Abstract
Oncoproteins from DNA tumor viruses associate with critical cellular proteins to regulate cell proliferation, survival, and differentiation.Human papillomavirus (HPV) E6 oncoproteins have been previously shown to associate with a cellular HECT domain ubiquitin ligase termed E6AP (UBE3A). Here we show that the E6-E6AP complex associates with and targets the degradation of the protein tyrosine phosphatase PTPN3 (PTPH1) in vitro and in living cells. PTPN3 is a membrane-associated tyrosine phosphatase with FERM, PDZ, and PTP domains previously implicated in regulating tyrosine phosphorylation of growth factor receptors and p97 VCP (valosin-containing protein, termed Cdc48 in Saccharomyces cerevisiae) and is mutated in a subset of colon cancers. Degradation of PTPN3 by E6 requires E6AP, the proteasome, and an interaction between the carboxy terminus of E6 and the PDZ domain of PTPN3. In transduced keratinocytes, E6 confers reduced growth factor requirements, a function that requires the PDZ ligand of E6 and that can in part be replicated by inhibiting the expression of PTPN3. This report demonstrates the potential of E6 to regulate phosphotyrosine metabolism through the targeted degradation of a tyrosine phosphatase.
Papillomaviruses are causative agents of benign epithelial tumors in vertebrates. A subset of these benign epithelial tumors may develop into epithelial malignancies, and the progression to malignancy is associated with particular papillomavirus types. The subset of human papillomavirus (HPV) types associated with cancer is termed “high risk.” Most human genital cancers contain integrated high-risk HPV genomes that express the viral E6 and E7 oncoproteins (reviewed in reference 28); continued expression of E6 and E7 is required for cancer cell proliferation, and in certain HPV-expressing cancer cell lines, negative regulation of E6 and E7 expression results in the cessation of proliferation and entry of the cells into a terminal differentiation pathway (11, 14).
Papillomavirus E6 oncoproteins are small zinc-binding proteins with conserved overall structure but diverse activities, and considerable effort has been directed toward establishing their cellular targets (reviewed in reference 25). The cancer-associated E6 oncoprotein from HPV type 16 (HPV-16) (16E6) and bovine papillomavirus E6 (BE6) directly interact with cellular proteins by interaction with LXXLL peptide sequences on the target protein, and this interaction is required for cellular transformation (3, 41). 16E6 interacts with an LXXLL peptide sequence found on the cellular E3 ubiquitin ligase E6AP and together with E6AP binds to the p53 tumor suppressor protein (17), resulting in its ubiquitin-mediated degradation by the proteasome. The efficient in vivo degradation of p53 requires both E6AP and the E3 ubiquitin ligase activity of E6AP (7). E6 proteins have also been reported to target the degradation of other cellular proteins, initially identified through yeast two-hybrid interaction searches or candidate approaches. A group of cellular proteins that interact with cancer-associated E6 proteins contain PDZ domains and bind the carboxy-terminal five amino acids of E6 that constitute a PDZ ligand consensus sequence [XX(S/T)X(V/I)]. Cellular targets of E6 that include PDZ domains include DLG1 (human discs large homolog) (20, 23, 24) and Scribble (29) (that are tumor suppressors in Drosophila melanogaster), MUPP1 (23), MAGI-1 (13), MAGI-2, and MAGI-3 (39). In these cases, the association with E6 has been shown to result in instability of the PDZ-containing proteins in vitro. Targeted degradation of Scribble and DLG1 by E6 can be accomplished through their association with the PDZ ligand of E6 and the LXXLL association of E6 with the E6AP ubiquitin ligase (4, 26).
There are many PDZ-containing proteins, including adapter molecules (such as DLG1 and Scribble), MAGUK proteins (membrane-associated guanylate kinase homologs with inactive kinase domains), and tyrosine phosphatases. There are three tyrosine phosphatases that contain PDZ domains that might be targeted by E6: PTPN3, PTPN4, and PTPN13. We have begun to use a proteomic approach to determine those substrates of E6 together with E6AP that form in vivo. Those experiments have isolated proteins that associate with E6AP together with HPV-16 E6 (16E6) by tandem affinity purification and identified the proteins using mass spectrometry. We find that 16E6 recruits the tyrosine phosphatase PTPN3 to E6AP, resulting in the E6AP and proteasome-dependent degradation of PTPN3.
MATERIALS AND METHODS
Cells and tissue culture.
CV-1 and HaCat cells were maintained in Dulbecco modified Eagle medium supplemented with 10% newborn calf serum, glutamine, and antibiotics. NIKS cells are normal immortalized epidermal growth factor (EGF)-dependent human keratinocytes that are passaged on 3T3 feeder cells as described previously (1). E6AP null mouse fibroblasts and vaccinia virus expression are as previously described (7).
Tandem affinity purification.
Either empty EE-Flag vector, EE-Flag-E6AP-C843A, or EE-Flag-E6AP-C843A together with untagged 16E6 were expressed in confluent monkey CV-1 cells by lipofection and T7 polymerase-directed expression using the T7-expressing vaccinia virus Vtf-7 (9). Eight hours after infection, the medium was removed, and the cells were washed three times with phosphate-buffered saline and lysed on ice with 0.5× NP-40 lysis buffer (1× NP-40 lysis buffer is 150 mM NaCl, 50 mM Tris, pH 7.5, 50 mM NaF, 5 mM Na PPi, 1% IPEGAL, 0.01% phenylmethylsulfonyl fluoride, 1 mM sodium vanadate, and 1 μg/ml leupeptin/aprotinin). Lysates from approximately 5 × 108 CV-1 cells were centrifuged at 15,000 × g for 10 min and then incubated with 1.0 mg EE monoclonal antibody covalently coupled to protein A-Sepharose beads for 1 h. The EE beads were washed extensively with NP-40 lysis buffer, and bound proteins were eluted by three successive elutions with 10 μg EE peptide each. The eluted proteins were applied to 20 μg Flag M2 monoclonal antibody coupled to Sepharose beads (Sigma-Aldrich Chemicals). After incubation on ice for 2 hours, the beads were washed three times with NP-40 lysis buffer, and bound proteins were eluted with three successive incubations of 2 μg Flag peptide (Sigma-Aldrich Chemicals) in 0.25× NP-40 lysis buffer. Eluted proteins were freeze-dried, resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, and applied to 5 to 20% Tris-glycine SDS-polyacrylamide gels (Cambrex). Gels were stained first with Coomassie blue and then restained with silver to visualize protein bands. Excised bands were reduced, alkylated, and digested with trypsin in the gel. The peptides formed in the digestion were extracted, concentrated, and characterized by capillary column liquid chromatography-tandem mass spectrometry. Database searches using the program SEQUEST were used to identify the protein by matching the collision-induced dissociation spectra to the PTPN3 sequence. These matching spectra were verified by manual inspection of the collision-induced dissociation spectra.
