Abstract
Smith-Magenis syndrome (SMS) is a multiple congenital anomaly/mental retardation syndrome associated with behavioral abnormalities and sleep disturbance. Most patients have the same ∼4 Mb interstitial genomic deletion within chromosome 17p11.2. To investigate the molecular bases of the SMS phenotype, we constructed BAC/PAC contigs covering the SMS common deletion interval and its syntenic region on mouse chromosome 11. Comparative genome analysis reveals the absence of all three ∼200-kb SMS-REP low-copy repeats in the mouse and indicates that the evolution of SMS-REPs was accompanied by transposition of adjacent genes. Physical and genetic map comparisons in humans reveal reduced recombination in both sexes. Moreover, by examining the deleted regions in SMS patients with unusual-sized deletions, we refined the minimal Smith-Magenis critical region (SMCR) to an ∼1.1-Mb genomic interval that is syntenic to an ∼1.0-Mb region in the mouse. Genes within the SMCR and its mouse syntenic region were identified by homology searches and by gene prediction programs, and their gene structures and expression profiles were characterized. In addition to 12 genes previously mapped, we identified 8 new genes and 10 predicted genes in the SMCR. In the mouse syntenic region of the human SMCR, 16 genes and 6 predicted genes were identified. The SMCR is highly conserved between humans and mice, including 19 genes with the same gene order and orientation. Our findings will facilitate both the identification of gene(s) responsible for the SMS phenotype and the engineering of an SMS mouse model.
Smith-Magenis syndrome (SMS) is a contiguous gene-deletion syndrome (Greenberg et al. 1991), in which a distinct and consistent phenotype is associated with deletion of a portion of chromosome band 17p11.2 (Smith et al. 1986; Stratton et al. 1986). The clinical features of SMS patients include mental retardation, delayed speech and motor development, behavior problems, sleep disturbance, minor craniofacial abnormalities, short stature, and brachydactyly (Greenberg et al. 1991). Less common features include otolaryngological abnormalities, hearing impairment, opthalmological abnormalities, and renal and cardiac abnormalities (Chen et al. 1996; Greenberg et al. 1996). The incidence of SMS is ∼1:25,000 births, which is likely underestimated given the often subtle clinical features, particularly early in life (Greenberg et al. 1991). A heterozygous interstitial deletion of ∼4 Mb in chromosome 17p11.2 was identified in >90% of SMS patients, whereas the remaining patients have rare smaller- or larger-sized deletions (Greenberg et al. 1991; Juyal et al. 1996; Chen et al. 1997).
To narrow the critical interval responsible for the SMS phenotype, STS content mapping was performed on somatic cell hybrids refining the deleted chromosome from 62 SMS patients. A common deletion region was defined between markers D17S58 and cDI17-498 (Juyal et al. 1996). Ten patients with a deletion distinct from the common deletion region were identified, and molecular analyses of these patients delineated an SMS critical interval between D17S29 and cCI17-638 (Elsea et al. 1997). Three copies of a low-copy repeat (SMS-REPs) were identified within the common deletion region, and homologous recombination and unequal crossing over between the flanking SMS-REPs were shown to comprise the mechanism responsible for the genomic deletion (Chen et al. 1997). The predicted reciprocal duplication of the SMS common deletion region dup(17)(p11.2p11.2), which was identified recently, causes a subtle clinical syndrome (Potocki et al. 2000).
The first gene identified within the SMS common deletion region, snU3, encodes a small nuclear RNA U3 (Chevillard et al. 1993). Since then, more than 15 genes have been mapped. In addition, a large number of ESTs with no homology to known genes were identified (Seranski et al. 1999). However, a potential role for any of these genes in the SMS phenotype through haploinsufficiency effects remains unclear.
The shaker-2 (sh2) mouse represents a mouse model for human deafness (DFNB3), resulting from mutation of MYO15A (Wang et al. 1998; Liburd et al. 2001). A physical map of the sh2 region revealed that 11 genes mapping within the SMS common deletion region have murine homologs in the sh2 region (Probst et al. 1998). The gene order was not completely conserved secondary to two independent genomic inversions. However, the order of the seven genes within the critical region was apparently conserved between humans and mice.
To identify potential causative gene(s) of the SMS phenotype and to facilitate the construction of an SMS mouse model, we constructed a large insert clone contig of the SMS common deletion region and its syntenic region in the mouse. Here we report the comparative genomic analysis between humans and mice, comparison of the human genetic and physical maps, delineation of the SMS critical region (SMCR), and the identification and characterization of genes in both human SMCR and the mouse syntenic region.
RESULTS
Construction of the BAC/PAC Contig of the SMS Common Deletion and Its Syntenic Region in the Mouse
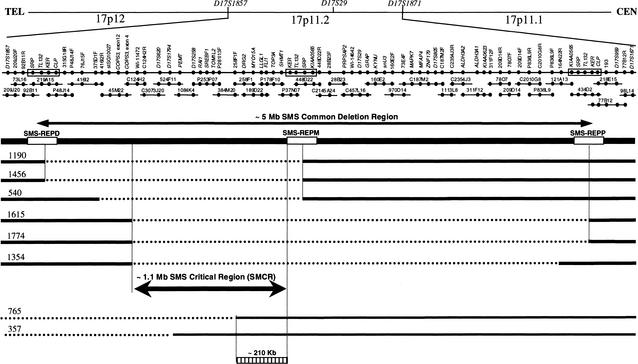
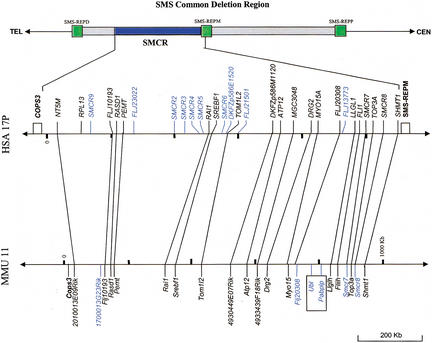
We constructed a BAC/PAC contig covering the SMS common deletion region with a minimal tilting path of 30 clones (Fig. 1). Three low-copy repeat gene clusters were identified previously inside and flanking the SMS deletion region (Chen et al. 1997). Clones specific to an individual SMS-REP were identified based on cis-morphisms, sequence differences among repeats on the same chromosome (Park et al. 2002). From the DNA sequence of these clones, we deduced that the size of the common deletion region including the three SMS-REPs is ∼3.7 Mb and the sizes of the SMS-REPs are between 176 and 256 kb. The middle SMS-REP is inverted with respect to the direct orientation of the proximal and distal copies (Park et al. 2002).
Figure 1.
The Smith-Magenis critical region (SMCR) refined by breakpoint studies of patients with deletions in 17p11.2. The SMS common deletion region falls between D17S959 and D17S1857, including SMS-REPs. Above are shown genetic markers and the cytogenetic bands on 17p. TEL represents telomeric orientation, and CEN represents centromeric orientation. The minimum BAC/PAC tiling path of the SMS common deletion region is shown toward the top of the figure, with STS-content markers represented by dots and BAC/PAC clones represented by horizontal bars. Clones without a prefix are BACs from RPCI-11; those with prefix P are PAC clones; those with prefix C are CTD clones. BAC end sequences were used as markers with R representing the BAC end sequence derived from the Sp6 primer, and F representing the sequence from the T7 primer. Above the BAC contig are listed the individual genes and genetic markers from this region. Below, patients are identified by number. The deleted region is indicated by dashed lines, whereas bold lines refer to genomic sequences retained. The distal breakpoints of patients 357 and 765 are outside the SMS common deleted region. The refined ∼1.1-Mb SMCR (double-edged bold arrow) and the ∼210 kb (hatched box) inside the SMCR, but not deleted in patient 765, are indicated.
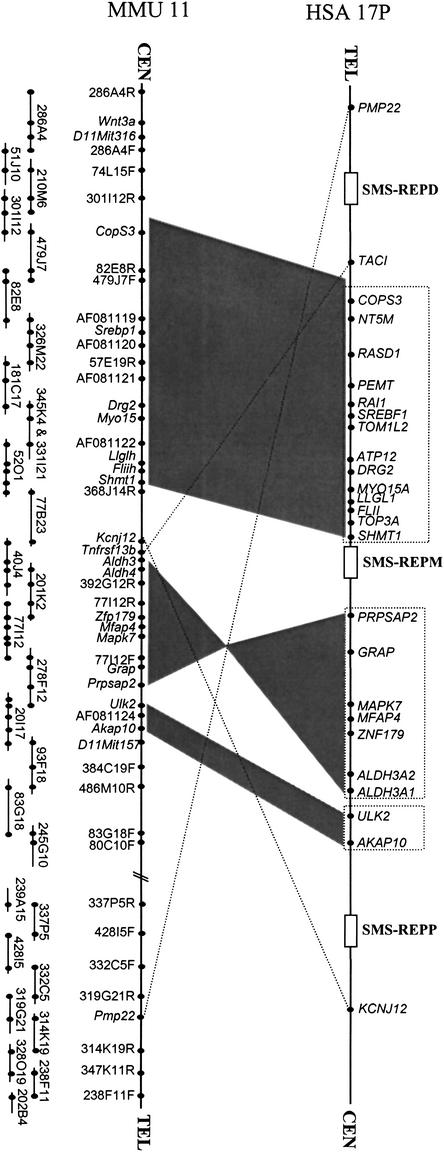
We also constructed a complete BAC contig covering the syntenic mouse region (Fig. 2). The DNA sequence from these BACs indicated that the SMS-REPs are not present, consistent with previous hybridization-based observations (Chen et al. 1997; Probst et al. 1999). The 28 genes that mapped within or around the common deletion region of SMS have homologs on the syntenic region in mouse chromosome 11. Comparisons between humans and mice (Fig. 2) confirmed previous observations that gene order is conserved within the genomic interval flanked by the distal and middle SMS-REPs (Probst et al. 1999). In addition, we found that the order of genes ULK2 and AKAP10 is also conserved. The gene order between COPS3 and AKAP10 is conserved with the exception of an inversion between PRPSAP2 and ALDH3A1. In contrast, the gene TACI is located centromeric to distal SMS-REP, whereas KCNJ12 is centromeric to proximal SMS-REP. In the mouse, these genes are adjacent and located in the middle of the syntenic SMS region. PMP22, the gene responsible for Charcot-Marie-Tooth disease type 1A (CMT1A) and hereditary neuropathy with liability to pressure palsies (HNPP; Lupski 1999), mapped telomeric to distal SMS-REP, but it is located in an inverted position with respect to the syntenic SMS region in mice. Thus, during evolution disruptions of linkage conservation apparently occurred in the genomic region around the SMS-REP low-copy repeats.
