Abstract
Maraviroc is a CCR5 antagonist in clinical development as one of a new class of antiretrovirals targeting human immunodeficiency virus type 1 (HIV-1) coreceptor binding. We investigated the mechanism of HIV resistance to maraviroc by using in vitro sequential passage and site-directed mutagenesis. Serial passage through increasing maraviroc concentrations failed to select maraviroc-resistant variants from some laboratory-adapted and clinical isolates of HIV-1. However, high-level resistance to maraviroc was selected from three of six primary isolates passaged in peripheral blood lymphocytes (PBL). The SF162 strain acquired resistance to maraviroc in both treated and control cultures; all resistant variants were able to use CXCR4 as a coreceptor. In contrast, maraviroc-resistant virus derived from isolates CC1/85 and RU570 remained CCR5 tropic, as evidenced by susceptibility to the CCR5 antagonist SCH-C, resistance to the CXCR4 antagonist AMD3100, and an inability to replicate in CCR5 Δ32/Δ32 PBL. Strain-specific mutations were identified in the V3 loop of maraviroc-resistant CC1/85 and RU570. The envelope-encoding region of maraviroc-resistant CC1/85 was inserted into an NL4-3 background. This recombinant virus was completely resistant to maraviroc but retained susceptibility to aplaviroc. Reverse mutation of gp120 residues 316 and 323 in the V3 loop (numbering from HXB2) to their original sequence restored wild-type susceptibility to maraviroc, while reversion of either mutation resulted in a partially sensitive virus with reduced maximal inhibition (plateau). The plateaus are consistent with the virus having acquired the ability to utilize maraviroc-bound receptor for entry. This hypothesis was further corroborated by the observation that a high concentration of maraviroc blocks the activity of aplaviroc against maraviroc-resistant virus.
The entry of human immunodeficiency virus (HIV) into CD4+ cells is a complex multistep process highlighting rational targets for potential new classes of antiretroviral drugs. Drugs directed against CD4 attachment, coreceptor (CCR5 or CXCR4) binding, or membrane fusion have the advantage that they act extracellularly. Therefore, in contrast to protease inhibitors and reverse transcriptase (RT) inhibitors, HIV type 1 (HIV-1) entry inhibitors do not need to cross the cell membrane and, unlike nucleoside RT inhibitors, they do not require intracellular processing in order to exert their activity. In addition, there is no cross-resistance between entry inhibitors and agents that act on intracellular targets (6, 18). The fusion inhibitor enfuvirtide is the first entry inhibitor to be licensed for clinical use, and a number of agents targeting the interaction between the virus envelope glycoprotein gp41/gp120 and either its cellular receptor CD4 (12, 16) or the coreceptor CCR5 or CXCR4 (7, 18, 26) are under development. Maraviroc is an orally administered small-molecule CCR5 antagonist that has demonstrated a promising efficacy and safety profile in phase IIa clinical trials (6, 8).
Antiretroviral drug resistance has been, and continues to be, a major barrier to the successful long-term treatment of HIV infection. The high replication rate of HIV, with the low fidelity and lack of proofreading activity of its RT, means that the virus has a high potential for mutations and developing resistance to antiviral agents. Since the envelope gene (Env) is the most sequence variable of all the HIV-1 genes (9), the possibility also exists that viruses with reduced susceptibility to entry inhibitors may exist in patients who are naive to this group of agents. Therefore, a clear understanding of the mechanisms of resistance and the potential for cross-resistance with other antiretrovirals is important for any new HIV-1 entry inhibitor in development. Of additional concern for CCR5 antagonists is the possibility that selective pressure from these drugs could result in the emergence of CXCR4-tropic viruses. Generally, only CCR5-tropic viruses are detected at the time of HIV-1 infection, with CXCR4-tropic viruses emerging in some patients in the later stages of infection. CXCR4-tropic virus is seen to emerge in around 60% of patients with disease progression and is associated with CD4 cell decline and rapid progression (13, 14, 25). However, it is unknown whether emergence of CXCR4 tropism is a cause or a consequence of severe immune system impairment.
Preclinical data suggest that natural resistance to maraviroc is likely to be rare. Maraviroc inhibited a large panel of 43 CCR5-tropic primary HIV-1 isolates of subtype B and non-B origin (geometric mean 90% inhibitory concentration [IC90], 2.0 nM; 95% confidence interval, 1.8 to 2.4 nM) in an acute infection assay using activated peripheral blood lymphocytes (PBL) (6). In a single-cycle replication assay using Env pseudotyped viruses, maraviroc was active against 200 clinically derived strains, of subtype B and non-B origin, from antiretroviral treatment-naive and treatment-experienced patients (geometric mean IC90, 13.7 nM; 95% confidence interval, 12.3 to 15.1 nM) (6). Furthermore, mutations associated with resistance to enfuvirtide map to the gp41 region of the viral envelope complex (31), so cross-resistance between enfuvirtide and CCR5 antagonists is unlikely. However, since most CCR5 antagonists under development bind to approximately the same region of CCR5, cross-resistance between agents within this class might be expected. The aim of this study was to generate maraviroc-resistant viruses by serial passage with a range of CCR5-tropic clinical isolates and to characterize the resultant viruses in order to better understand the molecular mechanisms of resistance to this agent and the potential for cross-resistance with other entry inhibitors.