Plasmids.
The transient expression of 16E6 and E6AP is as previously described (7). 16E6ΔC is deleted of the last two amino acids of 16E6 in the carboxy-terminal PDZ ligand. A cDNA expression plasmid with 5′ EE and Flag monoclonal antibody epitope tags was constructed by standard molecular biology techniques in the pcDNA3 plasmid. E6AP-C843A (a ubiquitin ligase-defective mutant of E6AP) was cloned into this plasmid for expression and purification of EE-Flag-E6AP-C843A by tandem affinity purification. Full-length cDNA for PTPN3 was the gift of Nicholas Tonks (Cold Spring Harbor Laboratories) and was subcloned as either native protein or fusions to myc, EE, or Flag epitope. A PTPN3 mutant with an in-frame 6-amino-acid insertion at amino acid 525 within the PDZ domain (FNLGKKV changed to FNLSMPWHVKGGV) was created by standard techniques. For retroviral transduction of mammalian cells, HPV-16 E6 and E7 genes cloned into pLXSN were a gift from Denise Galloway (University of Washington, Seattle), and retrovirus was packaged by transient transfection of Phoenix Ampho cells (provided by Gary Nolan, Stanford University). All mutants were sequenced to verify the mutation and were without second-site mutations. A short hairpin RNA (shRNA) expression retrovirus (p7194A) with defective 3′ long terminal repeats, U6 promoter shRNA expression cassette, and hygromycin selection was constructed by standard techniques and was based upon the retroviral plasmid pSiren-RetroX (BD Biosciences). shRNA to PTPN3 was created by cloning oligonucleotides corresponding to nucleotides 773 to 793 (AAGTTTCTATCCTTGGGTGAA for shRNA-2)in the coding region of human PTPN3 as hairpins into p7194A retroviral vector with a connecting loop containing an XhoI site (AAGTTTCTATCCTTGGGTGAAACACTCGAGTTCACCCAAGGATAGAAACTT). shRNA directed against luciferase was purchased from BD Biosciences.
In vitro protein expression, binding assays, and degradation assays were performed in rabbit reticulocyte lysate as previously described (3, 7, 41). Briefly, in vitro-coupled transcription and translation were performed utilizing standard nuclease-treated reticulocyte lysate (Promega) according to the manufacturer's recommendations, supplemented with 1.5 mM MgCl2, 0.5 mM (each) nucleotide triphosphate,and 25 units T7 RNA polymerase (Gibco-BRL) per 50-μl translation reaction mixture. For in vitro binding assays, 25 μl reticulocyte lysate programmed to express the indicated proteins was incubated for 30 min at 4°C, then 175 μl of 0.5 × NP-40 lysis buffer containing precipitating antibody and protein A-Sepharose or bead-immobilized glutathione S-transferase (GST) fusion was added, and binding was allowed to proceed at 4°C with rocking for 1 h. Beads were washed three times with 1.5 ml 0.5 × NP-40 lysis buffer. Retained proteins were eluted with SDS sample buffer, resolved by SDS-PAGE, transferred to polyvinylidene difluoride membranes, or fluorographed with salicylate (6), and subjected to autoradiography and quantification by beta counting with a Packard Instant Imager. In vitro degradation reactions were performed as previously described (7, 18).
Western blot analysis.
Cell lysates were equalized for protein content as determined with a commercial kit (Bio-Rad) before electrophoresis; equalized proteins boiled in complete SDS sample buffer were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Rabbit polyclonal antibody to PTPN3 was raised against human PTPN3 amino acids 311 to 510. Monoclonal antibodies and their sources are as follows: Ab-8 specific for human and not mouse p53 (Oncogene Science), DLG1 (BD Biosciences), vinculin and tubulin (Sigma). Epitope tags were obtained from Sigma Chemicals (M2 Flag), the Developmental Studies Hybridoma Bank (myc clone 9E10), EE (Gernot Walter, University of California, San Diego). Monoclonal antibody specific for human and not mouse PTPN3 was a generous gift from Nicholas Tonks (Cold Spring Harbor Laboratories), and monoclonal antibody 6F4 to 16E6 was provided by Gilles Trave (21).
RESULTS
Tandem affinity purification of PTPN3 associated with E6AP together with 16E6.
