Abstract
The importance of HLA class I-restricted CD8 T-cell responses in the control of human immunodeficiency virus (HIV) infection is generally accepted. While several studies have shown an association of certain HLA class I alleles with slower disease progression, it is not fully established whether this effect is mediated by HIV-specific CD8 T-cell responses restricted by these alleles. In order to study the influence of the HLA class I alleles on the HIV-specific CD8 T-cell response and on viral control, we have assessed HIV-specific epitope recognition, plasma viral load, and expression of HLA class I alleles in a cohort of HIV-seropositive bar workers. Possession of the HLA class I alleles B5801, B8101, and B0702 was associated with a low median viral load and simultaneously with a broader median recognition of Gag epitopes compared to all other HLA alleles (twofold increase) (P = 0.0035). We further found an inverse linear relationship between the number of Gag epitopes recognized and the plasma viral load (R = −0.36; P = 0.0016). Particularly, recognition of multiple epitopes within two regions of Gag (amino acids [aa] 1 to 75 and aa 248 to 500) was associated with the maintenance of a low steady-state viremia, even years after acute infection.
It is generally accepted that the HLA class I-restricted CD8 T-cell responses contribute substantially to the control of human immunodeficiency virus type 1 (HIV-1) replication in infected individuals. The appearance of HIV-specific CD8 T cells is temporally associated with the decline of plasma viremia after acute infection (5, 20), and their presence is associated with the control of viral replication during the chronic phase of infection (17, 28). Nonetheless, the parameters underlying the efficient control of viral replication by CD8 T cells remain controversial, since several recent studies have not found any correlation between the magnitude or breadth of the HIV-specific CD8 T-cell response and the plasma viral load in chronically infected individuals (1, 4, 6). Interestingly, CD8 T cells recognizing different HIV proteins may vary in their respective antiviral efficiencies (23, 33). Responses targeting the proteins encoded within the gag open reading frame have occasionally been shown to be associated with viral control (10, 23, 27, 29, 30), whereas responses to Nef or Env had no effect on viral control or even positively correlated with the plasma viral load (4, 23, 27).
CD8 T cells recognize virus-derived peptides in the context of HLA class I molecules. Three highly polymorphic genes, HLA-A, -B, and -C, encode these HLA class I molecules, and the CD8 T-cell specificities found within one individual are largely dependent on the HLA class I alleles expressed. Particular HLA-B alleles are strongly associated with different rates of disease progression and have been shown to restrict the majority of HIV-specific CD8 T-cell responses (18). These observations suggest that the association observed between a particular HLA-B allele and the corresponding viral load may be mediated by HIV-specific CD8 T cells restricted by these alleles. HLA-B alleles such as B5701/03, B5801, and B4201 (2, 16, 18) have been associated with a low viral load and also with slower disease progression. Conversely, HLA-B alleles such as B5802, B4501, and B1510 are associated with a high viral load and more rapid disease progression (18). Despite these important findings, it is not completely established whether disparities in the HIV-specific CD8 T-cell responses restricted by these alleles are contributing to these different rates of disease progression.
In order to study the mechanism underlying the beneficial effect of protective HLA class I alleles, seropositive study subjects from a high-risk cohort in Southwest Tanzania were HLA typed (54 of 56 subjects), and the plasma viral loads and CD4 counts were determined. At the same time, the HIV-specific CD8 T-cell response of each subject was examined for responses to Gag, Nef, and Env on a single-peptide level using multiple sets of 15-mer overlapping peptides representative of the locally occurring subtypes A, C, and D.
MATERIALS AND METHODS
Study subjects.
The 56 individuals in this study are part of a larger high-risk HIV cohort of female bar workers enrolled in a prospective study of HIV-1 superinfection (HIV Superinfection Study [HISIS]) in the Mbeya region of Southwest Tanzania. Between September and December 2000, 600 women were recruited after giving informed consent, and each participant provided blood samples at enrollment and every 3 months after for a period of up to 4 years. During the study, all participants received health care that included treatment of all acute infectious diseases, screening and treatment for sexually transmitted diseases, and, since 2003, cotrimoxazole prophylaxis for opportunistic infections for women with CD4 T-cell counts below 200 cells/μl. Since 2005, antiretroviral treatment has been available for all participants with AIDS-defining symptoms or CD4 counts below 200 cells/μl. During the course of this study, all individuals were antiretroviral naïve. Their HIV-1 status was determined using two diagnostic HIV enzyme-linked immunoassay tests (Enzygnost Anti HIV1/2 Plus [Dade Behring, Liederbach, Germany] and Determine HIV 1/2 [Abbott, Wiesbaden, Germany]). Discordant results were resolved by using a Western blot assay (Genelabs Diagnostics, Geneva, Switzerland). Plasma HIV-1 RNA levels were measured by using the Amplicor HIV-1 Monitor assay (Roche Diagnostics, Indianapolis, IN). In April 2003, 56 of the HIV-1 positive participants were enrolled in the HISIS-Cytotoxic T-Lymphocyte (CTL) substudy. The study was reviewed and approved by the ethics committees of all partners in compliance with national guidelines and institutional policies.
HLA typing.