MATERIALS AND METHODS
Compounds, cells, and viruses.
Enfuvirtide was synthesized by NeoMPS (Strasbourg, France). Maraviroc (UK427,857), SCH-C, aplaviroc, efavirenz, and saquinavir were synthesized at Pfizer Ltd., United Kingdom.
PBL were isolated by Ficoll-Paque gradient separation (Amersham Pharmacia Biotech, Piscataway, NJ) from pooled buffy coats (South London Blood Transfusion Services, United Kingdom) from three or four HIV- and hepatitis B virus-seronegative donors per batch. PBL were stimulated with phytohemagglutinin (PHA; Murex, Abbott Laboratories) at 1.5 μg/ml for 3 days before use. PBL homozygous for the CCR5 Δ32/Δ32 polymorphism were kindly provided by Rolf Kaiser (Cologne University). These cells carry a 32-bp deletion in the CCR5 gene that results in the production of CCR5 molecules that are not expressed on the surface of the cell. The ability of the CCR5 Δ32/Δ32 PBL to support HIV-1 infection under the conditions used was confirmed by using CXCR4-tropic laboratory-adapted strain NL4-3 (obtained from the Centralized Facility for AIDS Reagents, National Institute for Biological Standards and Control [NIBSC], Potters Bar, Hertfordshire, United Kingdom). The CD4+ T-cell clone PM1 was obtained from the Centralized Facility for AIDS Reagents (NIBSC, Potters Bar, Hertfordshire, United Kingdom) (17). All cells were maintained at 37°C and 5% CO2 in RPMI 1640 medium containing 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, 1 U/ml penicillin, and 0.1 mg/ml streptomycin. Interleukin-2 was included in PBL cultures at a final concentration of 10 to 15 ng/ml.
CCR5-tropic laboratory-adapted strain Ba-L and primary HIV-1 isolates 92BR017, 92BR018, 92BR023, SF162 (all subtype B), and RU570 (subtype G) were obtained from the Centralized Facility for AIDS Reagents. Primary HIV-1 isolate CC1/85 (subtype B) was kindly donated by John Moore (Cornell University).
Virus passage.
For experiments with laboratory-adapted strain Ba-L, PM1 cells or PHA-stimulated PBL (1 × 107 cells) were infected with 3.88 × 103 or 4.85 × 103 tissue culture infective doses, respectively, of virus. Infected PM1 cells and PBL were cultured at approximately 1 × 105 cells/ml and 2 × 105 cells/ml, respectively, in 2-ml cultures containing 0.04 nM maraviroc. Every 7 days, virus supernatant was passaged onto fresh cells, at the same cell density, with fresh maraviroc-containing medium. Virus levels were monitored weekly by measuring RT activity (Quan-T-RT assay; Amersham Biosciences/GE Healthcare). The concentration of maraviroc was increased upon subsequent passage whenever evidence of viral replication was observed.
For experiments with the six clinical isolates, PHA-stimulated PBL (1 × 107 cells) were infected with a standardized input of virus as determined by RT activity (equivalent to 75,000 cpm). Infected cultures were maintained at approximately 3 × 105 cells/ml for 1 week to establish virus growth, and these were then used to infect fresh, uninfected, PHA-stimulated PBL by cocultivation (10 ml of infected cell suspension plus 6 × 106 uninfected cells resuspended in 40 ml of medium). Cocultivated cells were plated in 4-ml cultures containing maraviroc at one of two concentrations, i.e., the previously determined IC90 for the virus or a twofold higher concentration. Virus growth was monitored after 6 days by measuring supernatant p24 levels (HIV MAB p24 ELISA; Murex, Abbott Diagnostics). The cultures were split on day 7 by transferring 1.25 ml of the resuspended PBL to replicate wells of a six-well plate containing 3.75 ml fresh PHA-stimulated PBL (cell density, 2 × 105/ml). After 4 days, maraviroc was added at final concentrations twice and/or equal to that of the presplit concentration, depending on whether the p24 levels had increased or not, respectively. After a further 2 days of incubation, the p24 levels were measured and the cultures were split on day 7 as described above. This was continued on a weekly basis.
For all experiments with Ba-L and clinical isolates, maraviroc-free passages were set up in parallel to control for changes in sensitivity to maraviroc upon prolonged culture of virus. For all maraviroc-associated passages when the virus no longer appeared to be inhibited by increasing amounts of compound, virus stocks were generated for further analysis.
Phenotypic analysis of tropism and susceptibility to CCR5 antagonists.
