Abstract
The small envelope protein (E) plays a role of central importance in the assembly of coronaviruses. This was initially established by studies demonstrating that cellular expression of only E protein and the membrane protein (M) was necessary and sufficient for the generation and release of virus-like particles. To investigate the role of E protein in the whole virus, we previously generated E gene mutants of mouse hepatitis virus (MHV) that were defective in viral growth and produced aberrantly assembled virions. Surprisingly, however, we were also able to isolate a viable MHV mutant (ΔE) in which the entire E gene, as well as the nonessential upstream genes 4 and 5a, were deleted. We have now constructed an E knockout mutant that confirms that the highly defective phenotype of the ΔE mutant is due to loss of the E gene. Additionally, we have created substitution mutants in which the MHV E gene was replaced by heterologous E genes from viruses spanning all three groups of the coronavirus family. Group 2 and 3 E proteins were readily exchangeable for that of MHV. However, the E protein of a group 1 coronavirus, transmissible gastroenteritis virus, became functional in MHV only after acquisition of particular mutations. Our results show that proteins encompassing a remarkably diverse range of primary amino acid sequences can provide E protein function in MHV. These findings suggest that E protein facilitates viral assembly in a manner that does not require E protein to make sequence-specific contacts with M protein.
Coronaviruses are a family of enveloped, positive-sense RNA viruses that infect numerous mammalian and avian species and have gained recent widespread attention due to the emergence of severe acute respiratory syndrome (SARS) (37, 48). Virion assembly of coronaviruses is the culmination of a series of interactions among a minimal set of four structural proteins and the viral genome at the site of budding in the endoplasmic reticulum-Golgi intermediate compartment (12). Three of the structural proteins are membrane bound and become incorporated into the virion envelope. These are the spike protein (S), which initiates infection through attachment to host cell receptors and fusion with host membranes; the membrane protein (M), the major constituent of the envelope; and the small envelope protein (E). In the interior of the virion, the fourth component, the nucleocapsid protein (N), forms a helical nucleocapsid with the RNA genome.
Much of our knowledge about coronavirus assembly has come from studies of virus-like particles (VLPs) produced by coexpression of virion structural proteins (12). The earliest investigations employing such systems demonstrated that coexpression of mouse hepatitis virus (MHV) M and E proteins was necessary and sufficient for VLP formation and release from cells (3, 47). Prior to this discovery, the significance of E protein had not been realized. It was subsequently shown that multiple other coronaviruses—bovine coronavirus (BCoV) (2), transmissible gastroenteritis virus (TGEV) (2), infectious bronchitis virus (IBV) (4, 6), and SARS coronavirus (SARS-CoV) (39)—conformed to the same rule. The only apparent exception to this pattern has been a report that the M and N proteins were necessary and sufficient for SARS-CoV VLP formation (23). M protein is a 25-kDa protein containing three transmembrane segments, with a short amino-terminal ectodomain and a large carboxy-terminal endodomain. M is the most abundant viral structural protein, and envelope formation is thought to largely be driven by M-M monomer interactions. The role of E protein is much less clear.
The E protein is a small polypeptide, ranging from 76 to 109 amino acids (8.4 to 12 kDa), and is a minor constituent of virions. E proteins all have large predicted hydrophobic domains, and it has been shown for MHV (47), IBV (4), and SARS-CoV (31) that E is an integral membrane protein, although it does not contain a cleavable signal sequence (43). The E proteins of IBV (5) and SARS-CoV (31) are palmitoylated on one or more cysteine residues, but it is currently unclear whether the TGEV or MHV E proteins share this modification (17, 43, 54). Also unresolved is the membrane topology of E. An early report suggested a C-exo, N-endo membrane orientation for the TGEV E protein (17). Contrary to this, the IBV E protein was observed to adopt an N-exo, C-endo orientation (4, 55). Different still, evidence obtained with the MHV E protein is consistent with an N-endo, C-endo topology (36, 43), and such a hairpin transmembrane configuration has also been proposed for the SARS-CoV E protein (1, 26, 55). It is thus possible that the E proteins of different coronavirus species do not have a uniform membrane topology or that the orientation of E varies with the protein's level of expression or oligomerization.
Our laboratory has been using genetic methods to study the MHV E protein in the intact virus. In previous work we found that certain charged-to-alanine mutations constructed in the E gene resulted in defective virus growth and in thermolability. The assembled virions of one such mutant had striking defects—elongated and pinched shapes—that were seldom seen among wild-type virions (14). This phenotype was clearly consistent with the critical role for E protein in virion assembly indicated by the VLP studies. It was therefore surprising that we were later able to generate a viable, albeit extremely defective, MHV recombinant (ΔE) in which the E gene, as well as the accessory genes 4 and 5a, were entirely deleted (30). This confirmed that although the E protein is important for MHV assembly, it is not absolutely essential. By contrast, for TGEV it was shown by two independent reverse genetic approaches that if the E gene was knocked out, then viable virus could be recovered only if E protein was provided in trans (8, 41).
In the present study, we investigated whether E proteins from different coronavirus species could functionally replace that of MHV. We found that heterologous E proteins with remarkably divergent amino acid sequences were able to substitute for MHV E. Our results suggest that most coronavirus E proteins play similar roles in virion assembly, without the sequence-specific constraints that would be expected if E protein were required to directly interact with M protein.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The MHV strain A59 wild type and mutants were grown in mouse 17 clone 1 (17Cl1) or L2 cells; plaque assays and plaque purifications were carried out with L2 cells. The feline-murine interspecies chimeric virus fMHV.v2 (18) was grown in feline FCWF cells.
For the generation of antibodies specific for the MHV E protein, the carboxy-terminal tail of E (amino acids 38 through 83) was connected to an amino-terminal hexahistidine tag in the vector pET-28a(+) (Novagen). The bacterially expressed fusion protein was isolated by Ni-nitrilotriacetic acid resin affinity purification (Novagen) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and was used to immunize rabbits. To test antiserum specificity, the MHV E and M genes were inserted into the Semliki Forest virus (SFV) replicon vector pSFV1 (32) for protein expression in L2 cells. Rabbit antipeptide antibodies specific for the IBV E protein (amino acids 96 through 109; PANFQDVQRDKLYS) were provided by Carolyn Machamer (Johns Hopkins University School of Medicine) (4). Rabbit antipeptide antibodies directed against the TGEV E protein (amino acids 61 through 82; AYDAYKNFMRIKAYNPDGALLA) were obtained from Washington Biotechnology, Inc. Monoclonal antibodies J.3.3 and J.1.3 (15), specific for the MHV N and M proteins, respectively, were provided by John Fleming (University of Wisconsin, Madison).
Plasmid constructs.
Transcription templates for viral mutant construction were derived from pMH54 (28), which contains cDNA corresponding to the 5′-most 0.5 kb of the MHV genome fused to the 3′-most 8.6 kb of the MHV genome (see Fig. 1A). All mutations and substitutions were generated via splicing overlap extension-PCR (22) and were inserted into pMH54 by exchange of the segment between the SbfI site following the S gene and either the EcoRV site at the end of the E gene or the EagI site near the start of the M gene. In pLK101, the open reading frame (ORF) for the E protein was knocked out with nine point mutations that disrupted the start codon and created stop codons in all three reading frames immediately downstream.
FIG. 1.