HLA-A, -B, and -C class I alleles were typed by DNA sequencing using an ABI 3700 sequencer. Blood samples of 200 μl were extracted using a QIAGEN blood extraction kit for yields of high-quality DNA. Exons 2 and 3 (encoding the peptide binding groove) were amplified in a first-round PCR as previously described (31). The initial amplicon of exons 2 and 3 then served as a template for the nested and/or heminested amplification of exon 2. Exon 3 was similarly amplified in a nested fashion. Both the first-round and nested PCR primers are located in the introns flanking the amplified exon(s) (31). The nested PCR primers are alternately tailed with an M13 Universal 10 DNA sequencing primer sequence. Bidirectional M13 cycle sequencing of exon 2 and exon 3 was executed with BigDye V3.1. HLA DNA sequence typing analysis was completed with Assign SBT software (Conexio Genomics). DNA sequence ambiguities were resolved using the PEL-FREEZ SSP kit.
Synthetic peptides and peptide pools.
Seven sets of peptides consisted of 15-mer peptides overlapping by 11 amino acids covering the entire Gag, Nef, and Env protein sequences of primary isolates 90CF402 (subtype A Gag) (GenBank accession no. AAB38823), DU422 (subtype C Gag) (accession no. CAD62240), 98UG57143 (subtypes D Gag and D Env) (accession no. AF484514), 92UG037 (subtype A Nef) (accession no. AAC97549), DU151 (subtype C Nef) (accession no. AAL05314), and 94UG114 (subtype D Nef) (accession no. AAC97574). Subtype C Env peptides consisted of 18-mers and were based on primary isolate Du179 (accession no. AX457092). Peptides were synthesized using 9-fluorenylmethoxy carbonyl chemistry and standard solid-phase techniques with free amino termini. All peptides were >80% pure as determined by high-performance liquid chromatography, mass spectrophotometry, amino acid analysis, and N-terminal sequencing. Peptides were synthesized at the Natural and Medical Sciences Institute (University of Tuebingen, Germany), the Henry M. Jackson Foundation (Rockville, MD), and Anaspec Incorporated (San Jose, CA). The Gag peptide sets were closely related to HIV-1 isolates from the Mbeya Region (data not shown). Initial screening for T-cell responses was performed using the peptides in a matrix format. Subtype A, C, and D Gag peptide sets were pooled in an 11-by-11 format, subtype A and D Nef peptide sets were pooled in a 7-by-7 format, and the subtype C Nef peptide set was pooled in a 10-by-5 format.
ELISPOT assays.
During follow-up 13 of the HISIS, freshly isolated peripheral blood mononuclear cells (PBMC) (viability of <95%) were screened for HIV-specific T-cell responses by stimulation with overlapping peptide pools representing Gag, Nef, and Env from isolates of subtypes A, C, and D. (Follow-up 13 is the 13th follow-up of the study, roughly 39 months after the beginning of the study.) Gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays were performed by using Nova-Red substrate (Vector, Burlingame, CA) as previously described (24). The assay consistency was monitored on each plate using quality control PBMC recovered from frozen stocks and stimulated with a peptide pool containing optimal peptides from cytomegalovirus, Epstein-Barr virus, and influenza virus (CEF peptide pool). Confirmation of individual peptide responses, viral loads, and CD4 counts was done at follow-up 14, 3 months after the initial screening with peptide matrices. Recognition of a single peptide or pair of adjacent overlapping peptides was defined as a single epitope response. Responses with at least 70 spot-forming cells/106 PBMC and at least three times the negative control were scored as positive.
Whole-blood-format intracellular cytokine staining assay.
Intracellular IFN-γ staining in whole blood was performed to determine whether the responses were mediated by CD4 or CD8 T cells. Briefly, 250 μl of heparinized whole blood was added into polypropylene tubes and stimulated with either individual peptides (2 μg/ml [final]), staphylococcal enterotoxin B (10 μg/ml [final]) (Sigma) as a positive control, or no added stimulant as a negative control. A solution containing 25 μg/ml of anti-CD28 and anti-CD49d monoclonal antibodies (Pharmingen, San Diego, CA), 0.5% dimethyl sulfoxide, and 25 μg/ml of brefeldin A (Sigma) was added to each tube. Tubes were incubated for 6 h at 37°C in the presence of 4.5% CO2. Samples were then processed by lysing, fixation, permeabilization, and staining according to the BD Bioscience standard whole-blood assay analysis. Cells were stained with a four-color panel of antibodies: anti-IFN-γ-fluorescein isothiocyanate (clone 4S.B3; Pharmingen), anti-CD4-phycoerythrin (clone RPA-T4; Pharmingen), anti-CD8-PerCp-Cy5.5 (clone SK1; BD Biosciences), and anti-CD3-allophycocyanin (clone UCHT1; Pharmingen). Data acquisition was performed using a FACScalibur flow cytometer (Becton Dickinson, San Jose, CA), and data sets were analyzed using FlowJo software (version 4) (TreeStar, Cupertino, CA). Responses of at least three times the negative control and at least 0.05% of CD8+ T cells were scored as positive.
Analysis of single peptide responses.
Associations between particular peptide responses and HLA class I allele expression were described previously (14). Optimal epitopes within the targeted peptides were inferred after analysis of the peptide sequence using the HIV Molecular Immunology Database (http://www.hiv.lanl.gov/content/hiv-db/ELF/epitope_analyzer.html). The potential B8101 epitope within peptide Gag83 was inferred using the HLA B8101 binding motif with proline as a second position anchor and methionine as the C-terminal anchor.
Statistical analysis.
Data analyses were carried out using either GraphPad Prism software or SPSS software. Comparisons of two groups were performed using the Mann-Whitney test. The relationship between viral load or CD4 counts and CD8 T-cell responses was analyzed using Spearman's rank correlation test and linear regression analysis. Tests used for statistical analysis are described in the figure legends.