Drug susceptibility assays with PBL and PM1 cells were carried out as previously described (6). Briefly, cells were infected for 1 h at 37°C with a predetermined volume of virus calculated to give an equal amount of RT activity per virus stock. Infected cells were washed and added to duplicate wells of 24-well plates (3.6 × 105 cells/well for PBL, 2 × 105 cells/well for PM1 cells) or triplicate wells of 96-well plates (7.2 × 104 cells/well for PBL only) containing serial dilutions of test compound in culture medium. After 5 to 7 days of incubation, the cultures were examined visually with a microscope for evidence of cytotoxicity and viral replication was quantified by measuring RT activity or supernatant p24 (for samples which replicated to lower levels).
Drug susceptibility and coreceptor tropism were also determined by Monogram Biosciences Inc. (South San Francisco, CA) with an in vitro pseudovirus assay (PhenoSense HIV Entry assay) (3, 29). The HIV-1 envelope coding sequence was amplified from virus samples by PCR and ligated into a pCXAS expression vector to create an envelope expression vector library. Virus particles were produced by transfecting HEK293 cells with the purified envelope expression vector library and an HIV-1 genomic vector lacking the envelope-encoding region and containing a firefly luciferase gene. The ability of the pseudoviruses to infect U87CD4+ cells overexpressing either CCR5 or CXCR4 in the presence or absence of various concentrations of inhibitors was assessed by measuring luciferase-generated relative light units (RLU). Tropism was assigned on the basis of RLU readings above the background level and was confirmed by the inhibition of viral replication in each cell type by a CCR5 or a CXCR4 antagonist. For clonal analyses, 100 to 200 individual Env clones from each sample were prescreened for viability and tropism in single-well cultures of CCR5- or CXCR4-expressing cells. At each time point, at least 12 viable clones were selected for further phenotypic and genotypic analysis.
For both phenotypic assays, 50% inhibitory concentrations (IC50) were calculated from plots of percent inhibition of viral replication versus drug concentration. For the PhenoSense HIV Entry Assay, plateaus in maximal percent inhibition were estimated visually from the inhibition curves. For PBL assays, sigmoidal dose-response inhibition curves were fitted to the antiviral data with a proprietary curve-fitting program (Excel add-in LabStats), and the maximum percent inhibition was reported by the curve-fitting algorithm as the upper asymptote.
Genotypic analysis.
Sequencing of the entire Env gene was performed by using Big Dye chain terminator chemistry (PE Biosystems, Foster City, CA) and an ABI 3700/3730XL sequencer. Sequences were aligned by using the CLUSTAL program, versions W and X (27), and then edited with GENEDOC (K. B. Nicholas and H. B. Nicholas, Jr., GeneDoc: a tool for editing and annotating multiple sequence alignments, 1997 [distributed by the authors]) to produce a final alignment. Mutations were numbered according to the HXB2 reference sequence (K03455).
Construction of recombinant molecular clones and SDM.
A 2.56-kb fragment encompassing the entire Env gene was amplified from DNA isolated from PBL infected with pre- and post-maraviroc passage stocks of primary isolate CC1/85. The respective Env genes were inserted into the EcoRI and XhoI restriction sites of pNL4-3 to produce molecular clones that we have named MVCsens (prepassage) and MVCres (postpassage). Site-directed mutagenesis (SDM) was performed on the MVCres Env sequence with the QuikChange XL system (Stratagene, La Jolla, CA), following its transfer into a shuttle vector (pSP72; Promega). The Env-recombinant NL4-3 viruses were recovered in the tissue culture supernatant of HEK293T cells 48 h posttransfection.
Nucleotide sequence accession numbers.
The following HIV-1 envelope sequences have been submitted to GenBank: 7 sequences for SF162-derived viruses (accession no. EF367173 through EF367179); 28 sequences for CC1/85-derived viruses (accession no. EF367180 through EF367207); and 27 sequences for RU570-derived viruses (accession no. EF367208 through EF367234).
RESULTS
Laboratory-adapted strain Ba-L remained sensitive to maraviroc following 30 passages in the presence of compound.
Initial in vitro selection experiments were carried out with laboratory-adapted CCR5-tropic HIV-1 strain Ba-L passaged weekly in either PBL or PM1 cells in the presence of increasing concentrations of maraviroc. Passage was started at a maraviroc concentration either 5- or 10-fold below the IC50 for Ba-L in PM1 and PBL cells, respectively, to allow the cultures to become established. After 30 weeks of passage in PBL and 32 weeks of passage in PM1 cells, the concentration of maraviroc had been increased to more than 100 times the IC50, reaching maxima of 20 and 80 nM, respectively. However, evidence of viral replication in the passage cultures was erratic; at most passages, RT levels in the maraviroc culture supernatants were either lower than in the control cultures or undetectable. Consistent with this observation, only small shifts in the maraviroc IC50 were observed (Table 1). While they may be biologically significant, these shifts in susceptibility to maraviroc were within the expected range of variability of the assay. The PM1 virus was sequenced, and no amino acid changes were observed in the V3 loop region between pre- and postpassage virus (data not shown).