Figure 2.
Comparison of the gene order in the human SMS common deletion region and its mouse syntenic region. Genes within the human SMS region on 17p11.2 are shown above. Open boxes represent the SMS-REPs. Below is shown the minimum BAC tiling path of the mouse syntenic region of the SMS common deletion interval and its flanking region. Each BAC clone is represented by a horizontal bar with STS-content markers represented by dots. Blocks of genes that show linkage conservation (i.e., identical gene order) in humans and mice are boxed and connected via gray shading.
Comparison between the Physical and Genetic Maps
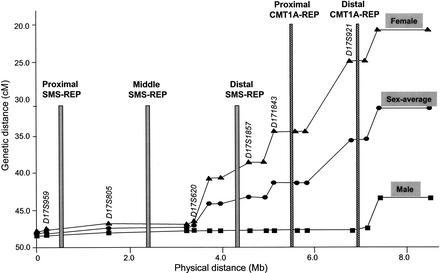
We previously identified a striking difference in recombination rates between the sexes in the CMT1A genomic region (Inoue et al. 2001), with reduced recombination noted for the male meiotic map. De novo CMT1A duplication occurs 10 times more frequently in male gametogenesis than in female germ cells (Palau et al. 1993). We hypothesized that reduced recombination may increase unequal crossing over resulting in an increased propensity to generate unequal reciprocal recombination products (Inoue et al. 2001).
We extended the genetic/physical map correlation over an ∼8.0-Mb region including both the CMT1A and SMS regions (Fig. 3). Reduced recombination is observed in both genders for most of the SMS region. Interestingly, no parent-of-origin frequency differences are observed for de novo SMS deletion (Greenberg et al. 1991; Juyal et al. 1996).
Figure 3.
Comparison between genetic and physical maps. The STR genetic markers from Marshfield are aligned to the sequence-based physical map. The marker order is as following: D17S1871, D17S959, D17S805, D17S1794, D17S620, D17S740, D17S2196, D17S1857, D17S953, D17S1843, D17S793, D17S918, D17S921, D17S1856, D17S947, and D17S1803 (markers within the SMS region are underlined). The three SMS-REPs are indicated by gray bars, the two CMT1A-REPs by hatched bars. Reduced recombination in both sexes was observed for most of the SMS region.
Refining the Smith-Magenis Syndrome Critical Region (SMCR)
Somatic cell hybrid analysis revealed that the breakpoints in most patients with SMS were located within the distal and proximal SMS-REPs (Chen et al. 1997; data not shown). We used an SMS junction fragment identified by PFGE analysis to distinguish SMS patients with the common deletion from those with an unusual-sized deletion. The breakpoints for these latter SMS patients were analyzed by FISH of patient lymphoblasts using BAC/PAC DNA probes; the results are summarized in Figure 1.
Smaller-sized deletions within 17p11.2 were identified in six patients who have typical behavioral and physical features consistent with SMS (Table 1). The deleted region for two SMS patients, 1190 and 1456, was mapped between the distal and middle SMS-REPs (Fig. 1). Furthermore, the distal deletion breakpoints for SMS patients 1615, 1774, and 1354 were mapped proximal to COPS3, a gene located within the genomic interval between the distal and middle SMS-REPs (Fig. 1). We therefore conclude that the genomic interval between COPS3 and the middle SMS-REP is the critical region for the major SMS features of mental retardation, craniofacial and behavioral abnormalities, and sleep disturbance. We propose that the dosage-sensitive genes responsible for the SMS phenotype are located inside this newly defined SMCR.
Table 1.
Clinical Features of Patients with Atypical Deletions within 17p11.2
| Featuresa | SMSb | Not SMS | ||||||
|---|---|---|---|---|---|---|---|---|
| 540 | 1190 | 1456 | 1354 | 1615 | 1774 | 765 | 357 | |
| Craniofacial | ||||||||
| Midfacial hypoplasia | + | + | + | + | + | + | + | − |
| Brachycephaly | − | + | − | − | + | + | + | − |
| Broad face | − | + | + | + | + | + | − | − |
| Abnormal ears | − | + | + | − | − | + | − | − |
| Down-turned mouth | + | + | + | + | + | + | − | − |
| Skeletal | ||||||||
| Short, broad hands | + | + | − | + | + | − | + | − |
| Short stature | + | + | + | + | − | + | − | − |
| Neuro/behavioral | ||||||||
| Mental retardation | + | + | + | + | + | + | + | + |
| Speech delay | + | + | + | + | + | + | + | + |
| Motor delay | − | + | + | + | + | + | + | + |
| Sleep disturbance | + | + | + | + | + | + | − | − |
| Self-destructive | + | + | − | + | + | + | − | − |
| Aggressive/tantrums | + | + | + | + | + | + | − | − |
| Self-hugging or hand clasp | ? | + | − | + | ? | − | − | − |
| Otolaryngologic | ||||||||
| Middle ear | + | − | + | + | − | + | − | − |
| Palatal anomaly/dysfunction | − | − | + | + | − | − | − | NA |
| Age at evaluation | 15 y | 26 y | 7 y | 7 y | 6 y | 4 y | 17 y | 6 y |
| Gender | M | M | F | F | F | M | M | M |
Found in >70% of patients.
+, feature present; −, feature absent; NA, not assessed.
Molecular analyses of three patients, 540, 357, and 765, have been reported (Juyal et al. 1996; Elsea et al. 1997). By our clinical analysis, patients 765 and 357 do not have SMS because they do not show self-destructive behavior, sleep disorder, or characteristic SMS facies (Table 1). Mapping of the deletion breakpoints in these patients by PCR analysis of hybrid DNA revealed that the deleted region for patient 765 is located distal to DRG2. Thus, the genomic region responsible for the SMS phenotype may be only ∼210 kb in size (Fig. 1). However, this conclusion is based on the absence of an SMS phenotype in one patient (765) only.
Sequences of the SMCR and Its Syntenic Region in the Mouse
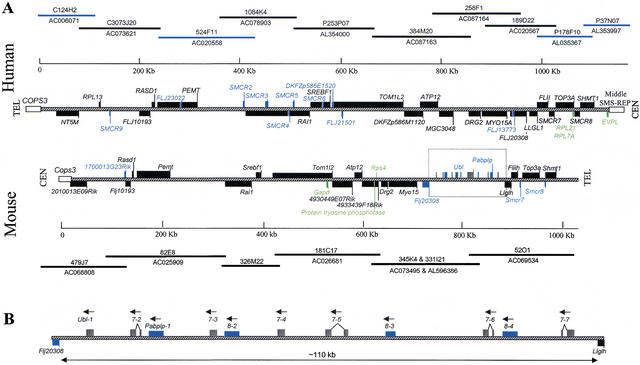
Ten BAC/PAC clones cover the entire SMCR (Fig. 4). The location of individual clones was confirmed by FISH on lymphoblast chromosomes derived from SMS patients with common deletions. The genomic sequence assembly using public (NCBI, http://www.ncbi.nlm.nih.gov) and private (Celera, http://www.celera.com) genome databases revealed that the size of entire SMCR from the putative distal end of the middle SMS-REP to the promoter of COPS3 is ∼1.1 Mb.
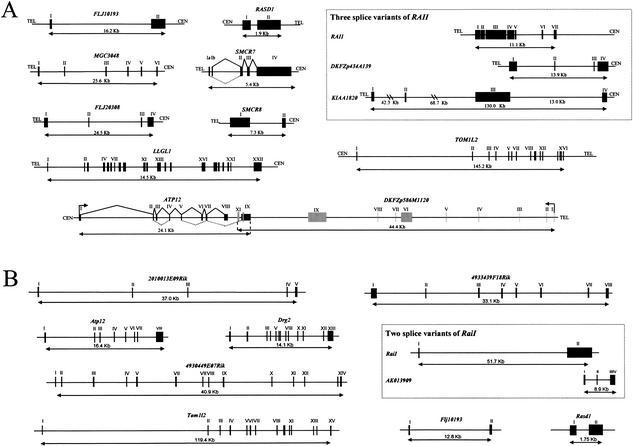
Figure 4.
Transcript map of the SMCR and its mouse syntenic region. The BAC and PAC clones are identified by name and accession number. (A) Clones without a prefix are BACs from RPCI-11 (human) or from RPCI-23 (mouse); those with prefix P are PAC clones; those with prefix C are CTD clones. The hatched line represents the SMCR, and the open boxes represent the flanking sequences of the SMCR: COPS3 and the middle SMS-REP. The position 0 on the size scale is assigned to the nucleotide proximal to the putative promoter of the COPS3 gene. Genes inside SMCR are represented by black boxes, predicted genes by blue boxes, and pseudogenes by green boxes. The predicted genes Ubl and Pabplp are located between Flj20308 and Llglh, and both are present as a gene cluster. To distinguish from Pabplp, gray boxes were used to represent the Ubl. Human genes transcribed from centromere to telomere are located above the central line, and genes transcribed from telomere to centromere are below the line. The mouse genes are drawn in the reciprocal manner (i.e., genes transcribed from telomere to centromere are above the central line). (B) The structure of the ∼110-kb gene clusters between Flj20308 and Llghl that is boxed in A. Seven copies of Ubl and four copies of Pabplp are intermixed. Solid lines connecting exons represent the splicing pattern. All copies are potentially transcribed in the same direction (arrows). The Ubl gene appears to have four copies with spliced variants.
RepeatMasker identified interspersed repeats that account for 42.19% of the SMCR. The repetitive elements include 20.54% Alu sequences and 6.19% LINE1 sequences, similar to that of chromosome 22, but different from chromosome 21, which contains 9.48% Alu and 15.51% LINE1 sequences, and the CMT1A/HNPP region in 17p12, that contains 9.97% Alu and 13.43% LINE1 elements (Dunham et al. 1999; Hattori et al. 2000; Inoue et al. 2001).
Seven BAC clones cover the mouse region syntenic to SMCR (Fig. 4; NCBI). Genome sequences from the private mouse database (Celera) were initially used to assemble some unordered pieces in the public database. However, subsequent analysis of the public sequence database revealed more robust sequence (fewer small gaps) than the Celera database. The mouse genome sequence is ∼1.0 Mb with interspersed repeats accounting for only 27.6%. This ∼15% decrease in repetitive sequences in the mouse genome as compared to the human has also been observed in other genomic regions (Amid et al. 2001).