Construction of E protein knockout and substitution mutants of MHV. (A) Transcription vectors used for the synthesis of donor RNA were derived from pMH54 (28), which contains a 5′ segment of the MHV genome (denoted by [1]) fused to a partial hemagglutinin (HE) gene and all genes downstream of HE. The T7 RNA polymerase start site is denoted by an arrow; the locations of restriction sites relevant to plasmid construction (SbfI, EcoRV, and EagI) or template linearization (PacI) are indicated. Shown above the pMH54 schematic are mutations (lowercase) incorporated into pLK101 to create the E knockout mutant. Changes in the first 17 bases of the E ORF eliminated the start codon and generated stop codons in all three reading frames (underlined). Shown beneath the pMH54 schematic are the sequences of the junctions of heterologous E genes substituted for the MHV E gene in transcription vectors pLK99 (BCoV E gene), pLK105 (SARS-CoV E gene), pLK107 (TGEV E gene), and pLK103 (IBV E gene). The 5′ end of every substituted E ORF was a direct AUG-for-AUG exchange. At the 3′ end of every substitution, the last 12 bases of the MHV E ORF were left intact in order to retain the upstream context of TRS6, as detailed in Materials and Methods. TRS6 and the EcoRV site are indicated with solid and broken lines, respectively. (B) Method of selection of E protein mutants by targeted RNA recombination between the interspecies chimeric virus fMHV.v2 (18) and donor RNAs transcribed from transcription vectors shown in panel A. The fMHV.v2 genome encodes the ectodomain region of the FIPV S gene (shaded rectangle) in place of that of the MHV S gene; consequently, fMHV.v2 grows in feline cells but not in murine cells. A single crossover event within the HE gene generates a recombinant that has simultaneously reacquired the MHV S ectodomain and the mutated E gene (solid rectangle). The rearranged order of genes following the S gene in fMHV.v2 rules out the occurrence of undesired secondary crossovers downstream of the S gene (18). Recombinants are selected as progeny that have reacquired the ability to grow in murine cells.
Heterologous E ORFs were substituted in pMH54 by an exact exchange of start codons, at their upstream ends, and in such a manner at their downstream ends as to preserve the context of the transcription-regulating sequence (TRS6) for the M gene. PCR templates for the substitution mutants were cDNA clones obtained from David Brian (University of Tennessee) for the BCoV and TGEV E genes and from Carolyn Machamer (Johns Hopkins University School of Medicine) for the IBV E gene. The SARS-CoV E gene was amplified from a cDNA clone made from infected cell RNA provided by Jill Taylor (Wadsworth Center). The MHV-A59 E ORF sequence in this study was identical to that found under GenBank accession number AY700211. The BCoV strain Mebus E ORF was the same as that under GenBank accession number U00735, except that we altered the last residue (V84I) in plasmid pLK99 in order to retain a nucleotide sequence context upstream of TRS6 identical to that in the MHV genome (see Fig. 1A). In addition, synonymous changes were made in the penultimate codon (GAC to GAU) and the stop codon (UAG to UAA). The SARS-CoV strain Urbani E ORF was the same as that under GenBank accession number AY278741, except for a single residue difference (L37H), which corresponds to the consensus sequence of our laboratory stock of SARS-CoV Urbani. The TGEV strain Purdue E ORF was identical to that under GenBank accession number AJ271965. The IBV strain M42 E ORF was the same as that found under GenBank accession number AY692454, except for a single residue difference (L73I). For the substituted SARS-CoV, TGEV, and IBV E genes in pLK105, pLK107, and pLK103, respectively, the E ORF was placed upstream of the EcoRV site in order to preserve 10 nucleotides (nt) of MHV genomic sequence upstream of TRS6 (see Fig. 1A).
Oligonucleotides for PCR and DNA sequencing were obtained from Integrated DNA Technologies. For every constructed vector, overall plasmid composition was monitored by restriction analysis, and the DNA sequences of all ligation junctions and all regions created by PCR amplification were verified by automated sequencing.
Mutant construction by targeted RNA recombination.
Mutants of MHV containing E gene modifications were isolated by targeted RNA recombination, using a host range-based selection described previously (see Fig. 1B) (27, 28, 38). In short, monolayers of feline FCWF cells were infected with the interspecies chimeric coronavirus fMHV.v2, which is an MHV recombinant containing the ectodomain of the S protein of feline infectious peritonitis virus (FIPV) (18). Infected cells were then transfected with capped synthetic donor RNA by electroporation (Gene Pulser II; Bio-Rad). Donor RNAs were synthesized by in vitro transcription with T7 RNA polymerase (mMessage mMachine; Ambion) using PacI-truncated plasmid templates. The infected and transfected feline cells were plated into 10-cm2 wells, and released progeny virus was harvested at 30 h postinfection at 37°C. Recombinant candidates were selected and purified by two rounds of plaque titration on mouse L2 cell monolayers at 37°C.
For analysis of recombinant candidates, total RNA was extracted from infected 17Cl1 cell monolayers with the Ultraspec reagent (Biotecx), and reverse transcription (RT) of RNA was carried out with a random hexanucleotide primer (Roche) and avian myeloblastosis virus reverse transcriptase (Life Sciences). To confirm the presence of incorporated mutations or genes, PCR amplifications of cDNAs were performed with AmpliTaq polymerase (Roche) using primer pairs flanking the E-gene region. RT-PCR products were analyzed directly by agarose gel electrophoresis and were purified with Quantum-prep columns (Bio-Rad) prior to automated sequencing.
TGEV E protein gain-of-function mutants were isolated as intermediate- to large-plaque variants that arose following two to four passages of independent plaques of the original TGEV E-gene substitution mutant. The loci of new mutations were determined by sequencing of RT-PCR products spanning the entire M and E genes of each mutant. A number of candidate gain-of-function mutations were reconstructed by splicing overlap extension-RT-PCR amplification of E genes from infected cell RNA, followed by cloning of the resulting product between the SbfI and EagI sites of pMH54. Substituted TGEV E genes containing candidate gain-of-function mutations were then transferred to MHV via targeted RNA recombination.
Virus purification.
Wild-type and IBV E substitution mutant viruses were purified exactly as described previously (52) by polyethylene glycol precipitation followed by two cycles of equilibrium centrifugation on preformed gradients of 0 to 50% potassium tartrate osmotically counterbalanced with 30 to 0% glycerol, in a buffer of 50 mM Tris-maleate (pH 6.5) and 1 mM EDTA. For further purification, a modification of a previously reported sucrose flotation procedure was used (28, 47). Virus in 0.5 ml of phosphate-buffered saline (PBS) was mixed with 1.25 ml of 70% sucrose in TM buffer (20 mM Tris-HCl, 20 mM MgCl2, pH 7.2) to give a final concentration of 50% sucrose. Each virus-sucrose mixture was transferred to the bottom of an SW60 centrifuge tube, and above it layers of 1 ml of 48% sucrose in TM, 0.5 ml of 40% sucrose in TM, and 0.5 ml 30% sucrose in TM were added. Samples were centrifuged at 133,000 × g in a Beckman SW60 rotor at 4°C for 46 h. The top 1 ml of each tube was collected and diluted with PBS, and virus was pelleted by centrifugation at 151,000 × g in a Beckman SW41 rotor at 4°C for 3 h; the virus pellet was then dissolved in SDS-PAGE sample buffer.
Western blotting.