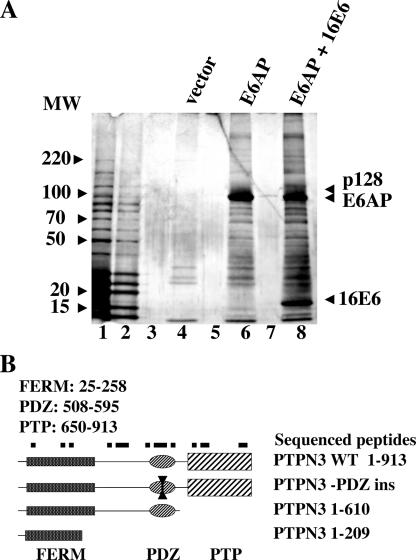
16E6 acts as an adapter protein in the targeted degradation of p53 by interacting with the cellular ubiquitin ligase E6AP, as neither E6AP nor 16E6 strongly associates with p53 alone (34). The E6AP mutant E6AP-C843A is defective for ubiquitin ligase activity and can act as a substrate-trapping mutant (35). E6AP-C843A epitope tagged with EE and FLAG epitopes was transiently expressed alone or together with native 16E6 in CV-1 epithelial cells. EE-FLAG-E6AP-C843A was purified by successive immune precipitation and peptide elution (tandem affinity purification) and analyzed by SDS-PAGE. Figure 1A demonstrates that only a few weakly stained proteins are obtained by tandem affinity purification from cells expressing the empty vector (lane 4). Several high-molecular-weight proteins were associated with EE-FLAG-E6AP-C843A (lane 6), but an additional 128-kDa protein was associated with E6AP-C843A only when 16E6 was coexpressed (lane 8). Analysis of the p128 band identified it as PTPN3 by peptide sequencing of 18 peptides mapping to each domain of PTPN3 and 22% of the entire PTPN3 primary sequence (Fig. 1B). PTPN3 is a member of a small group of membrane-associated nonreceptor tyrosine phosphatases that contain FERM domains (for 4.1, ezrin, radixin, and moesin) at the amino terminus and one or more PDZ domains, and is illustrated in Fig. 1B.
FIG. 1.
16E6 recruits the cellular tyrosine phosphatase PTPN3 protein to E6AP. (A) 16E6 recruits a 128-kDa protein to the E6AP ubiquitin ligase. Ubiquitin ligase-defective E6AP (E6AP-C843A) tagged at the amino terminus with the EE and Flag epitopes was transiently expressed with or without untagged 16E6 in CV-1 cells. Cleared lysates were purified by tandem affinity purification with EE and Flag antibodies followed by peptide elution and analysis on 5 to 20% SDS-polyacrylamide silver-stained gels. Lanes 1 and 2, molecular weight markers; lane 4, tandem affinity-purified (TAP), vector-transfected cells; lane 6, TAP, EE-Flag-E6AP-C843A-transfected cells; lane 8, TAP EE-Flag-E6AP-C843A expressed together with untagged 16E6; lanes 3, 5, and 7, loaded with sample buffer only. Peptides of PTPN3 sequenced from p128 band numbered by GenBank NP_002820: FFIPDPNTLQQEQTR (amino acids [aa] 109 to 115), VESLHEQHSGLK (aa 188 to 199), TLDFYGVELHSGR (aa 213 to 225), EHIVAFNMLNYR (aa 281 to 292), SCVEHHTFFQAK (aa 300 to 311), LLPQEK (aa 313 to 318), NVLSQYWTMGSR (aa 319 to 330), ITPDEDGKFGFNLK (aa 513 to 526), MPLVVSR (aa 533 to 539), INPESPADTCIPK (aa 540 to 552), LNEGDQIVLINGR (aa 553 to 565), ELALVIR (aa 589 to 595), GLESGTVLIQFEQLYR (aa 640 to 655), KKPGLAITFAK (aa 656 to 666), LPQNLDKNR (aa 667 to 675), YKDVLPYDTTR (aa 657 to 686), MRDQRAMMVQTSSQYK (aa 877 to 892), and FVCEAILR (aa 893 to 900). The positions of molecular weight standards (in thousands) are shown to the left of the gel. (B) Domain structure of PTPN3. A diagram of PTPN3 illustrates the relative locations of FERM, PDZ, and phosphatase domains. Black squares illustrate the approximate location of sequenced peptides from mass spectrometry of p128. Below are diagrammed plasmids used in this study with the amino acid segments of PTPN3. WT, wild type; ins, inserted.
PTPN3 is targeted for degradation in vitro by oncogenic HPV E6 proteins.
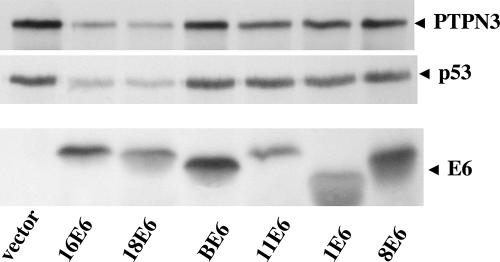
In order to determine whether 16E6 might functionally interact with PTPN3, E6 genes from high-risk oncogenic human mucosal HPVs (16E6 and 18E6), benign human mucosal (11E6) or cutaneous papillomas (human types 1E6 and 8E6, and bovine BE6) were in vitro translated in rabbit reticulocyte lysate and incubated together with in vitro-translated PTPN3 or in vitro-translated p53. Rabbit reticulocyte lysate contains ubiquitin-conjugating enzymes and active proteasomes and supports the degradation of p53 by 16E6 (36). A clear reduction in the amount of both p53 and PTPN3 was observed when in vitro translations of PTPN3 or p53 were incubated together with oncogenic 16E6 and HPV-18 E6, but not with other E6 types (Fig. 2). Although it is not apparent in the portion of the gel shown in Fig. 2, for both p53 and PTPN3 degradation, there is an accumulation of high-molecular-weight smeared protein at the top of the gel near the gel wells that is characteristic of polyubiquitinated proteins that have not yet been degraded.
FIG. 2.
In vitro degradation of PTPN3 by high-risk mucosal E6 proteins. PTPN3, p53, and the indicated 16E6 types from cancer-associated HPV (16E6 and 18E6), non-cancer-associated mucosal HPV (11E6), non-cancer-associated cutaneous HPV (1E6), cancer-associated cutaneous HPV (8E6), and bovine papillomavirus type 1 (BE6) were in vitro translated in reticulocyte lysate containing 35S-labeled amino acids and incubated together for 60 min at room temperature to assay for in vitro degradation of 35S-labeled p53 and PTPN3. Samples were analyzed by SDS-PAGE, with the segments containing p53, PTPN3, and E6 proteins indicated (broadening of some E6 bands is due to comigration with globin from the reticulocyte lysate).
The PTPN3 PDZ domain interacts with the 16E6 PDZ ligand.