Transcript Map of Human SMCR and Its Syntenic Region in the Mouse
Potential genes inside the SMCR and the mouse syntenic region were identified through a combination of sequence similarity searches and sequence analysis using gene prediction programs. The potential genes were categorized into three groups: (I) genes, (II) predicted genes, and (III) pseudogenes, using definitions that we employed previously (Inoue et al. 2001).
Within the ∼1.1-Mb SMCR, 30 genes were identified, including 20 genes (Group I), 10 predicted genes (Group II), and 3 pseudogenes (Group III; Fig. 4; Table 2). Thus, the gene density (1 gene per 37 kb) is much higher than the average calculated for the complete human genome (1 gene per 90 kb). The human SMCR genes are unevenly distributed; 23 genes are in the ∼730-kb interval between the middle SMS-REP and SMCR2, and only 7 genes are in the remaining ∼400-kb segment (Fig. 4).
Table 2.
Putative Genes Identified within the SMCR
| Group | Gene name | Accession number | Exons | mRNA (bp) | ORF (aa) | Domains | Expression profiles | Referencea |
|---|---|---|---|---|---|---|---|---|
| I. Genes | NT5M | NM_020201 | 5 | 1617 | 228 | NA | N/1.4, heart, brain, and muscle | Rampazzo et al. 2000 |
| RPL13 | NM_000977 | 1 | 1110 | 211 | NA | RT/ubiquitous | ||
| FLJ10193 | NM_018019 | 2 | 2222 | 146 | NA | N/2.4, ubiquitous | ||
| RASD1 | NM_016084 | 2 | 1740 | 281 | RAS, etc. | N/5.0, all, predominantly in heart | Tu et al. 1999 | |
| PEMT | NM_007169 | 9 | 1007 | 199 | NA | N/1.0, liver | Shields et al. 2001 | |
| RA11 | NM_030665 | 7 | 5667 | 1862 | NA | N/8.0, all, 1.5-placenta, 10.0-weak | Seranski et al. 2001 | |
| SREBF1 | NM_004176 | 22 | 4154 | 1147 | HLH | N/4.0, all, predominantly in liver, adrenal gland | Yokoyama et al. 1993 | |
| TOM1L2 | AF467441 | 16 | 1903 | 475 | VHS, GAT | N/6.0, 2.4, all, predominantly in heart, sk. mus.; RT/fetus (+) | ||
| DKFZp586M1120 | AL136926 | 11 | 5822 | 225 | LRR | N/1.8, predominantly in kidney; RT/fetus (+) | ||
| ATP12 | AF052185 | 8 | 1320 | 289 | NA | N/1.4, all, weak in brain, placenta, lung; RT/fetus (+) | ||
| MGC3048 | NM_024052 | 6 | 816 | 217 | NA | N/4.4, ubiquitous | ||
| DRG2 | NM_001388 | 13 | 1880 | 365 | GTP-OBG, etc. | N/1.4, 2.0, all, high level in sk. mus., heart, and kidney | Li et al. 2000 | |
| MYO15A | NM_016239 | 66 | 11876 | 3530 | MyTH4, etc. | N/12.0, pituitary gland | Liang et al. 1999 | |
| FLJ20308 | AK000315 | 4 | 2371 | 378 | NA | N/3.5, 4.0, ubiquitous; RT/fetus (+) | ||
| LLGL1 | D50550 | 22 | 3806 | 1032 | NA | N/4.4, all, more abundantly in brain and testis | Koyama et al. 1996 | |
| FL11 | NM_002018 | 30 | 14131 | 1269 | GEL, LRR | N/4.4, predominantly in sk. mus. | Campbell et al. 1997 | |
| SMCR7 | AF467443 | 4 | 2455 | 454 | NA | N/2.4, 3.4, all, predominantly in heart, sk. mus., RT/fetus (+) | ||
| TOP3A | U43431 | 19 | 3755 | 1001 | TOP | N/7.2, 6, 4, ubiquitous | Fritz et al. 1997 | |
| SMCR8 | AF467440 | 2 | 3206 | 787 | LBP/BPI/CETP | N/2.8, 3.0, 6.5, all; RT/fetus (+) | ||
| SHMT1 | XM_039898 | 13 | 1867 | 483 | SHMT | N/2.3, 2.1, predominantly in liver, kidney | Girgis et al. 1998 | |
| II. Predicted genes | SMCR9 | NA | 3 | 543 | 180 | PDZ | RT/adult (+), fetus (+) | |
| FLJ23022 | NM_025051 | 1 | 2145 | 147 | NA | RT/adult (+), fetus (+) | ||
| SMCR2 | AI821758 | 4 | 2255 | NA | NA | RT/adult brain, leukemia, fetus (+) | ||
| SMCR3 | AA609047 | 1 | 3235 | NA | NA | RT/adult (+), fetus (+) | ||
| SMCR4 | BG992883 | 3 | 502 | NA | NA | RT/adult (+), fetus (+) | ||
| SMCR5 | AF467442 | 1 | 2843 | 140 | NA | RT/ubiquitous | ||
| SMCR6 | BF511382 | 3 | 588 | NA | NA | RT/ubiquitous | ||
| DKFZp586E1520 | AL133641 | 1 | 1328 | 136 | NA | RT/ubiquitous | ||
| FLJ21501 | AK025154 | 1 | 3203 | NA | NA | RT/ubiquitous | ||
| FLJ13773 | AK023835 | 1 | 4512 | NA | NA | RT/adult (+), fetus (+) | ||
| III. Pseudogenes | EVPL | |||||||
| RPL7A | ||||||||
| RPL21 |
References for gene expression profile. Alternative splicing was observed for genes PEMT, RAII, SREBF1, ATP12, SMCR7, DRG2, and SHMT1. The gene description for them is about the major transcripts. NA, not available. For the gene expression profile, N means data from Northern blotting, and the numbers after the N are the sizes of transcripts in kilobases; RT means data from RT-PCR analyses.
We identified 16 genes (Group I), 6 predicted genes (Group II), and 3 pseudogenes (Group III) in the mouse region syntenic to SMCR (Fig. 4; Table 3); 8 fewer than in the human. Similar to the gene distribution in the human SMCR, the gene density in the region between Shmt1 and Rai1 is higher than in the remaining portion. Comparison of human and mouse sequences indicated that 19 genes in the human SMCR have orthologs in the mouse syntenic region, with conservation of both the order and the orientation, and the same numbers of pseudogenes are present (Fig. 5). Homology is higher across the exons, but extends to some introns and intergenic regions.
Table 3.
Putative Genes Identified within the Mouse Syntenic Region of the SMCR
| Group | Gene names | Accession number | Exons | mRNA (bp) | ORF (aa) | Expression profiles | Referencea | Human homologs |
|---|---|---|---|---|---|---|---|---|
| I. Genes | 2010013E09Rik | AK008226 | 5 | 1271 | 111 | N/1.8, all; RT embryo (+) | NT5M | |
| Flj10193 | AF467888 | 2 | 740 | 142 | N/4.0, 2.5, 1.3, all; RT/embryo (+) | FLJ10193 | ||
| Rasd1 | NM_009026 | 2 | 1623 | 280 | N/2.1, heart, brain, kidney, and liver | Kemppainen and Behrend 1998 | RASD1 | |
| Pemt | NM_008819 | 7 | 600 | 200 | N/1.0, only liver after birth | Walkey et al. 1996 | PEMT | |
| Rai1 | NM_009021 | 2 | 7222 | 1840 | RT/adult (+), before embryo 10 dpc (+) | RAII | ||
| Srebf1 | AF374266 | 18 | 3570 | 1076 | RT/ubiquitous | SREBF1 | ||
| Tom112 | AF467887 | 15 | 1678 | 507 | N/all, with 7 splice variants; RT/embryo (+) | TOM1L2 | ||
| 4930449E07Rik | AK015430 | 14 | 1860 | 523 | N/2.2, predominantly in testis; RT/embryo at 15 dpc (+) | DKFZp586M1120 | ||
| Atp12 | BC013607 | 8 | 1813 | 289 | RT/ubiquitous | ATP12 | ||
| 4933439F18Rik | AK017120 | 8 | 1717 | 217 | N/2.0, all except spleen; RT/embryo (+) | MGC3048 | ||
| Drg2 | NM_021354 | 13 | 1713 | 364 | N/2.0, low in sk. mus. | Li et al. 2000 | DRG2 | |
| Myo15 | NM_010862 | 66 | 11769 | 3511 | N/1.2, pituitary gland | Liang et al. 1999 | MYO15A | |
| LIglh | NM_008502 | 22 | 4279 | 1034 | RT/adult (+), embryo (+) | Tomotsune et al. 1993 | LLGLI | |
| Fliih | NM_022009 | 31 | 4012 | 1271 | RT/adult (+), embryo (+) | FLII | ||
| Top3a | AB006074 | 20 | 3741 | 1003 | N/3.8, predominantly in testis | Seki et al. 1998 | TOP3A | |
| Shmtl | AF237702 | 13 | 1771 | 478 | N/2.1, 3.2, liver | Liu et al. 2001 | SHMT1 | |
| II. Predicted genes | 1700013G23Rik | AK005951 | 2 | 293 | NA | RT/testis | ||
| Flj20308 | BI412421 | 3 | 783 | NA | N/6.0, 3.5; ubiquitous | FLJ20308 | ||
| Ubl | NA | 2 | cluster | NA | RT/ubiquitous | |||
| Pabplp | NA | 2 | cluster | NA | RT/testis, weak in embryos 10 dpc and 15 dpc | |||
| Smcr7 | BF582554 | 1 | 694 | NA | N/2.6, heart, liver; RT/embryo after 10 dpc (+) | SMCR7 | ||
| Smcr8 | AK010198 | 1 | 1050 | NA | N/7.5, 3.8-all except sk. mus.; RT/embryo (+) | SMCR8 | ||
| III. Pseudogene | Rps4 | |||||||
| Protein tyrosine phosphatase 1F2 | ||||||||
| Gapd |
References for gene expression profile. NA, not available. For the gene expression profile, N means data from Northern blotting, and the numbers after the N are the sizes of transcripts in kilobases; RT means data from RT-PCR analyses. Reference is for gene expression profile.
Figure 5.