To generate a sufficiently high-titer stock of the TGEV E-gene substitution mutant for subsequent infections for Western blot analysis, we grew this mutant and a subset of reconstructed TGEV E gain-of-function mutants in 17Cl1 cells expressing the MHV E protein. This cell line was generated using the plasmid pTetON-IRES-MHV-E, in which the MHV E gene was linked to the encephalomyocarditis virus internal ribosome entry site (IRES) and inserted into the vector pTet-On (Clontech). pTetON-IRES-MHV-E, which also contains the neomycin/kanamycin resistance gene, was provided by Volker Thiel (Cantonal Hospital, St. Gallen, Switzerland). Cells constitutively expressing the MHV E protein were selected on the basis of resistance to Geneticin (Invitrogen).
For the preparation of cell lysates, confluent 25-cm2 monolayers of regular (not E-expressing) 17Cl1 cells were mock infected or were infected with wild-type MHV or one of the constructed mutants. Cells were then incubated at 37°C for 12 to 14 h. Monolayers were washed twice with PBS and then lysed by addition of 600 μl of 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.0% Nonidet P-40, 0.7 μg/ml pepstatin, 1.0 μg/ml leupeptin, 1.0 μg/ml aprotinin, and 0.5 mg/ml Pefabloc SC (Roche). Lysates were held for 5 to 15 min on ice and were then clarified by centrifugation. Samples of infected-cell lysates were separated by SDS-PAGE through 15% or 17.5% polyacrylamide and were transferred to polyvinylidine difluoride membranes. Blots were then probed with various antibodies, and bound primary antibodies were visualized using a chemiluminescence detection system (ECL; Amersham).
RESULTS
Expression of E protein is critical for MHV replication.
In a prior study, we reported the generation of a viable MHV recombinant, ΔE, in which the E gene, together with accessory genes 4 and 5a, was deleted (30). Our isolation of the ΔE mutant demonstrated that the E protein is not absolutely essential for MHV replication, as had been previously assumed. However, the ΔE mutant grew very poorly, reaching maximal titers that were at least three orders of magnitude lower than those of wild-type MHV, and the ΔE virus produced very tiny plaques when plated onto mouse L2 cell monolayers (30). We concluded that this extremely defective phenotype was the result of the E-gene deletion, since earlier work by a number of laboratories had shown that mutation, deletion, or replacement of genes 4 and 5a had no significant effect on viral phenotype in tissue culture (11, 13, 40, 49, 53). Nevertheless, it could not be entirely ruled out that the characteristics of the ΔE mutant were partially due to the size of the genomic deletion, the concomitant loss of one or both of the accessory genes, or the new context that had been created upstream of TRS6, which governs transcription of the M gene.
To ascertain whether the highly defective phenotype observed for the ΔE mutant did indeed result from ablation of the E gene, we generated a recombinant virus in which E protein expression was specifically abolished. For this purpose, we designed an E gene knockout plasmid, pLK101, in which 9 point mutations within the first 17 bases of the E ORF changed the start codon from AUG to ACG and instituted stop codons in all 3 reading frames immediately downstream (Fig. 1A). The remainder of the E gene, as well as genes 4 and 5a, was kept intact, except that an additional amino acid was appended to ORF 5a as a result of one of the introduced E ORF stop codons. These mutations were transferred to the MHV genome by the technique of targeted RNA recombination, using host-range-based selection with the chimeric virus fMHV.v2 (Fig. 1B) (18, 28, 38), and the presence of all intended nucleotide changes was confirmed by direct sequencing of RT-PCR products. The phenotype of the resulting E knockout mutant, E-KO, was identical to that of ΔE. When plated onto mouse L2 cells, the E-KO mutant produced plaques that were indistinguishable from those of the ΔE mutant and were tiny by comparison to plaques of the wild type (Fig. 2A). Moreover, the E-KO mutant grew very slowly in murine 17Cl1 cells, with exceptionally low peak titers compared to those of the wild type. The wild-type MHV used throughout this study was Alb240, a well-characterized isogenic recombinant that had been previously reconstructed from fMHV and pMH54 donor RNA (30).
FIG. 2.
Phenotype of the E-KO mutant. (A) Plaques of the E-KO mutant (Alb517) compared with those of the ΔE mutant (Alb291) (30) and an isogenic wild-type control (Alb240) (30) that was reconstructed with pMH54 donor RNA. Plaque titrations were carried out with L2 cells at 37°C. Monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later. Mock-infected cells are shown at the left. (B) Specificity of rabbit polyclonal antiserum raised against the carboxy-terminal tail of the MHV E protein (amino acids 38 through 83). Antibodies were used to probe a Western blot of the following: lanes 1 to 4, lysates of mock-transfected L2 cells or L2 cells transfected with empty SFV vector, SFV vector encoding MHV M protein, or SFV vector encoding MHV E protein; lanes 5 to 7, lysates of mock-infected, wild-type MHV-infected, or ΔE-infected 17Cl1 cells; lanes 8 and 9, lysates of E. coli expressing the carboxy-terminal tail of E or full-length E. (C) Western blots of lysates from L2 cells that were either mock infected or infected with wild-type MHV (Alb240) or with two independent isolates each of the ΔE mutant (Alb290 and Alb291) or the E-KO mutant (Alb517 and Alb518). Blots were probed with polyclonal anti-MHV E antiserum or with monoclonal anti-N antibody J.3.3 (15).
To examine E protein expression in wild-type, ΔE, and E-KO viruses, we generated antiserum specific for the E protein of our laboratory strain of MHV-A59. We (30) and others (11) have previously found that antisera raised against the E proteins of other laboratory strains of MHV (43, 54) fail to recognize our E protein in Western blots, owing to sequence differences of as few as two amino acids. We therefore bacterially expressed and purified an amino-terminal hexahistidine-tagged fragment of the MHV E protein (E-tail), corresponding to amino acids 38 through 83, and this polypeptide was used to immunize rabbits. The specificity of the collected antiserum was demonstrated by Western blotting with samples from a number of sources. Anti-MHV E antiserum was found to react with bacterially expressed full-length, hexahistidine-tagged E protein (10.7 kDa) and with the original E-tail immunogen (6.2 kDa) (Fig. 2B). The antiserum also specifically reacted with E protein that was expressed from an SFV RNA expression vector in mouse L2 cells (9.7 kDa) but not with lysates from mock-transfected cells or cells transfected with empty SFV vector or SFV expressing MHV M protein. Similarly, the antiserum reacted with a 9.7-kDa band in wild-type MHV-infected 17Cl1 cells but not with lysates from mock-infected or ΔE-infected cells (Fig. 2B).
Further Western blot analyses showed that no E protein was expressed by either the E-KO mutant or the ΔE mutant. As shown in Fig. 2C, there was no detectable signal corresponding to E protein in lysates of L2 cells infected with two independent isolates each of E-KO or ΔE, whereas wild-type-infected controls exhibited a strong band for E protein. Both E-KO-infected and ΔE-infected lysates exhibited the same nonspecific background of higher-molecular-weight bands as was seen in wild-type-infected and mock-infected controls. Prolonged exposure did not reveal a detectable specific E protein signal in either of the two mutants. Moreover, the lack of E expression in either E-KO-infected or ΔE-infected cells was unlikely to have resulted from their slow virus growth relative to that of the wild type, since all infected-cell lysates were harvested at similar stages of infection, when nearly 90% syncytium formation was observed. Consistent with this, a duplicate blot of the same samples probed with anti-N monoclonal antibody, as an internal control for the extent of infection, showed similar levels of N expression in all infected-cell lysates (Fig. 2C). It should be noted that since the anti-MHV E antiserum was raised against the carboxy terminus of E protein, we would also have been able to detect an amino-terminally truncated E protein fragment initiated from a downstream methionine at codon 25 in the E-KO mutant if such a species had been synthesized. Our results with the E-KO mutant thus confirmed our previous conclusion that the defects observed in the ΔE mutant directly resulted from the absence of E protein expression. The combined findings from both E mutants established that E protein expression, while not essential in MHV, is critical for the virus to carry out a productive infection.
