Abstract
This work demonstrates a central role for the retinoblastoma (Rb) family in driving the transcriptional program of induced and replicative senescence. HeLa cervical carcinoma cells rapidly undergo senescence when the human papillomavirus (HPV) type 18 E7 gene in these cells is repressed by the bovine papillomavirus (BPV) E2 protein. This senescence response requires the endogenous Rb pathway but not the p53 pathway. Microarray analysis 6 days after BPV E2 introduction into HeLa cells identified 224 cellular genes induced by E7 repression and 354 repressed genes. Many repressed genes were involved in cell cycle progression, and numerous induced genes encoded lysosomal proteins. These gene expression changes were blocked by constitutive expression of the wild-type HPV16 E7 or adenovirus E1A gene, but not by E7 or E1A mutants defective for Rb binding. Short hairpin RNAs targeting the Rb family also inhibited these gene expression changes and blocked senescence. Therefore, surprisingly, the transcriptional response to BPV E2 expression was entirely dependent on E7 repression and activation of the Rb family, and the BPV E2 protein did not directly affect the expression of cellular genes. Activation of the Rb family repressed E2F-responsive genes and stimulated transcriptional activators, thereby mobilizing multiple signals, such as repression of B-MYB and DEK, that were independently sufficient to induce senescence. There was extensive overlap between the transcriptional profiles of senescent, late-passage primary human fibroblasts and senescent cervical carcinoma cells, suggesting that this Rb family-mediated transcriptional cascade also plays a central role in replicative senescence.
Replicative senescence, an irreversible growth-arrested state attained by primary somatic cells after extensive passage in culture, reflects an important tumor suppressor mechanism in vivo and is a model for various aspects of cellular and organismal aging (10, 13, 16, 76). Senescence is characterized by a constellation of features including irreversible growth arrest, elevated senescence-associated β-galactosidase (SAβ-gal) activity, and increased autofluorescence, cell size, and cellular granularity. Although the p53 and retinoblastoma (Rb) tumor suppressor pathways appear to be important mediators of replicative senescence (37, 77), it has been difficult to characterize the genetic and biochemical basis of replicative senescence because it is a heterogeneous process that occurs after months in cell culture. Senescence-like states can also be acutely induced by oxidative stress, introduction of an activated ras oncogene or tumor suppressor genes, or various other treatments (reviewed in reference 78). The rapid induction of senescence by these diverse stimuli implies that senescence is a genetically programmed cellular process analogous to cell cycle progression or apoptosis.
We and others have described a model of induced senescence in cervical cancer cells (36, 57, 88). Cervical carcinogenesis is initiated by persistent infection of cervical keratinocytes with a high-risk human papillomavirus (HPV), such as HPV16 or HPV18. Cervical carcinoma cells continuously express the HPV E6 and E7 oncogenes, resulting in sustained inactivation of the p53 and Rb pathways, respectively (54, 58). Repression of the E6 and E7 genes in cervical carcinoma cell lines such as HeLa cells by expression of the bovine papillomavirus (BPV) E2 transcriptional repressor reactivates the endogenous p53 and Rb pathways and induces a robust senescence response that proceeds rapidly and synchronously and becomes irreversible by 5 days after E2 expression (20, 24, 36, 41, 42, 45, 57, 88). Constitutive expression of the HPV16 E6 and E7 genes in HeLa cells can prevent BPV E2-induced senescence, demonstrating that HPV repression is required for this response (18, 27, 33, 34, 36, 45, 63, 64, 68). In contrast, high-level expression of HPV E2 proteins can result in cellular effects independently of E6/E7 repression (9, 19, 28, 66, 84).
To initiate a genetic dissection of the pathways leading to senescence in response to HPV repression, we engineered HeLa cells to constitutively express an exogenous HPV16 E6 gene (18). Repression of the endogenous HPV18 E7 gene in these cells by E2 expression activates the Rb but not the p53 pathway, which remains dormant because of enforced expression of HPV16 E6. Nevertheless, E7 repression efficiently induces senescence in these cells, demonstrating that p53 induction is not required for senescence (18). Genetic studies strongly suggest that this induced senescence response requires activation of the Rb family (63, 70, 88). In addition, inactivation of the Rb pathway in primary keratinocytes is required to bypass replicative senescence (22, 48) and Rb-dependent heterochromatic foci are a hallmark of cells undergoing replicative senescence (60). These findings indicate that the Rb family also plays a major role in regulating replicative senescence.
The best characterized E7 targets are p105Rb, p107, and p130, which comprise the Rb family. Rb family members regulate transcription primarily by modulating the activity of the E2F family of transcription factors (30). Complexes composed of E2F and Rb family members actively repress transcription of numerous genes required for cellular DNA replication and cell cycle progression by recruitment of chromatin-modifying enzymes, including histone deactylases, to the promoters of these genes (11). Expression of the E7 protein disrupts Rb/E2F complexes and stimulates degradation of the Rb family, relieving repression and converting E2F family members into transcriptional activators, resulting in cell cycle progression (58). Rb family members can also interact with a number of transcriptional activators, including c-jun and chromatin remodeling proteins (e.g., see references 26, 62, 65, 80, and 94).
The E7 protein binds numerous other proteins in addition to members of the Rb family (58), but the functional consequences of these interactions are poorly characterized. Therefore, in order to understand the mechanistic basis of induced senescence in cervical cancer cells, it is important to determine which of the transcriptional consequences of HPV E7 repression are caused by activation of the Rb pathway and which do not require Rb activation, and to explore the molecular basis for the transcriptional response to Rb activation. In addition as noted above, in some situations, papillomavirus E2 proteins can exert effects independently of HPV repression (9, 19-21, 28, 66, 84), presumably by binding and influencing the activity of cellular genes or proteins (e.g., see references 56 and 72). Therefore, although BPV E2-induced senescence requires E7 repression, it is also important to determine whether changes in gene expression that occur following E2 expression are solely due to repression of the endogenous HPV oncogenes or also require other effects of the E2 protein.
To address these issues, we conducted RNA microarray analysis on senescing HeLa cells in the absence of p53 activation. Our results revealed that few, if any, cellular genes are directly regulated by the BPV E2 protein or by responses to E7 repression independent of Rb family activation. Rather, the entire transcriptional response to the BPV E2 protein is mediated by E7 repression and activation of the Rb pathway. Our studies also provided insight into the mechanistic basis for the resulting transcriptional profile and senescent phenotype and revealed substantial overlap between Rb family-induced senescence and replicative senescence, suggesting that common transcriptional events may underlie both processes.
MATERIALS AND METHODS
Cell culture and viruses.
The HeLa/16E6-LXSN-1, HeLa/16E6-16E7-21, and HeLa/16E6-16E7Δ21-24-28 stable cell lines referred to here as the E6, E6E7, and E6E7Δ21-24 cells, respectively, were previously described (70). HeLa cells constitutively expressing the HPV16 E6 gene and either the wild-type adenovirus 5 13S E1A gene (E6/E1A cells) or E1A with a point mutation (C124G) in the Rb-binding domain (E6/E1A-C124G cells) were generated by infecting HeLa/16E6-5K cells (single-cell cloned from HeLa/16E6-5 cells) (18) with retroviral stocks generated from pBabe-13S E1A or pBabe-13S E1A-C124G. Cells were pooled after 2 days of selection in 0.4 μg/ml puromycin. To generate HeLa cells constitutively expressing the HPV16 E6 gene and DEK (E6/DEK) or B-MYB (E6/BM), full-length DEK cDNA (Invitrogen Mammalian Gene Collection [IMGC], clone 5122743) was cloned into the pLPCX retroviral vector and full-length B-MYB (IMGC, clone 3162656) was cloned into the pBabe-Puro retroviral vector. Concentrated retrovirus was used to infect HeLa/16E6-5K cells. Cells were plated at low density and selected for 5 to 6 days in medium containing 0.4 μg/ml puromycin. After further incubation in the absence of selection, individual colonies were expanded to generate clonal cell lines. Lines maintaining a high level of expression of each transcription factor mRNA after expression of the E2 protein, as assessed by quantitative real-time PCR (qRT-PCR), were selected for further analysis. To generate HeLa/E6 cells constitutively expressing the TAM67 dominant-negative deletion mutant of c-jun, the TAM67 c-jun gene was released from pLRT-TAM67 (46) with BamHI and cloned into the pBabe-Puro retroviral vector to generate pBabe-Puro-TAM67. Concentrated control Babe-Puro or Babe-Puro-TAM67 retrovirus was used to infect HeLa/16E6-5K cells, and cells were pooled after 2 days of selection in puromycin (0.4 μg/ml) to generate the E6/pBabe and E6/TAM67 cells, respectively. Primary human foreskin fibroblasts were harvested at passage 8 or serially passaged 1:4 until passage 26, at which time they had reached senescence. All cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 10 mM HEPES (pH 7.3), penicillin, and streptomycin.
Retroviruses were generated by cotransfecting 293T cells with the retroviral plasmids of interest, pVSVg (Clontech) and pCL-ECO (61). Retroviral supernatants were harvested in OptiMEM every 24 h for 3 to 4 days, pooled, and concentrated with Centricon Ultracel PL-30 columns (Microcon). Cells were infected at a multiplicity of infection of 2 to 3 and selected in puromycin. The wild-type BPV E2 protein or E2 with a point mutation in the DNA-binding domain (E2C340R) was expressed by infection at a multiplicity of infection of 20 with high-titer recombinant simian virus 40 (SV40) viruses (E2 virus or E2 mutant virus), amplified and with the titer determined as previously described (35, 59).
Microarray analysis.
A total of 1 × 106 E6, E6E7, or E6E7Δ21-24 cells were plated per 150-mm-diameter dish and mock infected or infected with the mutant or wild-type E2 virus the following day. Two days later, inhibition of DNA synthesis by expression of the wild-type E2 protein was confirmed by measuring bromodeoxyuridine incorporation using a cell proliferation enzyme-linked immunosorbent assay bromodeoxyuridine kit (Roche), and repression of the endogenous HPV18 E7 gene was confirmed by qRT-PCR. Three days after mock infection or infection with a virus expressing the wild-type or mutant E2 protein, cells were trypsinized and 1 × 106 cells were replated per 150-mm dish. Because expression of the E2 protein from the nonreplicating vector is diluted over time in proliferating cells, the E6E7 cells were reinfected with the E2 or E2 mutant virus at 4 days after the initial infection. Six days after the initial infection, total RNA was prepared using Qiashredder columns, the RNeasy Midi kit, and the RNase-free DNase set (QIAGEN). Three biological replicates of each cell sample were analyzed, one of which was hybridized twice. Labeled cDNA probes were prepared and microarrays were hybridized using protocols provided by the Keck Microarray Resource (KMR) at Yale University. Briefly, 50 μg of RNA concentrated using Microcon YM-30 columns (Millipore) was primed with random hexamers and oligo(dT) and reverse transcribed at 42°C for 2 h using Superscript II reverse transcriptase (Invitrogen) in the presence of 500 μM each dATP, dCTP, and dGTP and 200 μM each dTTP and amino-allyl dUTP. Reaction mixtures were treated with sodium hydroxide at 65°C for 20 min to remove the RNA template, neutralized, and purified using the CyScribe GFX purification kit (Amersham Biosciences) according to manufacturer's instructions, except columns were washed with 80% ethanol and samples were eluted with 0.1 M sodium bicarbonate, pH 9.0. Samples were labeled with Cy3 or Cy5 monoreactive dye (Amersham Biosciences) for 90 min in the dark. Reactions were quenched with ethanolamine, and the labeled cDNA probes were purified using the CyScribe GFX purification kit (Amersham Biosciences). Three micrograms each of differentially labeled E2-infected or control samples were mixed for each hybridization reaction, with the dyes used to label the E2-infected and control samples swapped in half of the replicates. Microarray slides were prepared by the KMR and contained approximately 28,000 unique oligonucleotide probes derived from the Human Genome Oligo set V2.0 and V3.0 (Operon) representing approximately 24,000 genes. Slides were covered with 22- by 60-mm LifterSlips (Erie Scientific), prehybridized at 55°C for 1 h, hybridized at 55°C overnight in Gene Machines hybridization chambers, washed, and scanned in an Axon GenePix 4000A scanner. Using the GenePix Pro3 software (Axon), spots were automatically detected and then the position and quality of each spot were manually confirmed. The Cy3 and Cy5 fluorescent intensities of each spot were quantified. Data for spots of good quality in which the fluorescence intensity was ≥3× above background in at least one channel were uploaded to the GeneSpring program (Silicone Genetics), and those genes which satisfied these criteria in at least half of the replicates were defined as expressed. A lowess normalization was applied, and the ratio of fluorescent intensities in the E2-infected samples and control samples was determined for each gene. Significant expression changes in HeLa cells were defined as a twofold or greater change in expression with a t test P value of ≤0.001. The gene ontology feature of GeneSpring and the online Database for Annotation, Visualization and Integrated Discovery 2006 (DAVID 2006) were used to classify genes according to function and subcellular location.