TABLE 1.
Overview of maraviroc passage experiments using CCR5-tropic viruses in PM1 cells or PBL
| Virus strain (reference)a | Subtypeb | Maraviroc IC50/IC90 (nM)c | Passage experiments
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Cell type | Concn (nM) at:
|
Duration (wk) | End IC50 (nM) | FCd | End tropism | ||||
| Start | End | ||||||||
| Ba-L | B (United States) | 0.2/2.0 | PM1 | 0.04 | 80 | 32 | 0.6-1.4 | 2.6-6.6 | CCR5 |
| 0 | 0 | 32 | 0.5 | 2.3 | CCR5 | ||||
| 0.5/6.4 | PBL | 0.04 | 20 | 30 | 1.8-4.1 | 3.6-8.3 | CCR5 | ||
| 0 | 0 | 30 | 0.6 | 1.1 | CCR5 | ||||
| 92BR017 (29)e | B (Brazil) | 0.3/1.4c | PBL | 2.0 | 32 | 11 | NDg | ND | ND |
| 0 | 0 | 11 | ND | ND | ND | ||||
| 92BR018 (29)e | B (Brazil) | 0.3/1.3c | PBL | 2.0 | 64 | 20 | ND | ND | ND |
| 0 | 0 | 20 | ND | ND | ND | ||||
| 92BR023 (29) | B (Brazil) | 0.7/4.1c | PBL | 8.0 | 32 | 9 | ND | ND | ND |
| 0 | 0 | 9 | ND | ND | ND | ||||
| SF162 (1) | B (United States) | 2.3/15.2 | PBL | 16 | 8,000 | 12 | >50,000 | >21,000 | CXCR4 |
| 0 | 0 | 12 | >50,000 | >21,000 | CXCR4 | ||||
| CC1/85 (3)f | B (United States) | 3.0/12.2 | PBL | 2 | 4,000 | 16 | >5,600 | >1,526 | CCR5 |
| 0 | 0 | 16 | 1.9 | 0.65 | CCR5 | ||||
| RU570 | G (Russia) | 2.5/149 | PBL | 32 | 16,000 | 18 | >50,000 | >20,000 | CCR5 |
| 0 | 0 | 18 | 0.59 | 0.24 | CCR5 | ||||
All isolates were obtained from the Centralized Facility for AIDS Reagents (NIBSC, Potters Bar, United Kingdom), except for CC1/85 (kindly provided by John Moore). Isolates were chosen for inclusion on the basis of a relatively high maraviroc IC90 as determined by Dorr et al. (6) (unless otherwise specified).
Subtype and starting coreceptor usage were assigned by NIBSC.
The IC50 and IC90 quoted for Brazilian isolates were determined as described by Dorr et al. (6) and are means of four to eight determinations. For other isolates, the values quoted were determined in drug susceptibility assays where the start and passaged viruses were tested in parallel. In most cases, these values were in good agreement with those determined by Dorr et al. (6); the IC90 for the RU570 isolate was 5- to 10-fold higher than previously reported, but a complete dose-response curve was obtained.
FC, fold change in IC50 relative to the prepassage virus tested in parallel.
Chosen on the basis of good growth in PBL in vitro.
Chosen on the basis of prior use to generate escape mutants to other CCR5 antagonists (14, 18, 27).
ND, not done.
Maraviroc-resistant variants were selected in three of six primary HIV-1 isolates following serial passage in PBL.
Following the failure to generate maraviroc-resistant virus with laboratory-adapted HIV-1 strain Ba-L, we initiated a new series of experiments with primary clinical isolates of HIV-1 grown in PBL as detailed in Table 1.
No resistance was observed with CCR5-tropic primary isolates 92BR017, 92BR018, and 92BR023 over 11, 20, and 9 weeks of culture, respectively (Table 1). During the early passages of these viruses, p24 levels remained in step with levels in the control passage through increasing concentrations of maraviroc. However, as maraviroc concentrations increased, virus replication appeared to be inhibited and cultures were terminated when there was no longer any evidence of ongoing virus replication, as determined by an absence of p24 antigen in consecutive passages (Fig. 1). In contrast, virus replication was detected in cultures of isolates CC1/85, RU570, and SF162 when maraviroc concentrations were increased hundreds of times above the starting concentration (Fig. 1). For all three viruses, cultures were continued until increasing the concentration of maraviroc no longer suppressed virus replication, and the p24 production rates paralleled that of the control culture.
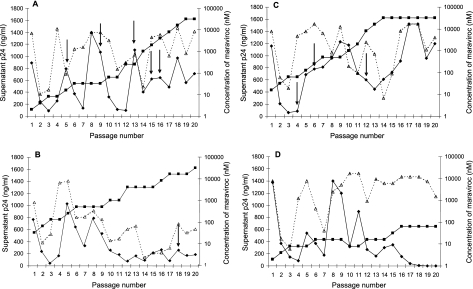
FIG. 1.