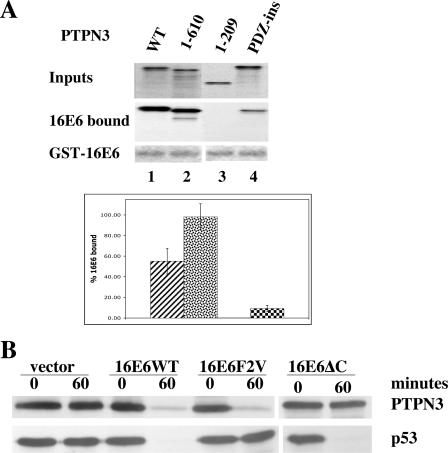
As noted in the introduction, 16E6 has been shown to target the degradation of other PDZ-containing cellular proteins through interaction of a 5-amino-acid peptide at the end of 16E6 with the PDZ domains of the targeted cellular proteins. In order to determine whether the PDZ domain of PTPN3 is required for binding to 16E6, fragments of PTPN3 were in vitro translated and assayed for binding to 16E6. Also, a PTPN3 mutant containing an in-frame insertion was generated in the first GLGF conserved loop that stabilizes the ligand-binding pocket of the PDZ domain. As predicted, only PTPN3 molecules containing an intact PDZ domain bound efficiently to GST-16E6 (Fig. 3A). The in-frame insertion mutant in the GLGF loop of the PDZ domain (PTPN3-PDZ-ins) was reduced for binding to GST-16E6. In order to determine whether PTPN3 was targeted for degradation through association of its PDZ domain with the carboxy terminus of 16E6, in vitro-translated 16E6 or 16E6 mutants were incubated together with in vitro-translated PTPN3. One 16E6 mutant, 16E6F2V, has a substitution of valine for phenylalanine at amino acid position 2, which reduces association with and degradation of p53 (7). The other mutant, 16E6ΔC, has a deletion of carboxy-terminal two amino acids of 16E6, disrupting the PDZ ligand motif (7, 20, 24). Figure 3B demonstrates that the degradation of PTPN3 by 16E6 requires an intact PDZ ligand at the carboxy terminus of 16E6 but is unaffected by the 16E6F2V mutation that reduces degradation of p53. Figure 3C demonstrates that fragments of PTPN3 that contain an intact PDZ domain are degraded by 16E6, while fragments deleted of the PDZ domain are not degraded. Mutation of the PTPN3 PDZ domain prevents degradation by 16E6 in vitro. Thus, the binding and degradation assays in Fig. 3 correspond. We have also tested a mutant of 16E6 that fails to interact with E6AP and therefore fails to target degradation of p53; as expected, this mutant (16E6_Y79N) also fails to target the degradation of PTPN3 in vitro (data not shown). In order to compare the in vitro degradation of PTPN3 to p53 and another PDZ domain-containing target of 16E6, in vitro-translated PTPN3, p53, and DLG1 were incubated together with 16E6 and 18E6. Figure 3D shows that while PTPN3 and p53 were efficiently degraded by both 16E6 and 18E6, DLG1 was degraded efficiently only by 18E6 and not appreciably by 16E6. As in Fig. 3A, degradation of PTPN3 by 18E6 also required an intact PDZ ligand at the carboxy terminus of 18E6.
FIG. 3.
Role of PDZ interactions in the degradation of PTPN3 by 16E6. (A) An intact PDZ domain on PTPN3 is required for efficient binding to 16E6 in vitro. Input and GST-16E6 bound in vitro translation products of PTPN3 and the indicated mutants of PTPN3 (Fig. 1B) were resolved on SDS-polyacrylamide gels fluorographed and quantified by beta counting. GST-16E6 is shown from Coomassie blue staining. Bound counts resulting from four independent binding reactions are graphed below with standard deviation error bars. WT, wild type. (B) 16E6 degradation of PTPN3 requires the carboxy-terminal PDZ-binding peptide of 16E6. In vitro-translated p53, PTPN3, 16E6 or the indicated 16E6 mutants were mixed together as indicated and incubated for 60 min at 25°C. 16E6F2V refers to substitution of valine for phenylalanine at amino acid position 2, and 16E6ΔC contains a deletion of the carboxy-terminal 2 amino acids of 16E6. Aliquots removed at 0 and 60 min were analyzed by SDS-PAGE followed by autoradiography. (C) An intact PDZ domain in PTPN3 is required for in vitro degradation by 16E6. In vitro-translated p53, PTPN3, or the indicated PTPN3 mutants were incubated with mock-translated lysate,16E6, or 16E6ΔC for 60 min. (D) Comparison of in vitro degradation of PTPN3, p53, and DLG1. PTPN3, p53, and DLG1 were in vitro translated in reticulocyte lysate with 35S-labeled amino acids and incubated with in vitro-translated 16E6, 18E6, or 18E6ΔC for 0 or 60 min before analysis by SDS-PAGE and autoradiography. 16E6 bands are distorted by reticulocyte-derived globin in the gel.
16E6 expression reduces PTPN3 levels in vivo.