RNA for the microarray analysis of replicative senescence was harvested from the same strain of primary human foreskin fibroblasts at passage 8 and passage 26. Preparation of RNA, labeling of cDNA probes, hybridization of differentially labeled probes from senescent and proliferating cell samples to microarray slides, and data analysis were performed as described above for HeLa cells, except significant expression changes were defined as a twofold or greater change in expression with a t test P value of ≤0.01.
qRT-PCR.
RNA for qRT-PCR was prepared 6 days after mock infection or wild-type E2 infection as described above for the microarrays. RNA was converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad), and qRT-PCR was performed with Hot-Start iTaq DNA polymerase using 40 ng of cDNA and the iQ SYBR green supermix (Bio-Rad) in a single-color i-Cycler (Bio-Rad). Gene-specific primers were designed using the Universal ProbeLibrary ProbeFinder software (Roche). Results were normalized to expression of glyceraldehyde 3-phosphate dehydrogenase, which did not change in response to the E2 protein.
Western blots.
A total of 1 × 106 cells were lysed in 2× sample buffer (4% sodium dodecyl sulfate, 200 mM dithiothreitol, 300 mM Tris-HCl, pH 6.8, 20% glycerol, 5% β-mercaptoethanol, 0.04% bromophenol blue). Lysates were cleared by centrifugation, and Western blot analysis was performed as previously described (70) using c-jun (D) primary antibody (Santa Cruz Biotechnology, Inc.) and horseradish peroxidase-conjugated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch).
Gene knockdown.
Short hairpin RNAs (shRNAs) specifically targeting the Rb family members (p105Rb, p107, and p130), DEK, and B-MYB were designed using the BLOCK-iT RNA interference designer (Invitrogen). The following shRNAs were used for each gene: p105 sh#3, 5′-GGAAAGGACATGTGAACTTATCGAAATAAGTTCACATGTCCTTTCC-3′; p107 sh#1, 5′-GCACCAAGTGACCAACTTATACGAATATAAGTTGGTCACTTGGTGC-3′; p130 sh#1, 5′-GCATAGCTTGAGTCGTCTTCACGAATGAAGACGACTCAAGCTATGC-3′; DEK sh#1, 5′-GGAAGGCTAAGCGAACCAAATCGAAATTTGGTTCGCTTAGCCTTCC-3′; DEK sh#2, 5′-GCCTCGAAAGTGTCATCAATTCGAAAATTGATGACACTTTCGAGGC-3′; DEK sh#3, 5′-GCTTTCAGAGTTTGGTTAATCCGAAGATTAACCAAACTCTGAAAGC-3′; MYB sh#1, 5′-GAACTTCCAGTCCTGCTGTCCACGAATGGACAGCAGGACTGGAAG-3′; and MYB sh#2, 5′-GAAGTTCAGAAACTGGGAGGGCCGAAGCCCTCCCAGTTTCTGAAC-3′. These shRNAs were cloned into the pSIREN-RetroQ retroviral vector (BD Biosciences), in which expression of the shRNA was driven by the human U6 promoter. The negative control vector expresses a scrambled sequence that does not form a hairpin. To generate E6/shRbFam cells, serial infections with concentrated retrovirus were used to express p105 sh#3, p107 sh#1, and p130 sh#1 in HeLa/16E6-5K cells. After each infection, cells were selected for 2 to 3 days in medium containing 0.4 μg/ml puromycin, expanded, and screened by Western blotting or qRT-PCR for Rb family member knockdown. To inhibit expression of DEK and B-MYB in primary fibroblasts, concentrated retrovirus was used to infect proliferating primary human foreskin fibroblasts at passage 16. Cells were selected in 0.6 μg/ml puromycin for 4 to 5 days and stained 6 days later for SAβ-gal activity at pH 6.0, as previously described (23).
RESULTS
Microarray analysis of induced senescence.
To characterize induced senescence, transcriptional profiling was carried out in the HeLa/16E6 cell line (E6 cells) (18). In addition to the endogenous HPV18 E6 and E7 genes, these cells express the HPV16 E6 protein from a promoter that does not respond to the BPV E2 protein. Expression of the BPV E2 protein in E6 cells represses the HPV18 genes, but HPV16 E6 expression is maintained. E7 repression activates the Rb pathway and induces the cells to undergo Rb-dependent senescence, but the p53 pathway remains dormant because of constitutive expression of the HPV16 E6 protein. The BPV E2 gene was delivered by infection with a replication-defective SV40-based virus that infects essentially every cell in the population, so the senescence response unfolds uniformly throughout the entire population (36).
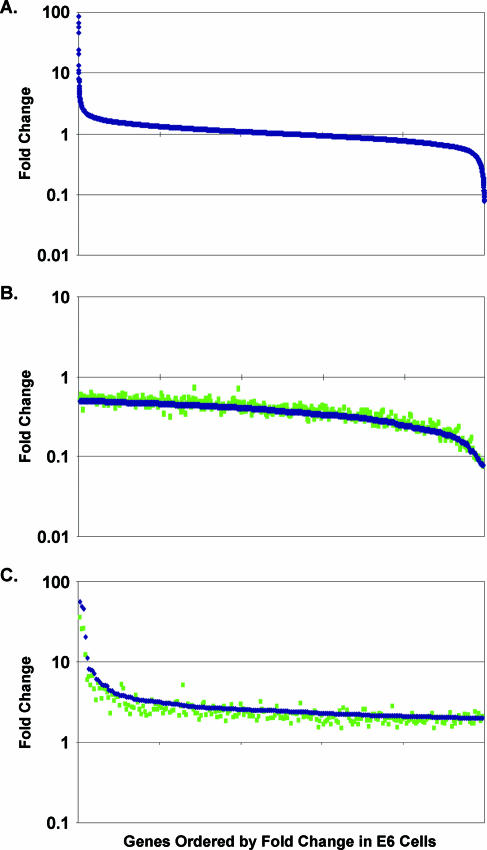
RNA was isolated in three independent experiments from E6 cells 6 days after mock infection or infection with the BPV E2 virus. This time point was chosen because preliminary studies showed that the vast majority of gene expression changes increased progressively following BPV E2 expression, and this time point allowed us to capture these changes with high statistical confidence. The RNA was reverse transcribed into cDNA and fluorescently labeled, and differentially labeled cDNAs from control cells and cells expressing the E2 protein were mixed and hybridized to arrays representing approximately 24,000 genes, 13,635 of which were expressed in E6 cells. The transcriptional response of these genes to E2 expression is shown in Fig. 1A, in which the relative expression of each gene is represented by a symbol. Using a P value cutoff of ≤0.001 applied to genes regulated twofold or more by E2 expression, 224 genes were induced and 352 genes were repressed. Thus, approximately 4% of the expressed genes were regulated twofold or more by the introduction of the BPV E2 gene into E6 cells.
FIG. 1.
E2 expression induces many changes in cellular gene expression in E6 cells. Each symbol represents a different gene, and genes are ordered along the x axis by change in expression in E6 cells in response to the E2 protein compared to mock-infected cells. Change in expression at 6 days after E2 expression is graphed on the y axis on a logarithmic scale. Blue symbols represent change after E2 expression in E6 cells when mock infection is used as a control. Green symbols represent change after E2 expression in E6 cells when infection with the E2 mutant is used as a control. (A) All 13,635 genes expressed in E6 cells. (B) A total of 352 genes repressed ≥2× in E6 cells in comparison to mock infection (P ≤ 0.001). (C) A total of 224 genes induced ≥2× in E6 cells in comparison to mock infection (P ≤ 0.001).
We conducted experiments to determine whether this pattern of cellular gene expression was due entirely to expression of a DNA binding-competent BPV E2 protein or whether the viral vector used to introduce the E2 gene directly affected transcription. In previous microarray analysis of E2-induced senescence, cells infected with an adenovirus vector expressing a full-length or chimeric E2 protein were compared to cells infected with the vector lacking E2 expression (82, 87). This design would fail to reveal changes caused by the vector itself, which might nonetheless contribute to senescence. We compared mock infection and infection with a virus expressing a BPV E2 DNA-binding mutant (E2C340R) as control samples. Any genes whose expression is directly affected by virus infection would show differential expression when wild-type E2-infected cells are compared to mock-infected cells but not to cells infected with the E2 mutant virus.
In fact, the wild-type BPV E2 protein generated essentially the same transcriptional profile when either control sample was used. Figure 1B shows genes repressed twofold or more in E6 cells by the wild-type E2 protein, and Fig. 1C shows genes induced twofold or more. Similar levels of repression and induction of all of these genes occurred when either infection with the E2 mutant (green symbols) or mock infection (blue symbols) was used as a control sample. The repression of two poorly expressed genes (bottom 7th percentile of expressed genes) appeared partially blocked when the E2 mutant was used as a control, but qRT-PCR analysis demonstrated that both genes were repressed to similar extents in comparison to either control. Therefore, although it is possible that direct effects of virus infection occur at very early time points, virus infection per se does not affect the expression of any cellular genes at 6 days after infection. Furthermore, the DNA binding activity of the BPV E2 protein is required for its transcriptional effects.
Confirmation of regulated gene expression during senescence.
To confirm the induction and repression of selected genes, independent RNA samples isolated from E6 cells at 6 days after infection with the wild-type or mutant E2 virus were analyzed by Northern blotting. This analysis confirmed that the HPV18 oncogenes and a major repressed cellular gene (FOXM1) were highly expressed in mutant-infected cells and displayed a substantial repression in response to the wild-type E2 protein and that a major induced gene (PSCA) was expressed at a low level in cells infected with the virus expressing the E2 mutant and underwent a substantial induction in response to the wild-type E2 protein (data not shown).
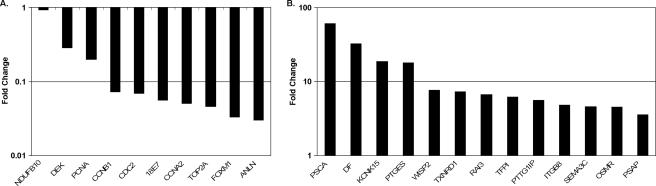
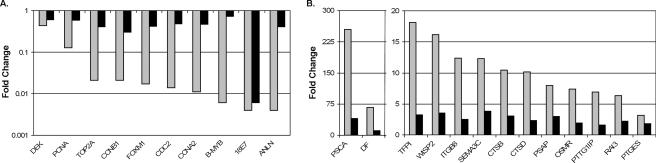
qRT-PCR analysis was used to examine the expression of a larger number of genes in a quantitative fashion. This analysis revealed excellent agreement with the microarray data, although in most cases the amplitude of the change was greater according to qRT-PCR. The NADH dehydrogenase (ubiquinone) 1β gene (NDUFB10), a gene that did not change in expression by microarray analysis, similarly showed no change when expression was assessed by qRT-PCR (Fig. 2A). In contrast, all 20 genes examined that were repressed by microarray analysis were also repressed when assessed by qRT-PCR (Fig. 2A) (data not shown). Similarly, we confirmed the induction of 38 out of 39 (>97%) of the induced genes (Fig. 2B) (data not shown). Therefore, the microarray results are an accurate reflection of gene expression changes in response to the E2 protein.