Growth of HIV-1 isolates passaged through PBL in the presence of maraviroc. Filled diamonds represent p24 values in wells containing virus passaged with maraviroc, open triangles represent p24 values in untreated control wells, and filled squares represent the concentrations of maraviroc in the treated cultures. The arrows indicate the passages at which virus supernatants were harvested for further analysis. Virus appeared to escape maraviroc inhibition in cultures of CC1/85 (A), RU570 (B), and SF162 (C), as shown by the fact that p24 levels in the maraviroc-containing cultures approximately matched the p24 levels in the drug-free control cultures by the end of the experiment. In contrast, escape variants were not recovered from cultures of 92BR017, 92BR018, and 92BR023. Results for 92BR018 are shown as an example in panel D. Although virus appeared to break through in the presence of 16 nM maraviroc at passage 8, this virus subsequently failed to replicate and there was no detectable p24 by passage 20 (not shown). A frozen aliquot of passage 10 supernatant was recovered and passaged in 8 nM maraviroc. This culture showed evidence of breakthrough again (passage 11) but was subsequently unable to replicate in higher concentrations of maraviroc.
Virus stocks for further analysis were expanded from passage 16 (4,000 nM maraviroc) for CC1/85, passage 18 (16,000 nM maraviroc) for RU570, and passage 12 (8,000 nM maraviroc) for SF162. To control for the possibility that the resistance phenotype might revert during the expanded culture, stocks were prepared both in the presence and in the absence of maraviroc. Virus stocks were tested for susceptibility to maraviroc, the CXCR4 antagonist AMD3100, and the protease inhibitor saquinavir in PBL. Table 2 summarizes the results obtained for virus stocks expanded in the absence of maraviroc. Almost identical results were obtained with stocks expanded in the presence of maraviroc. Therefore, in later experiments only the stock expanded in the absence of maraviroc was used in order to minimize the potential effect of maraviroc being carried over in the virus inoculum. All viruses showed similar levels of susceptibility to the control compound saquinavir, indicating that no shift in sensitivity had occurred during prolonged in vitro passage, either in the presence or in the absence of maraviroc.
TABLE 2.
Susceptibilities to maraviroc, AMD3100, saquinavir, SCH-C, and enfuvirtide of pre- and postpassage virus derived from primary isolates CC1/85, RU570, and SF162 in PBL and U87CD4+ cells
| Virus, drug, passage no. | IC50 of virus stocks in PBL (nM)
|
IC50 in PhenoSense HIV Entry assay (U87CD4+CCR5+ cells, nM)c
|
|||||
|---|---|---|---|---|---|---|---|
| Maraviroca | AMD3100a | Maraviroc + AMD3100b | Saquinavira | Maraviroc | SCH-C | Enfuvirtide | |
| CC1/85, start | 3.0 | >1,000 | NDd | 4.4 | 7.0 | 222 | 171 |
| CC1/85, control, 16 | 1.9 | >1,000 | 0.53 | 4.6 | 6.2 | 165 | 111 |
| CC1/85, maraviroc, 16 | >5,600 | >1,000 | >3,300 | 4.8 | 3.7 | 161 | 129 |
| RU570, start | 2.5 | >1,000 | 0.43 | 3.7 | 3.4 | 63 | 11 |
| RU570, control, 18 | 0.59 | >1,000 | 0.28 | ND | 4.1 | 82 | 6.0 |
| RU570, maraviroc, 18 | >50,000 | >1,000 | >3,300 | 2.0 | 2.0 | 20 | 3.0 |
| SF162, start | 2.3 | >1,000 | ND | 3.2c | ND | ND | ND |
| SF162, control, 12 | >50,000 | 202 | 27 | 2.1c | ND | ND | ND |
| SF162, maraviroc, 12 | >50,000 | 24 | 30 | ND | ND | ND | ND |
Results for CC1/85 are the mean of two separate experiments. Results for RU570 and SF162 are from single experiments unless otherwise specified.
Maraviroc and AMD3100 were titrated in the same dose range. Thus, the IC50 relates to both compounds being present at the same concentration.
Mean of two experiments.
ND, not done.
Sequential passage of SF162 resulted in a tropism shift irrespective of the presence of maraviroc.
SF162-derived viruses passaged either with or without maraviroc were both highly resistant to maraviroc and susceptible to AMD3100 in the PBL assay (Table 2), suggesting that a CXCR4-using variant was selected in both the maraviroc and control passage cultures. This was confirmed by the tropism assay (Table 3), in which dual- and mixed-tropic viruses were detected by passage 4 in both the maraviroc and control cultures. Sequencing of the Env regions of SF162-derived viruses revealed changes from isoleucine to arginine at position 309 (I309R) and from alanine to valine at position 316 (A316V) in both cultures (Table 3). The virus passaged in the presence of maraviroc harbored an additional change from alanine to threonine at position 319 (A319T).