RESULTS
Expression of “protective” HLA class I B alleles correlates with a broad, Gag-specific CD8 T-cell response.
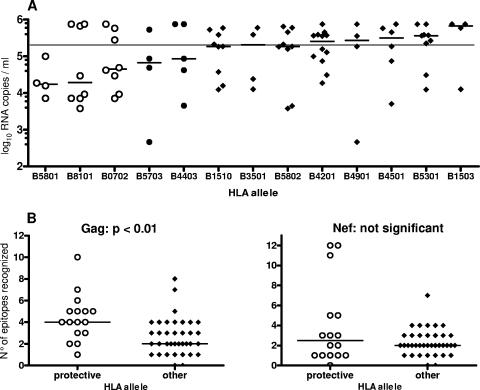
The most common HLA-B allele within the HISIS-CTL cohort was B4201 (n = 12), followed by B5301 (n = 10), B5802 (n = 9), B8101 (n = 9), B0702 (n = 8), and B1510 (n = 8) (data not shown). The alleles B5801 and B5703, which have been associated with a low viral load in previous studies, were less common (n = 4). The association between HLA-B alleles and viral load is shown in Fig. 1A. B5801-expressing subjects had the lowest viral load (median, 17,250 RNA copies/ml), followed by individuals expressing B8101 (median, 19,360 RNA copies/ml), B0702 (median, 45,350 RNA copies/ml), B5703 (median, 67,250 RNA copies/ml), and B4403 (median, 85,800 RNA copies/ml). On the contrary, expression of B1503, B4501, B5301 was associated with median plasma viral loads that were well above 200,000 RNA copies/ml.
FIG. 1.
Expression of “protective” HLA class I B alleles correlates with a broad Gag-specific CD8 T-cell response. HLA class I expression and plasma viral load in chronically infected study subjects (n = 54) are shown in panel A. The line indicates the median viral load of the HISIS-CTL cohort (198,500 viral RNA copies/ml). (B) Shown is the number of recognized epitopes for Gag and Nef from study subjects who express either HLA-B alleles B0702, B8101, or B5801 (n = 16) (open circles) or other HLA-B alleles (n = 37) (closed circles). Gag peptide responses were determined with an IFN-γ ELISPOT assay. Recognition of two consecutive peptides was counted as one epitope response. The Mann-Whitney test was used to analyze the breadth of the Gag- and Nef-specific T-cell responses associated with these alleles.
Next, we examined whether subjects expressing HLA alleles that were associated with a median plasma viral load of below 50,000 RNA copies/ml (B5801, B8101, and B0702, referred to as “protective” HLA alleles for simplicity) differed from other subjects in their HIV-specific CD8 T-cell responses. The recognition of the immunodominant HIV proteins Gag, Nef, and Env was analyzed with an IFN-γ ELISPOT assay using fresh PBMC and HIV subtype A, C, and D overlapping peptide matrices. Cytokine flow cytometric analysis for IFN-γ production confirmed the peptide responses as detected with the ELISPOT assay and revealed that 89% of peptide-specific responses were from CD8+ T cells (data not shown).
Subjects (n = 16) expressing “protective” HLA-B alleles had a broader median recognition of Gag peptides, twofold higher than that of individuals expressing other HLA-B alleles (P = 0.0035) (Fig. 1B). The magnitude of Gag-specific CD8 T cells was not significantly different between these groups, although the median magnitude was twofold higher (2,840 versus 1,420 spot-forming cells/106 PBMC) (data not shown) for subjects expressing a “protective” HLA allele. Interestingly, five of the six outlier subjects, expressing B8101 or B0702 but with viral loads above 200,000 RNA copies/ml, coexpressed either B4501 (two subjects), B1801 (two subjects), or B1503 (one subject), all HLA-B alleles that have previously been shown to be associated with rapid disease progression in a subtype C-dominated epidemic (11, 18). On the contrary, the coexpression of two protective B alleles was associated with low viral loads (mean, 13,243 copies/ml) and a broad Gag-specific T-cell response. For Nef- and Env-specific CD8 T-cell responses, there were no significant differences between the two groups (Fig. 1B and data not shown). The HLA class I allele B57 was not associated with broad Gag or Nef epitope recognition (data not shown). In summary, individuals expressing the “protective” HLA class I alleles B5801, B8101, and B0702 have a significantly broader recognition of Gag epitopes, but the protective effect may be abrogated in individuals coexpressing B alleles associated with rapid disease progression.
Inverse linear relationship between the number of Gag epitopes recognized and the plasma viral load.
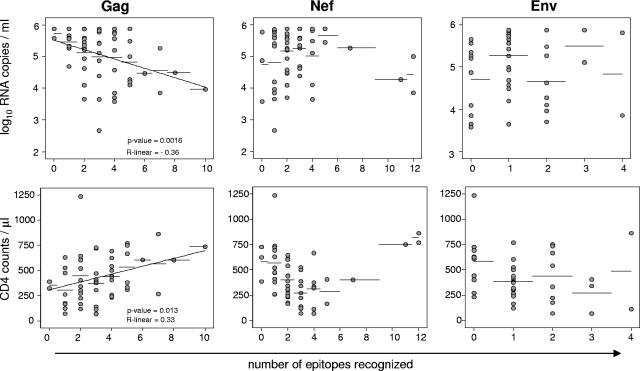
To determine whether a broad recognition of Gag epitopes protects HIV-infected individuals from rapid disease progression, we compared the breadth of epitope recognition and the total magnitude of protein-specific CD8 T-cell responses with the plasma viral load and the CD4 T-cell count. As shown in Fig. 2, there was a negative linear relationship between the number of recognized epitopes within Gag and the plasma viral load (P = 0.0016; R linear = −0.36) and a positive linear relationship between the number of recognized epitopes within Gag and the CD4 T-cell count (P = 0.013; R linear = 0.33). Neither Nef- nor Env-specific CD8 T-cell responses correlated with a low plasma viral load or with the CD4 cell counts. There was no significant inverse linear relationship between the total magnitude of Gag-specific CD8 T cells and the plasma viral load (data not shown).