FIG. 2.
Confirmation of significantly induced and repressed genes in E6 cells by quantitative real-time PCR. Change in expression at 6 days after E2 infection of E6 cells compared to mock infection, as detected by qRT-PCR normalized to glyceraldehyde-3-phosphate dehydrogenase expression, is graphed for selected repressed (A) and induced (B) genes. NDUFB10 is a gene that did not change in expression on the array and is shown as a control in panel A.
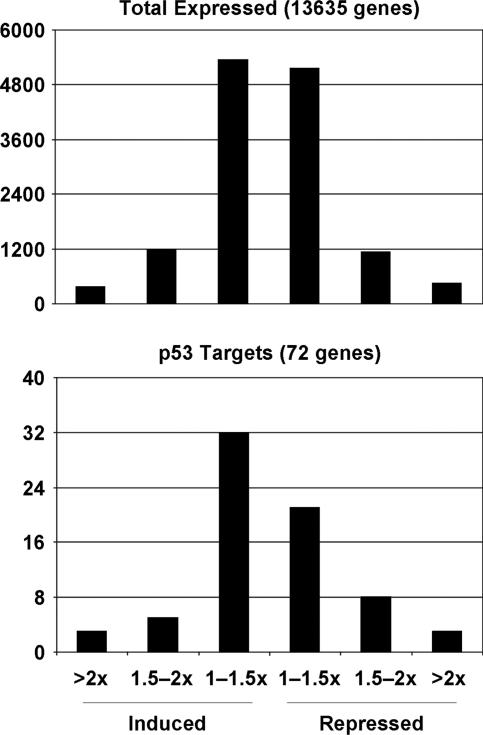
To confirm that the p53 pathway was not activated during senescence in this system, we examined the expression of genes known to be direct targets of p53 (85, 92). Seventy-two p53-responsive genes were expressed in E6 cells. Only three of these genes (4%) were induced twofold or more after expression of the E2 protein, and another three were repressed twofold or more (Fig. 3, bottom panel). Notably, expression of the vast majority of p53-dependent genes was not regulated to a significant extent by the E2 protein in these cells, and there was little if any enrichment of p53-regulated genes in the genes that were induced or repressed by the E2 protein (compare top and bottom panels of Fig. 3). These results confirmed that the BPV E2 protein did not activate the p53 pathway in E6 cells and that regulated expression of p53-responsive genes is not required for induced senescence in these cells.
FIG. 3.
p53 pathway is not activated during induced senescence in E6 cells. Genes expressed in E6 cells were divided into six bins according to the magnitude of expression changes at 6 days after expression of the E2 protein compared to mock infection. The total number of genes in each bin is graphed for all 13,635 expressed genes (top panel) and the 72 p53-responsive genes expressed in E6 cells (bottom panel).
Global transcriptional response to BPV E2 expression requires E7 repression.
Because there are many consensus E2 DNA binding sites in the cellular genome, it is possible that the E2 protein directly regulates the expression of some cellular genes. In addition, as well as binding Rb family members, the high-risk HPV E7 protein binds to several other proteins, including transcription factors. Therefore, it is important to determine the role of E7 repression and activation of the Rb family in the transcriptional response to E2 expression during induced senescence. For this purpose, we analyzed two additional cell lines. The HeLa/16E6-16E7 cell line (E6E7 cells) constitutively expresses both HPV16 E6 and HPV16 E7 from retrovirus vectors (18). Expression of the E2 protein in these cells represses the endogenous HPV18 oncogenes, but the cells continue to proliferate because expression of HPV16 E6 and E7 persists. In these cells, enforced expression of the HPV16 E7 protein prevents changes in cellular gene expression due to repression of the HPV18 E7 protein. The HeLa/16E6-16E7Δ21-24 cell line (E6E7Δ21-24 cells) constitutively expresses the HPV16 E6 gene and an HPV16 E7 mutant that is defective for binding and degrading Rb family members (70). This mutant does not block E2-induced senescence (70), but retains other E7 activities (3, 39, 40, 52). Therefore, any E2-induced changes in gene expression that are blocked by expression of the mutant E7 gene in E6E7Δ21-24 cells are likely Rb family-independent responses to E7 repression, whereas expression changes dependent on the Rb family should not be blocked by this mutant. As predicted, transient assays with an E2F-responsive reporter gene demonstrated that HPV18 E7 repression activated the Rb/E2F repressor pathway in E6 cells and that activation of the Rb pathway was blocked by constitutive expression of the wild-type HPV16 E7 protein in the E6E7 cells but not the E7 mutant in the E6E7Δ21-24 cells (data not shown).
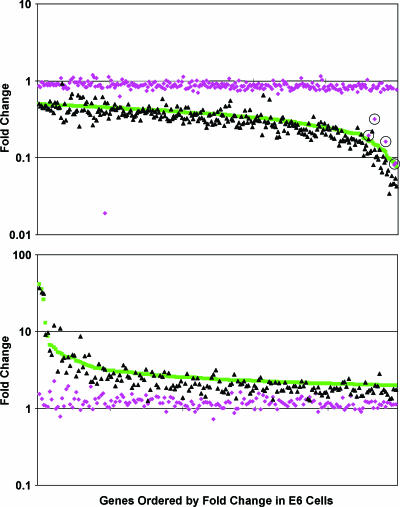
Microarray analysis was used to examine the global effect of the BPV E2 protein in the face of constitutive expression of the HPV16 E7 protein (Fig. 4). The repression and induction of essentially all of the genes significantly affected by the wild-type E2 protein in E6 cells (green symbols) were substantially inhibited by the wild-type HPV16 E7 protein (purple symbols). The five symbols representing genes significantly repressed by the E2 protein in E6E7 cells corresponded to the endogenous HPV18 DNA (circled symbols in Fig. 4, top panel). One gene appeared repressed by the E2 protein to a far greater extent in E6E7 cells, but because of the quality of the data for this gene, repression was not statistically significant (P = 0.2). Thus, gene induction and repression by the E2 protein were blocked by constitutive expression of the wild-type E7 protein in the E6E7 cells, indicating that transcriptional effects of the BPV E2 protein 6 days after E2 introduction require E7 repression.
FIG. 4.
Comparison between the transcriptional profiles of E6 cells (green), E6E7 cells (purple), and E6E7Δ21-24 cells (black) after expression of the E2 protein. Change in expression at 6 days after E2 infection is graphed for the genes repressed (top panel) or induced (bottom panel) twofold or more (P ≤ 0.001) in E6 cells compared to infection with the E2 mutant. Because the magnitude of gene expression changes was, in general, slightly decreased when infection with the E2 mutant virus rather than mock infection was used as a control, according to these criteria, the E2 protein induced 175 genes and repressed 294 genes. Each symbol indicates a different gene, ordered along the x axis by change in expression in E6 cells. Circled symbols correspond to integrated HPV DNA.
Microarray analysis was also used to examine gene expression 18 h after infection of E6 cells with the E2 virus. At this early time point, induction of the SV40 capsid genes (expressed from the viral vector) and repression of the endogenous HPV18 E7 gene were the only significant expression changes observed in response to the E2 protein (data not shown). Thus, repression of the endogenous E7 oncogene preceded any sustained changes in cellular transcription. Because direct targets of the E2 protein would presumably show changes in gene expression early after E2 expression, these results supported the surprising conclusion that the BPV E2 protein does not directly bind to and regulate the expression of any cellular genes at 6 days after infection.
Global transcriptional response to BPV E2 expression requires Rb family activation.
To determine whether there was a component of the transcriptional response to E7 repression that was independent of Rb family activation, we examined E2-mediated gene expression changes in E6E7Δ21-24 cells. As shown in Fig. 4, the transcriptional responses to BPV E2 expression were essentially identical in the E6 cells (green symbols) and the E6E7Δ21-24 cells (black symbols), but clearly divergent from the pattern in E6E7 cells (purple symbols). In fact, the vast majority (∼98%) of genes were induced or repressed to similar extents in E6 cells and E6E7Δ21-24 cells. The expression of the handful of genes that appeared divergent in these two cell lines was assayed by qRT-PCR analysis of independent RNA samples. This analysis revealed that four of these induced genes did in fact display Rb family-dependent regulation and that the expression of the other outliers was either undetectable or was not regulated by E2 expression in E6 cells (data not shown). Thus, we did not identify a single gene that was regulated in an Rb family-independent manner. The inability of the HPV16 E7 mutant to block the transcriptional response to HPV18 E7 repression strongly suggests that the major expression changes during senescence induced by E7 repression were dependent on activation of the Rb family.
As an independent assessment of the role of the Rb family in generating the transcriptional profile after E7 repression, we analyzed E6 cells expressing either a wild-type adenovirus type 5 13S E1A protein, which binds and inactivates Rb family members, or 13S E1A-C124G, which contains a mutation in the LXCXE motif that prevents binding to p105Rb and p130, but retains a reduced ability to bind p107 (17). Other than the sequences required for the binding and degradation of the Rb family, the sequences of E7 and E1A are largely unrelated (69). The wild-type and mutant E1A proteins were expressed at similar levels, and the wild-type but not the mutant E1A protein prevented E2-mediated senescence (data not shown). RNA was isolated from these cells 6 days after mock infection or infection with the E2 virus and analyzed by qRT-PCR. Genes identified as being repressed or induced by microarray analysis of E6 cells were also repressed or induced in cells expressing the E1A mutant deficient for binding Rb family members, but regulation of these genes was markedly inhibited by expression of wild-type E1A, despite effective repression of the endogenous E7 gene in these cells (Fig. 5A and B). These results provide independent evidence that induction and repression of the vast majority of genes during induced senescence are dependent on activation of the Rb family.
FIG. 5.
Effect of adenovirus E1A on gene induction and repression by the E2 protein. Expression of selected repressed (A) and induced (B) genes was examined by qRT-PCR at 6 days after repeated E2 infection of E6/E1A cells (black) or E6/E1A-C124G cells (gray), compared to mock infection, and normalized for glyceraldehyde-3-phosphate dehydrogenase expression.
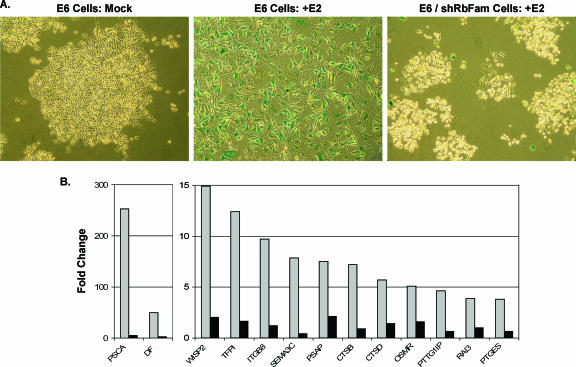
As a final test of the role of the Rb family in senescence in E6 cells, short hairpin interfering RNAs (shRNAs) were designed to specifically target p105Rb, p107, and p130. Rb family shRNAs were expressed individually or in combination in E6 cells by infection with high-titer retroviruses, and knockdown of p105Rb, p107, and p130 was confirmed by qRT-PCR and/or Western blotting (data not shown). E6 cells expressing each Rb family shRNA individually still senesced in response to expression of the BPV E2 protein (data not shown), suggesting that no specific Rb family member was required to initiate a senescence response in E6 cells. However, reduction in expression of individual Rb family members caused compensatory increases in other family members (data not shown). Therefore, we used serial infection with concentrated shRNA retroviral stocks to generate E6/shRbFam cells in which the expression of all three Rb family members was repressed as assessed by qRT-PCR (data not shown). As shown in Fig. 6A, 10 days after repeated infection with the E2 virus, parental E6 cells acquired a senescent morphology and positive staining for SAβ-gal activity, but E2-infected E6/shRbFam cells continued to proliferate and formed small colonies lacking SAβ-gal activity, confirming that E2-mediated senescence in E6 cells requires activation of the Rb family. Because E6/shRbFam cells grew poorly in culture, microarray analysis of these cells was not performed. Rather, qRT-PCR analysis was conducted on RNA harvested from E6/shRbFam cells and parental E6 cells 6 days after mock infection or E2 infection. Even though the E2 protein repressed the endogenous HPV18 E7 gene 38-fold in E6/shRbFam cells (data not shown), E2-mediated induction and repression of cellular genes were markedly inhibited by expression of shRNAs targeting the Rb family (Fig. 6B) (data not shown). These results provide additional direct evidence that gene regulation during senescence triggered by E7 repression requires activation of the Rb family.