Heterologous E genes substituted from other coronaviruses have various levels of functionality in MHV.
The dramatic impairment of the ΔE and E-KO mutants provided a basis for us to examine the constraints on E protein sequence and structure by determining whether E genes from other coronaviruses can functionally replace that of MHV. Coronaviruses are classified into three phylogenetic groups (37), which are likely to be accorded the status of genera (19); MHV is the prototype member of group 2. E protein sequences diverge widely among the three coronavirus groups. In pairwise amino acid sequence comparisons, the MHV E protein ranges from a maximum of 65% identity with its closest group 2 relative, the BCoV E protein, to a mere 22% identity with the E protein of the group 3 IBV E protein (Fig. 3A). Similarly, pairwise sequence identities among a set of coronavirus E proteins, other than that of MHV, fall between 28% and 16% (Fig. 3A). A more comprehensive analysis of pairwise sequence identities found that the E protein has the highest intergroup divergence of the four canonical coronavirus structural proteins (19). A multiple alignment of a set of E proteins from all three coronavirus groups shows only four completely conserved residues and considerable heterogeneity in the size of this small protein (Fig. 3B). Despite such extensive variability, common characteristics can be discerned for the different E proteins. Each has a short hydrophilic amino terminus followed by a relatively large transmembrane domain. The remainder of the polypeptide is a carboxy-terminal tail, constituting one-half or more of the molecule. Two to four cysteine residues are found within the transmembrane domain or in the membrane-proximal portion of the carboxy-terminal tail. As yet, however, the exact boundaries of the transmembrane domain have not been well defined for any E protein. Moreover, membrane protein topology prediction programs other than the one that we have used (7) position the transmembrane domains somewhat differently than is shown in Fig. 3B, and in many cases, a second transmembrane segment is predicted within the carboxy-terminal tail.
FIG. 3.
Replacement of the MHV E protein with heterologous coronavirus E proteins. (A) Percent amino acid identities for pairwise alignments between representative members of all three coronavirus groups. Pairwise alignments were generated with the gap program of the Genetics Computer Group sequence analysis package (16). (B) Multiple alignment of the coronavirus E proteins used in this study. The alignment was produced manually from comparison of the pairwise alignments. Residues that are common to three, four, or all five sequences are marked by open, gray, or black circles, respectively. The solid bar represents the limits of the transmembrane domains of each of the sequences, as assigned by the dense alignment surface method (7) (http://www.sbc.su.se/∼miklos/DAS/); the thicker section of the bar indicates positions predicted to be transmembrane residues in all five E proteins. (C) Plaques of heterologous E gene substitution mutants, compared with plaques of wild-type MHV and the ΔE mutant. Plaque titrations were carried out on L2 cells at 37°C. Monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later.
To substitute E genes from other coronaviruses, we in each case swapped the MHV E ORF with a heterologous E ORF by making an exact exchange of start codons and by making an exact, or nearly exact, exchange of the stop codon position, as shown in Fig. 1A and described in detail in Materials and Methods. Because the MHV E ORF is not preceded by its own TRS, it consequently follows ORF 5a in subgenomic mRNA5. The translation of the MHV E protein has been proposed to be governed by an IRES that occupies part of the immediately upstream gene 5a (25, 45). Since the IRES could conceivably affect both the level and timing of E protein expression, we made an AUG-for-AUG replacement at the upstream end of each substitution in order to preserve the E ORF genomic context as closely as possible (Fig. 1A). Similarly, at the downstream end of each substitution, we made the ORF replacement such that the immediately upstream context of TRS6 was retained in order to avoid unintended effects on the expression of the downstream M gene. In the case of the BCoV E ORF, the latter criterion allowed us to position the stop codon of the heterologous ORF at the same nucleotides as those of its MHV counterpart. This positioning was done at the expense of altering the last residue of the BCoV E protein from valine to isoleucine, a conservative change that turned out to have no effect. For the SARS-CoV, TGEV, and IBV E ORFs, the stop codon was moved 12 nucleotides upstream of the position of its MHV counterpart, thereby leaving unchanged the identity of the 10 nucleotides preceding TRS6 (Fig. 1A).
Recombinants containing heterologous coronavirus E genes were generated by targeted RNA recombination. Multiple recombinant viruses were obtained for each E replacement, and two independent isolates of each substitution mutant were purified, confirmed by sequencing of the E gene and its boundaries, and used in further analyses. We first examined the plaque size phenotype of each chimeric virus. As expected, the recombinant containing the highly homologous BCoV E gene in place of the MHV E gene was indistinguishable from wild-type MHV. It produced wild-type-sized plaques on mouse L2 cells (Fig. 3C) and grew as efficiently as and to similar titers as wild-type MHV. Similarly, the E gene derived from SARS-CoV was almost completely able to functionally replace its MHV counterpart. The recombinant containing the SARS-CoV E gene produced plaques nearly equivalent in size to those of the wild type. This finding is in line with the classification of SARS-CoV as an outlying member of the group 2 coronaviruses, more closely related to MHV and BCoV than to other members of the family (21). It must be noted that the SARS-CoV E gene used in our study differs from the reported SARS-CoV strain Urbani sequence (44) by a single transversion that results in a change of codon 37 from CUU (leucine) to CAU (histidine). However, since the sequence containing this change is the only sequence detected in our laboratory stock of SARS-CoV, it must represent a functional E gene for that virus. We therefore do not think that the L37H change accounts for the slightly diminished plaque size in the SARS-CoV E substitution mutant.
In contrast to the intragroup E gene substitutions, the E gene from the group 1 coronavirus TGEV was not able to functionally replace the MHV E gene. The recombinant containing the TGEV E gene formed very tiny plaques that were not measurably different from those of the ΔE mutant (Fig. 3C). One explanation for this result might have been that the transplanted TGEV E gene was not expressed in the MHV genome, but this possibility was later ruled out (see below). Therefore, unlike the BCoV or SARS-CoV E protein, the TGEV E protein was inert with respect to facilitation of MHV virion assembly. This outcome, the incompatibility of group 1 and group 2 structural proteins in virion morphogenesis, appears to echo previous results in which VLPs could not be formed by coexpression of BCoV M protein and TGEV E protein or, conversely, by coexpression of TGEV M protein and BCoV E protein (2).
The most surprising result with the substitution mutants was that the E gene from IBV, a group 3 coronavirus, was completely functional in MHV. This finding was unexpected, since the IBV E protein has the lowest degree of sequence homology with the MHV E protein and has a size difference of 26 amino acids (Fig. 3A and B). The chimeric MHV mutant containing the IBV E gene formed wild-type-sized plaques on L2 cells (Fig. 3C), and it grew efficiently in 17Cl1 cells, to high titers equivalent to those of wild-type MHV (data not shown). Viral protein expression in the IBV E substitution mutant was assessed by Western blot analysis comparing lysates from mutant-infected cells with those from wild-type-infected and mock-infected controls (Fig. 4). This confirmed that IBV E protein was expressed in two independent isolates of the substitution mutant, whereas no trace of MHV E was detected in the same mutants on a duplicate blot probed with anti-MHV E antiserum. The latter result ruled out the possibility that a functional copy of the MHV E gene was somehow retained elsewhere in the substitution mutants. Additionally, the expression of two other MHV structural proteins, N and M, showed little or no alteration as a consequence of the IBV E gene substitution; the same lysates probed with monoclonal antibodies against MHV N or M gave rise to nearly equivalent signals for wild-type MHV and the substitution mutant (Fig. 4). These data indicate that the IBV E protein can substitute for the MHV E protein in MHV replication.