FIG. 2.
Linear relationship between viral load and CD4 counts with the number of recognized epitopes within Gag but not within Nef or Env. Shown are (A) the linear regression analysis of the viral load and (B) the CD4 count versus the number of epitopes recognized per subject for Gag (left panels), Nef (middle panels), and Env (right panels). Recognition of two consecutive peptides was counted as on epitope response. Statistical analysis was performed using the Spearman rank test.
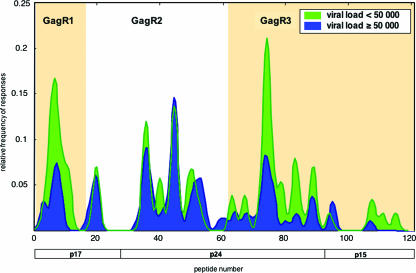
The majority of subjects with efficient control of viral replication recognize multiple epitopes within two specific regions of Gag.
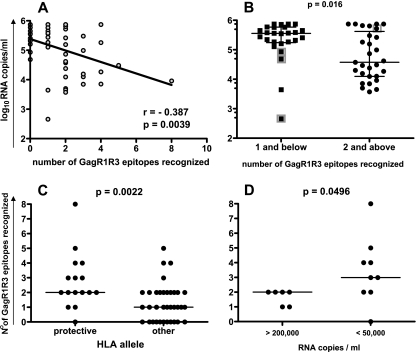
To identify particular regions within the Gag protein that are rich in potentially protective epitopes, we first stratified the cohort into two groups accounting for approximately one-third and two-thirds of the studied cohort, respectively: the low-viral-load group (LVL group) (<50,000 RNA copies/ml; n = 19) and the high-viral-load group (HVL group) (>50,000 RNA copies/ml; n = 36). We then compared the Gag responses between the two groups (Fig. 3). There were striking differences in the CD8 T-cell specificities between these two groups. The average LVL subject not only recognized more peptides within the whole Gag protein (1.4-fold greater) but also differed in the specific Gag regions targeted compared to the HVL group. In comparison to HVL subjects, LVL subjects recognized peptides within the N-terminal region of Gag (GagR1, amino acids [aa] 1 to 75, peptides 1 to 16) and within the C-terminal Gag region (GagR3, aa 248 to 500, peptides 62 to 120) almost twice as often (1.8-fold and 2.0-fold, respectively). Peptides within the middle region of Gag (GagR2, aa 76 to 247, peptides 17 to 61) were recognized equally between the groups. We found an inverse linear relationship between the number of recognized GagR1 and GagR3 (GagR1R3) epitopes and the viral load (Fig. 4A). Furthermore, recognition of more than one epitope within the GagR1R3 regions was strongly associated with a low viral load (P = 0.015) (Fig. 4B) and a higher CD4 T-cell count (P = 0.016) (data not shown). The importance of targeting multiple epitopes within the GagR1R3 regions at a population level is further supported by the fact that 16 of 19 subjects (86%) with a viremia below 50,000 RNA copies/ml targeted at least two epitopes within GagR1R3. Expression of the protective HLA class I alleles B5801, B8101, and B0702 was associated with a broad recognition of GagR1R3 epitopes (Fig. 4C), and breadth was significantly associated with a low viral load within subjects expressing these alleles (Fig. 4D).
FIG. 3.
Gag-specific CD8 T-cell epitope recognition of subjects with a plasma viral load below (n = 19) or above (n = 36) 50,000 RNA copies/ml. Shown is the magnitude of responses after smooth local average with Gaussian kernel weights and a standard deviation (bandwidth) of 1 peptide.
FIG. 4.
Relationship of the recognition of multiple epitopes within Gag regions at aa 1 to 075 and aa 248 to 500 (GagR1R3) with efficient viral control and protective HLA class I B alleles. Shown are (A) the linear regression analysis of the viral load and the number of GagR1R3 epitopes recognized and (B) the viral loads of subjects with a broad (n = 29) or narrow (n = 26) CD8 T-cell response against GagR1R3. The cutoff value was 2 epitopes recognized. The respective median is indicated. HLA B5703-expressing subjects are indicated as gray squares. (C) The numbers of recognized epitopes for GagR1R3 from study subjects who express HLA-B alleles B0702, B8101, or B5801 (n = 16) (“protective”) and from those expressing other HLA-B alleles (n = 37,other) and within these subjects (D) show a comparison of the number of recognized GagR1R3 epitopes of subjects with a high viral load above 200,000 RNA copies/ml (n = 6) and those with a viral load below 50,000 RNA copies/ml (n = 9). Statistical analysis for B, C, and D was performed using the Mann-Whitney test (two tailed).