FIG. 6.
Short hairpin RNAs targeting the Rb family block E2-induced senescence and gene induction. (A) E6 cells or E6/shRbFam cells were stained for senescence-associated β-galactosidase activity 10 days after mock infection or repeated E2 infection. (B) Expression of selected induced genes was examined by qRT-PCR analysis of RNA harvested 6 days after repeated E2 infection of E6 cells (gray) or E6/shRbFam cells (black), compared to mock infection, and normalized to glyceraldehyde-3-phosphate dehydrogenase expression.
Functional classification of genes regulated during induced senescence.
The 20 genes showing the greatest induction and repression during induced senescence are listed in Tables 1 and 2, respectively. For a complete list of regulated genes, see Tables S1A and S1B in the supplemental material. Classes of repressed genes include many histones and other cell cycle-related genes. A total of 336 of the 352 genes repressed twofold or more (P ≤ 0.001) were present in the DAVID 2006 database. Nineteen percent of these 336 genes were classified as cell cycle-related genes, and the P value for this classification (a measure of overrepresentation of a functional category compared to its representation in the Homo sapiens proteome) was 2.7E-29. Twenty-six genes involved in DNA repair and 33 genes involved in mitosis, some of which were also in the cell cycle class, were repressed (P values of 3.6E-14 and 2.1E-27, respectively).
TABLE 1.
Top 20 significantly induced genes in HeLa/16E6 cells
| Fold inductiona | Entrez gene no. | Gene symbol | Description |
|---|---|---|---|
| 56.5 | 84460 | ZMAT1 | Zinc finger, matrin type 1 |
| 48.8 | 80183 | C13orf18 | Clone RP11-247M1 on chromosome 13 |
| 46.0 | 84725 | PLEKHA8 | Pleckstrin homology domain containing, family A member 8 |
| 20.4 | 23387 | KIAA0999 | cDNA FLJ25293 fis, clone STM07384 |
| 11.2 | 90120 | LOC90120 | Homo sapiens cDNA FLJ11396 fis, clone HEMBA1000604 |
| 8.1 | 1675 | DF | D component of complement (adipsin) |
| 7.9 | 22890 | ZBTB1 | Zinc finger and BTB domain containing 1 |
| 7.9 | 8334 | HIST1H2AC | Histone 1, H2ac |
| 7.2 | 3906 | LALBA | Lactalbumin, alpha |
| 6.1 | 3696 | ITGB8 | Integrin, beta 8 |
| 6.0 | 51406 | NOL7 | 27-kDa nucleolar protein |
| 5.6 | 8000 | PSCA | Prostate stem cell antigen |
| 5.3 | 10512 | SEMA3C | Semaphorin 3C |
| 5.2 | 9635 | CLCA2 | Chloride channel, calcium activated, family member 2 |
| 5.1 | 8349 | HIST2H2BE | H2B histone family, member Q |
| 5.0 | 8714 | ABCC3 | ATP-binding cassette, subfamily C (CFTR/MRP), member 3 |
| 4.5 | AF017104 | Chromosome 7 common fragile site | |
| 4.4 | 440533 | PSG8 | Pregnancy-specific beta-1-glycoprotein 8 |
| 4.3 | AC009708 | Chromosome 8, clone RP11-318G5 | |
| 4.1 | 60598 | KCNK15 | Potassium channel, subfamily K, member 15 (TASK-5) |
Induction in E6 cells 6 days after expression of the E2 protein compared to mock infection.
TABLE 2.
Top 20 significantly repressed genes in HeLa/16E6 cells
| Fold repression in E6 cellsa | Fold repression in fibroblastsb | Entrez gene no. | Gene symbol | Description | Rb/E2F targetc |
|---|---|---|---|---|---|
| 12.8 | 4.5 | 259266 | ASPM | Abnormal spindle-like microcephaly-associated protein | |
| 12.6 | 3.3 | 9232 | PTTG1 | Pituitary tumor-transforming 1 | X |
| 10.9 | 4.2 | 983 | CDC2 | Cell division cycle 2, G1 to S and G2 to M | X |
| 10.7 | 3.5 | 7153 | TOP2A | Topoisomerase (DNA) II alpha (170 kDa) | X |
| 10.0 | 2.9 | 55872 | PBK | PDZ binding kinase | X |
| 9.4 | 2.9 | 54443 | ANLN | Anillin, actin binding protein | X |
| 9.2 | 4.8 | 259266 | ASPM | Abnormal spindle-like microcephaly-associated protein | |
| 8.6 | 3.4 | 51203 | NUSAP1 | Nucleolar and spindle-associated protein 1 | X |
| 8.6 | 2.8 | 195828 | ZNF367 | Zinc finger protein 367 | |
| 8.6 | 4.0 | 10744 | PTTG2 | Pituitary tumor-transforming 2 | |
| 8.6 | 3.7 | 6241 | RRM2 | Ribonucleotide reductase M2 polypeptide | |
| 7.7 | 3.8 | 26255 | PTTG3 | Pituitary tumor-transforming 3 | |
| 7.1 | 3.5 | 890 | CCNA2 | Cyclin A2 | X |
| 6.9 | 2.9 | 29028 | ATAD2 | ATPase family, AAA domain containing 2 | |
| 6.7 | 2.8 | 3070 | HELLS | Helicase, lymphoid specific | |
| 6.4 | 3.0 | 55635 | DEPDC1 | DEP domain containing 1 | X |
| 6.1 | 3.7 | 64151 | HCAP-G | Chromosome condensation protein G | X |
| 6.1 | 2.8 | 9133 | CCNB2 | Cyclin B2 | X |
| 6.1 | 3.1 | 6941 | TCF19 | Transcription factor 19 (SC1) | X |
| 6.0 | 2.9 | 79968 | WDR76 | WD repeat domain 76 | X |
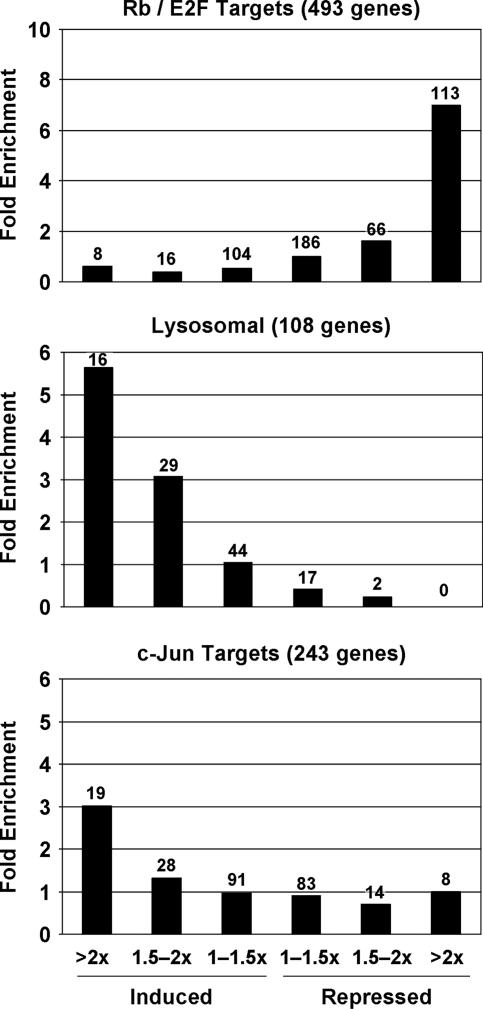
To assess whether genes in particular functional classes were preferentially induced or repressed relative to all 13,635 genes expressed in these cells, each gene was placed in one of six bins according to its change in expression in response to the E2 protein. Expression of the vast majority of genes did not change and hence was not associated with significant P values. Therefore, we did not apply a P value cutoff for these assignments. For each functional class, the distribution of genes within these bins was then compared to the distribution of all expressed genes. A total of 493 Rb/E2F target genes, identified by chromatin immunoprecipitation against E2F4 and Rb family members followed by hybridization to human promoter arrays (4, 12, 73, 86), were expressed in E6 cells. Although only 3.3% of all expressed genes were repressed twofold or more, 113 (23%) of the Rb/E2F target genes were repressed, representing a sevenfold enrichment (Fig. 7, upper panel). Indeed, 16 of the top 25 repressed cellular genes were known E2F target genes (Table 2) (data not shown). These results demonstrated that, as expected, mobilization of an Rb/E2F transcriptional inhibitory response was a major consequence of E7 repression in E6 cells.
FIG. 7.
Regulation of specific functional classes of genes during induced senescence. Genes expressed in E6 cells were divided into six bins according to the magnitude of expression changes at 6 days after expression of the E2 protein compared to mock infection. Enrichment for a given functional class was calculated by dividing the percentage of genes in that functional class in each bin by the percentage of total expressed genes in that bin. The number of genes in each bin is shown above the bars.
Even though gene induction was also dependent on Rb family activation, induced genes included few known Rb/E2F targets. Only eight (1.6%) of the known Rb/E2F target genes were induced twofold or more, while 2.6% of the total genes were induced to this extent. The major class of induced genes encoded proteins involved in lysosome function (Table 3). Sixteen lysosomal genes were induced greater than twofold, representing a five- to sixfold enrichment, another 29 lysosomal genes were induced greater than 1.5-fold, and none were repressed greater than 2-fold (Fig. 7, middle panel). The P value for overrepresentation of lysosomal genes in the DAVID database among genes induced twofold or more (P ≤ 0.001) was highly significant (P value of 8.7E-9). Additional functional classes of induced genes that were significantly enriched (P ≤ 0.01) were the plexin/semaphorin/integrin class, the electron transport and oxidoreductase activity classes, the transporter activity class, the endoplasmic reticulum class, the calcium binding class, the protease class, and the complement pathway class (see Table S2 in the supplemental material).
TABLE 3.
Induced lysosomal genes in HeLa/16E6 cells
| Fold inductiona | Entrez gene no. | Gene symbol | Description |
|---|---|---|---|
| 3.8 | 427 | ASAH1 | N-Acylsphingosine amidohydrolase (acid ceramidase) |
| 3.1 | 1509 | CTSD | Cathepsin D (lysosomal aspartyl protease) |
| 3.1 | 54414 | CSE-C | Cytosolic sialic acid 9-O-acetylesterase homolog |
| 2.6 | 4126 | MANBA | Mannosidase, beta A, lysosomal |
| 2.6 | 1508 | CTSB | Cathepsin B |
| 2.4 | 6609 | SMPD1 | Sphingomyelin phosphodiesterase 1, acid lysosomal (acid sphingomyelinase) |
| 2.4 | 414 | ARSD | Arylsulfatase D |
| 2.3 | 1200 | CLN2 | Ceroid-lipofuscinosis, neuronal 2, late infantile (Jansky-Bielschowsky disease) |
| 2.3 | 5660 | PSAP | Prosaposin |
| 2.3 | 51449 | PCYOX1 | Prenylcysteine oxidase 1 |
| 2.1 | 537 | ATP6S1 | ATPase, H+ transporting, lysosomal (vacuolar proton pump), subunit 1 |
| 2.0 | 57192 | MCOLN1 | Mucolipin 1 |
| 2.0 | 967 | CD63 | CD63 antigen (melanoma 1 antigen) |
Induction in E6 cells 6 days after expression of the E2 protein compared to mock infection.