Comparison of the order of the putative genes in the SMCR and the mouse syntenic region. The open boxes represent the flanking sequences of the SMCR: COPS3 and the middle SMS-REP. Genes are in black, and predicted genes are in blue. Genes in the SMCR are connected with their mouse homologs by lines. The region that contains two mouse gene clusters that are not inside the SMCR is boxed. The location of the SMCR in the common deletion region is indicated at the top.
Differences in the gene content between the human SMCR and the mouse syntenic region were also observed. RPL13 and 10 predicted genes within the human SMCR are not present within the mouse syntenic region. In the SMCR no genes were identified in the ∼20-kb genomic sequence between LLGL1 and FLJ20308 (Fig. 4); as defined by RepeatMasker, this region consists of 56% repetitive sequences. In contrast, in the mouse, an ∼110-kb genomic region separates Flj20308 and Llglh, two genes, Ubl and Pabplp, were predicted within this interval (Fig. 4).
Genes Ubl and Pabplp are homologous to ubiquitin and poly(A)-binding protein, respectively. The human ubiquitin gene subfamily consists of primarily processed pseudogenes (Baker and Board 1992). The poly(A)-binding protein is a conserved protein that binds to the 3′ poly(A) tail on mRNAs in eukaryotic cells (Kleene et al. 1994). Both genes are present as intermixed gene repeating units with seven copies for Ubl and four copies for Pabplp (Fig. 4B). Because three copies of each Ubl and Pabplp could be translated without a premature stop codon, we placed both genes in Group II. In addition, another predicted mouse gene, 1700013G23Rik, is also not located within the human SMCR (Fig. 4).
Genes in the SMCR (Group I)
We identified 20 genes within the human SMCR (Table 2). Of the 12 genes mapped in 17p11.2, the genomic structures have been described previously for the following 9: NT5M, PEMT, SREBF1, RAI1, DRG2, MYO15A, FLII, TOP3A, and SHMT1. Here we describe the genomic structures of the other 3 genes: LLGL1, TOM1L2, and ATP12. In addition, two known genes: RASD1 and RPL13, and six unknown genes: FLJ10193, DKFZp586M1120, MGC3048, FLJ20308, SMCR7, and SMCR8, are newly mapped to this region (Fig. 6).
Figure 6.
Genomic structure of the genes in the SMCR (A) and in the mouse syntenic region (B). Exons of DKFZp586M1120 are represented by gray boxes; exons of other genes are represented by black boxes. Alternative splicing is observed for RAI1, ATP12, and SMCR7. Dashed lines connecting exons represent the alternative splicing. Exon 10 of DKFZp586M1120 is located inside exon 6 of the ATP12 splice variant. Arrows indicate the orientation of transcription. The introns 2 and 3 of KIAA1820 are not to scale.
Previously, only seven genes in mice: Pemt, Srebp1, Drg2, Myo15, Llglh, Fliih, and Top3a, were mapped within the SMS syntenic region. Of these, the genomic structure of Drg2 is not known. We describe the gene structure of Rasd1, Rai1, and Drg2, and assemble six unknown mouse genes homologous to human NT5M, FLJ10193, TOM1L2, DKFZp586M1120, MGC3048, and FLJ20308 (Fig. 6).
FLJ10193
The human and mouse FLJ10193 have the same two-exon gene structure (Fig. 6), but the proteins that they encode have only 64% homology, which is the lowest homology among all genes identified in the SMCR. FLJ10193 is expressed ubiquitously in human and mouse tissues and has a predicted proline-rich region (Fig. 7; Tables 4 and 5).
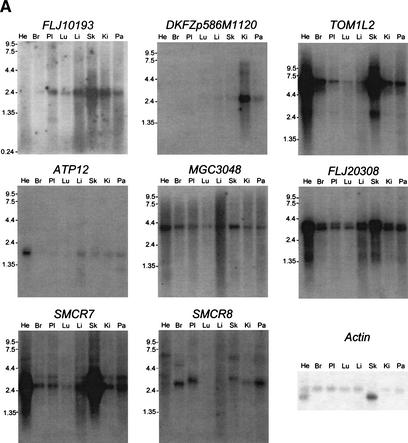
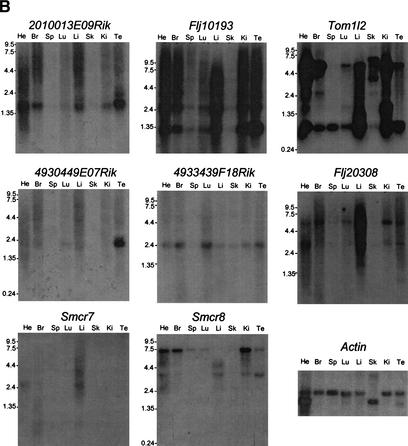
Figure 7.
Northern blotting of the genes in the SMCR (A) and the genes in its mouse syntenic region (B). Tissues are indicated at the top of each lane, and the size markers in kilobases (kb) are the numbers beside the blots. (He) heart; (Br) brain; (Pl) placenta; (Lu) lung; (Li) liver; (Sk) skeletal muscle; (Ki) kidney; (Pa) pancreas; (Te) testis. For mRNA quantity, β-actin was probed as a control. Ubiquitous expression was observed for FLJ10193, MGC3048, FLJ20308, and SMCR7. Tissue-specific various-sized transcripts were observed for TOM1L2 and SMCR8. DKFZp586M1120 is predominantly expressed in the kidney, whereas 4930449E07Rik is in the testis.
Table 4.
RT-PCR Analysis of the Genes within the SMCR
| Gene name | Adult | Fetal | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | H | K | Li | Lu | Pa | Pl | Sm | C | Le | O | Pr | Si | Sp | Te | Th | B | H | K | Li | Lu | Sm | Sp | Th | |
| SMCR9 | + | + | + | + | + | + | + | + | + | − | w | + | + | + | w | w | + | + | + | w | w | w | − | w |
| FLJ10193 | + | + | + | + | + | + | − | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FLJ23022 | + | + | + | + | + | + | − | + | w | + | + | + | + | + | + | + | w | w | + | + | w | w | − | − |
| SMCR2 | + | − | − | − | − | − | − | − | − | + | − | − | − | − | w | − | − | − | − | − | w | − | w | w |
| SMCR3 | − | + | + | w | + | + | w | w | − | + | w | w | − | − | + | − | w | w | + | w | w | w | − | w |
| SMCR4 | − | + | + | w | − | + | − | − | − | − | − | w | − | w | w | + | + | + | + | w | − | − | w | w |
| SMCR5 | + | + | + | + | + | + | w | + | + | + | + | + | + | + | + | + | + | w | + | w | w | w | + | + |
| TOM1L2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w | w |
| SMCR6 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FLJ21501 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | w |
| DKFZp586M1120 | − | − | + | + | + | + | − | − | − | − | w | w | − | − | − | − | w | w | + | − | + | − | w | w |
| ATP12 | + | + | + | + | + | + | + | + | + | + | + | − | + | + | − | + | + | + | + | + | + | + | + | + |
| MGC3048 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FLJ20308 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| FLJ13773 | − | + | + | w | + | + | − | − | w | + | + | + | + | + | + | + | + | + | + | + | + | − | + | w |
| SMCR7 | + | + | + | + | + | + | + | + | + | + | + | w | + | + | w | + | + | + | + | + | + | + | + | + |
| SMCR8 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + |
Adult or fetal tissues used are indicated on the top line. B, brain; H, heart; K, kidney; Li, liver; Lu, lung; Pa, pancreas; Pl, placenta; Sm, smooth muscle; C, colon; Le, leukemia; O, ovary; Pr, prostate; Si, small intestine; Sp, spleen; Te, testis; Th, thymus; Sp, spleen; +, positive expression; w, very weak expression; −, no expression.
Table 5.
RT-PCR Analysis of the Genes within the Mouse Syntenic Region of the SMCR
| Gene name | Adult | Embryo | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | H | K | Li | Lu | Sm | Sp | Te | E7 | E10 | E15 | E17 | |
| 2010013E09Rik | + | + | + | + | + | + | w | + | + | + | + | + |
| 1700013G23Rik | w | w | w | − | w | w | w | + | − | − | − | − |
| Flj10193 | + | + | + | + | + | + | w | + | + | + | + | + |
| Rasd1 | w | w | w | − | w | w | − | + | − | − | − | − |
| RaiI | w | − | w | w | w | w | w | w | + | + | − | − |
| Srebfl | + | + | + | + | + | + | + | + | + | w | w | w |
| Tom1l2 | + | − | + | + | + | + | + | + | + | + | + | + |
| 4930449E07Rik | w | w | − | − | + | − | − | + | − | − | + | + |
| Atp12 | + | + | + | + | + | + | + | + | + | + | + | + |
| 4933439F18Rik | + | + | + | + | + | w | + | + | − | + | + | + |
| Flj20308 | + | + | + | + | + | + | w | + | − | + | + | + |
| Ubl | + | + | + | + | + | + | + | + | − | + | + | + |
| Pabplp | − | − | − | − | − | − | − | + | − | w | w | − |
| Llglh | + | + | − | + | + | + | + | + | + | + | + | + |
| Fliih | + | + | + | + | − | + | + | + | − | + | + | + |
| Smcr7 | + | + | w | + | + | + | − | + | − | + | w | + |
| Smcr8 | + | + | + | + | + | + | + | + | + | + | + | + |
B, brain; H, heart; K, kidney; Li, liver; Lu, lung; Sm, smooth muscle; Sp, spleen; Te, testis; E7, E10, E15, E17, embryo at 7, 10, 15, 17 dpc. +, positive expression; w, very weak expression; −, no expression.
RASD1
Mouse Rasd1 is a member of the RAS superfamily, which was induced rapidly by dexamethasone in AtT-20 cells (Kemppainen and Behrend 1998). RASD1 and Rasd1 have the same genomic structure of two exons separated by a <1-kb intron (Fig. 6).
RAI1
The genomic structure of RAI1 was described recently (Seranski et al. 2001). We found that there are two additional transcripts: KIAA1820 (AB058723) and DKFZp434A139 (AL133649) in an ∼130-kb region, which share part of their coding regions with RAI1 (AJ271790; Fig. 6A). A polyglutamine was observed in RAI1 and KIAA1820 but not in DKFZp434A139. Mouse Rai1 contains two exons corresponding to the exons 2 and 3 of transcript KIAA1820 (Fig. 6B). Mouse transcript AK013909, the homolog of DKFZp586M1120, was also identified (Fig. 6B).