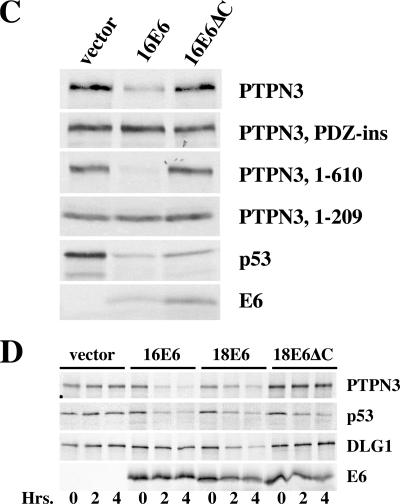
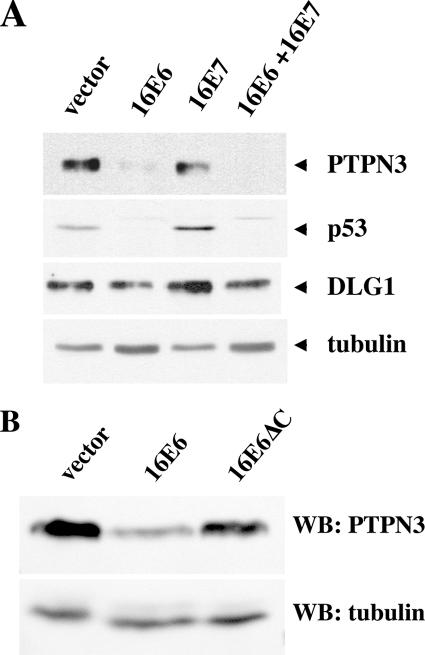
In keratinocytes that express cancer-associated E7 oncoproteins, there is an induction of p53 protein due to stabilization of p53 (8, 37); E6 degradation of p53 compensates for the induction of p53 by E7. Cancer-associated E7 proteins are reported to induce p73 (5), and PTPN3 has been described as an induced target of p73 (10). In Fig. 4A, E7 induced p53 in NIKS keratinocytes as expected but did not markedly enhance the expression of either PTPN3 or DLG1; PTPN3 was efficiently reduced by 16E6 expression, and DLG1 was slightly reduced by 16E6 in the presence of E7. Retrovirally transduced HaCat keratinocytes showed a clear reduction of PTPN3 levels by 16E6 but not by a 16E6 mutant deleted of the C-terminal PDZ ligand (16E6ΔC), confirming the in vivo degradation of PTPN3 seen in NIKS cells and demonstrating the requirement for the PDZ ligand of 16E6 (Fig. 4B). In all the experiments with intact cells, the reduction of PTPN3 by 16E6 was clear but not complete, and residual amounts of residual PTPN3 were observed. In separate experiments, we have determined that the expression levels of 16E6 and 16E6ΔC in transduced NIKS cells are quite similar (data not shown). Quantitative reverse transcription-PCR for PTPN3 RNA showed no reduction in RNA levels between vector-transduced NIKS cells and 16E6-transduced cells (data not shown). Expression levels of PTPN3 in vivo are low, and clear localization of cellular PTPN3 could not be observed with either monoclonal antibodies or affinity-purified polyclonal antibodies (data not shown).
FIG. 4.
16E6 reduces the in vivo levels of PTPN3 in stably transduced cells. (A) 16E6 reduces PTPN3 expression in NIKS keratinocytes. NIKS human keratinocytes stably retrovirally transduced by the indicated oncogenes were removed from feeder cells, reattached to collagen type 1-coated plates overnight in 3T3 feeder cell-conditioned medium, and lysed in SDS lysis buffer the next day, and equalized amounts of protein were analyzed by sequential probing of a Western blot for the expression of PTPN3, p53, DLG1, and tubulin. (B) In vivo reduction of PTPN3 by 16E6 requires an intact PDZ motif on 16E6. HaCat keratinocytes stably transduced with the indicated retroviral plasmids were lysed in SDS sample buffer, and equal amounts of protein were analyzed with antibodies to PTPN3 (top panel) or tubulin (bottom panel). The blot exposures were quantitated by densitometry scanning to determine that the residual amounts of PTPN3 in lane 2 were 22% of lane 1 and 79% in lane 3. WB, Western blotting.
Efficient degradation of PTPN3 by 16E6 in vivo requires E6AP and the proteasome.
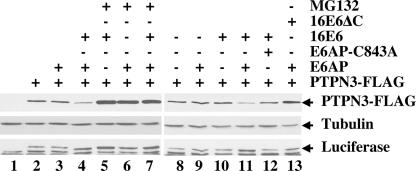
The association of PTPN3 with 16E6 and E6AP ubiquitin ligase (Fig. 1) and the reduction of PTPN3 levels in response to oncogenic E6 proteins imply that an E6AP- and proteasome-dependent process degrades PTPN3, as is the case with p53. To confirm this hypothesis, murine fibroblasts null for E6AP were transiently transfected with E6AP, 16E6, 16E6 mutants, and epitope-tagged PTPN3 (Fig. 5). Expression of PTPN3 protein was reduced by coexpression of E6AP together with 16E6, but not by either 16E6 or E6AP alone. Mutation of the carboxy-terminal PDZ ligand of 16E6 (16E6ΔC) restored expression of PTPN3. Addition of proteasome inhibitor MG132 allowed higher accumulation of PTPN3 under different conditions. An ubiquitin ligase-defective mutant of E6AP (E6AP-C843A) did not reduce PTPN3 levels when coexpressed with 16E6, demonstrating the requirement of wild-type E6AP, 16E6, and intact proteasome for the degradation of PTPN3 in vivo.
FIG. 5.
Role of E6AP in proteasome-mediated degradation of PTPN3 by 16E6. E6AP null mouse embryo fibroblasts were transfected with the indicated plasmids together with a constant amount of internal luciferase expression plasmid (starting in lane 2) and empty vector DNA to make the amount of transfected DNA constant. Cells were lysed in SDS sample buffer 20 h later and analyzed sequentially by immunoblotting with antibodies to FLAG, tubulin, and luciferase. PTPN3 was tagged at the carboxy-terminal end with the FLAG epitope. The results of one representative experiment of five experiments are shown.