FIG. 4.
Western blots of lysates from 17Cl1 cells that were mock infected or were infected with wild-type MHV (Alb240) or with two independent isolates of the IBV E substitution mutant (Alb499 and Alb500). Blots were probed with polyclonal anti-IBV E antiserum (4), polyclonal anti-MHV E antiserum, monoclonal anti-N antibody J.3.3 (15), or monoclonal anti-M antibody J.1.3 (15).
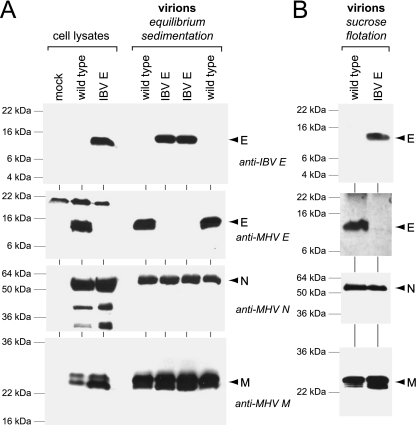
To determine whether IBV E protein was incorporated into virions of the substitution mutant, we purified viruses released from infected 17Cl1 cells by two cycles of equilibrium sedimentation on tartrate-glycerol gradients (52). Western blot analysis of purified virions revealed that IBV E protein was incorporated into virions that were otherwise entirely composed of MHV components (Fig. 5A). Because the IBV and MHV E proteins were detected with different antisera with different affinities for their respective antigens, we could not determine whether equivalent molar amounts of the two E proteins were incorporated into virions. However, the quantity of each E protein incorporated into virions was a similar fraction of the total E protein expressed in infected cells. To further test the stringency of the virus preparation, gradient-isolated virions were additionally purified by a sucrose flotation procedure (28, 47). This confirmed that IBV E protein was a component of virions of the substitution mutant (Fig. 5B).
FIG. 5.
Incorporation of IBV E protein into virions. (A) Wild-type MHV (Alb240) and an IBV E substitution mutant (Alb499) were grown in 17Cl1 cells and purified by two cycles of equilibrium sedimentation on tartrate-glycerol gradients, as described in Materials and Methods. (B) Portions of isolated virus were further purified by sucrose flotation. Virions and cell lysate controls were analyzed by Western blots probed with polyclonal anti-IBV E antiserum (4), polyclonal anti-MHV E antiserum, monoclonal anti-N antibody J.3.3 (15), or monoclonal anti-M antibody J.1.3 (15).
Specific mutations in either the transmembrane domain or the carboxy-terminal tail of the TGEV E protein render it functional in MHV.
The phenotype of the TGEV E substitution mutant, which was essentially the same as that of the ΔE mutant, presented us with the opportunity to search for mutations that might facilitate better growth of this virus. To accomplish this, we started cultures from multiple individual plaques of the TGEV E substitution mutant and serially passaged each of them two to four times in L2 cells, frequently monitoring the spread of infection in the monolayer. When accelerated growth was detected for a given passage, viral supernatants were titered on L2 cells. From this procedure we identified and purified plaques that were markedly larger than those of the original mutant, although none had fully attained the size of plaques of wild-type MHV. A total of 30 viruses with enhanced growth, representing 26 independent mutants of the original TGEV E substitution recombinant, were isolated in this fashion through two separate searches.
To map genetic changes in each of these TGEV E gain-of-function mutants, we sequenced RT-PCR products that spanned the entirety of the E and the M genes. The results of this analysis are summarized in Fig. 6. For each mutant, we found at least one base change in the TGEV E gene. By contrast, no changes were seen in the M gene of any mutant. All identified mutations resulted in coding changes in the TGEV E ORF; there were no silent nucleotide changes. Most mutations were found in more than one mutant, which suggests that our searches were saturating. With two exceptions, all changes were point mutations, and in almost all cases, these occurred as a single point mutation per mutant. The paucity of multiple mutations in members of this set was likely a reflection of our approach, in which we searched for large-plaque variants at the earliest passage in which an upward shift in viral growth rate was observed. Most strikingly, the mutations were found to cluster almost exclusively in two small regions of the TGEV E protein, one in the transmembrane domain, between residues 25 and 32, and the other within the carboxy-terminal tail, between residues 50 and 60 (Fig. 6).
FIG. 6.
Summary of the sequence analysis of TGEV E gain-of-function mutants. Two separate searches for intermediate- or large-plaque mutants of the TGEV E substitution recombinant were performed following serial passaging of independent plaques. Mutants that were obtained in the first search are denoted by open circles; mutants that were obtained in the second search are denoted by filled circles. All mutants are independent except for the following pairs: TE1 and TE26; TE23 and TE30; and TE27 and TE28. The entire E and M genes of all mutants were sequenced. The wild-type TGEV E protein is represented linearly at the top; the solid rectangle indicates the predicted transmembrane domain. Positions within the E protein at which potential gain-of-function mutations were found are connected by arrows to the wild-type residue, under which is indicated changes found in each mutant. All coding changes are shown; there were no noncoding changes. No changes were found in the M gene of any of the revertants. Boxed residues, or pairs of residues, are those that were chosen for further analysis. At the bottom are details about an insertion and a deletion, respectively, found in mutants TE29 and TE30.
Two of the TGEV E mutants harbored changes that were not point mutations. Mutant TE29 contained a 15-nt insertion that resulted in the replacement of I55 with six amino acids, MSLMKF. It is notable that this alteration also occurred in one of the two regions where point mutations were found to cluster. Curiously, the insertion in mutant TE29, 5′GUCGUUGAUGAAGUU3′, is identical to nucleotides 17,472 to 17,486 of the MHV genome, which are within the nsp13 segment of the pp1ab ORF. This suggests that the insertion arose from a nonhomologous recombination event. A second mutant, TE30, contained a 103-nt deletion that fused a thereby truncated ORF 5a to codon 15 of the TGEV E ORF. We have not yet determined whether the potential 5a-E fusion protein is expressed. The mutations in TE29 and TE30 recall other nonhomologous rearrangements that we have found in revertants of M and N protein mutants that had extremely defective phenotypes (24, 29). We believe that coronavirus RNA synthesis must produce such rearrangements at a constant low rate and that they become visible only when revealed by a sufficiently strong selective pressure.