TOM1L2
We assembled TOM1L2 and Tom1l2, the human and mouse homologs of chicken Tom1B, respectively, by multiple EST alignments. Human TOM1L2 shares a similar genomic structure with its mouse homolog (Fig. 6). VHS and GAT domains, which are involved in vesicular trafficking (Lohi and Lehto 1998; Puertollano et al. 2001), are present in both the human and mouse hypothetical proteins. A major 6.0-kb and a minor 2.4-kb transcript were identified in all human tissues examined, with a significantly higher level in the heart and skeletal muscle (Fig. 7A). However, when mouse tissues were analyzed, seven transcripts were observed, and the alternative splicing patterns displayed tissue-specific variations (Fig. 7B).
ATP12
ATP12 is a human homolog of a yeast nuclear gene required for the assembly of the mitochondrial F1-ATPase (Wang et al. 2001). Alternative splicing is observed for exon 3, exon 6, and the last exon (Fig. 6A). The 3′-UTR of ATP12 overlaps with exon 10 of DKFZp586M1120, which encodes the 3′-UTR (Fig. 6A). However, the corresponding mouse genes are not overlapping.
DKFZp586M1120
DKFZp586M1120 encodes a putative 225-amino-acid protein (Table 2), in which five leucine-rich repeats (LRR) were identified. LRR-containing proteins often are involved in protein–protein interactions and cellular adhesion (Rothberg et al. 1990). The putative protein shares 61% homology with PPP1R7, the human homolog of yeast sds22, a mitotic regulator of protein phosphatase-1. A 1.8-kb transcript was observed predominantly in human kidneys and mouse testes (Fig. 7).
MGC3048
A putative 217-amino-acid protein encoded by human MGC3048 has the same size as that encoded by its mouse homolog. The two proteins share significant homology with 93% identity. PipMaker analysis revealed conservation of sequence not only in all exons but also throughout the first intron, which may potentially contain regulatory elements for MGC3048 (data not shown). Ubiquitous expression was observed in both human and mouse tissues (Fig. 7).
FLJ20308
FLJ20308 contains 4 exons encoding a putative 378-amino-acid transmembrane protein (Fig. 6). This gene is well conserved in the mouse with conservation in all exons, introns, and the promoter region (data not shown). Northern blotting identified two transcripts in both human and mouse tissues (Fig. 7). The expression is observed in all tissues, with relatively higher expression in heart and skeletal muscle.
LLGL1
Lethal(2) giant larvae is a Drosophila tumor-suppressor gene. Both human and mouse homologs have been described (Tomotsune et al. 1993; Koyama et al. 1996). The overlapping of the 3′ end of LLGL1 with the 3′ end of FLII is similar to that observed for ATP12 and DKFZp586M1120, but in contrast the LLGL1 and FLII overlap is found in both humans and mice (Campbell et al. 1997, 2000). LLGL1 spans 14.5 kb with 22 exons and is transcribed toward the centromere (Fig. 6A).
SMCR7
SMCR7 was represented by a cluster of more than 20 ESTs, and two splicing variants with different 5′-UTRs were identified (Fig. 6A). The putative 353-amino-acid transmembrane protein shows no homology to proteins with defined functions. PipMaker analysis revealed sequence conservation throughout the coding region, indicating the presence of the mouse homolog in the syntenic region. We have not obtained the full-length mouse gene, but several mouse ESTs have been identified. Two transcripts were detected by Northern blotting on human tissues with a similar expression pattern, a major 2.8-kb transcript and a minor 3.4-kb transcript, confirming alternative splicing for SMCR7 (Fig. 7A). Both transcripts were expressed ubiquitously with a relatively higher expression level in heart and skeletal muscle.
SMCR8
A novel gene, SMCR8, was identified between SHMT1 and TOP3A, with its putative promoter only 286 bp proximal to the promoter of TOP3A. SMCR8 contains two exons encoding a putative 787-amino-acid protein that encompasses an N-terminal domain of the LBP/BPI/CETP family involved in lipid binding (Fig. 6; Beamer et al. 1997). More than 75% homology between human and mouse sequences in its coding region was identified. SMCR8 is expressed in all tissues examined (Tables 4 and 5), and several transcripts were observed in both human and mouse tissues (Fig. 7).
Predicted Genes in the SMCR (Group II)
Ten predicted genes were identified within the human SMCR (Table 2). The existence of these genes is indicated by the presence of multiple ESTs, poly(A), or intron structure, as well as expression in RT-PCR analysis. Each of the predicted genes has no homologs identified in the syntenic region of the mouse. Seven genes are located inside introns of other genes. This potentially explains why these genes have not been identified by gene prediction programs (Fig. 4). RT-PCR showed that each predicted gene is expressed in almost all adult and fetal tissues examined except SMCR2, which was expressed in the brain, fetal brain, spinal cord, and trachea (Table 4; data not shown). SMCR9 contains two putative PDZ domains, a protein–protein interaction domain likely involved in protein clustering and scaffolding (Sheng and Sala 2001).
Pseudogenes (Group III)
Three pseudogenes were identified within the SMCR, including EVPL and two adjacent ribosome protein genes, RPL17 and RPL7A. EVPL is located adjacent to the middle SMS-REP and distal to SHMT1, a region with no conservation to the mouse syntenic region as shown by PipMaker. Also, three pseudogenes, Rps4, Gapd, and Protein tyrosine phosphatase IF2, were identified in the mouse syntenic region.
DISCUSSION
Chromosome 17p11.2 is an unstable genomic region harboring several low-copy repeats associated with genomic disorders (Lupski 1998; Stankiewicz and Lupski 2002). Our comparative analysis between the genomic sequence of the SMS region and the syntenic region on mouse chromosome 11 revealed information regarding genomic architecture. Moreover, we refined the critical region responsible for SMS to a minimum interval. New genes were identified within this refined SMCR as well as its mouse syntenic region.
Physical and Genetic Map Comparisons
We previously hypothesized that the reduced male recombination frequency at the CMT1A locus may increase the unequal crossing over. This was proposed to result from an extended region of allelic chromosomes without synapse formation to provide an anchor and prevent chromosomal slipping. Indeed, the SMS region shows reduced recombination in both sexes, as might be expected because there is no parent-of-origin preference for the de novo deletion. In the SMS case, the reduced recombination may also reflect interference owing to proximity to the centromere. Nevertheless, it will be interesting to determine if reduced recombination is a general feature of genomic regions that undergo nonallelic homologous recombination.
Genomic Architecture Revealed through Comparative Genomics
The percentage of low-copy repeats (LCRs), also termed segmental duplications, in the human genome is greater than in other sequenced genomes, such as the fly and worm (Lander et al. 2001; Venter et al. 2001). Three low-copy repeats SMS-REPs on human chromosome 17p11.2 mediate DNA rearrangements (deletions and duplications) associated with genomic disorders, by nonallelic homologous recombination (Chen et al. 1997; Potocki et al. 2000; Park et al. 2002). Sequence analyses show that all three SMS-REPs within the SMS common deletion are not present in the mouse syntenic region (Fig. 2). Apparently, except for a chromosome inversion of the region between the middle and proximal SMS-REP syntenic region in mouse, the gene order between SMS-REPs is conserved (Fig. 2). Interestingly, transposition occurred for the TACI and KCNJ12 genes adjacent to the SMS-REPs. This rearrangement of gene order might have occurred during the evolution of the SMS-REPs, indicating that segmental duplications might transpose surrounding genes.
Another genome architectural feature revealed by our human/mouse comparative genome analysis is a region containing two intermixed gene repeat units (Fig. 4B). Four of the Ubl copies retain a putative intron. The four Pabplp copies are interspersed among the Ubl copies. How this complex array evolved is not immediately obvious, but this entire segment is absent in the human genome.
Gene Density and Evolution
The gene density of the human SMCR is higher than the estimated average in the human genome. Within an ∼1.1-Mb interval, 19 genes are also present in the same order and orientation in the mouse chromosome (Fig. 5). Highly conserved orthologous regions can contain a high gene density. The number, order, and orientation of all 17 genes in a gene-rich cluster at human 12p13 are conserved between humans and mice (Ansari-Lari et al. 1998). Conservation between these two species was also shown for the distal 700 kb of the Cat eye syndrome (tetrasomy of 22q11.2) critical region and for human chromosome 11p15.3, both of which are gene-rich regions (Amid et al. 2001; Footz et al. 2001). According to the mosaic model of genomic evolution, different portions of the genome evolve at different rates (Koop 1995). Thus, gene-rich regions in the genome might have evolved much more slowly than the gene-poor regions.
None of the 10 predicted genes in the SMCR was found in mice. Most of these predicted genes match multiple homologous ESTs. Their existence was further confirmed by their expression in RT-PCR analysis. These genes might have important functions for silencing gene expression as an antisense RNA (Nellen and Lichtenstein 1993). However, the fact that seven predicted genes are located inside introns of other genes indicates that these predicted genes could represent differentially spliced exons or untranslated exons. Therefore, the absence of these predicted genes in mice could be due to less alternative splicing. In humans, an alternative-spliced last exon of ATP12 overlaps DKFZp586M1120; however, no alternative splicing was observed for mouse Atp12, and the two corresponding mouse homologs are separated from each other.
A Newly Defined SMS Critical Interval
Analyses of the deleted intervals of the SMS patients with smaller-sized deletions enabled refinement of the SMS critical region (Fig. 1). Six SMS patients—540, 1190, 1456, 1354, 1615, and 1774—were determined to harbor unusual deletions in 17p11.2. The same deletions were identified for patients 1190 and 1456; but some SMS features such as brachycephaly; short, broad hands; and self-destructive and self-hugging behavior are present in 1190 but not in 1456. Slightly different phenotypes were also observed for patients 1615 and 1774 with the same deletions. Thus, genetic background and/or stochastic factors may contribute to penetrance of the phenotype. However, because the major SMS features including craniofacial, skeletal, and neuro/behavioral phenotypes were present in two or more patients (Table 1), we determined that the smallest overlap among these patients is the SMCR and that the genes that contribute to the craniofacial, skeletal, and neuro/behavioral features are likely located within this genomic region.
Two patients, 357 and 765, with an unusual deletion in 17p11.2 have atypical clinical features. The deletions for both 357 and 765 include part of the region between the distal SMS-REP and the middle SMS-REP, with the proximal breakpoints located inside the SMCR. Further study of the breakpoint of patient 765 using a somatic hybrid cell line indicated that it is located inside RP11-258F1, ∼210 kb away from the middle SMS-REP. Because patient 765 does not manifest SMS, it is possible that genes responsible for SMS are located within this ∼210 kb (Fig. 1).