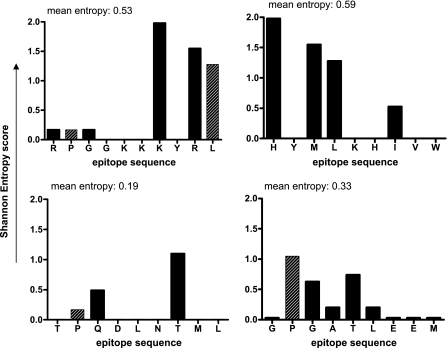
Interestingly, the conservation of some important epitopes within GagR1R3 is relatively limited. For two epitope responses that were most strongly associated with low viral load, RPGGKKKYML (aa 22 to 31, presented by HLA B0702 and B4201) and the inferred B8101 epitope GPGATLEEM (aa 338 to 346), one of the HLA anchor residues for each epitope (shown in boldface type) is highly variable (Fig. 5). Mutations affecting anchor residues are very rare in the more frequently recognized epitopes within GagR2 (http://www.hiv.lanl.gov/content/hiv-db/ELF/epitope_analyzer.html). Another frequently recognized epitope, HYMLNHIVW (aa 28 to 36), is the most variable of the immunodominant epitopes, and recognition of this epitope is overrepresented in the LVL group.
FIG. 5.
Variability of important immunodominant Gag epitopes. Shown is the Shannon entropy score (y axis) for each amino acid position of the epitopes (A) RL10 (aa 22 to 31), (B) HW9 (aa 28 to 36), (C) TL9 (aa 180 to 188), and (D) GM9 (aa 338 to 346). The entropy at amino acid anchor residues is indicated as striped bars. The epitope sequence included in the best recognized peptide variant is shown on the x axis. The entropy score analysis is based on HIV-1 Gag sequences of primary isolates from the Mbeyan Region (n = 41).
Recognition of multiple epitopes within Gag R1R3 is associated with the long-term maintenance of a low viral load after seroconversion.
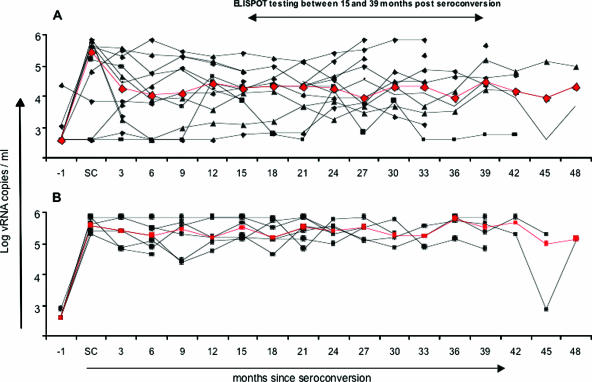
It was of further interest to determine whether the recognition of multiple epitopes within GagR1R3 could be associated with the maintenance of low steady-state viral load levels. To address this question, the course of plasma viremia was studied in 19 subjects who seroconverted during the HISIS (three HLA B57-expressing subjects were excluded, because B57 is often associated with the maintenance of a very low viral set point, low viral load, and an exceptional disease course) (2). In subjects (n = 12) recognizing two or more GagR1R3 epitopes during follow-up 14, the course of plasma viremia is typical for HIV infection (Fig. 6A). During the acute phase, the viral load rises to a median peak level of 286,000 RNA copies/ml and subsequently drops down to a steady-state level of 10,000 to 20,000 RNA copies/ml for the next 24 months. Only 2 of 12 subjects had a viral load above 50,000 RNA copies/ml at 24 months after seroconversion. The course of plasma viremia of B57-negative subjects recognizing zero or one epitope within GagR1R3 (n = 7) is strikingly different (Fig. 6B). The plasma viral load rises to peak levels with a median of 403,000 RNA copies/ml during acute infection and subsequently stays at a steady-state level of viremia of 150,000 to 350,000 RNA copies/ml over the next 24 months. At 24 months after seroconversion, none of this group had a viral load below 50,000 RNA copies/ml, and only one subject was below 100,000 RNA copies/ml. It appears that viral replication is never brought under efficient control in these subjects. This difference in viral load is likely to affect disease progression, and indeed, there was a strong tendency supporting this assumption. At 24 to 27 months after seroconversion, 6 of 12 subjects recognizing multiple GagR1R3 epitopes had CD4 T-cell counts above 500 cells/μl, whereas 6 of 7 subjects with narrow recognition of GagR1R3 had CD4 T-cell counts below 500 cells/μl (data not shown). These results indicate that long after the acute phase of peak viremia, the recognition of multiple epitopes within GagR1R3 may still contribute substantially to the maintenance of a low steady-state viremia.
FIG. 6.
Dynamics of the plasma viral load after seroconversion. Shown is the viral load since seroconversion of study subjects who (A) recognized two or more epitopes (n = 12) or (B) zero or one epitope (n = 7) within the Gag regions at aa 1 to 75 and aa 297 to 500. The median viral load is indicated in red. Gag peptide responses were determined between 15 and 39 months (follow-up 14) after seroconversion with an IFN-γ ELISPOT assay. Three HLA-B5703-expressing subjects were excluded from the analysis. Recognition of two consecutive peptides was counted as one epitope response. vRNA, viral RNA.
DISCUSSION
The main objective of this study was to reveal antiviral determinants of the HIV-specific CD8 T-cell response and to examine the influence of the host genetic (HLA) background on the HIV-specific CD8 T-cell response. The HISIS-CTL cohort is composed of 56 chronically infected female bar workers highly exposed to a mixed-subtype epidemic where subtype C predominates (14). Within the HISIS-CTL cohort, HLA-B alleles B5801, B8101, and B0702 were associated with a low viral load. This finding is consistent with a larger study in a cohort of subtype C-infected individuals previously conducted in South Africa (18). We have expanded upon these observations to show that within this cohort of high HIV-1 exposure, it is T-cell responses restricted by these alleles, particularly those targeting certain regions of the Gag protein, that mediate significantly better control of viral load.