In addition to its effects on E2F function, p105Rb can bind and stimulate the transcriptional activity of c-jun, a component of the AP1 transcriptional activator. Furthermore, qRT-PCR analysis revealed a >10-fold Rb family-dependent induction of c-jun mRNA itself following E7 repression, consistent with the known autoregulatory activity of c-jun on its own promoter (1) (data not shown). A total of 243 c-jun target genes were expressed in E6 cells (excluding genes also known to be Rb/E2F targets) (38). Nineteen of these genes were induced twofold or more, representing a threefold enrichment (Fig. 7, bottom panel).
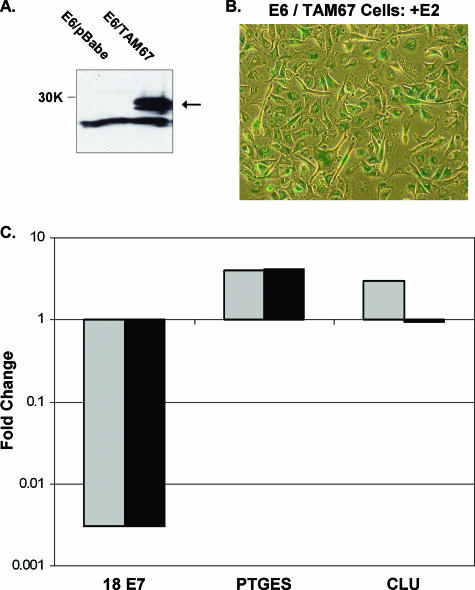
To analyze the role of c-jun activation in Rb family-dependent gene induction, we generated E6 cells expressing the TAM67 transactivation domain deletion mutant of c-jun, which functions as a dominant negative (46) (E6/TAM67 cells). Expression of the c-jun mutant in these cells was confirmed by Western blotting (Fig. 8A). E6/TAM67 cells efficiently entered senescence in response to expression of the E2 protein (Fig. 8B) (data not shown), indicating that c-jun activity was not required for senescence in this system. qRT-PCR was used to analyze RNA harvested from E6/TAM67 cells and control cells 6 days after mock infection or repeated E2 infection (Fig. 8C) (data not shown). All of the repressed genes examined, including the endogenous HPV18 E7 gene, and many of the induced genes, including the prostaglandin E synthase (PTGES) gene, were regulated by the E2 protein to a similar extent in the E6/TAM67 cells and in the control cells. PTGES lacks a recognizable AP1 site and is not a known c-jun target. However, expression of the dominant-negative c-jun mutant blocked the E2-mediated induction of several genes, most dramatically the gene coding for clusterin, a known target of c-jun (38) that contains an exact AP1 consensus site in its promoter. Inhibition of clusterin induction by TAM67 in three independent experiments was significant (P ≤ 0.007). Therefore the activation of c-jun transcriptional activity appears to be in part responsible for Rb family-dependent gene induction in response to E7 repression.
FIG. 8.
Expression of a dominant-negative c-jun mutant blocks E2-mediated induction of a known c-jun target gene. (A) Western blot of the c-jun TAM67 mutant. E6 cells transduced with an empty vector or E6/TAM67 cells are shown. An arrow indicates the TAM67 doublet at the predicted molecular size of 29 kDa (30K marker on the left). (B) E6/TAM67 cells that were stained for senescence-associated β-galactosidase activity 10 days after repeated E2 infection. (C) RNA was isolated from E6 cells containing an empty vector (gray) or E6/TAM67 cells (black) 6 days after mock infection or two rounds of E2 infection and analyzed by qRT-PCR for expression of HPV18 E7 (18 E7), PTGES, or clusterin (CLU). Data are plotted as described in the legend to Fig. 2.
Mobilization of multiple signals that can induce senescence.
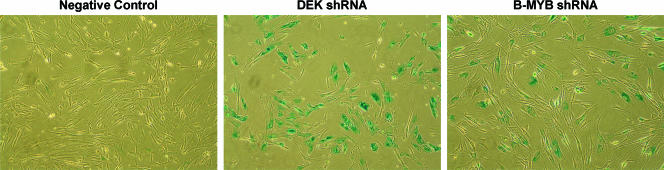
Activation of the Rb family repressed 19 transcriptional regulators twofold or more during induced senescence in E6 cells, 8 of which are known E2F targets, including DEK and B-MYB (designated MYBL2) (Table 4) (14, 50). Published studies indicate that DEK and B-MYB may play a role in cell growth control (55, 89, 90). To determine if repression of DEK and B-MYB was sufficient to induce senescence in the absence of HPV oncogenes, concentrated retrovirus stocks were used to introduce DEK and B-MYB shRNAs into mid-passage primary human fibroblasts. Knockdown of the expression of DEK (up to 100-fold) and B-MYB (up to 12-fold) was confirmed by qRT-PCR (data not shown). Inhibition of the expression of either DEK or B-MYB in fibroblasts induced senescence, as assessed by growth arrest, cell morphology, increased autofluorescence, and SAβ-gal activity (Fig. 9) (data not shown). The same result was obtained with multiple shRNAs for each gene. Cells infected at the same passage number with a control retrovirus containing a scrambled insert did not senesce. Therefore, the repression of either DEK or B-MYB is sufficient to induce senescence. Previous studies reported that B-MYB or DEK repression can inhibit cell growth or survival, but did not explicitly demonstrate the acquisition of senescence markers (2, 31, 71, 75, 89).
TABLE 4.
Repressed transcription factors in HeLa/16E6 cells
| Fold repressiona | Entrez gene no. | Gene symbol | Description | Rb/E2F targetb |
|---|---|---|---|---|
| 12.6 | 9232 | PTTG1 | Pituitary tumor transforming 1 | X |
| 8.6 | 195828 | ZNF367 | Zinc finger protein 367 | |
| 6.1 | 6941 | TCF19 | Transcription factor 19 (SC1) | X |
| 3.9 | 58500 | ZNF250 | Zinc finger protein 250 | |
| 3.4 | 7913 | DEK | DEK oncogene | X |
| 3.3 | 2305 | FOXM1 | Forkhead box M1 | |
| 3.1 | 4678 | NASP | Nuclear autoantigenic sperm protein (histone binding) | X |
| 3.0 | 4609 | MYC | v-myc myelocytomatosis viral oncogene homolog (avian) | X |
| 2.8 | 3181 | HNRPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | X |
| 2.7 | 3148 | HMGB2 | High-mobility group box 2 | |
| 2.7 | 4605 | MYBL2 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | X |
| 2.5 | 10151 | HNRPA3 | Heterogeneous nuclear ribonucleoprotein A3 | |
| 2.5 | 388275 | HNRPA1 | Heterogeneous nuclear ribonucleoprotein A1 | X |
| 2.3 | 253782 | LASS6 | LAG1 longevity assurance homolog 6 (Saccharomyces cerevisiae) | |
| 2.3 | 3187 | HNRPH1 | Heterogeneous nuclear ribonucleoprotein H1 (H) | |
| 2.2 | 3189 | HNRPH3 | Heterogeneous nuclear ribonucleoprotein H3 (2H9) | |
| 2.2 | 3159 | HMGA1 | High-mobility group AT-hook 1 | |
| 2.1 | 5036 | PA2G4 | Proliferation-associated 2G4, 38 kDa | |
| 2.1 | 6628 | SNRPB | Small nuclear ribonucleoprotein polypeptides B and B1 |
FIG. 9.
Inhibition of expression of DEK and B-MYB induces senescence in primary fibroblasts. Short hairpin interfering RNAs (shRNAs) specifically targeting DEK or B-MYB (DEK sh#2 or MYB sh#2) or a control vector was expressed in mid-passage primary human foreskin fibroblasts, and cells were stained in situ 10 days later for senescence-associated β-galactosidase activity.
We also determined whether repression of DEK or B-MYB was required for senescence in E6 cells. Retrovirus infection was used to constitutively express these genes in E6 cells, and clonal lines maintaining a high level of expression of each transcription factor after E2 expression were identified by qRT-PCR analysis. Wise-Draper and colleagues reported that overexpression of DEK using an adenovirus expression vector provides partial protection against senescence induced by HPV E6 and E7 repression in HeLa cells (90). In contrast, in our experiments, E6 cells constitutively expressing DEK or B-MYB efficiently senesced in response to the BPV E2 protein (data not shown). Thus, although repression of DEK or B-MYB was sufficient to induce senescence in primary cells, maintenance of the expression of these genes did not block E2-induced senescence in HeLa cells. Taken together, these results indicated that E7 repression and activation of the Rb pathway mobilized multiple signals that were independently capable of inducing senescence. Even if some of these signals were blocked, the remaining signals were sufficient to trigger the senescence response.
Comparison between induced and replicative senescence.
To compare the transcriptional profile of induced senescence to the most commonly studied system of replicative senescence, we used microarray analysis to identify differences in gene expression in late-passage primary human foreskin fibroblasts that had undergone replicative senescence compared to the same strain of fibroblasts at early passage. Because the amplitude of changes in gene expression was, in general, lower in the fibroblasts than in the senescent HeLa cells and displayed more variability between the biological replicates, the expression changes in the fibroblasts tended to display lower statistical significance than the changes in the HeLa cells. Therefore, a P value cutoff of 0.01 was used to identify 248 genes induced twofold or greater in fibroblasts during replicative senescence and 284 repressed genes. For the complete list of genes regulated during replicative senescence in primary fibroblasts, see Tables S1C and S1D in the supplemental material. In senescent fibroblasts, there was a 6.2-fold enrichment for induction of p53-responsive genes and a 7.7-fold enrichment for repression of Rb/E2F-responsive genes, indicating that as expected both the p53 and Rb pathways were activated during replicative senescence (data not shown). There was substantial overlap between the results of these microarray experiments and similar published studies of replicative senescence in late-passage human fibroblasts (e.g., reference 93) (data not shown).
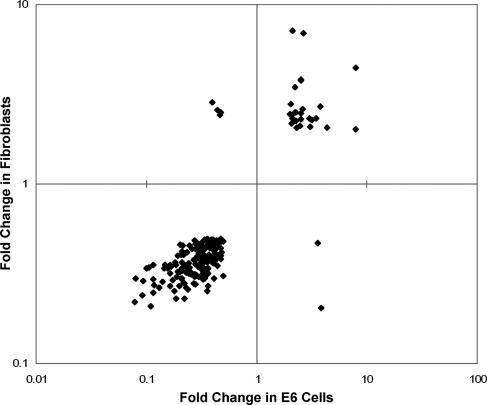
A large number of genes were regulated in common in senescing HeLa cells and fibroblasts. A total of 207 genes were regulated twofold or more in both E6 cells (P ≤ 0.001) and primary fibroblasts (P ≤ 0.01), representing 39% of the genes regulated during replicative senescence. Twenty-seven of these 207 genes were induced in both cell types during senescence, and 174 were repressed in both cell types, and the expression of only 6 genes changed in different directions (Fig. 10 and see Table S3 in the supplemental material). Therefore, 97% of the genes regulated twofold or more in both systems of senescence were regulated in the same direction, while only 3% showed divergent regulation. The 27 genes induced in both senescent HeLa cells and fibroblasts include c-jun targets and the lysosomal genes acid ceramidase (ASAH1) and prenylcysteine oxidase 1 (PCYOX1) (Table 5). Strikingly, all 20 of the top repressed genes in senescent HeLa cells were also repressed in fibroblasts and are primarily cell-cycle-related genes (Table 2). Thus, there is considerable overlap in the transcriptional profiles of Rb family-mediated induced senescence in HeLa cells and replicative senescence in fibroblasts. This similarity establishes the critical role for the Rb family in driving the transcriptional profile of replicative senescence.
FIG. 10.
Comparison between the transcriptional profiles of replicative senescence and induced senescence. The 207 genes regulated twofold or more during senescence in both fibroblasts (P ≤ 0.01) and E6 cells (P ≤ 0.001, comparing E2 infected to mock infected) are shown. Change in fibroblasts is graphed on the y axis, and change in E6 cells is graphed on the x axis.