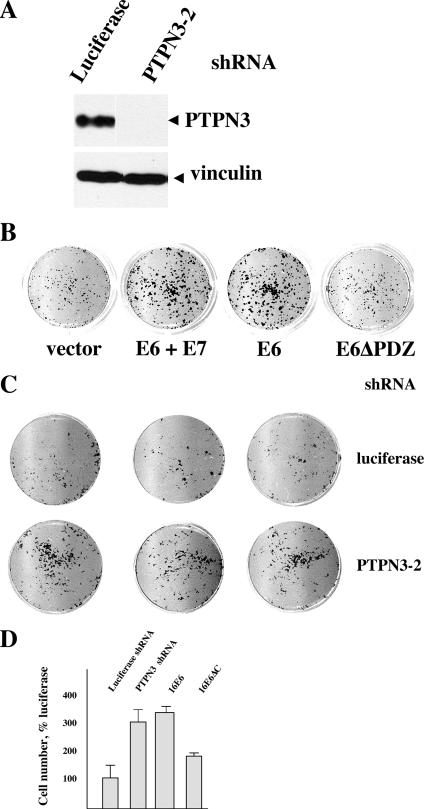
Tyrosine phosphatases have diverse functions but are thought to primarily act to reduce phosphotyrosine-based signal transduction initiated by growth factor receptors. It is possible that targeted degradation of PTPN3 by E6 might be manifested by reduced growth factor requirements in E6-transduced cells. To test this possibility, NIKS keratinocytes were transduced with empty retroviral vector, 16E6, 16E6ΔC, shRNA vector directed against luciferase, or shRNAs directed against PTPN3. Eight shRNAs were directed against various parts of PTPN3, but only one gave rise to robust inhibition of PTPN3 protein expression (PTPN3-2 shRNA [Fig. 6A and data not shown]). Figure 6A shows that shRNA directed against PTPN3 (shRNA PTPN3-2, directed to the region in-between the FERM and PDZ domains) reduced PTPN3 expression to undetectable levels by Western blotting. E6 transduction enhanced the accumulation of keratinocytes cultured in 0.25% serum without added EGF or insulin, and deletion of the PDZ ligand of E6 reduced this E6 phenotype (Fig. 6B). Interestingly, the effect of addition of E7 to E6 was similar in this assay to that of E6 alone at enhancing growth in the absence of EGF and insulin. Compared to shRNA directed against luciferase, shRNA directed against PTPN3 also enhanced the accumulation of keratinocytes cultured in reduced serum without added EGF or insulin (Fig. 6C and D).
FIG. 6.
Role of the PDZ ligand of E6 in reducing the growth factor requirements of keratinocytes. (A) shRNA to PTPN3 reduces PTPN3 expression. Epithelial cells transduced with the indicated retroviruses were lysed in SDS sample buffer and analyzed by immunoblotting for PTPN3 expression. Each lane shows the result of independently transduced cell cultures arising from pooled drug-resistant cell colonies. (B) Role of the E6 PDZ ligand in the acquisition of reduced growth factor requirements. A total of 5 × 103 NIKS keratinocytes transduced by the indicated retroviruses were seeded into wells with mitomycin C-treated feeder cells in complete medium for 48 h and then changed to medium with 0.25% serum and no added EGF or insulin. Nine days later, the plates were fixed, stained with crystal violet, and scanned. (C) Effect of reduced PTPN3 upon cell accumulation during culture in reduced growth factors. NIKS keratinocytes retrovirally transduced with shRNA to luciferase or to PTPN3 were transduced and seeded into tissue culture dishes as in panel B above, then stained, and scanned. (D) E6 and shRNA to PTPN3 alter cell accumulation in NIKS keratinocytes. Crystal violet from stained cells was solubilized in 1% acetic acid and quantified by absorbance at 600 nm. Error bars represent the standard deviations of 12 wells of cells and are representative of three independent experiments. E6 and shRNA directed against PTPN3 (shRNA PTPN3-2) enhances cell accumulation, while deletion of the E6 PDZ ligand results in a loss of accumulated cells compared to E6.
DISCUSSION
PTPN3 was purified in a complex with 16E6 and E6AP-C843A. It is possible that other proteins might have been associated with E6AP together with 16E6 in our experiment, but they were too scarce in the complex for visualization, masked by background staining, comigrated with E6AP or E6AP fragments, or were lost during prolonged washing and peptide elutions over 8 h. Although the E6AP-E6-PTPN3 complex was isolated from CV-1 cells, PTPN3 is present in cultured keratinocytes as well. The stable introduction of 16E6 into keratinocytes resulted in a decline of PTPN3 protein expression that required the PDZ ligand of E6 (Fig. 4). In vitro degradation assays established the requirement for cancer-associated HPV types (Fig. 2), as well as an intact PDZ ligand of E6 and an intact PDZ domain of PTPN3 for both the association with E6 and the targeted degradation of PTPN3 by E6 (Fig. 3). Using E6AP null fibroblasts, we determined that the degradation of PTPN3 by 16E6 was enhanced by coexpression of E6AP, required E6AP ubiquitin ligase activity, and required the proteasome (Fig. 5).
PTPN3 is one of only three tyrosine phosphatase groups containing FERM domains at the amino terminus and one of three tyrosine phosphatases containing one or more PDZ domains; PTPN3 is most closely related to PTPMEG1, and PTPN3 and PTPMEG1 together comprise a distinct group of tyrosine phosphatases that are highly conserved in vertebrates, flies, and worms (2). Interestingly, PTPN3 has recently been shown to be mutated in a minor fraction of colon cancers (42). PTPN13 also contains PDZ domains and is (like PTPN3) also mutated in colon cancers (42). In preliminary experiments, PTPN13 expression is reduced in keratinocytes in which 16E6 is expressed but not in cells in which 16E6ΔC is expressed. While we did not observe a clear band the size of PTPN13 in our silver-stained gel, it is possible that E6 may target the degradation of more than one tyrosine phosphatase.
Substrate-trapping experiments from vanadate-treated cell lysates identified VCP as a PTPN3 substrate (44). VCP is the mammalian form of the Saccharomyces cerevisiae protein Cdc48 that is essential for cell cycle progression in all phases of the cell cycle (12, 27) and is an abundant AAA-ATPase associated with many essential cellular functions (reviewed in reference 33). A defect in degradation of polyubiquitinated proteins may underlie these phenotypes, as both yeast Cdc48 cells at the nonpermissive temperature and mammalian cells treated with RNA interference to VCP accumulate polyubiquitinated proteins (43). However, detection of VCP tyrosine phosphorylation in vivo has been elusive. PTPN3 has been shown to dephosphorylate VCP in vitro, but it is unproven if other phosphatases may be able to perform this function in vivo as well. As yet, we have failed to detect clear differences in the tyrosine phosphorylation of VCP between normal cells compared to either cells expressing E6 or in cells where PTPN3 has been reduced by shRNA in the presence or absence of vanadate (our unpublished observations). Recent experiments indirectly implicated PTPN3 as a candidate phosphatase in the dephosphorylation of the growth hormone receptor (32) and the T-cell receptor zeta chain after ligand stimulation (16, 38). In summary, the role of PTPN3 in regulating the in vivo phosphorylation of VCP or other cellular substrates is not yet clearly defined.