Candidate Genes for the SMS Phenotype
To delineate genes that contribute to the SMS features, we identified and characterized genes within the SMCR. SMS patients manifest abnormalities in multiple tissues/organs including: neural (100% mental retardation, 100% self-hugging, 75% peripheral neuropathy, and 69% sleep disorder), eyes (68% iris abnormalities), ears (81% hearing impairment), hearts (29%), kidneys (28%), and skeletal (93% midface hypoplasia and 85% brachydactyly) (Chen et al. 1996). The SMS-causing genes are therefore likely expressed in multiple tissues. With the exception of PEMT and MYO15A, all genes mapped within the SMCR are widely expressed in multiple tissues.
The genes that are responsible for the SMS phenotype are located within the SMCR and are probably dosage-sensitive. One way to evaluate whether a gene manifests haploinsufficiency effects is by the evaluation of animal models, for example, mice. Until now, targeted disruption of five mouse genes: Pemt, Srebp1, Myo15, Top3a, and Fliih, within SMCR has been reported. The normal heterozygous mutant mice indicated that these genes are not haploinsufficient (Walkey et al. 1996; Shimano et al. 1997; Li and Wang 1998; Probst et al. 1998; H. Campbell, pers. comm.) and therefore are less likely to contribute to the SMS phenotypes. However, dosage sensitivity for a given gene in humans does not necessarily correlate with haploinsufficiency in mice. For example, haploinsufficiency of GATA3 is the underlying mechanism for the hypoparathyroidism, sensorineural deafness, and renal anomalies (HDR) syndrome. Mice heterozygous for the Gata3 mutation appear to be normal (Pandolfi et al. 1995; Van Esch and Devriendt 2001). We can also exclude MYO15A because the heterozygous individual carrying this recessive deafness gene does not show SMS (Wang et al. 1998; Liburd et al. 2001).
One potential SMS candidate gene is LLGL1, the human homolog of lethal giant larvae ( Lgl), a tumor-suppressor gene in Drosophila. LLGL1 is expressed ubiquitously, with the most abundant expression in the brain and testis (Koyama et al. 1996). Moreover, Lgl in Drosophila is essential for asymmetric cortical localization of basal determinants in mitotic neuroblasts, and is required for neural fate decisions (Ohshiro et al. 2000; Peng et al. 2000). Another SMS candidate is RAI1. Mouse Rai1 has neuron-specific expression in the brain and is induced by retinoic acid, which is involved in craniofacial development (Imai et al. 1995; Helms et al. 1997; Padmanabhan and Ahmed 1997). Therefore, haploinsufficiency of RAI1 may account for both neuro/behavioral abnormalities and facial abnormalities in SMS. Two GTP-binding proteins were identified within the SMCR: DRG2 and RASD1. DRG2 is a developmentally regulated protein, and is closely related to DRG1 (Li and Trueb 2000). RASD1 is a brain-enriched G protein that is strongly and rapidly induced during treatment with dexamethasone (Kemppainen and Behrend 1998). Both the LRR in DKFZp586M1120 and the PDZ domain in SMCR9 are associated with protein–protein interaction. Genes involved in signal transduction or protein–protein interactions have the potential to show haploinsufficiency effects, because half dosage may affect the balance of protein complexes, and then interfere with a series of related events required for normal development. The three genes FLJ20308, SMCR7, and SMCR8 located within the ∼210-kb region also remain promising SMS candidates, although we have no clues as to their functions. Targeted mutation of these genes will help unravel their potential roles in SMS.
One or More SMS-Causing Genes?
It is unclear whether haploinsufficiency of either one single gene or several contiguous genes causes the characteristic features of SMS as classically proposed (Schmickel 1986; Shaffer et al. 2001). The causative genes for several microdeletion syndromes have been identified. In some syndromes, haploinsufficiency of a single gene is responsible for the entire phenotype. The Rubinstein-Taybi syndrome (RTS), associated with microdeletion of 16p13.3, consists of facial abnormalities, broad thumbs, broad big toes, and mental retardation. Loss of one functional copy of a transcriptional coactivator cyclic AMP response-element-binding protein, (CREB)-binding protein (CBP), underlies all the developmental abnormalities in RTS (Petrij et al. 1995). This was clearly documented by identifying patients with CBP null alleles caused by frameshift or nonsense mutations. Another example of a monogenic microdeletion syndrome is Alagille syndrome, sometimes associated with the deletion of 20p12. The causative gene for all the features of this syndrome is JAG1, which encodes a ligand for Notch1 (Li et al. 1997; Oda et al. 1997).
Because deletions of genomic regions inside 17p11.2 are identified in almost all SMS patients, it is possible that haploinsufficiency of more than one gene underlies the SMS phenotype. In some contiguous gene-deletion syndromes, mutation in one gene accounts for only a portion of the phenotype (Shaffer et al. 2001). For example, deletion of the elastin gene, ELN, leads to vascular stenoses, one of the clinical features of Williams syndrome (WS), a developmental disorder caused by a deletion of band 7q11.23 (Ewart et al. 1993). The specific gene(s) for other features of WS including growth retardation, hypercalcemia, renal anomalies, and mental retardation are still unknown (Ewart et al. 1993). Miller-Dieker syndrome (MDS) is a multiple malformation syndrome characterized by lissencephaly and facial abnormalities (Dobyns et al. 1984). LIS1, the gene inside the MDS critical region, encodes a subunit of the brain platelet-activating factor acetylhydrolase (PAFAH), and is responsible only for brain malformation (Hirotsune et al. 1998; Pilz et al. 1998). The additional features such as facial abnormalities are thought to be caused by other unknown genes. Therefore, to identify the genes involved in manifesting phenotypes of a contiguous gene deletion syndrome, an important step is to determine the finished sequence of the genomic region corresponding to the deleted interval, and identify all the genes within that region.
Conclusions
We have refined the critical SMS interval to an ∼1.1-Mb genomic region and performed gene identification and characterization for this newly defined critical region and its syntenic region in the mouse. Our data provide insights into genome architecture and evolution, and new genomic information for comparative analysis between humans and mice, indicate potential SMS candidate genes, facilitate the identification of the haploinsufficient genes involved in this syndrome, and provide information necessary for engineering a mouse model of SMS.
METHODS
Construction of BAC/PAC Contig
CTD and PAC clones were selected from the NCBI database (http://www.ncbi.nlm.nih.gov) based on their sequence homology with the STS markers mapped inside the SMS common deletion region. The BAC clones were isolated by radioactive filter hybridization of human RPCI-11 and mouse RPCI-23 BAC libraries (BACPAC Resources) using probes to STS markers. BAC end sequences were determined either from the database (http://www.tigr.org) or by direct sequencing. The extent of the genomic clones was determined by STS content mapping. Gaps were filled by multipoint walking based on end sequences of several selected BACs.
Up to 20 oligonucleotide overgo probes (Cai et al. 1998) were included in one hybridization, and positive clones were identified by PCR using primers derived from the parental clone ends. The location of each BAC in the SMCR was further confirmed by FISH on cells from SMS patient lymphoblasts. Human BACs for the minimum tiling paths were submitted to the Whitehead Institute for Biomedical Research at the Massachusetts Institute of Technology. Mouse BACs were submitted to the Human Genome Sequencing Center at Baylor College of Medicine or the Sanger Center for sequencing.
Refining the SMCR
The SMCR was refined by FISH on lymphoblast cell lines derived from SMS patients with unusual smaller deletions. BAC/PAC DNA was prepared using the PSI Clone BAC DNA kit (Princeton Separations, Inc.) or the Plasmid Midi Kit (QIAGEN) according to manufacturers' instructions. The DNA probe (1 μg) was labeled by nick-translation using biotin (Life Technologies–GIBCO BRL) or digoxigenin (Boehringer Mannheim). Biotin was detected with FITC-avidin DCS (Vector labs), and digoxigenin was detected with rhodamine-anti-digoxigenin antibodies (Sigma). Chromosomes were counterstained with DAPI diluted in Vectashield antifade (Vector Labs). Cells were viewed under a Zeiss Axioskop fluorescence microscope.
Informatics
Sequencher 3.1 software (Gene Codes) was used for sequence alignment, DNA translation, and annotation. Human and mouse interspersed repeat sequences were detected and masked using RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker). DNA sequences were separated into ∼50-kb segments and analyzed using the NIX analysis (http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix); an integrated Web-based multiple DNA analysis bioinformatics tool including GRAIL, Fex, Hexon, MZEF, Genemark, GeneFinder, FGENES, Polyah, RepeatMasker, tRNAscan, and BLAST, that searches many databases. Potential genes were further analyzed individually using FGENES (http://genomic.sanger.ac.uk/gf/gfb.html) for gene structure and ORF Finder (http://www.ncbi.nlm.nih.gov/gorf) for translation and ORFs. The putative proteins were analyzed using Pfam 6.6 (http://pfam.wustl.edu), InterPRO (http://www.ebi.ac.uk/interpro) for domains, and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) for transmembrane regions.
DNA and protein sequence similarity was analyzed with BLAST (http://www.ncbi.nlm.nih.gov/BLAST) against the nr, EST, and htgs databases using the default parameters. Human noncontinuous DNA sequences proximal to COPS3 and distal to the middle SMS-REP, corresponding to the SMCR, were repeat-masked and compared with mouse noncontinuous DNA sequences of five BACs (a gap between RP23-82E8 and RP23-181C17 was filled using Celera's genome sequences) using the PipMaker program (http://bio.cse.psu.edu/pipmaker/).PipMaker computes alignments of similar regions in two DNA sequences, and the resulting alignments are summarized with a percent identity plot (PIP; Schwartz et al. 2000).
RT-PCR Analyses and Northern Blotting
Gene expression profiles in human and mouse tissues were analyzed by Northern blotting (Fig. 7) and/or RT-PCR (Tables 4 and 5). RT-PCR analyses were performed using the first-strand cDNA from various adult and fetal tissues (Clontech). Primers were designed using Primer3 (http://www-genome.wi.mit.edu/genome_software/other/primer1.html). Hotstart DNA polymerase (QIAGEN) was used to reduce the amplification of nonspecific PCR products; PCR conditions consisted of 95°C for 15 min, 1 cycle; 95°C for 30 sec, 60°C for 30 sec, 72°C for 1 min, 32 cycles; and a final extension cycle at 72°C. There was no amplification of genomic untranscribed sequences consistent with the absence of genomic DNA contamination in both human and mouse cDNAs. For Northern blotting, probes were designed to DNA sequences of the 3′-UTR of each gene. Radioactive hybridization was performed on multiple tissue blots following the manufacturer's instructions (Clontech).