The CD8 T-cell specificities found within an HIV-infected individual are largely dependent on the HLA class I alleles expressed, the sequence of the infecting virus (8, 11), and, partly, the stage of infection (1, 15). Additionally, HIV-specific CD4 T cells may also help to maintain CD8 T-cell responses of broad HIV specificity (22). It was first shown in 1995 that the Gag-specific CD8 T-cell response may play an important role in slowing down progression to AIDS (30). However, recent reports are contradictory with respect to a role for the Gag-specific CD8 T-cell response in controlling viral load. Some studies supported an association of a strong Gag-specific CD8 T-cell response with the control of HIV replication (10, 27-30, 34), whereas others showed no such association (1, 4, 6, 12). Our results suggest that the breadth of epitope recognition within specific regions of Gag strongly contributes to the antiviral efficiency of the CD8 T-cell response during the chronic phase of infection. The expression of certain HLA-B alleles, namely, B0702, B5801, and B8101, was associated with this particular pattern of CD8 T-cell recognition. This finding, and the inverse linear relationship between the number of recognized Gag (or GagR1R3) epitopes and plasma viral load, further supports the notion that the viral load-HLA associations are mediated by the CD8 T-cell response restricted by these HLA alleles. The hypothesis that the breadth of GagR1R3 recognition is an important correlate of the expression of these HLA B alleles is reinforced by the examination of six individuals that have a high viral load despite expressing the protective allele B5801, B8101, or B0702. The breadth of GagR1R3 recognition was significantly reduced in these individuals compared to the other subjects expressing these B alleles (Fig. 4D).
Is the antigen specificity of CD8 T cells important for the maintenance of antiviral efficiency? In contrast to the Gag response, there was a trend of a broad Nef-specific response with higher viral loads, resembling observations of previous studies (4, 23, 27). The observed differences between Gag- and Nef-specific responses support the notion that antigen specificity plays a role in maintaining antiviral efficiency of HIV-specific CD8 T cells during the chronic phase of infection.
The observation that the magnitude of Gag- or GagR1R3-responding CD8 T cells, measured by cumulative IFN-γ ELISPOT responses, did not correlate with a low viral load contrasts with most of the recent reports that have found a protective role of strong Gag-specific T-cell responses. One possible explanation for this difference is that the majority of the HISIS-CTL substudy subjects were infected for extended time periods and had high viral loads. An alternative explanation is that the usage of three different Gag peptide sets permitted a better detection of epitope breadth compared to other studies. We have shown previously that that the Gag and Nef peptide sets used during this study did afford better detection of the breadth of the T-cell response (7, 14).
Importantly, “protective” HLA B alleles, B0702, B5801, and B8101, were associated with a median plasma viral load below 50,000 RNA copies/ml, constituting a fourfold reduction in the median viral load observed within the whole HISIS-CTL cohort. This comparatively high median viral load of 198,500 RNA copies per ml most likely reflects the long time since primary HIV infection in the majority of the study subjects, 63% of which (35 of 56) were infected for more than 3.5 years. The remaining 21 subjects seroconverted during the study and had a median viral load of 40,600 RNA copies/ml at 24 months after seroconversion, which is in the range observed in most other studies (10, 18, 23, 27). Together, these results underline that all study subjects were in the chronic phase of HIV infection: 21 were in the postacute or early chronic phase (9 months to 3.5 years of infection), and 35 were in later stages of the chronic phase of HIV infection (>3.5years of infection).
It is also noteworthy that most previous studies (and the present study) used IFN-γ-based assays to detected HIV-specific CD8 T cells. IFN-γ is a sensitive marker for HIV-specific CD8 T-cell activation; however, virus-specific cytolytic activity and IFN-γ secretion upon restimulation do not necessarily correlate during chronic HIV infection (3, 9, 19). This dislinkage of cytolytic activity and IFN-γ secretion has been ascribed to a phenomenon known as the “partial functional exhaustion” of T cells. In a mouse model of chronic LCMV infection, CD8 T cells of different epitope specificities can vary in their respective functional exhaustion, which is partly dependent on the antigenic load and the time from infection (32). A similar phenomenon may explain the differences between the present study and previous studies of HIV-1 infection.
One example of a very strong, yet potentially ineffective, CD8 T-cell response during chronic HIV infection is the targeting of the Gag epitope TL9. This epitope is presented by the class I alleles B0702, B4201, and B8101 and is targeted by 41% of the subjects in this study cohort, and responses are among those with the highest magnitude (14). This epitope is highly conserved within a particular subtype, and there is no evidence supporting viral escape by an epitope point mutation. Additionally, the high magnitude of responding cells and the lack of mutational changes in the immediate epitope proximity within subtype C argue against abrogated epitope processing. Therefore, we believe that neither the epitope itself nor its presentation is changed in the course of chronic infection. Despite all of these parameters, previously assumed to be favorable, we did not find any evidence supporting a substantial role for this immunodominant response in viral control during the chronic phase of infection. It is noteworthy that the three subjects that possess protective allele B8101, but with high viral loads, also targeted this epitope. The persistence of unaltered yet targeted epitopes during the course of chronic HIV infection has also been demonstrated in two recent studies (9, 19).