To determine whether the mutations found in the TGEV E gain-of-function mutants were indeed responsible for the enhanced growth of the mutants, we reconstructed many TGEV E substitution recombinants, each containing one of a subset of these mutations (those that are boxed in Fig. 6). This permitted us to specifically gauge the effects of individual mutations in the absence of possible mutations elsewhere in the genome. From the cluster of mutations in the transmembrane region, we separately reconstructed TGEV E substitution recombinants with L31V, L32R, and I17V-plus-L32R mutations. In all three cases, the reconstructed mutants produced considerably larger plaques than did the original TGEV E substitution mutant (Fig. 7), and the efficiency of virus replication was greatly enhanced. Likewise, we reconstructed TGEV E substitution recombinants with mutations from the carboxy-terminal tail cluster: G50A, I55S, I55T, V56G, V56A, P57L, H60Y, and I17V plus H60Y. These reconstructed mutants also produced plaques that, to various degrees, were significantly larger than those of the TGEV E substitution mutant (Fig. 7). Plaques of all reconstructed mutants were indistinguishable from those of the original isolates in which the mutations had been identified. In addition, reconstructed mutants exhibited the same extents of enhanced growth as did the original gain-of-function isolates. Passage 1 stocks begun from single plaques of the TGEV E substitution mutant had titers on the order of 102 PFU per ml. By contrast, passage 1 stocks of either the original or the reconstructed TGEV E gain-of-function mutants ranged from 104 to 105 PFU per ml. These results allowed us to conclude that the mutations are directly responsible for the larger plaque size and enhanced growth phenotypes of the gain-of-function mutants. The greatest increases in plaque size were observed with the L31V or L32R change in the transmembrane domain and with the I55S, I55T, V56G, or P57L change in the carboxy-terminal tail of TGEV E. These positions may thus define the core residues of each cluster, where changes can exert the greatest effect. Intermediate effects on plaque size were found to be caused by mutations at residues more distant from either of these two loci. For two of the core residues, I55 and V56, two different mutations each were reconstructed. TGEV E substitution recombinants with I55S and I55T mutations produced equally large plaques, whereas the recombinant with V56G produced larger plaques than did the recombinant with V56A (Fig. 7). Two reconstructed recombinants with double mutations, I17V plus L32R and I17V plus H60Y, were not significantly different from recombinants with the respective single mutations, L32R and H60Y. This suggests that the I17V mutation did not function synergistically with either of the other mutations with which it had been isolated in double mutants. We do not yet know the effects, if any, of pairing other mutations that were not originally found together. In sum, our reconstructions confirmed that any one of several single mutations in either of two crucial regions of the molecule was sufficient to render the TGEV E protein functional in MHV.
FIG. 7.
Plaques of a reconstructed subset of 11 gain-of-function mutations, representing 20 of the TE mutants, compared with plaques of the wild type and the original TGEV E substitution mutant. Plaque titrations were carried out on L2 cells at 37°C. Monolayers were stained with neutral red at 72 h postinfection and were photographed 18 h later.
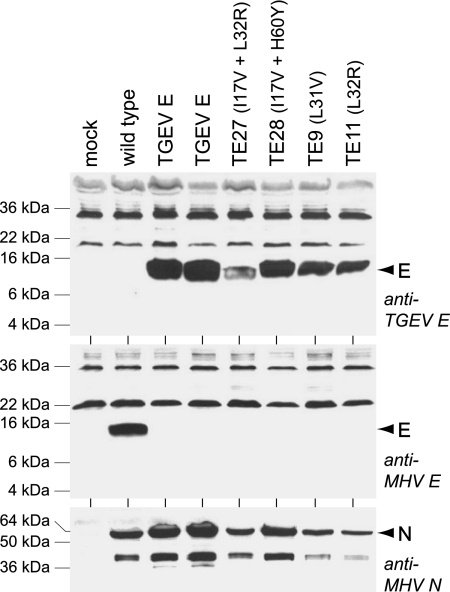
In addition, we examined protein expression by the original TGEV E substitution mutant and by four of the reconstructed gain-of-function mutants. For this purpose, an antipeptide antiserum was raised against the carboxy-terminal 22 amino acids of the TGEV E protein. This antiserum was used in Western blots of lysates from cells infected with the TGEV E substitution mutant or with four of the reconstructed gain-of-function mutants, along with lysates from wild-type-infected and mock-infected controls (Fig. 8). This analysis revealed equivalent expression of TGEV E protein by two independent isolates of the original substitution mutant and the reconstructed mutants TE9 (L31V), TE11 (L32R), TE27 (I17V plus L32R) and TE28 (I17V plus H60Y). The anti-TGEV E antiserum did not cross-react with MHV E protein, and conversely, the anti-MHV E antiserum did not cross-react with TGEV E protein. Also, the levels of MHV N protein, probed as an internal infection control, were comparable among all lysates, indicating that the expression of TGEV E protein in the original substitution mutant did not impair the expression of another MHV structural protein. These results demonstrated that the failure of the original (wild-type) TGEV E gene to functionally substitute for that of MHV was not due to failure of the original substitution mutant to express the TGEVE protein. Therefore, the gain-of-function mutations acted through qualitative, rather than quantitative, effects on the TGEV protein.
FIG. 8.
Western blots of lysates from 17Cl1 cells that were mock infected or were infected with wild-type MHV (Alb240), two independent isolates of the TGEV E substitution mutant (Alb515 and Alb516), or four reconstructed gain-of-function mutants (TE27, TE28, TE9, or TE11). Blots were probed with polyclonal anti-TGEV E antiserum, polyclonal anti-MHV E antiserum, or monoclonal anti-N antibody J.3.3 (15).
DISCUSSION
To obtain insights into the role played by E protein in coronavirus assembly, we examined the extent to which heterologous E proteins could replace that of MHV. As a prerequisite to this study, however, we needed to determine whether the ΔE mutant that we had previously isolated (30) truly represented the E-null phenotype in MHV. This had not been unequivocally demonstrated, because the ΔE mutant also contained complete deletions of genes 4 and 5a. Although prior work with a number of strains of MHV had shown that genes 4 and 5a are nonessential for growth in tissue culture (11, 13, 40, 49, 53), a Δ45a mutant had exhibited slightly reduced kinetics of growth and had been found to be attenuated in vivo (11). Therefore, to clarify the status of the ΔE mutant, we constructed an E-KO mutant in which only expression of the E gene was disrupted through the introduction of point mutations (Fig. 1). The E-KO mutant entirely recapitulated the severe growth defect of the ΔE mutant (Fig. 2A), and the ablation of E expression in both mutants was ascertained by Western blot analysis (Fig. 2B and C). We can thus conclude that knockout of the E gene is responsible for the observed defect and that the absence of genes 4 and 5a did not make a detectable contribution to the phenotype of ΔE. The combined results establish that E protein, although not essential, is extremely important for the growth of MHV.
The exact nature of this importance, nevertheless, is currently obscure. Some evidence appears to point to a requirement for direct physical interactions between E and M proteins in virion morphogenesis. In coexpression systems, particular interspecies combinations of E and M proteins are not competent for VLP assembly (2, 12). Similarly, the physical proximity of the E and M proteins in IBV has been demonstrated by their chemical cross-linking (6) or coimmunoprecipitation (33) in infected or cotransfected cells, although these properties did not always correlate with the capabilities of mutant E proteins to form VLPs with M protein (6). Conversely, many previous results could be taken to indicate that E protein acts at a distance, independently of it having to come into contact with M protein. The isolated expression of either IBV or MHV E protein leads to the formation and cellular export of membranous vesicles (4, 35). Likewise, the expression of MHV E protein has been shown to give rise to intracellular clusters of convoluted membrane structures (43) that strongly resemble those seen in coronavirus-infected cells (9). These observations suggest that the principal role of E protein is to induce membrane curvature in the budding compartment.