TABLE 3.
Coreceptor tropism and V3 loop amino acid sequences of pre- and postpassage SF162
| Virus, drug, and passage no. | Coreceptor tropisma | gp120 V3 loop sequenceb |
|---|---|---|
| SF162, start | R5 | CTRPNNNTRKSITIGPGRAFYATGDIIGDIRQAHC |
| SF162, control | ||
| 4 | DM | -------------r----v---------------- |
| 6 | DM | -------------r----v---------------- |
| 12 | DM | -------------r----v---------------- |
| SF162, maraviroc | ||
| 4 | DM | -------------R-------t------------- |
| 6 | DM | -------------R----v--T------------- |
| 12 | DM | -------------R----V--T------------- |
Tropism was assigned phenotypically by using the PhenoSense HIV Entry assay. R5, CCR5 tropic; DM, dual or mixed tropic.
Sequences were obtained by population sequencing. Amino acid positions are numbered according to HXB2. Changes shown in lowercase indicate positions where a mixture was observed between the starting sequence and the mutated sequence. Amino acids 309 and 316 are underlined in the topmost sequence.
Maraviroc-resistant variants derived from CC1/85 and RU570 remained CCR5 tropic.
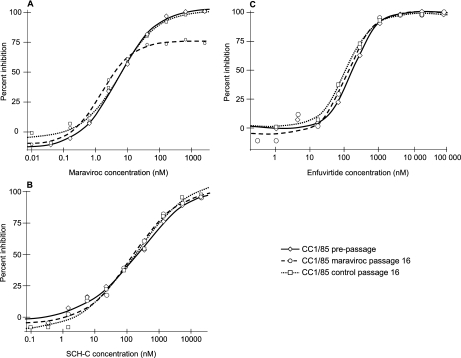
Viruses derived from CC1/85 and RU570 passaged with maraviroc were highly resistant to maraviroc, giving almost flat dose-response curves in PBL (not shown) from which an IC50 could not be calculated (Table 2). In contrast, stocks from the control passages for these two viruses remained fully susceptible to maraviroc. The maraviroc-resistant viruses were not susceptible to AMD3100 or to the combination of maraviroc and AMD3100 in PBL (Table 2), indicating that resistance was not associated with the use of CXCR4 as a coreceptor. This was confirmed by the fact that none of the CC1/85- and RU570-derived viruses were able to replicate in PBL from a CCR5 Δ32/Δ32 homozygous donor but replicated well in PBL from a CCR5 wt/wt donor (not shown). In addition, in the PhenoSense HIV Entry assay, all the CC1/85- and RU570-derived pseudoviruses selectively infected U87CD4+ cells expressing CCR5 but not those expressing CXCR4 and were therefore classified as CCR5 tropic. When maraviroc-resistant CC1/85 and RU570 were tested for susceptibility to maraviroc in the PhenoSense HIV Entry assay, no shifts in IC50 between pre- and postpassage virus were observed (Table 2). Rather, incomplete dose-response curves were obtained, characterized by a plateau at less than 90% inhibition (Fig. 2). To determine whether these plateaus were a consequence of a mixed population of viruses with different susceptibilities to maraviroc, individual Env clones were tested. Again there were no shifts in IC50, and incomplete susceptibility curves, with plateaus at 90% or less inhibition, were obtained for 8 of 12 CC1/85-derived clones and 7 of 11 RU570-derived clones (Table 4). Since the majority of individual Env clones gave a plateau in maximum percent inhibition, it was concluded that the plateau phenotype observed in the pooled sample could not have arisen due to a mixture of a fully sensitive virus (i.e., a virus that gives a dose-response inhibition curve from 0% to 100%) with a fully resistant virus (i.e., a virus that shows no dose-dependent inhibition across all concentration ranges).
FIG. 2.
Susceptibility of pseudoviruses derived from CC1/85 to maraviroc (A), SCH-C (B), and enfuvirtide (C) in the PhenoSense HIV Entry assay (U87CD4+ cells expressing CCR5). Similar results were obtained for RU570-derived viruses.
TABLE 4.