Why may the breadth of Gag or GagR1R3 but not Nef or Env recognition be important for viral control during the chronic stage of HIV infection? One possible explanation is that a broad Gag or GagR1R3 response is more likely to select for viral escape mutants that are characterized by a reduced replicative capacity. Responding T cells would at the same time poorly recognize the mutant epitope(s), and a reduced T-cell receptor stimulation may in turn slow down the functional exhaustion of virus-specific T cells. Indeed, some of the most “protective” epitopes within GagR1R3 are highly variable, and it has been shown previously that mutational escape from an epitope-specific T-cell response can cripple the replicative capacity of the mutant virus (25), despite it being less well recognized by virus-specific T cells. Episodic epitope reversion may then contribute to the persistence of wild-type epitope-specific T cells (13, 21) and at the same time “conserve” the antiviral efficiency of these T cells.
It is commonly accepted that the steady-state viral load is a major parameter affecting the rate of disease progression (26). Within this context, perhaps the most striking observation during this study was the course of the viral load in 19 B5703-negative individuals after seroconversion. Most subjects targeting multiple epitopes within GagR1R3 had a “normal” course of viral load, whereas subjects with a very narrow GagR1R3 response had an unusually high median viral load steady-state level. Therefore, this pattern of HIV-specific T-cell recognition is likely to be associated with slow/normal disease progression, and to the contrary, its absence is likely to be associated with a more rapid progression. Importantly, the latter finding may affect not only survival but also HIV transmission rates. Taken together, our findings highlight the association between the host genetic (HLA) background and the quality of the HIV-specific CD8 T-cell response during the chronic phase of HIV infection. The results may help to explain differences in disease progression on a population level. During evaluations of candidate HIV-1 vaccines, it may be useful to include vaccine immunogens that are capable of stimulating broad and targeted Gag responses in individuals with certain HLA class I alleles in the population.
Acknowledgments
This work was supported by the European Commission, DG XII, INCO-DC (grant ICA-CT-2002-10048), and by a cooperative agreement between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense. This study was conducted at the Mbeya Medical Research Programme.
We thank the excellent staff at the Mbeya Medical Research Programme that conducted the HISIS, especially Vera Kleinfeldt, Frowin Nichombe, Weston Assisya, and Clemence Konkamkula, and all of the HISIS participants. Furthermore, we thank Clive Gray (NICD, Johannesburg, South Africa) for critical advice on some experimental procedures and the subtype C peptides. Additionally, we thank Patricia D'Souza (Division of AIDS, NIH), Christian Brander (Massachusetts General Hospital), Steve Cate, and William Hildebrand (University of Oklahoma Health Science Center) for facilitating the HLA typing of the specimens.
The views and opinions expressed herein do not necessarily reflect those of the U.S. Army or the Department of Defense.
Footnotes
Published ahead of print on 20 December 2006.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, et al. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altfeld, M., M. M. Addo, E. S. Rosenberg, F. M. Hecht, P. K. Lee, M. Vogel, et al. 2003. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17:2581-2591. [DOI] [PubMed] [Google Scholar]
- 3.Appay, V., D. F. Nixon, S. M. Donahoe, G. M. Gillespie, T. Dong, A. King, et al. 2000. HIV-specific CD8(+) T cells produce antiviral cytokines but are impaired in cytolytic function. J. Exp. Med. 192:63-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Betts, M. R., D. R. Ambrozak, D. C. Douek, S. Bonhoeffer, J. M. Brenchley, J. P. Casazza, R. A. Koup, and L. J. Picker. 2001. Analysis of total human immunodeficiency virus (HIV)-specific CD4+ and CD8+ T-cell responses: relationship to viral load in untreated HIV infection. J. Virol. 75:11983-11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao, J., J. McNevin, S. Holte, L. Fink, L. Corey, and M. J. McElrath. 2003. Comprehensive analysis of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-secreting CD8+ T cells in primary HIV-1 infection. J. Virol. 77:6867-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Currier, J. R., U. Visawapoka, S. Tovanabutra, C. J. Mason, D. L. Birx, F. E. McCutchan, and J. H. Cox. 2006. CTL epitope distribution patterns in the Gag and Nef proteins of HIV-1 from subtype A infected subjects in Kenya: use of multiple peptide sets increases the detectable breadth of the CTL response. BMC Immunol. 7:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Draenert, R., T. M. Allen, Y. Liu, T. Wrin, C. Chappey, C. L. Verrill, et al. 2006. Constraints on HIV-1 evolution and immunodominance revealed in monozygotic adult twins infected with the same virus. J. Exp. Med. 203:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draenert, R., C. L. Verrill, Y. Tang, T. M. Allen, A. G. Wurcel, M. Boczanowski, A. Lechner, A. Y. Kim, T. Suscovich, N. V. Brown, M. M. Addo, and B. D. Walker. 2004. Persistent recognition of autologous virus by high-avidity CD8 T cells in chronic, progressive human immunodeficiency virus type 1 infection. J. Virol. 78:630-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards, B. H., A. Bansal, S. Sabbaj, J. Bakari, M. J. Mulligan, and P. A. Goepfert. 2002. Magnitude of functional CD8+ T-cell responses to the Gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 76:2298-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frahm, N., P. Kiepiela, S. Adams, C. H. Linde, H. S. Hewitt, K. Sango, et al. 2006. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat. Immunol. 7:173-178. [DOI] [PubMed] [Google Scholar]