To learn more about the requirements imposed on E protein sequence relative to the possible specificity of interactions between E protein and M protein, we attempted to replace the MHV E gene with E genes from a range of phylogenetically conserved and divergent coronaviruses covering all three groups within the coronavirus family. This was carried out by creation of ORF-for-ORF exchanges, by which heterologous E proteins could be expressed in the same manner as native MHV E protein. The most conservative substitution made was that of the E protein of BCoV, the group 2 coronavirus that is the closest relative of MHV. The E gene of BCoV was fully functional when installed in place of the MHV E gene, and the resulting recombinant behaved identically to a wild-type MHV control (Fig. 3C). This result was not unexpected, since substitutions of similarly homologous domains of the BCoV M or N protein have been found to be able to replace the counterpart domains in MHV (10, 42). More striking was our finding that the E protein of SARS-CoV, the outlying member of the group 2 coronaviruses (21), was almost fully able to replace the MHV E protein, notwithstanding the much lower degree of sequence identity between these two E proteins.
Most remarkably, however, we observed that the MHV mutant with an E gene obtained from the group 3 coronavirus IBV was indistinguishable from wild-type MHV. The IBV E substitution mutant produced plaques identical in size to those of the wild type, and it grew efficiently, to titers equivalent to those of the wild type. This interchangeability was not hindered by the large disparity in primary sequence between the two E proteins. Not only does IBV E have the most limited homology to its MHV counterpart of any coronavirus (22% amino acid sequence identity), but it is also 26 residues longer than the 83-amino-acid MHV E protein (Fig. 3B). This result, together with the substitution of the SARS-CoV E protein, supports the notion that E proteins function without having to make sequence-specific contacts with other viral components, including, most notably, M protein. We did observe, however, that IBV E protein was incorporated into virions of the IBV E substitution mutant (Fig. 5), just as MHV E protein is incorporated into MHV (54). Although this could be taken as evidence for a physical interaction between the E and M proteins, we currently think that it merely reflects that these two proteins are colocalized in infected cells and that M-M interactions during budding do not entirely exclude E protein.
At present, the precise mechanism by which IBV E is able to entirely replace MHV E is undetermined. Most likely, the two E proteins provide identical functionality during infection, despite their sequence divergence. It cannot be ruled out, however, that IBV E offers an alternate, but equally efficient, option for MHV assembly in the absence of MHV E. Elegant expression studies of the IBV E protein have shown that the carboxy-terminal tail of IBV E interacts with the IBV M protein, and it also contains a Golgi-targeting signal (5, 6). Both of these characteristics were found to be important, and probably sufficient, for VLP formation. Further detailed study will be required to delineate how these results are commensurate with the mechanism by which IBV E compensates for the loss of MHV E in a complete viral infection. An attractive approach would be to examine the functionality within MHV of hybrid E proteins that mix domains taken from IBV E and from MHV E.
Contrary to results with the substitutions of group 2 and group 3 E genes, the E gene from the group 1 coronavirus TGEV was inactive in MHV (Fig. 3C), even though the TGEV E protein was clearly expressed by the substitution mutant (Fig. 8). This finding strongly resembles the outcome of prior coexpression work showing that interspecies combinations of the M and E proteins of TGEV and BCoV fail to yield VLPs (2). Negative results were also obtained in VLP experiments that mixed the M and E proteins of FIPV and MHV (12). In both cases, as well as with our TGEV E substitution mutant, the incompatibilities that were found occurred between components from group 1 coronaviruses (TGEV and FIPV) and group 2 coronaviruses (MHV and BCoV). At first, these observations appeared at variance with the idea that E protein acts independently of a physical association with M protein. However, we were able to isolate numerous gain-of-function mutants of the TGEV E substitution recombinant, in which single amino acid changes in the TGEV E protein allowed it to assist MHV replication. Since the gain-of-function mutant E proteins remain extremely divergent in sequence from MHV E protein, we believe that such a sequence difference is consistent with a mechanism of E acting at a distance from M. On the other hand, the fact that the TGEV E protein required modification before it could replace its MHV counterpart suggests that some fundamental disparity exists in the details of how the TGEV and MHV E proteins operate during infections by their respective viruses. This disparity most likely resides at the level of interactions among viral components rather than in the ability of the TGEV E protein to function in mouse cells. All available evidence indicates that once the barrier of S protein-receptor recognition is overcome, there is no significant block to coronavirus replication in different species of cells. In particular, the interspecies chimera mFIPV (12, 38), which is an FIPV recombinant containing the ectodomain of the MHV S protein, grows well in mouse cells. This indicates that the group 1 FIPV E protein (which has 94% sequence identity with the TGEV E protein) is fully functional in mouse cells in conjunction with its cognate viral structural proteins.
All of the gain-of-function mutants that we selected in two separate searches mapped in the (TGEV) E gene (Fig. 6), and reconstruction of many of these mutants proved that the responsible mutations had been correctly identified (Fig. 7). No changes were found in the (MHV) M genes of any of the gain-of-function mutants, supporting the idea that it was not possible to obtain modifications of M protein able to compensate for the deficiency of the wild-type TGEV E protein in MHV. In accord with this result, we previously found that revertants of clustered charged-to-alanine mutants of the MHV E protein mapped solely in the E gene and not in the M gene (14). Similarly, we have been able to isolate many intergenic second-site revertants for extremely defective mutants of the M (29) and N (24) proteins of MHV, but none of these revertants mapped in the E gene.
Almost all of the TGEV E gain-of-function mutations were clustered at two loci, one in the transmembrane domain (with the strongest effects resulting from changes in residues L31 and L32) and the other in the carboxy-terminal tail (with the strongest effects at residues I55, V56, and P57). The mutations that occurred at each of these positions actually reduced, rather than increased, the homology between TGEV E and MHV E. The nature of the mutations in the transmembrane region is particularly intriguing. Most of these, such as L25V, I27V, I27L, and L31V, are fairly conservative substitutions, perhaps indicative of subtle modifications in the interactions among hydrophobic residues in oligomers of E (46). By contrast, the L32R mutation, which arose independently in three distinct mutants (TE11, TE26, and TE27), might be expected to significantly alter the topology or orientation of the transmembrane domain.
The distribution of the TGEV E gain-of-function mutations into two clusters appears to highlight two important domains of the molecule, but it is yet indeterminate whether these domains govern separate functions or whether they interact with one another. Analyses of the IBV E protein have similarly indicated two separable functional domains for that molecule and have suggested that the transmembrane domain is not specifically required for either VLP formation or Golgi localization (5, 6). Multiple roles of E protein in viral infection may also be implied by the lack of correspondence between the lethality of the E-KO mutants of TGEV (8, 41) and the viability of the ΔE and E-KO mutants of MHV (30; this work). It is conceivable that one function of E protein is to potentiate the kinetics of virion assembly, but this role is not essential. A hypothetical second function may be essential for TGEV, but either it is not required by MHV or else it is supplied redundantly by some other MHV protein.
It was recently demonstrated that the E protein of SARS-CoV can form membrane channels having a selectivity for monovalent cations (51). Moreover, a peptide corresponding to the amino-terminal 40 amino acids of SARS-CoV E (comprising the hydrophilic amino terminus and the transmembrane domain) had the same properties as did full-length E. This result accords with reports that the expression of MHV or SARS-CoV E protein enhanced the permeability of bacterial or mammalian cells (34, 31); such a capability has led some to classify E among a set of small viral proteins termed viroporins (20). Very recently, the channel-forming ability of E protein was shown to extend to the E proteins of human coronavirus 229E, MHV, and IBV (50). This property therefore generalizes to all three coronavirus groups. Interestingly, channels formed by E proteins of group 2 (MHV and SARS-CoV) and group 3 (IBV) coronaviruses exhibited a selectivity for sodium ions over potassium ions (50, 51). By contrast, the E protein of human coronavirus 229E, which, like TGEV, is a member of group 1, formed channels that were selective for potassium ions over sodium ions. This observation may explain the inactivity of the wild-type TGEV E protein in MHV infection, and it may further provide a basis for understanding the nature of the transmembrane gain-of-function mutations.