TABLE 5.
Genes induced in both HeLa/16E6 cells and fibroblasts
| Fold induction in E6 cellsa | Fold induction in fibroblastsb | Entrez gene no. | Gene symbol | Description | Classc |
|---|---|---|---|---|---|
| 7.9 | 2.0 | 22890 | ZBTB1 | Zinc finger and BTB domain containing 1 | |
| 7.9 | 4.4 | 8334 | HIST1H2AC | Histone 1, H2ac | |
| 4.4 | 2.1 | 440533 | PSG8 | Pregnancy-specific beta-1-glycoprotein 8 | |
| 3.8 | 2.7 | 427 | ASAH1 | N-Acylsphingosine amidohydrolase (acid ceramidase) | L |
| 3.5 | 2.3 | 6840 | SVIL | Supervillin | CB |
| 3.2 | 2.3 | 2247 | FGF2 | Fibroblast growth factor 2 (basic) | CJ |
| 3.1 | 2.1 | 26060 | APPL | Adaptor protein, pH and PTB domains, leucine zipper motif | |
| 3.0 | 2.3 | 84140 | cDNA FLJ13305 fis, clone OVARC1001399 | ||
| 2.7 | 6.9 | 23766 | GABARAPL3 | GABA(A) receptors associated protein like 3 | |
| 2.6 | 2.6 | 134145 | Chromosome 5 clone CTD-2199O4 | ||
| 2.5 | 2.5 | 390 | RND3 | Rho family GTPase 3 | |
| 2.5 | 3.8 | 440449 | Hypothetical gene supported by AF086204 | ||
| 2.5 | 2.3 | 51706 | CYB5R1 | Cytochrome b5 reductase 1 | ET, TA |
| 2.5 | 3.8 | 718 | C3 | Complement component 3 | CP |
| 2.5 | 2.1 | 57088 | PLSCR4 | Phospholipid scramblase 4 | TA, CB |
| 2.3 | 2.1 | 26020 | LRP10 | Low-density lipoprotein receptor-related protein 10 | |
| 2.3 | 2.5 | 51449 | PCYOX1 | Prenylcysteine oxidase 1 | L, O |
| 2.3 | 2.3 | 9052 | RAI3 | Retinoic acid-induced 3 | |
| 2.2 | 3.5 | 7474 | WNT5A | Wingless-type mouse mammary tumor virus integration site family, member 5A | |
| 2.2 | 2.5 | 81533 | ITFG1 | Integrin alpha FG-GAP repeat containing 1 | |
| 2.2 | 2.2 | 64065 | PERP | TP53 apoptosis effector; p53-induced protein PIGPC1 | |
| 2.1 | 7.2 | 23710 | GABARAPL1 | GABA(A) receptor-associated protein-like 1 | |
| 2.1 | 2.3 | 5654 | PRSS11 | Protease, serine, 11 (IGF binding) | P |
| 2.1 | 2.2 | 2745 | GLRX | Glutaredoxin (thioltransferase) | CJ, ET, TA |
| 2.1 | 2.4 | 1601 | DAB2 | Disabled homolog 2, mitogen-responsive phosphoprotein | |
| 2.1 | 2.8 | 114294 | LACTB | Lactamase, beta | |
| 2.0 | 2.5 | 1026 | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
Induction in E6 cells 6 days after expression of the E2 protein compared to mock infection.
Induction in senescent fibroblasts compared to proliferating fibroblasts.
L, lysosomal; CB, calcium binding; CJ, c-jun target; ET, electron transport activity; TA, transporter activity; CP, complement pathway; O, oxidoreductase activity; P, protease.
DISCUSSION
Repression of the HPV18 E7 gene in HeLa cervical carcinoma cells by expression of the BPV E2 protein induces a senescence response that requires activation of endogenous Rb family members but not the p53 pathway. At 6 days after introduction of the BPV E2 gene, we did not detect any transcriptional effects due to virus infection per se or to DNA binding-independent effects of the BPV E2 protein. Strikingly, the BPV E2 transcription factor did not directly affect the expression of any cellular genes in this system, but rather the entire gene expression profile required E7 repression. At face value, this finding conflicts with reports that E2 proteins can exert biological effects independently of HPV repression. However, HPV-independent effects of E2 were demonstrated for the most part with high-risk HPV E2 proteins, which appear to have additional biological activities compared to the BPV E2 protein or the low-risk HPV E2 proteins (6, 9, 19, 28, 66, 84). In addition, many of these other studies utilized adenovirus infection, transfection, induction of E2 expression with heavy metals, or fusion to reporter proteins, which may influence the cellular response to the E2 protein.
Even though consensus E2 binding sites occur thousands of times in the human genome, our results indicate that the BPV E2 protein acts remarkably specifically on the integrated HPV18 genome. There are four conserved E2 binding sites in the viral long control region, including two separated by only 4 bp immediately upstream of the E6/E7 promoter. This arrangement may facilitate specific, cooperative binding and action of the BPV E2 protein at the viral DNA. It is also possible that cellular E2 sites are inaccessible to E2 binding because they are methylated or in heterochromatin. Methylation of CpG dinucleotides in the E2 binding sites blocks the ability of the E2 protein to bind (81) and affect transcription (47). Finally, the consensus E2 DNA binding site was determined in in vitro studies using naked DNA, but in vivo the E2 protein may prefer its natural sites in chromatin and in the presence of cellular proteins.
In addition to Rb family members, the E7 protein interacts with numerous other binding partners, including cdk inhibitors p21 and p27, several transcription factors, transcriptional coactivators, and proteins involved in chromatin remodeling (58), and Rb family-independent functions of E7 have been described (5, 8, 25). Nevertheless, the gene expression changes in E6 cells 6 days after expression of the BPV E2 protein were blocked by constitutive expression of wild-type 16 E7, but not by expression of an E7 mutant defective for binding and degradation of Rb family members. This E7 mutant is not globally disrupted but retains several biological activities, including the ability to bind the p600 Rb-associated protein, pCAF acetyltransferase, and the BRG1 component of SWI-SNF chromatin remodeling complexes and the ability to abrogate p21-induced cell cycle arrest (3, 39, 40, 52). In fact, interaction domains for most cellular E7-binding proteins are within the carboxyl terminus of E7, distal to the Rb binding motif (58). However, interaction between E7 and some cellular proteins other than Rb family members involves the Rb binding domain (7, 67), and the E7Δ21-24 mutation can affect p21 binding in vitro (44). Therefore, we used two additional approaches to independently confirm the Rb family dependence of the transcriptional response to E7 repression. Induction and repression of all the genes examined were also inhibited by expression of shRNAs targeting all three Rb family members or by expression of the wild-type adenovirus type 5 E1A protein, but not by an E1A mutant defective for binding Rb family members. Overall, different patterns of gene expression were not detected when various methods were used to ablate the Rb family pathway. Similarly, because we were not able to repress Rb family members individually and because the E1A mutant displayed only a partial defect in p107 binding, our results did not permit us to assign specific activities to different Rb family members. Taken together, these results establish that the transcriptional response to E7 repression in HeLa cells is dependent on activation of the Rb family and that interactions of the E7 protein with other binding partners are not sufficient to exert transcriptional effects in the absence of activation of the Rb family.
In our experiments, approximately one-third of the repressed genes were known targets of E2F4 or Rb family members. Furthermore, almost one-quarter of E2F target genes expressed in E6 cells were significantly repressed during induced senescence, consistent with activation of the Rb/E2F pathway. Typically, only a fraction of targets identified by global chromatin immunoprecipitation studies demonstrate regulated expression in complementary expression studies (e.g., see reference 38). Therefore, repression of E2F target genes, many of which are involved in cell cycle progression, is likely a direct effect of complex formation between Rb family members and repressor E2F family members and is undoubtedly responsible for cell cycle arrest. These results are consistent with the results of Jackson et al., who detected recruitment of Rb family members to promoters of cell cycle-related genes in MCF-7 breast carcinoma cells induced to senesce by doxorubicin treatment (43). Furthermore, stable repression of E2F-responsive genes in senescent human fibroblasts has been attributed to the Rb-dependent formation of senescence-associated heterochromatic foci, involving the recruitment of p105Rb to the promoters of these genes (60). Finally, our earlier studies demonstrated that repression of cdc25A expression by E7 repression in HeLa cells required an E2F site in the cdc25A promoter and was accompanied by the assembly of p105Rb/E2F4 complexes able to bind this site in vitro (91).
E7 repression and activation of the Rb family members also induced more than 200 genes, notably including genes encoding proteins involved in lysosome function. These results suggest that there is a transcriptional basis for the increased lysosome content characteristic of senescent cells, which may contribute to many features of the senescence phenotype, including increased autofluorescence, cellular granularity, and SAβ-gal activity, the widely used marker of senescence, which is due to increased activity of lysosomal β-galactosidase (23, 49, 51).
The ability of Rb family members to directly stimulate gene expression by interacting with various positively acting transcription factors and chromatin remodeling proteins may account for much of the gene induction observed. Direct support for this model is provided by our finding that induction of an AP1 target gene, coding for clusterin, was blocked by a dominant-negative version of c-jun. Induction of jun-responsive genes may involve not only direct activation of c-jun itself, but also the consequent autostimulation of the c-jun promoter, which contains an AP1 site. In addition, p105Rb associates with and activates the CBFA-1 transcription factor (83). Pituitary tumor-transforming 1 interacting protein (PTTG1IP), coded for by a gene induced in response to E7 repression, is a target of CBFA-1 (79). Similarly, p105Rb binds and activates GATA-1, C/EBPα (53), and C/EBPβ (15). Genes significantly induced by E7 repression include targets of GATA-1, such as those coding for cyclin-dependent kinase inhibitor 2B [p15 (CDKN2B)] and cellular repressor of E1A-stimulated genes (CREG) (74), as well as targets of C/EBPα and/or C/EBPβ, such as genes coding for complement component 3 (C3) and S100 calcium binding protein A10 (29, 32).
Transcription factors repressed in an Rb family-dependent manner after E7 repression in E6 cells include the known Rb/E2F targets DEK and B-MYB. Some of the gene expression changes observed during induced senescence are likely to be downstream effects of the regulated expression of these transcription factors. In addition, inhibition of expression of either DEK or B-MYB by RNA interference induced senescence in primary cells. Thus, activation of the Rb family results in the delivery of multiple signals, several of which are independently sufficient to elicit senescence. The simultaneous delivery of reinforcing senescence-inducing signals may account for the profound growth inhibition caused by E7 repression and for our inability to block senescence by the expression of DEK, B-MYB, or dominant-negative c-jun.
Two previous microarray-based studies examined transcription in HeLa cells following E2-induced repression of both E6 and E7. Wells et al. described a transcriptional profile that included the induction of p53-responsive genes and repression of cell cycle-regulated genes (87). Two sets of regulated genes highlighted by Wells et al., the RAB vesicular transport genes and genes encoding the GAGE tumor-associated antigens, did not significantly change expression in our study, in which only E7 was repressed. Thierry et al. concluded that gene induction by a chimeric green fluorescent protein-HPV18 E2 protein was largely due to activation of the p53 pathway in response to E6 repression, while gene repression was attributed to the assembly of active Rb/E2F complexes in response to E7 repression, with mitotic genes representing a major repressed class (82). Both of these studies expressed the E2 protein from adenovirus vectors. Unlike these prior studies, we showed that the gene expression changes observed were not due to HPV-independent effects of E2 expression. We also eliminated effects of E6 repression, including p53-dependent gene expression changes, from the transcriptional profile of induced senescence. Furthermore, we demonstrated directly that the observed changes in gene expression were a consequence of HPV E7 repression and activation of the Rb family, identified lysosomal genes as the major induced class, and revealed a role of c-jun in mediating the transcriptional response.