The biological significance of particular cellular PDZ targets of the E6 oncoproteins also remains uncertain. The PDZ ligand of E6 is important, as the development of eye lens hyperplasias or skin hyperplasia in mice that express 16E6 requires an intact PDZ-binding peptide at the carboxy terminus of E6 (30, 31). Further, deletion of the PDZ ligand of E6 in the context of the full HPV-31 genome resulted in transfected cells that were significantly reduced in their growth rates and reduced in their viral copy numbers compared to keratinocytes transfected with wild-type genomes (22). Since deletion of the carboxy terminus of E6 eliminates the interaction with all of its PDZ targets, the full significance of any particular PDZ-containing target of 16E6 in the viral life cycle or the development of cancer remains open for investigation. One might suspect that the biologically significant targets of E6 would be common to all the oncogenic types. While 16E6 expression causes hyperplasia of mouse skin and the eye lens, and insertional inactivation of DLG1 also causes hyperplasia of the eye lens, 16E6 does not significantly target the degradation of DLG1 in vitro (Fig. 3D) (also shown in reference 40) and has a modest effect upon overall expression levels in vivo (Fig. 4A), indicating that 16E6 interactions with DLG1 may be more subtle than simply reducing the overall level of DLG1 expression. PTPN3 is one of a limited number of PDZ proteins that is efficiently targeted for degradation by both 16E6 and 18E6 (Fig. 2 and 3D).
One hallmark of the progression from normal to malignant cells is a reduced requirement for exogenously supplied growth factors (15). Keratinocytes proliferate in response to exogenously supplied growth factors but also produce and respond to growth factors in an autocrine fashion (19). Because primary keratinocytes have limited growth potential after retroviral transduction and selection, we utilized NIKS cells that are growth factor dependent yet immortalized. We showed that E6 reduced the requirement of NIKS cells for growth factors and that mutation of the PDZ ligand of E6 reduced this effect (Fig. 6). It could be that the accumulation of cells conferred by E6 under these conditions is due to either an increase in proliferation rate or a decrease in cell death or terminal differentiation; these studies are under way. It is reasonable to suppose that a reduced requirement for exogenous growth factors could facilitate the initial establishment or maintenance of an epithelioma. It is possible that E6 degradation of PTPN3 contributes to such a phenotype, as shRNA to PTPN3 also gave a similar but less pronounced phenotype than E6 (Fig. 6); this correlates with the finding that PTPN3 is mutated in a small fraction of colon cancers (42). As yet, it is unclear if the effect of PTPN3 upon growth factor requirements will be direct or indirect. It is possible that the reduced growth factor requirement of E6-transduced cells could also be conferred by degradation either alone or in combination of additional E6 targets (besides PTPN3) that interact with the PDZ ligand of E6. These possibilities are currently under investigation.
Acknowledgments
This research was supported by NIH CA80931 and CA69292 to S.B.V.P.
We thank Charles Lyons and the University of Virginia Department of Pathology Collaborative Mass Spectrometry Facility for additional analysis of protein samples. We thank David Brautigan and Janet Cross for critical reading of the manuscript.
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Allen-Hoffmann, B. L., S. J. Schlosser, C. A. Ivarie, C. A. Sattler, L. F. Meisner, and S. L. O'Connor. 2000. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J. Investig. Dermatol. 114:444-455. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. N., O. H. Mortensen, G. H. Peters, P. G. Drake, L. F. Iversen, O. H. Olsen, P. G. Jansen, H. S. Andersen, N. K. Tonks, and N. P. Moller. 2001. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol. Cell. Biol. 21:7117-7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohl, J., K. Das, B. Dasgupta, and S. B. Vande Pol. 2000. Competitive binding to a charged leucine motif represses transformation by a papillomavirus E6 oncoprotein. Virology 271:163-170. [DOI] [PubMed] [Google Scholar]
- 4.Brimer, N., C. Lyons, and S. B. Vande Pol. 3. October 2006. Association of E6AP (UBE3A) with human papillomavirus type 11 E6 protein. Virology doi: 10.1016/j.virol.2006.08.038. [DOI] [PMC free article] [PubMed]
- 5.Brooks, L. A., A. Sullivan, J. O'Nions, A. Bell, B. Dunne, J. A. Tidy, D. J. Evans, P. Osin, K. H. Vousden, B. Gusterson, P. J. Farrell, A. Storey, M. Gasco, T. Sakai, and T. Crook. 2002. E7 proteins from oncogenic human papillomavirus types transactivate p73: role in cervical intraepithelial neoplasia. Br. J. Cancer 86:263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamberlain, J. P. 1979. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal. Biochem. 98:132-135. [DOI] [PubMed] [Google Scholar]
- 7.Cooper, B., S. Schneider, J. Bohl, Y. Jiang, A. Beaudet, and S. Vande Pol. 2003. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology 306:87-99. [DOI] [PubMed] [Google Scholar]
- 8.Eichten, A., M. Westfall, J. A. Pietenpol, and K. Munger. 2002. Stabilization and functional impairment of the tumor suppressor p53 by the human papillomavirus type 16 E7 oncoprotein. Virology 295:74-85. [DOI] [PubMed] [Google Scholar]
- 9.Elroy-Stein, O., T. R. Fuerst, and B. Moss. 1989. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc. Natl. Acad. Sci. USA 86:6126-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontemaggi, G., I. Kela, N. Amariglio, G. Rechavi, J. Krishnamurthy, S. Strano, A. Sacchi, D. Givol, and G. Blandino. 2002. Identification of direct p73 target genes combining DNA microarray and chromatin immunoprecipitation analyses. J. Biol. Chem. 277:43359-43368. [DOI] [PubMed] [Google Scholar]