- 12.Frahm, N., B. T. Korber, C. M. Adams, J. J. Szinger, R. Draenert, M. M. Addo, M. E. Feeney, K. Yusim, K. Sango, N. V. Brown, D. SenGupta, A. Piechocka-Trocha, T. Simonis, F. M. Marincola, A. G. Wurcel, D. R. Stone, C. J. Russell, P. Adolf, D. Cohen, T. Roach, A. StJohn, A. Khatri, K. Davis, J. Mullins, P. J. R. Goulder, B. D. Walker, and C. Brander. 2004. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J. Virol. 78:2187-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, et al. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275-281. [DOI] [PubMed] [Google Scholar]
- 14.Geldmacher, C., J. R. Currier, M. Gerhardt, A. Haule, L. Maboko, D. Birx, et al. 2007. In a mixed subtype epidemic, the HIV-1 Gag-specific T-cell response is biased towards the infecting subtype. AIDS 21:135-143. [DOI] [PubMed] [Google Scholar]
- 15.Goulder, P. J., M. A. Altfeld, E. S. Rosenberg, T. Nguyen, Y. Tang, R. L. Eldridge, et al. 2001. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendel, H., S. Caillat-Zucman, H. Lebuanec, M. Carrington, S. O'Brien, J. M. Andrieu, et al. 1999. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J. Immunol. 162:6942-6946. [PubMed] [Google Scholar]
- 17.Jones, N. A., X. Wei, D. R. Flower, M. Wong, F. Michor, M. S. Saag, et al. 2004. Determinants of human immunodeficiency virus type 1 escape from the primary CD8+ cytotoxic T lymphocyte response. J. Exp. Med. 200:1243-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, et al. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-775. [DOI] [PubMed] [Google Scholar]
- 19.Koibuchi, T., T. M. Allen, M. Lichterfeld, S. K. Mui, K. M. O'Sullivan, A. Trocha, S. A. Kalams, R. P. Johnson, and B. D. Walker. 2005. Limited sequence evolution within persistently targeted CD8 epitopes in chronic human immunodeficiency virus type 1 infection. J. Virol. 79:8171-8181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 22.Lichterfeld, M., D. E. Kaufmann, X. G. Yu, S. K. Mui, M. M. Addo, M. N. Johnston, et al. 2004. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J. Exp. Med. 200:701-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masemola, A., T. Mashishi, G. Khoury, P. Mohube, P. Mokgotho, E. Vardas, M. Colvin, L. Zijenah, D. Katzenstein, R. Musonda, S. Allen, N. Kumwenda, T. Taha, G. Gray, J. McIntyre, S. Abdool Karim, H. W. Sheppard, and C. M. Gray 2004. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J. Virol. 78:3233-3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mashishi, T., and C. M. Gray. 2002. The ELISPOT assay: an easily transferable method for measuring cellular responses and identifying T cell epitopes. Clin. Chem. Lab. Med. 40:903-910. [DOI] [PubMed] [Google Scholar]
- 25.Matano, T., M. Kobayashi, H. Igarashi, A. Takeda, H. Nakamura, M. Kano, et al. 2004. Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J. Exp. Med. 199:1709-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. (Erratum, 275:14, 1997.) [DOI] [PubMed] [Google Scholar]
- 27.Novitsky, V., P. Gilbert, T. Peter, M. F. McLane, S. Gaolekwe, N. Rybak, I. Thior, T. Ndung'u, R. Marlink, T. H. Lee, and M. Essex. 2003. Association between virus-specific T-cell responses and plasma viral load in human immunodeficiency virus type 1 subtype C infection. J. Virol. 77:882-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, et al. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 29.Ramduth, D., P. Chetty, N. C. Mngquandaniso, N. Nene, J. D. Harlow, I. Honeyborne, et al. 2005. Differential immunogenicity of HIV-1 clade C proteins in eliciting CD8+ and CD4+ cell responses. J. Infect. Dis. 192:1588-1596. [DOI] [PubMed] [Google Scholar]
- 30.Riviere, Y., M. B. McChesney, F. Porrot, F. Tanneau-Salvadori, P. Sansonetti, O. Lopez, et al. 1995. Gag-specific cytotoxic responses to HIV type 1 are associated with a decreased risk of progression to AIDS-related complex or AIDS. AIDS Res. Hum. Retrovir. 11:903-907. [DOI] [PubMed] [Google Scholar]
- 31.Turner, S., M. E. Ellexson, H. D. Hickman, D. A. Sidebottom, M. Fernandez-Vina, D. L. Confer, and W. H. Hildebrand. 1998. Sequence-based typing provides a new look at HLA-C diversity. J. Immunol. 161:1406-1413. [PubMed] [Google Scholar]
- 32.Wherry, E. J., J. N. Blattman, K. Murali-Krishna, R. van der Most, and R. Ahmed. 2003. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77:4911-4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, O. O., P. T. Sarkis, A. Ali, J. D. Harlow, C. Brander, S. A. Kalams, and B. D. Walker. 2003. Determinant of HIV-1 mutational escape from cytotoxic T lymphocytes. J. Exp. Med. 197:1365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuñiga, R., A. Lucchetti, P. Galvan, S. Sanchez, C. Sanchez, A. Hernandez, H. Sanchez, N. Frahm, C. H. Linde, H. S. Hewitt, W. Hildebrand, M. Altfeld, T. M. Allen, B. D. Walker, B. T. Korber, T. Leitner, J. Sanchez, and C. Brander. 2006. Relative dominance of Gag p24-specific cytotoxic T lymphocytes is associated with human immunodeficiency virus control. J. Virol. 80:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]