In our experiments, the transcriptional profile of induced senescence driven by the Rb family in cervical carcinoma cells was compared to the profile of replicative senescence in primary fibroblasts triggered by telomere erosion. This analysis revealed substantial overlap between the transcriptional programs of these two senescence systems, indicating that activation of the Rb family plays a prominent role in driving the transcriptional events during replicative senescence. Furthermore, the common transcriptional response suggests that there is extensive mechanistic and phenotypic overlap between senescence in cancer cells and in primary cells. Therefore, this model of induced senescence, which is rapid, synchronous, and easily manipulated, can facilitate the genetic and biochemical characterization of senescence.
Supplementary Material
Acknowledgments
We thank Valerie Reinke, Kevin White, and Kristin Yates for helpful discussions. We also thank Janet Hager and Irina Tikhonova for assistance with the microarray experiments, Archibald Perkins for assistance with qRT-PCR, Kathi Fries and Kristin Yates for initial experiments with the E1A gene, and Jan Zulkeski for assistance with preparation of the manuscript. The E1A gene was the generous gift of Elizabeth Moran (Temple University), and the TAM67 c-jun mutant was the generous gift of Michael Birrer (NIH).
The DNA Microarray Resource of the Keck Biotechnology Center was supported by the Anna and Argall Hull Fund, the Swebilius Trust, and a grant from the NIH (DK58776). K.J. was supported in part by an MSTP training grant (GM07205) from NIGMS, and E.G. was supported in part by a grant (IRG 58-012-45) from the American Cancer Society. This work was supported by a grant from the NCI (CA16038) to D.D.
Footnotes
Published ahead of print on 20 December 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Angel, P., and M. Karin. 1991. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta 1072:129-157. [DOI] [PubMed] [Google Scholar]
- 2.Arsura, M., M. Introna, F. Passerini, A. Mantovani, and J. Golay. 1992. B-myb antisense oligonucleotides inhibit proliferation of human hematopoietic cell lines. Blood 79:2708-2716. [PubMed] [Google Scholar]
- 3.Avvakumov, N., J. Torchia, and J. S. Mymryk. 2003. Interaction of the HPV E7 proteins with the pCAF acetyltransferase. Oncogene 22:3833-3841. [DOI] [PubMed] [Google Scholar]
- 4.Balciunaite, E., A. Spektor, N. H. Lents, H. Cam, H. te Riele, A. Scime, M. A. Rudnicki, R. Young, and B. D. Dynlacht. 2005. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 25:8166-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balsitis, S. J., J. Sage, S. Duensing, K. Münger, T. Jacks, and P. F. Lambert. 2003. Recapitulation of the effects of the human papillomavirus type 16 E7 oncogene on mouse epithelium by somatic Rb deletion and detection of pRb-independent effects of E7 in vivo. Mol. Cell. Biol. 23:9094-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellanger, S., S. Blachon, F. Mechali, C. Bonne-Andrea, and F. Thierry. 2005. High-risk but not low-risk HPV E2 proteins bind to the APC activators Cdh1 and Cdc20 and cause genomic instability. Cell Cycle 4:1608-1615. [DOI] [PubMed] [Google Scholar]
- 7.Bernat, A., N. Avvakumov, J. S. Mymryk, and L. Banks. 2003. Interaction between the HPV E7 oncoprotein and the transcriptional coactivator p300. Oncogene 22:7871-7881. [DOI] [PubMed] [Google Scholar]
- 8.Bischof, O., K. Nacerddine, and A. Dejean. 2005. Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways. Mol. Cell. Biol. 25:1013-1024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Blachon, S., S. Bellanger, C. Demeret, and F. Thierry. 2005. Nucleo-cytoplasmic shuttling of high risk human papillomavirus E2 proteins induces apoptosis. J. Biol. Chem. 280:36088-36098. [DOI] [PubMed] [Google Scholar]
- 10.Braig, M., S. J. Lee, C. Loddenkemper, C. Rudolph, A. H. F. M. Peters, B. Schlegelberger, H. Stein, B. Dorken, J. Jenuwein, and C. A. Schmitt. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature 436:660-665. [DOI] [PubMed] [Google Scholar]
- 11.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:535-536. [DOI] [PubMed] [Google Scholar]
- 12.Cam, H., E. Balciunaite, A. Blais, A. Spektor, R. C. Scarpulla, R. Young, Y. Kluger, and B. D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16:399-411. [DOI] [PubMed] [Google Scholar]
- 13.Campisi, J. 2005. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 120:513-522. [DOI] [PubMed] [Google Scholar]
- 14.Carro, M. S., F. M. Spiga, M. Quarto, V. Di Ninni, S. Volorio, M. Alcalay, and H. Muller. 2006. DEK expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle 5:1202-1207. [DOI] [PubMed] [Google Scholar]
- 15.Chen, P.-L., D. J. Riley, S. Chen-Kiang, and W.-H. Lee. 1996. Retinoblastoma protein directly interacts with and activates the transcription factor NF-IL6. Proc. Natl. Acad. Sci. USA 93:465-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, Z., L. C. Trotman, D. Schaffer, H. K. Lin, Z. A. Dotan, M. Niki, J. A. Koutcher, H. I. Scher, T. Ludwig, W. Gerald, C. Cordon-Cardo, and P. P. Pandolfi. 2005. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436:636-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corbeil, H. B., and P. E. Branton. 1994. Functional importance of complex formation between the retinoblastoma tumor suppressor family and adenovirus E1A proteins as determined by mutational analysis of E1A conserved region 2. J. Virol. 68:6697-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeFilippis, R. A., E. C. Goodwin, L. Wu, and D. DiMaio. 2003. Endogenous human papillomavirus E6 and E7 proteins differentially regulate proliferation, senescence, and apoptosis in HeLa cervical carcinoma cells. J. Virol. 77:1551-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demeret, C., A. Garcia-Carranca, and F. Thierry. 2003. Transcription-independent triggering of the extrinsic pathway of apoptosis by human papillomavirus 18 E2 protein. Oncogene 22:168-175. [DOI] [PubMed] [Google Scholar]
- 20.Desaintes, C., C. Demeret, S. Goyat, M. Yaniv, and F. Thierry. 1997. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 16:504-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desaintes, C., S. Goyat, S. Garbay, M. Yaniv, and F. Thierry. 1999. Papillomavirus E2 induces p53-independent apoptosis in HeLa cells. Oncogene 18:4538-4545. [DOI] [PubMed] [Google Scholar]
- 22.Dickson, M. A., W. C. Hahn, Y. Ino, V. Ronfard, J. Y. Wu, R. A. Weinberg, D. N. Louis, F. P. Li, and J. G. Rheinwald. 2000. Human keratinocytes that express hTERT and also bypass a p16INK4a-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 20:1436-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dimri, G. P., X. Lee, G. Basile, et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dowhanick, J. J., A. A. McBride, and P. M. Howley. 1995. Suppression of cellular proliferation by the papillomavirus E2 protein. J. Virol. 69:7791-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duensing, S., and K. Munger. 2003. Human papillomavirus type 16 E7 oncoprotein can induce abnormal centrosome duplication through a mechanism independent of inactivation of retinoblastoma protein family members. J. Virol. 77:12331-12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 27.Francis, D. A., S. I. Schmid, and P. M. Howley. 2000. Repression of the integrated papillomavirus E6/E7 promoter is required for growth suppression of cervical cancer cells. J. Virol. 74:2679-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frattini, M. G., S. D. Hurst, H. B. Lim, S. Swaminathan, and L. A. Laimins. 1997. Abrogation of a mitotic checkpoint by E2 proteins from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumor suppressor protein. EMBO J. 16:318-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman, J. R., B. Larris, P. P. Le, T. H. Peiris, A. Arsenlis, J. Schug, J. W. Tobias, K. H. Kaestner, and L. E. Greenbaum. 2004. Orthogonal analysis of C/EBPb targets in vivo during liver proliferation. Proc. Natl. Acad. Sci. USA 101:12986-12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frolov, M. V., and N. J. Dyson. 2004. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J. Cell Sci. 117:2173-2181. [DOI] [PubMed] [Google Scholar]
- 31.Garcia, P., and J. Frampton. 2006. The transcription factor B-Myb is essential for S-phase progression and genomic stability in diploid and polyploid megakaryocytes. Cell Sci. 119:1483-1493. [DOI] [PubMed] [Google Scholar]
- 32.Gery, S., A. F. Gombart, W. S. Yi, C. Koeffler, W. K. Hofmann, and H. P. Koeffler. 2005. Transcription profiling of C/EBP targets identifies Per2 as a gene implicated in myeloid leukemia. Blood 106:2827-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodwin, E. C., and D. DiMaio. 2001. Induced senescence in HeLa cervical carcinoma cells containing elevated telomerase activity and extended telomeres. Cell Growth Differ. 12:525-534. [PubMed] [Google Scholar]
- 34.Goodwin, E. C., and D. DiMaio. 2000. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. USA 97:12513-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodwin, E. C., L. K. Naeger, D. E. Breiding, E. J. Androphy, and D. DiMaio. 1998. Transactivation-competent bovine papillomavirus E2 protein is specifically required for efficient repression of human papillomavirus oncogene expression and for acute growth inhibition of cervical carcinoma cell lines. J. Virol. 72:3925-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodwin, E. C., E. Yang, C.-J. Lee, H.-W. Lee, D. DiMaio, and E.-S. Hwang. 2000. Rapid induction of senescence in human cervical carcinoma cells. Proc. Natl. Acad. Sci. USA 97:10978-10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hara, E., H. Tsuri, S. Shinozaki, and K. Oda. 1991. Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of lifespan in human diploid fibroblasts. Biochem. Biophys. Res. Commun. 179:528-534. [DOI] [PubMed] [Google Scholar]
- 38.Hayakawa, J., S. Mittal, Y. Wang, K. S. Korkmaz, E. Adamson, C. English, M. Omichi, M. McClelland, and D. Mercola. 2004. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol. Cell 16:521-535. [DOI] [PubMed] [Google Scholar]
- 39.Helt, A.-M., J. O. Funk, and D. A. Galloway. 2002. Inactivation of both the retinoblastoma tumor suppressor and p21 by the human papillomavirus type 16 E7 oncoprotein is necessary to inhibit cell cycle arrest in human epithelial cells. J. Virol. 76:10559-10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huh, K.-W., J. DeMasi, H. Ogawa, Y. Nakatani, P. M. Howley, and K. Munger. 2005. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc. Natl. Acad. Sci. USA 102:11492-11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang, E.-S., L. K. Naeger, and D. DiMaio. 1996. Activation of the endogenous p53 growth inhibitory pathway in HeLa cervical carcinoma cells by expression of the bovine papillomavirus E2 gene. Oncogene 12:795-803. [PubMed] [Google Scholar]
- 42.Hwang, E.-S., D. J. Riese II, J. Settleman, L. A. Nilson, J. Honig, S. Flynn, and D. DiMaio. 1993. Inhibition of cervical carcinoma cell line proliferation by the introduction of a bovine papillomavirus regulatory gene. J. Virol. 67:3720-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jackson, J. G., and O. M. Pereira-Smith. 2006. Primary and compensatory roles for RB family members at cell cycle gene promoters that are deacetylated and downregulated in doxorubicin-induced senescence of breast cancer cells. Mol. Cell. Biol. 26:2501-2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang, H. T., C. J. Lee, E. J. Seo, Y. J. Bahn, H. J. Kim, and E. S. Hwang. 2004. Transition to an irreversible state of senescence in HeLa cells arrested by repression of HPV E6 and E7 genes. Mech. Ageing Dev. 125:31-40. [DOI] [PubMed] [Google Scholar]