- 11.Francis, D. A., S. I. Schmid, and P. M. Howley. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 74:2679-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frohlich, K. U., H. W. Fries, M. Rudiger, R. Erdmann, D. Botstein, and D. Mecke. 1991. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 114:443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodwin, E. C., E. Yang, C. J. Lee, H. W. Lee, D. DiMaio, and E. S. Hwang. 2000. Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. USA 97:10978-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, W. C., and R. A. Weinberg. 2002. Rules for making human tumor cells. N. Engl. J. Med. 347:1593-1603. [DOI] [PubMed] [Google Scholar]
- 16.Han, S., S. Williams, and T. Mustelin. 2000. Cytoskeletal protein tyrosine phosphatase PTPH1 reduces T cell antigen receptor signaling. Eur. J. Immunol. 30:1318-1325. [DOI] [PubMed] [Google Scholar]
- 17.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1993. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol. Cell. Biol. 13:4918-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jost, M., C. Kari, and U. Rodeck. 2000. The EGF receptor—an essential regulator of multiple epidermal functions. Eur. J. Dermatol. 10:505-510. [PubMed] [Google Scholar]
- 20.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagrange, M., S. Charbonnier, G. Orfanoudakis, P. Robinson, K. Zanier, M. Masson, Y. Lutz, G. Trave, E. Weiss, and F. Deryckere. 2005. Binding of human papillomavirus 16 E6 to p53 and E6AP is impaired by monoclonal antibodies directed against the second zinc-binding domain of E6. J. Gen. Virol. 86:1001-1007. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C., and L. A. Laimins. 2004. Role of the PDZ domain-binding motif of the oncoprotein E6 in the pathogenesis of human papillomavirus type 31. J. Virol. 78:12366-12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, S. S., R. S. Weiss, and R. T. Javier. 1997. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:6670-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantovani, F., and L. Banks. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874-7887. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto, Y., S. Nakagawa, T. Yano, S. Takizawa, K. Nagasaka, K. Nakagawa, T. Minaguchi, O. Wada, H. Ooishi, K. Matsumoto, T. Yasugi, T. Kanda, J. M. Huibregtse, and Y. Taketani. 2006. Involvement of a cellular ubiquitin-protein ligase E6AP in the ubiquitin-mediated degradation of extensive substrates of high-risk human papillomavirus E6. J. Med. Virol. 78:501-507. [DOI] [PubMed] [Google Scholar]
- 27.Moir, D., S. E. Stewart, B. C. Osmond, and D. Botstein. 1982. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics 100:547-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Munger, K., and P. M. Howley. 2002. Human papillomavirus immortalization and transformation functions. Virus Res. 89:213-228. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa, S., and J. M. Huibregtse. 2000. Human Scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20:8244-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen, M. L., M. M. Nguyen, D. Lee, A. E. Griep, and P. F. Lambert. 2003. The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required for E6's induction of epithelial hyperplasia in vivo. J. Virol. 77:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen, M. M., M. L. Nguyen, G. Caruana, A. Bernstein, P. F. Lambert, and A. E. Griep. 2003. Requirement of PDZ-containing proteins for cell cycle regulation and differentiation in the mouse lens epithelium. Mol. Cell. Biol. 23:8970-8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasquali, C., M. L. Curchod, S. Walchli, X. Espanel, M. Guerrier, F. Arigoni, G. Strous, and R. H. Van Huijsduijnen. 2003. Identification of protein tyrosine phosphatases with specificity for the ligand-activated growth hormone receptor. Mol. Endocrinol. 17:2228-2239. [DOI] [PubMed] [Google Scholar]
- 33.Sauer, R. T., D. N. Bolon, B. M. Burton, R. E. Burton, J. M. Flynn, R. A. Grant, G. L. Hersch, S. A. Joshi, J. A. Kenniston, I. Levchenko, S. B. Neher, E. S. Oakes, S. M. Siddiqui, D. A. Wah, and T. A. Baker. 2004. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell 119:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 35.Scheffner, M., U. Nuber, and J. M. Huibregtse. 1995. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373:81-83. [DOI] [PubMed] [Google Scholar]
- 36.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 37.Seavey, S. E., M. Holubar, L. J. Saucedo, and M. E. Perry. 1999. The E7 oncoprotein of human papillomavirus type 16 stabilizes p53 through a mechanism independent of p19ARF. J. Virol. 73:7590-7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sozio, M. S., M. A. Mathis, J. A. Young, S. Walchli, L. A. Pitcher, P. C. Wrage, B. Bartok, A. Campbell, J. D. Watts, R. Aebersold, R. H. Van Huijsduijnen, and N. S. Van Oers. 2004. PTPH1 is a predominant protein-tyrosine phosphatase capable of interacting with and dephosphorylating the T cell receptor ζ subunit. J. Biol. Chem. 279:7760-7769. [DOI] [PubMed] [Google Scholar]
- 39.Thomas, M., R. Laura, K. Hepner, E. Guccione, C. Sawyers, L. Lasky, and L. Banks. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, M., P. Massimi, C. Navarro, J. P. Borg, and L. Banks. 2005. The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24:6222-6230. [DOI] [PubMed] [Google Scholar]
- 41.Vande Pol, S. B., M. C. Brown, and C. E. Turner. 1998. Association of bovine papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene 16:43-52. [DOI] [PubMed] [Google Scholar]
- 42.Wang, Z., D. Shen, D. W. Parsons, A. Bardelli, J. Sager, S. Szabo, J. Ptak, N. Silliman, B. A. Peters, M. S. van der Heijden, G. Parmigiani, H. Yan, T. L. Wang, G. Riggins, S. M. Powell, J. K. Willson, S. Markowitz, K. W. Kinzler, B. Vogelstein, and V. E. Velculescu. 2004. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science 304:1164-1166. [DOI] [PubMed] [Google Scholar]
- 43.Wojcik, C., M. Yano, and G. N. DeMartino. 2004. RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Cell Sci. 117:281-292. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, S. H., J. Liu, R. Kobayashi, and N. K. Tonks. 1999. Identification of the cell cycle regulator VCP (p97/CDC48) as a substrate of the band 4.1-related protein-tyrosine phosphatase PTPH1. J. Biol. Chem. 274:17806-17812. [DOI] [PubMed] [Google Scholar]