Our work supports the conclusion that the E proteins of coronaviruses possess certain common functions that can be provided by a broad, and largely interchangeable, range of primary amino acid sequences. Our findings are most consistent with a scenario in which sequence-specific interactions of E protein with M protein are not required. We hope to use such a framework to obtain further insights into the in vivo functions of E protein and the mechanism of this protein's involvement in virion morphogenesis.
Acknowledgments
We are indebted to the following colleagues for generously providing materials essential to this study: Carolyn Machamer for the IBV E gene clone and antiserum to the IBV E protein; David Brian for clones of the BCoV and TGEV E genes; John Fleming for monoclonal antibodies to the MHV M and N proteins; Volker Thiel for the pTetON-IRES-MHV-E expression plasmid; Jill Taylor for RNA from SARS-CoV-infected cells; and Scott Goebel for a clone containing the SARS-CoV E gene. We also thank Victoria Derbyshire for valuable advice on the bacterial expression of the MHV E protein and the Molecular Genetics Core Facility of the Wadsworth Center for DNA sequencing.
This work was supported by Public Health Service grants AI064603 and AI060755 from the National Institutes of Health.
Footnotes
Published ahead of print on 20 December 2006.
REFERENCES
- 1.Arbely, E., Z. Khattari, G. Brotons, M. Akkawi, T. Salditt, and I. T. Arkin. 2004. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 341:769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos, E. C. W., W. Luytjes, H. van der Meulen, H. K. Koerten, and W. J. M. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corse, E., and C. E. Machamer. 2002. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 76:1273-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corse, E., and C. E. Machamer. 2003. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 312:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David-Ferreira, J. F., and R. A. Manaker. 1965. An electron microscope study of the development of a mouse hepatitis virus in tissue culture cells. J. Cell Biol. 24:57-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A. M., L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72:6838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan, C. A. M., P. S. Masters, X. Shen, S. Weiss, and P. J. M. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Haan, C. A. M., and P. J. M. Rottier. 2005. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64:165-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleming, J. O., S. A. Stohlman, R. C. Harmon, M. M. C. Lai, J. A. Frelinger, and L. P. Weiner. 1983. Antigenic relationships of murine coronaviruses: analysis using monoclonal antibodies to JHM (MHV-4) virus. Virology 131:296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Genetics Computer Group. 1996. Program manual for the Wisconsin package. Version 9.0. Genetics Computer Group, Madison, WI.
- 17.Godet, M., R. L'Haridon, J.-F. Vautherot, and H. Laude. 1992. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology 188:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez, J. M., P. Gomez-Puertas, D. Cavanagh, A. E. Gorbalenya, and L. Enjuanes. 2003. A comparative sequence analysis to revise the current taxonomy of the family Coronaviridae. Arch. Virol. 148:2207-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, M. E., and L. Carrasco. 2003. Viroporins. FEBS Lett. 552:28-34. [DOI] [PubMed] [Google Scholar]
- 21.Gorbalenya, A. E., E. J. Snijder, and W. J. M. Spaan. 2004. Severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J. Virol. 78:7863-7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton, R. M., and L. R. Pease. 1991. Recombination and mutagenesis of DNA sequences using PCR, p. 217-247. In M. J. McPherson (ed.), Directed mutagenesis, a practical approach. IRL Press, New York, N.Y.
- 23.Huang, Y., Z. Y. Yang, W. P. Kong, and G. J. Nabel. 2004. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: implications for assembly and vaccine production. J. Virol. 78:12557-12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurst, K. R., L. Kuo, C. A. Koetzner, R. Ye, B. Hsue, and P. S. Masters. 2005. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 79:13285-13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jendrach, M., V. Thiel, and S. Siddell. 1999. Characterization of an internal ribosome entry site within mRNA5 of murine hepatitis virus. Arch. Virol. 144:921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khattari, Z., G. Brotons, M. Akkawi, E. Arbely, I. T. Arkin, and T. Salditt. 2006. SARS coronavirus E protein in phospholipid bilayers: an X-ray study. Biophys. J. 90:2038-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koetzner, C. A., M. M. Parker, C. S. Ricard, L. S. Sturman, and P. S. Masters. 1992. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 66:1841-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao, Y., Q. Yuan, J. Torres, J. P. Tam, and D. X. Liu. 2006. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 349:264-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liljestrom, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Biotechnology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 33.Lim, K. P., and D. X. Liu. 2001. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 276:17515-17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madan, V., M. de Jesus Garcia, M. A. Sanz, and L. Carrasco. 2005. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 579:3607-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda, J., A. Maeda, and S. Makino. 1999. Release of E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeda, J., J. F. Repass, A. Maeda, and S. Makino. 2001. Membrane topology of coronavirus E protein. Virology 281:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masters, P. S., and P. J. M. Rottier. 2005. Coronavirus reverse genetics by targeted RNA recombination. Curr. Top. Microbiol. Immunol. 287:133-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortola, E., and P. Roy. 2004. Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett. 576:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ontiveros, E., L. Kuo, P.S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 41.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng, D., C. A. Koetzner, T. McMahon, Y. Zhu, and P. S. Masters. 1995. Construction of murine coronavirus mutants containing interspecies chimeric nucleocapsid proteins. J. Virol. 69:5475-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raamsman, M. J. B., J. Krijnse Locker, A. de Hooge, A. A. F. de Vries, G. Griffiths, H. Vennema, and P. J. M. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 45.Thiel, V., and S. G. Siddell. 1994. Internal ribosome entry in the coding region of murine hepatitis virus mRNA5. J. Gen. Virol. 75:3041-3046. [DOI] [PubMed] [Google Scholar]
- 46.Torres, J., J. Wang, K. Parthasarathy, and D. X. Liu. 2005. The transmembrane oligomers of coronavirus protein E. Biophys. J. 88:1283-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vennema, H., G.-J. Godeke, J. W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D.-J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weiss, S. R., P. W. Zoltick, and J. L. Leibowitz. 1993. The ns 4 gene of mouse hepatitis virus (MHV), strain A59 contains two ORFs and thus differs from ns 4 of the JHM and S strains. Arch. Virol. 129:301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, L., P. Gage, and G. Ewart. 2006. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353:294-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, L., C. McKinlay, P. Gage, and G. Ewart. 2004. SARS coronavirus E protein forms cation-selective ion channels. Virology 330:322-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye, R., C. Montalto-Morrison, and P. S. Masters. 2004. Genetic analysis of determinants for spike glycoprotein assembly into murine coronavirus virions: distinct roles for charge-rich and cysteine-rich regions of the endodomain. J. Virol. 78:9904-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yokomori, K., and M. M. C. Lai. 1991. Mouse hepatitis virus S RNA sequence reveals that nonstructural proteins ns4 and ns5a are not essential for murine coronavirus replication. J. Virol. 65:5605-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yu, X., W. Bi, S. R. Weiss, and J. L. Leibowitz. 1994. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology 202:1018-1023. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, Q., Y. Liao, J. Torres, J. P. Tam, and D. X. Liu. 2006. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 580:3192-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]