V3 loop amino acid sequences of prepassage, postpassage, and revertanta CC1/85 and RU570
| Virus, drug | Passage no. | Clone(s)b | gp120 V3 loop amino acid sequencec |
|---|---|---|---|
| CC1/85, startd | NAh | NA | CTRPNNNTRKSIHIGPGRAFYATGDIIGDIRQAHCi |
| CC1/85, controld | 16 | NA | ------Y------M-----L--------------- |
| CC1/85, maravirocd | 16 | NA | ------------------T--X---V--------- |
| CC1/85, maraviroce | 16 | 2, 4, 9, 12, 31, 37 | ------------------T------V--------- |
| 3, 5, 7 | ------------------T--S---V--------- | ||
| 1 | ------S-----------S------V--------- | ||
| 8 | ------------------T------V--G------ | ||
| 10 | ------------------S------V--------- | ||
| CC1/85, revertantd | 20a | -------------------X-----V--------- | |
| CC1/85, revertante,f | 20a | 2, 6, 9, 15, 17, 19, 22, 30, 31, 34, 35, 46 | -------------------------V--------- |
| RU570, startd | NA | NA | CTRPNNNTRKSISFGPGQAIYTTGNIIGDIRQAHCj |
| RU570, controld | 18 | NA | ---------------------K--D---------- |
| RU570, maravirocd | 18 | NA | ---------------X----------------k |
| RU570, maraviroce | 18 | 7g, 8, 10, 23, 31, 32, 38, 48 | ---------------------D----------k |
| 19, 41 | ------------------------D---------- | ||
| 25 | ------------------------D------R--- | ||
| RU570, revertantd | 20a | NA | ---------------------X--D---------- |
| RU570, revertante | 20a | 5, 27, 29, 40, 43, 46, 53, 57 | ---------------------K--D---------- |
| 16, 32, 44, 48 | ---------------------T--D---------- |
Viruses generated by passage in the presence of maraviroc were further passaged for 20 weeks in the absence of inhibitor to investigate the stability of the escape mutants. We have named the resultant variants revertants.
Individual clones were also tested for maraviroc susceptibility in the PhenoSense HIV Entry assay. The majority of clones resulted in incomplete inhibition curves with plateaus of maximum inhibition at 90% or less, similar to that seen in Fig. 2A. Clones that did give a complete dose-response curve, reaching 100% inhibition, are in boldface.
Amino acid positions are numbered according to HXB2. X indicates an unresolvable mixture.
Sequence obtained by population sequencing.
Sequence of an individual clone.
Plateaus of maximum inhibition were at 90 to 95%.
The presence of a plateau could not be clearly determined due to the low number of RLU.
NA, not applicable.
Amino acids 316 and 323 are underlined.
Amino acids 316 and 322 are underlined.
Blank spaces in the alignment represent a 3-amino-acid deletion.
Maraviroc resistance is associated with mutations in Env.
Distinct amino acid changes in the V3 loop were observed in maraviroc-free passage controls and maraviroc-resistant CC1/85- and RU570-derived viruses, compared with the prepassage sequences (Table 4). For maraviroc-resistant CC1/85, the population sequence and 12 of 12 clones showed changes at amino acid positions 316 and 323, while for maraviroc-resistant RU570, a three-amino-acid QAI deletion was observed in the population sequence and in 8 of 11 clones. Of note for RU570, the three clones that did not carry this deletion were the three that did not give plateaus in maximal inhibition of <90% in the PhenoSense HIV Entry assay, consistent with the QAI deletion conferring the resistance phenotype in this strain. Mutations outside the V3 loop were also observed in the CC1/85 and RU570 resistant variants. All 12 CC1/85-derived Env clones sequenced contained the substitutions T163K in V2, N355Y in C3, and S405A in V4. On the other hand, all 11 RU570-derived Env clones sequenced contained the substitutions D389N in V4, N442K in C4, and E509K in the gp120/gp41 junction. In addition, 9 of 11 Env clones contained the substitution S132N in V1. Virus derived from the control passage of CC1/85 carried a two-amino-acid SN deletion at amino acid positions 188 and 189 in V2. This virus appeared to infect cells more efficiently than the prepassage virus in that it consistently gave higher readings in the PhenoSense assay and was the only CC1/85 variant that replicated in PM1 cells, the latter of which express low levels of CD4 and CCR5 (data not shown).
Two amino acid substitutions in the V3 loop confer maraviroc resistance in CC1/85.
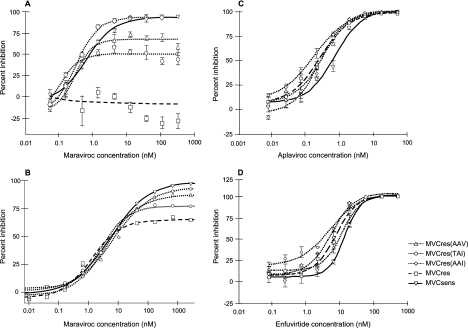
Maraviroc-resistant virus derived from CC1/85 was chosen for further characterization. The accumulation of mutations over time in CC1/85 passaged in the presence of maraviroc appeared to occur in an ordered fashion (Table 5). Of note, the I323V V3 mutation was established first. To determine which amino acid residues were associated with the resistance phenotype, the Env genes from prepassage and maraviroc-resistant CC1/85 were cloned into pNL4-3 to create the replication-competent Env recombinant clones that we have called MVCsens and MVCres, respectively. The Env gene from the MVCres clone was further modified within the V3 region by using SDM to “reverse mutate” amino acid residues 316 and 323 at either or both positions (Table 6). The maraviroc susceptibility of the SDM clones was assessed both in PBL and in the PhenoSense HIV Entry assay (Fig. 3). As expected, in the PBL assay, MVCsens remained susceptible to maraviroc while MVCres was fully resistant, confirming that the NL4-3 background did not alter the virus tropism or maraviroc susceptibility. In addition, both viruses were susceptible to another CCR5 antagonist (aplaviroc), confirming their reliance on CCR5 for cell entry (Table 6). Maraviroc susceptibility curves for MVCres, MVCres(TAI), and MVCres(AAV) were characterized by plateaus in maximum inhibition below 90% in both PBL and PhenoSense HIV Entry assays (Fig. 3). As a consequence, the maraviroc IC50 for these SDM were not increased in comparison with the MVCsens virus (Table 6). However, a trend could be observed in the level of the plateaus. In both assays, the maximum percent inhibition was greatest for MVCsens and MVCres(AAI) (>90%) and successively lower for MVCres(AAV), MVCres(TAI), and MVCres (carrying the TAV motif). Of note, the dose-response curves for MVCsens and MVCres(AAI) did not quite reach 100% inhibition (Fig. 3 and Table 6).