WEB SITE REFERENCES
http://bio.cse.psu.edu/pipmaker/; PipMaker to compare two or more noncontinuous DNA sequences.
http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker to detect and mask human and mouse interspersed repeat sequences.
http://genomic.sanger.ac.uk/gf/gfb.html; FGENES for gene prediction.
http://pfam.wustl.edu; Pfam 6.6 to analyze proteins.
http://www.celera.com; Celera private database.
http://www.ch.embnet.org/software/TMPRED_form.html; TMpred for transmembrane regions.
http://www.ebi.ac.uk/interpro; InterPRO for domains.
http://www-genome.wi.mit.edu/genome_software/other/primer1.html; Primer3 to design primers.
http://www.hgmp.mrc.ac.uk/Registered/Webapp/nix; NIX for DNA sequence analysis.
http://www.ncbi.nlm.nih.gov; NCBI public database.
http://www.tigr.org; BAC ends database.
Acknowledgments
We appreciate the critical reviews of B.A. Bejjani and N. Katsanis and the technical assistance of M. Withers. We thank B. Birren and K. Dewar of Whitehead Institute for Biomedical Research/MIT Center for Genome Research for contributing many thoughtful discussions. S.-S.P. was supported by a fellowship from the South Korean government, and K.I. by postdoctoral fellowships from the Charcot-Marie-Tooth Association and the Muscular Dystrophy Association. This research was supported in part by grants from the Muscle Dystrophy Association, the National Institute of Child Health and Human Development (P01 HD38420), the National Institute of Neurological Disorders and Stroke (R01 NS27042), and the National Cancer Institute (P01 CA75719).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
While our manuscript was under review, a paper regarding analysis of a human genomic contig and a transcription map of the SMS critical interval was published (Lucas et al. 2001). Comparison of the gene lists within our SMCR indicated that 15 genes or ESTs (NT5M, FLJ10193, RASD1, PEMT, RAI1, SREBF1, TOM1L2, ATP12, MGC3048, DRG2, MYO15A, LLGL1, FLII, TOP3A, and SHMT1) were identified in both papers, and 5 genes (RPL13, DKFZp586M1120, FLJ20308, SMCR7, and SMCR8) and 10 predicted genes (SMCR9, FLJ23022, SMCR2, SMCR3, SMCR4, SMCR5, SMCR6, DKFZp586E1520, FLJ21501, and FLJ13773) described in this paper are not mentioned in the Lucas paper. One gene (EEF1A3) and 4 ESTs (IB1187, stSG8339, stSG9692, and T78887) reported in the Lucas paper are not included in this paper because the information available was not sufficient to identify them as genes.
Footnotes
E-MAIL jlupski@bcm.tmc.edu; FAX (713) 798-5073.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.73702.
REFERENCES
- Amid C, Bahr A, Mujica A, Sampson N, Bikar S-E, Winterpacht A, Zabel B, Hankeln T, Schmidt ER. Comparative genomic sequencing reveals a strikingly similar architecture of a conserved syntenic region on human chromosome 11p15.3 (including gene ST5) and mouse chromosome 7. Cytogenet Cell Genet. 2001;93:284–290. doi: 10.1159/000056999. [DOI] [PubMed] [Google Scholar]
- Ansari-Lari MA, Oeltjen JC, Schwartz S, Zhang Z, Muzny DM, Lu J, Gorrell JH, Chinault AC, Belmont JW, Miller W, et al. Comparative sequence analysis of a gene-rich cluster at human chromosome 12p13 and its syntenic region in mouse chromosome 6. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- Baker RT, Board PG. The human ubiquitin/52-residue ribosomal protein fusion gene subfamily (UbA52) is composed primarily of processed pseudogenes. Genomics. 1992;14:520–522. doi: 10.1016/s0888-7543(05)80258-7. [DOI] [PubMed] [Google Scholar]
- Beamer LJ, Carroll SF, Eisenberg D. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science. 1997;276:1861–1864. doi: 10.1126/science.276.5320.1861. [DOI] [PubMed] [Google Scholar]
- Cai W-W, Reneker J, Chow C-W, Vaishnav M, Bradley A. An anchored framework BAC map of mouse chromosome 11 assembled using multiplex oligonucleotide hybridization. Genomics. 1998;54:387–397. doi: 10.1006/geno.1998.5620. [DOI] [PubMed] [Google Scholar]
- Campbell HD, Fountain S, Young IG, Claudianos C, Hoheisel JD, Chen K-S, Lupski JR. Genomic structure, evolution, and expression of human FLII, a gelsolin and leucine-rich-repeat family member: Overlap with LLGL. Genomics. 1997;42:46–54. doi: 10.1006/geno.1997.4709. [DOI] [PubMed] [Google Scholar]
- Campbell HD, Fountain S, Young IG, Weitz S, Lichter P, Hoheisel JD. Fliih, the murine homologue of the Drosophila melanogaster flightless I gene: Nucleotide sequence, chromosomal mapping and overlap with Llglh. DNA Seq. 2000;11:29–40. doi: 10.3109/10425170009033967. [DOI] [PubMed] [Google Scholar]
- Chen K-S, Potocki L, Lupski JR. The Smith-Magenis syndrome [del(17)p11.2]: Clinical review and molecular advances. MRDD Res Rev. 1996;2:122–129. [Google Scholar]
- Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- Chevillard C, Le Paslier D, Passage E, Ougen P, Billault A, Boyer S, Mazan S, Bachellerie JP, Vignal A, Cohen D, et al. Relationship between Charcot-Marie-Tooth 1A and Smith-Magenis regions. snU3 may be a candidate gene for the Smith-Magenis syndrome. Hum Mol Genet. 1993;2:1235–1243. doi: 10.1093/hmg/2.8.1235. [DOI] [PubMed] [Google Scholar]
- Dobyns WB, Stratton RF, Greenberg F. Syndromes with lissencephaly. I: Miller-Dieker and Norman-Roberts syndromes and isolated lissencephaly. Am J Med Genet. 1984;18:509–526. doi: 10.1002/ajmg.1320180320. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Elsea SH, Purandare SM, Adell RA, Juyal RC, Davis JG, Finucane B, Magenis RE, Patel PI. Definition of the critical interval for Smith-Magenis syndrome. Cytogenet Cell Genet. 1997;79:276–281. doi: 10.1159/000134742. [DOI] [PubMed] [Google Scholar]
- Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, Keating MT. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- Footz TK, Brinkman-Mills P, Banting GS, Maier SA, Riazi MA, Bridgland L, Hu S, Birren B, Minoshima S, Shimizu N, et al. Analysis of the Cat Eye Syndrome critical region in humans and the region of conserved synteny in mice: A search for candidate genes at or near the human chromosome 22 pericentromere. Genome Res. 2001;11:1053–1070. doi: 10.1101/gr.154901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz E, Elsea SH, Patel PI, Meyn MS. Overexpression of a truncated human topoisomerase III partially corrects multiple aspects of the ataxia-telangiectasia phenotype. Proc Natl Acad Sci. 1997;94:4538–4542. doi: 10.1073/pnas.94.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis S, Nasrallah IM, Suh JR, Oppenheim E, Zanetti KA, Mastri MG, Stover PJ. Molecular cloning, characterization and alternative splicing of the human cytoplasmic serine hydroxymethyltransferase gene. Gene. 1998;210:315–324. doi: 10.1016/s0378-1119(98)00085-7. [DOI] [PubMed] [Google Scholar]
- Greenberg F, Guzzetta V, Montes de Oca-Luna R, Magenis RE, Smith ACM, Richter SF, Kondo I, Dobyns WB, Patel PI, Lupski JR. Molecular analysis of the Smith-Magenis syndrome: A possible contiguous-gene syndrome associated with del(17)(p11.2) Am J Hum Genet. 1991;49:1207–1218. [PMC free article] [PubMed] [Google Scholar]
- Greenberg F, Lewis RA, Potocki L, Glaze D, Parke J, Killian J, Murphy MA, Williamson D, Brown F, Dutton R, et al. Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2) Am J Med Genet. 1996;62:247–254. doi: 10.1002/(SICI)1096-8628(19960329)62:3<247::AID-AJMG9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Hattori M, Fujiyama A, Taylor TD, Watanabe H, Yada T, Park H-S, Toyoda A, Ishii K, Totoki Y, Choi D-K, et al. The Chromosome 21 Mapping and Sequencing Consortium. The DNA sequence of human chromosome 21. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- Helms JA, Kim CH, Hu D, Minkoff R, Thaller C, Eichele G. Sonic hedgehog participates in craniofacial morphogenesis and is down-regulated by teratogenic doses of retinoic acid. Dev Biol. 1997;187:25–35. doi: 10.1006/dbio.1997.8589. [DOI] [PubMed] [Google Scholar]
- Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, Clark GD, Ledbetter DH, McBain CJ, Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- Imai Y, Suzuki Y, Matsui T, Tohyama M, Wanaka A, Takagi T. Cloning of a retinoic acid-induced gene, GT1, in the embryonal carcinoma cell line P19: Neuron-specific expression in the mouse brain. Mol Brain Res. 1995;31:1–9. doi: 10.1016/0169-328x(95)00020-s. [DOI] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res. 2001;11:1018–1033. doi: 10.1101/gr.180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juyal RC, Figuera LE, Hauge X, Elsea SH, Lupski JR, Greenberg F, Baldini A, Patel PI. Molecular analyses of 17p11.2 deletions in 62 Smith-Magenis syndrome patients. Am J Hum Genet. 1996;58:998–1007. [PMC free article] [PubMed] [Google Scholar]
- Kemppainen RJ, Behrend EN. Dexamethasone rapidly induces a novel ras superfamily member-related gene in AtT-20 cells. J Biol Chem. 1998;273:3129–3131. doi: 10.1074/jbc.273.6.3129. [DOI] [PubMed] [Google Scholar]
- Kleene KC, Wang M-Y, Cutler M, Hall C, Shih D. Developmental expression of poly(A) binding protein mRNAs during spermatogenesis in the mouse. Mol Reprod Dev. 1994;39:355–364. doi: 10.1002/mrd.1080390403. [DOI] [PubMed] [Google Scholar]
- Koop BF. Human and rodent DNA sequence comparisons: A mosaic model of genomic evolution. Trends Genet. 1995;11:367–371. doi: 10.1016/s0168-9525(00)89108-8. [DOI] [PubMed] [Google Scholar]