- 46.Kielosto, M., P. Nummela, R. Katainen, V. Leaner, M. J. Birrer, and E. Holtta. 2004. Reversible regulation of the transformed phenotype of ornithine decarboxylase- and ras-overexpressing cells by dominant-negative mutants of c-Jun. Cancer Res. 64:3772-3779. [DOI] [PubMed] [Google Scholar]
- 47.Kim, K., P. A. Garner-Hamrick, C. Fisher, D. Lee, and P. F. Lambert. 2003. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 77:12450-12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiyono, T., S. A. Foster, J. I. Koop, J. K. McDougall, D. A. Galloway, and A. J. Kleingelhutz. 1998. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature 396:84-88. [DOI] [PubMed] [Google Scholar]
- 49.Kurz, D. J., S. Decary, Y. Hong, and J. D. Erusalimsky. 2000. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J. Cell Sci. 113:3613-3622. [DOI] [PubMed] [Google Scholar]
- 50.Lam, E. W.-F., J. D. Bennett, and R. J. Watson. 1995. Cell-cycle regulation of human b-myb transcription. Gene 160:277-281. [DOI] [PubMed] [Google Scholar]
- 51.Lee, B. Y., J. A. Han, J. S. Im, A. Morrone, K. Johung, E. C. Goodwin, W. J. Kleijer, D. DiMaio, and E. S. Hwang. 2006. Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell 5:187-195. [DOI] [PubMed] [Google Scholar]
- 52.Lee, D., C. Lim, T. Seo, H. Kwon, H. Min, and J. Choe. 2002. The viral oncogene human papillomavirus E7 deregulates transcriptional silencing by Brm-related gene 1 via molecular interactions. J. Biol. Chem. 277:48842-48848. [DOI] [PubMed] [Google Scholar]
- 53.Liu, H., B. Dibling, B. Spike, A. Dirlam, and K. Macleod. 2004. New roles for the RB tumor suppressor protein. Curr. Opin. Genet. Dev. 14:55-64. [DOI] [PubMed] [Google Scholar]
- 54.Mantovani, F., and L. Banks. 2001. The human papillomavirus E6 protein and its contribution to malignant progression. Oncogene 20:7874-7887. [DOI] [PubMed] [Google Scholar]
- 55.Masselink, H., N. Vastenhouw, and R. Bernards. 2001. B-myb rescues ras-induced premature senescence, which requires its transactivation domain. Cancer Lett. 171:87-101. [DOI] [PubMed] [Google Scholar]
- 56.Massimi, P., D. Pim, C. Bertoli, V. Bouvard, and L. Banks. 1999. Interaction between the HPV-16 E2 transcriptional activator and p53. Oncogene 18:7748-7754. [DOI] [PubMed] [Google Scholar]
- 57.Moon, M. S., C. J. Lee, S. J. Um, J. S. Park, J. M. Yang, and E. S. Hwang. 2001. Effect of BPV1 E2-mediated inhibition of E6/E7 expression in HPV16-positive cervical carcinoma cells. Gynecol. Oncol. 80:168-175. [DOI] [PubMed] [Google Scholar]
- 58.Munger, K., J. R. Basile, S. Duensing, A. Eichten, S. L. Gonzalez, M. Grace, and V. L. Zacny. 2001. Biological activities and molecular targets of the human papillomavirus E7 oncoprotein. Oncogene 20:7888-7898. [DOI] [PubMed] [Google Scholar]
- 59.Naeger, L. K., E. C. Goodwin, E.-S. Hwang, R. A. DeFilippis, H. Zhang, and D. DiMaio. 1999. Bovine papillomavirus E2 protein activates a complex growth-inhibitory program in p53-negative HT-3 cervical carcinoma cells that includes repression of cyclin A and cdc25A phosphatase genes and accumulation of hypophosphorylated retinoblastoma protein. Cell Growth Differ. 10:413-422. [PubMed] [Google Scholar]
- 60.Narita, M., S. Nunez, E. Heard, M. Narita, A. W. Lin, S. A. Hearn, D. L. Spector, G. J. Hannon, and S. W. Lowe. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113:703-716. [DOI] [PubMed] [Google Scholar]
- 61.Naviaux, R. K., E. Costanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nead, M. A., L. A. Baglia, M. J. Antinore, J. W. Ludlow, and D. J. McCance. 1998. Rb binds c-Jun and activates transcription. EMBO J. 17:2342-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishimura, A., T. Nakahara, T. Ueno, K. Sasaki, S. Yoshida, S. Kyo, P. M. Howley, and H. Sakai. 2006. Requirement of E7 oncoprotein for viability of HeLa cells. Microbes Infect. 8:984-993. [DOI] [PubMed] [Google Scholar]
- 64.Nishimura, A., T. Ono, A. Ishimoto, J. J. Dowhanick, M. A. Frizzell, P. M. Howley, and H. Sakai. 2000. Mechanisms of human papillomavirus E2-mediated repression of viral oncogene expression and cervical cancer cell growth inhibition. J. Virol. 74:3752-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nishitani, J., T. Nishinaka, C.-H. Cheng, W. Rong, K. K. Yokoyama, and R. Chiu. 1999. Recruitment of the retinoblastoma protein to c-Jun enhances transcription activity mediated through the AP-1 binding site. J. Biol. Chem. 274:5454-5461. [DOI] [PubMed] [Google Scholar]
- 66.Parish, J. L., A. Kowalczyk, H.-T. Chen, G. E. Roeder, R. Sessions, M. Buckle, and K. Gaston. 2006. E2 proteins from high- and low-risk human papillomavirus types differ in their ability to bind p53 and induce apoptotic cell death. J. Virol. 80:4580-4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park, J.-S., E.-J. Kim, H.-J. Kwon, E.-S. Hwang, S.-E. Namkoong, and S.-J. Um. 2000. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. J. Biol. Chem. 275:6764-6769. [DOI] [PubMed] [Google Scholar]
- 68.Park, R. B., and E. J. Androphy. 2002. Genetic analysis of high-risk E6 in episomal maintenance of human papillomavirus genomes in primary human keratinocytes. J. Virol. 76:11359-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phelps, W. C., C. L. Yee, K. Munger, and P. M. Howley. 1988. The human papillomavirus type 16 E7 gene encodes transactivation and transformation functions similar to those of adenovirus E1A. Cell 53:539-547. [DOI] [PubMed] [Google Scholar]
- 70.Psyrri, A., R. A. DeFilippis, A. P. B. Edwards, K. E. Yates, L. Manuelidis, and D. DiMaio. 2004. Role of the retinoblastoma pathway in senescence triggered by repression of the human papillomavirus E7 protein in cervical carcinoma cells. Cancer Res. 64:3079-3086. [DOI] [PubMed] [Google Scholar]
- 71.Raschella, G., A. Negroni, A. Sala, S. Pucci, A. Romeo, and B. Calabretta. 1995. Requirement of b-myb function for survival and differentiative potential of human neuroblastoma cells. J. Biol. Chem. 270:8540-8545. [DOI] [PubMed] [Google Scholar]
- 72.Rehtanz, M., H.-M. Schmidt, U. Warthorst, and G. Steger. 2004. Direct interaction between nucleosome assembly protein 1 and the papillomavirus E2 proteins involved in activation of transcription. Mol. Cell. Biol. 24:2153-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rylski, M., J. J. Welch, Y.-Y. Chen, D. L. Letting, J. A. Diehl, L. A. Chodosh, G. A. Blobel, and M. J. Weiss. 2003. GATA-1-mediated proliferation arrest during erythroid maturation. Mol. Cell. Biol. 23:5031-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sala, A., and B. Calabretta. 1992. Regulation of BALB/c 3T3 fibroblast proliferation by B-myb is accompanied by selective activation of cdc2 and cyclin D1 expression. Proc. Natl. Acad. Sci. USA 89:10415-10419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sedivy, J. M. 1998. Can ends justify the means? Telomeres and the mechanisms of replicative senescence and immortalization in mammalian cells. Proc. Natl. Acad. Sci. USA 95:9078-9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shay, J. W., O. M. Pereira-Smith, and W. E. Wright. 1991. A role for both RB and p53 in the regulation of human cellular senescence. Exp. Cell Res. 196:33-39. [DOI] [PubMed] [Google Scholar]
- 78.Shay, J. W., and I. B. Roninson. 2004. Hallmarks of senescence in carcinogenesis and cancer therapy. Oncogene 23:2919-2933. [DOI] [PubMed] [Google Scholar]
- 79.Stock, M., H. Schafer, M. Fliegauf, and F. Otto. 2004. Identification of novel genes of the bone-specific transcription factor Runx2. J. Bone Miner. Res. 19:959-972. [DOI] [PubMed] [Google Scholar]
- 80.Strober, B. E., J. L. Dunaief, S. Guha, and S. P. Goff. 1996. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol. Cell. Biol. 16:1576-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thain, A., O. Jenkins, A. R. Clarke, and K. Gaston. 1996. CpG methylation directly inhibits binding of the human papillomavirus type 16 E2 protein to specific DNA sequences. J. Virol. 70:7233-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thierry, F., M. A. Benotmane, C. Demeret, M. Mori, S. Teissier, and C. Desaintes. 2004. A genomic approach reveals a novel mitotic pathway in papillomavirus carcinogenesis. Cancer Res. 64:895-903. [DOI] [PubMed] [Google Scholar]
- 83.Thomas, D. M., S. A. Carty, D. M. Piscopo, J.-S. Lee, W.-F. Wang, W. C. Forrester, and P. W. Hinds. 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell 8:303-316. [DOI] [PubMed] [Google Scholar]
- 84.Webster, K., J. Parish, M. Pandya, P. L. Stern, A. R. Clarke, and K. Gaston. 2000. The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. J. Biol. Chem. 275:87-94. [DOI] [PubMed] [Google Scholar]
- 85.Wei, C.-L., Q. Wu, V. B. Vega, K. P. Chiu, P. Ng, T. Zhang, A. Shahab, H. C. Yong, Y. Fu, Z. Weng, J. Liu, X. D. Zhao, J.-L. Chew, Y. L. Lee, V. A. Kuznetsov, W.-K. Sung, L. D. Miller, B. Lim, E. T. Liu, Q. Yu, H.-H. Ng, and Y. Ruan. 2006. A global map of p53 transcription-factor binding sites in the human genome. Cell 124:207-219. [DOI] [PubMed] [Google Scholar]
- 86.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H.-M. Huang, and P. J. Farnham. 2006. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wells, S. I., B. J. Aronow, T. M. Wise, S. S. Williams, J. A. Couget, and P. M. Howley. 2003. Transcriptome signature of irreversible senescence in human papillomavirus-positive cervical cancer cells. Proc. Natl. Acad. Sci. USA 100:7093-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wells, S. I., D. A. Francis, A. Y. Karpova, J. J. Dowhanick, J. D. Benson, and P. M. Howley. 2000. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21CIP-dependent pathways. EMBO J. 19:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wise-Draper, T. M., H. V. Allen, E. E. Jones, K. B. Habash, H. Matsuo, and S. I. Wells. 2006. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol. Cell. Biol. 26:7506-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wise-Draper, T. M., H. V. Allen, M. N. Thobe, E. E. Jones, K. B. Habash, K. Munger, and S. I. Wells. 2005. The human DEK proto-oncogene is a senescence inhibitor and an upregulated target of high-risk human papillomavirus E7. J. Virol. 79:14309-14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu, L., E. C. Goodwin, L. K. Naeger, E. Vigo, K. Galaktionov, K. Helin, and D. DiMaio. 2000. E2F-Rb complexes assemble and inhibit cdc25A transcription in cervical carcinoma cells following repression of human papillomavirus oncogene expression. Mol. Cell. Biol. 20:7059-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu, J., L. Zhang, P. M. Hwang, C. Rago, K. W. Kinzler, and B. Vogelstein. 1999. Identification and classification of p53-regulated genes. Proc. Natl. Acad. Sci. USA 96:14517-14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang, H., K.-H. Pan, and S. N. Cohen. 2003. Senescence-specific gene expression fingerprints reveal cell-type-dependent physical clustering of up-regulated chromosomal loci. Proc. Natl. Acad. Sci. USA 100:3251-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang, H. S., M. Gavin, A. Dahlya, A. A. Postigo, D. Ma, R. X. Luo, J. W. Harbour, and D. C. Dean. 2000. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell 101:79-89. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.