- Koyama K, Fukushima Y, Inazawa J, Tomotsune D, Takahashi N, Nakamura Y. The human homologue of the murine Llglh gene (LLGL) maps within the Smith-Magenis syndrome region in 17p11.2. Cytogenet Cell Genet. 1996;72:78–82. doi: 10.1159/000134167. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li B, Trueb B. DRG represents a family of two closely related GTP-binding proteins. Biochim Biophy Acta. 2000;1491:196–204. doi: 10.1016/s0167-4781(00)00025-7. [DOI] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- Li W, Wang JC. Mammalian DNA topoisomerase IIIα is essential in early embryogenesis. Proc Natl Acad Sci. 1998;95:1010–1013. doi: 10.1073/pnas.95.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Wang A, Belyantseva IA, Anderson DW, Probst FJ, Barber TD, Miller W, Touchman JW, Jin L, Sullivan SL, et al. Characterization of the human and mouse unconventional myosin XV genes responsible for hereditary deafness DFNB3 and shaker 2. Genomics. 1999;61:243–258. doi: 10.1006/geno.1999.5976. [DOI] [PubMed] [Google Scholar]
- Liburd N, Ghosh M, Riazuddin S, Naz S, Khan S, Ahmed Z, Riazuddin S, Liang YP, Menon PSN, Smith T, et al. Novel mutations of MYO15A associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with Smith-Magenis syndrome. Hum Genet. 2001;109:535–541. doi: 10.1007/s004390100604. [DOI] [PubMed] [Google Scholar]
- Liu X, Szebenyi DME, Anguera MC, Thiel DJ, Stover PJ. Lack of catalytic activity of a murine mRNA cytoplasmic serine hydroxymethyltransferase splice variant: Evidence against alternative splicing as a regulatory mechanism. Biochemistry. 2001;40:4932–4939. doi: 10.1021/bi002598t. [DOI] [PubMed] [Google Scholar]
- Lohi O, Lehto V-P. VHS domain marks a group of proteins involved in endocytosis and vesicular trafficking. FEBS Lett. 1998;440:255–257. doi: 10.1016/s0014-5793(98)01401-x. [DOI] [PubMed] [Google Scholar]
- Lucas RE, Vlangos CN, Das P, Patel PI, Elsea SH. Genomic organisation of the ∼1.5 Mb Smith-Magenis syndrome critical interval: Transcription map, genomic contig, and candidate gene analysis. Euro J Hum Genet. 2001;9:892–902. doi: 10.1038/sj.ejhg.5200734. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- ————— Charcot-Marie-Tooth polyneuropathy: Duplication, gene dosage, and genetic heterogeneity. Pediatr Res. 1999;45:159–165. doi: 10.1203/00006450-199902000-00001. [DOI] [PubMed] [Google Scholar]
- Nellen W, Lichtenstein C. What makes an mRNA anti-sense-itive? Trends Biochem Sci. 1993;18:419–423. doi: 10.1016/0968-0004(93)90137-c. [DOI] [PubMed] [Google Scholar]
- Oda T, Elkahloun AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Ohshiro T, Yagami T, Zhang C, Matsuzaki F. Role of cortical tumour-suppressor proteins in asymmetric division of Drosophila neuroblast. Nature. 2000;408:593–596. doi: 10.1038/35046087. [DOI] [PubMed] [Google Scholar]
- Padmanabhan R, Ahmed I. Retinoic acid-induced asymmetric craniofacial growth and cleft palate in the TO mouse fetus. Reprod Toxicol. 1997;11:843–860. doi: 10.1016/s0890-6238(97)00068-3. [DOI] [PubMed] [Google Scholar]
- Palau F, Lofgren A, De Jonghe P, Bort S, Nelis E, Sevilla T, Martin J-J, Vilchez J, Prieto F, Van Broeckhoven C. Origin of the de novo duplication in Charcot-Marie-Tooth disease type 1A: Unequal nonsister chromatid exchange during spermatogenesis. Hum Mol Genet. 1993;2:2031–2035. doi: 10.1093/hmg/2.12.2031. [DOI] [PubMed] [Google Scholar]
- Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, Engel JD, Lindenbaum MH. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11:40–44. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- Park S-S, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR. Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res. 2002;12:729–738. doi: 10.1101/gr.82802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CY, Manning L, Albertson R, Doe CQ. The tumour-suppressor genes lgl and dlg regulate basal protein targeting in Drosophila neuroblasts. Nature. 2000;408:596–600. doi: 10.1038/35046094. [DOI] [PubMed] [Google Scholar]
- Petrij F, Giles RH, Dauwerse HG, Saris JJ, Hennekam RCM, Masuno M, Tommerup N, van Ommen G-JB, Goodman RH, Peters DJM, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- Pilz DT, Matsumoto N, Minnerath S, Mills P, Gleeson JG, Allen KM, Walsh CA, Barkovich AJ, Dobyns WB, Ledbetter DH, et al. LIS1 and XLIS (DCX) mutations cause most classical lissencephaly, but different patterns of malformation. Hum Mol Genet. 1998;7:2029–2037. doi: 10.1093/hmg/7.13.2029. [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Park S-S, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, et al. Molecular mechanism for duplication 17p11.2: The homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, et al. Correction of deafness in shaker-2 mice by an unconventional myosin in a BAC transgene. Science. 1998;280:1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- Probst FJ, Chen K-S, Zhao Q, Wang A, Friedman TB, Lupski JR, Camper SA. A physical map of the mouse shaker-2 region contains many of the genes commonly deleted in Smith-Magenis syndrome (del17p11.2p11.2) Genomics. 1999;55:348–352. doi: 10.1006/geno.1998.5669. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Randazzo PA, Presley JF, Hartnell LM, Bonifacino JS. The GGAs promote ARF-dependent recruitment of clathrin to the TGN. Cell. 2001;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- Rampazzo C, Gallinaro L, Milanesi E, Frigimelica E, Reichard P, Bianchi V. A deoxyribonucleotidase in mitochondria: Involvement in regulation of dNTP pools and possible link to genetic disease. Proc Natl Acad Sci. 2000;97:8239–8244. doi: 10.1073/pnas.97.15.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg JM, Jacobs JR, Goodman CS, Artavanis-Tsakonas S. slit: An extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains. Genes & Dev. 1990;4:2169–2187. doi: 10.1101/gad.4.12a.2169. [DOI] [PubMed] [Google Scholar]
- Schmickel RD. Contiguous gene syndromes: A component of recognizable syndromes. J Pediatr. 1986;109:231–241. doi: 10.1016/s0022-3476(86)80377-8. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. PipMaker: A web server for aligning two genomic DNA sequences. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T, Seki M, Katada T, Enomoto T. Isolation of a cDNA encoding mouse DNA topoisomerase III which is highly expressed at the mRNA level in the testis. Biochim Biophys Acta. 1998;1396:127–131. doi: 10.1016/s0167-4781(97)00192-9. [DOI] [PubMed] [Google Scholar]
- Seranski P, Heiss NS, Dhorne-Pollet S, Radelof U, Korn B, Hennig S, Backes E, Schmidt S, Wiemann S, Schwarz CE, et al. Transcription mapping in a medulloblastoma breakpoint interval and Smith-Magenis syndrome candidate region: Identification of 53 transcriptional units and new candidate genes. Genomics. 1999;56:1–11. doi: 10.1006/geno.1998.5647. [DOI] [PubMed] [Google Scholar]
- Seranski P, Hoff C, Radelof U, Hennig S, Reinhardt R, Schwartz CE, Heiss NS, Poustka A. RAI1 is a novel polyglutamine encoding gene that is deleted in Smith-Magenis syndrome patients. Gene. 2001;270:69–76. doi: 10.1016/s0378-1119(01)00415-2. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Ledbetter DH, Lupski JR. Molecular cytogenetics of contiguous gene syndromes: Mechanisms and consequences of gene dosage imbalance. In: Scriver CR, et al., editors. The metabolic and molecular bases of inherited disease. 8th ed. New York: McGraw-Hill; 2001. pp. 1291–1324. [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Ann Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Shields DJ, Agellon LB, Vance DE. Structure, expression profile and alternative processing of the human phosphatidylethanolamine N-methyltransferase (PEMT) gene. Biochim Biophys Acta. 2001;1532:105–114. doi: 10.1016/s1388-1981(01)00122-6. [DOI] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ACM, McGavran L, Robinson J, Waldstein G, Macfarlane J, Zonona J, Reiss J, Lahr M, Allen L, Magenis E. Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am J Med Genet. 1986;24:393–414. doi: 10.1002/ajmg.1320240303. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements, and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stratton RF, Dobyns WB, Greenberg F, DeSana JB, Moore C, Fidone G, Runge GH, Feldman P, Sekhon GS, Pauli RM, et al. Interstitial deletion of (17)(p11.2p11.2): Report of six additional patients with a new chromosome deletion syndrome. Am J Med Genet. 1986;24:421–432. doi: 10.1002/ajmg.1320240305. [DOI] [PubMed] [Google Scholar]
- Tomotsune D, Shoji H, Wakamatsu Y, Kondoh H, Takahashi N. A mouse homologue of the Drosophila tumour-suppressor gene l(2)gl controlled by Hox-C8 in vivo. Nature. 1993;365:69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- Tu Y, Wu C. Cloning, expression and characterization of a novel human Ras-related protein that is regulated by glucocorticoid hormone. Biochim Biophys Acta. 1999;1489:452–456. doi: 10.1016/s0167-4781(99)00197-9. [DOI] [PubMed] [Google Scholar]
- Van Esch H, Devriendt K. Transcription factor GATA3 and the human HDR syndrome. Cell Mol Life Sci. 2001;58:1296–1300. doi: 10.1007/PL00000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Walkey CJ, Cui Z, Agellon LB, Vance DE. Characterization of the murine phosphatidylethanolamine N-methyltransferase-2 gene. J Lipid Res. 1996;37:2341–2350. [PubMed] [Google Scholar]
- Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, et al. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science. 1998;280:1447–1451. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]
- Wang Z-G, White PS, Ackerman SH. Atp11p and Atp12p are assembly factors for the F1-ATPase in human mitochondria. J Biol Chem. 2001;276:30773–30778. doi: 10.1074/jbc.M104133200. [DOI] [PubMed] [Google Scholar]
- Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. SREBP-1, a basic-helix–loop–helix–leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]