TABLE 5.
Accumulation of mutations in gp120 during passage of CC1/85 in increasing concentrations of maraviroc
| Condition | Mutation in gp120a
|
|||||
|---|---|---|---|---|---|---|
| Domain V2 | Domain V3
|
Domain C3
|
Domain V4
|
|||
| Amino acid 163 | Amino acid 316 | Amino acid 319 | Amino acid 323b | Amino acid 355 | Amino acid 405 | |
| CC1/85, start | T | A | A ≫ Tc | I ≫ Vd | N ≫ P> K = Ye | S ≫ A > T = E |
| Passage 5 (16)f | T | A | A | I | N > P | S |
| Passage 9 (32) | T > K | A | A | V | Y > N > S | S ≫ A |
| Passage 13 (128) | K > T | T > S | A | V | Y | A |
| Passage 15 (2,000) | K | T ≫ S | A > S | V | Y | A ≫ V |
| Passage 16 (4,000) | K | T ≫ S | A > S | V | Y | A |
Mutations were identified by sequencing the entire gp160 region of 36 envelope clones amplified from the start virus and 12 envelope clones amplified from the five other time points. Amino acid positions are numbered according to HXB2. Bold font indicates where the majority population carries a change from the prepassage virus.
This could be considered position 322 according to the placement of an insertion in the alignment.
A319T was seen in 4/36 clones.
I323V was seen in 1/36 clones.
N355Y was seen in 1/36 clones.
The concentration [nanomolar] of maraviroc in the selection culture is in parentheses.
TABLE 6.
Susceptibilities of Env-recombinant NL4-3 molecular clones created from maraviroc-resistant CC1/85 by SDM to entry inhibitors in PBL
| Clonej | Mutation in gp120a
|
Entry inhibitor susceptibility in PBLb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Domain V2 | Domain V3
|
Domain C3
|
Domain V4
|
Maraviroc
|
Aplaviroc IC50 (nM) | Enfuvirtide IC50 (nM) | ||||
| Amino acid 163 | Amino acid 316 | Amino acid 319 | Amino acid 323 | Amino acid 355 | Amino acid 405 | IC50 (nM) | Plateau (%)c | |||
| MVCres | K | T | A | V | Y | A | >333 | NDRi | 0.23 | 5.78 |
| MVCres(TAI) | K | T | A | I | Y | A | 0.35 | 67d | 0.13 | 3.28 |
| MVCres(AAV) | K | A | A | V | Y | A | 0.41 | 74d | 0.32 | 9.29 |
| MVCres(AAI) | K | A | A | I | Y | A | 0.50 | 95 | 0.23 | 10.1e |
| MVCsensh | T | A | A | I | N | S | 0.89f | 93f | 0.53f | 12.2g |
The 2.56-kb Env gene was cloned by PCR from maraviroc-resistant variants of CC1/85 and inserted into pNL4-3, to produce MVCres. A similar fragment was cloned from the start virus to produce MVCsens. SDM was performed on the MVCres clone to revert mutations in the V3 loop back to the wild type.
Drug susceptibility was assessed over 7 days in PBL. Viral replication was quantified by p24. Results shown are the geometric means of five experiments unless otherwise stated.
Percent inhibition for the plateau in maximal inhibition.
P < 0.05 versus MVCsens (analysis of variance).
Geometric mean of four experiments.
Geometric mean of three experiments.
Geometric mean of two experiments.
There were an additional eight amino acid differences in gp41 between MVCsens and MVCres.
NDR, no dose response (see Fig. 3A).
Boldface letters in clone designations indicate amino acid substitutions in MVCres.
FIG. 3.
Representative susceptibility curves for MVCsens, MVCres, and SDM Env-recombinant NL4-3 clones with maraviroc tested in PBL (A) and in the PhenoSense HIV Entry assay (B) and with aplaviroc (C) and enfuvirtide (D) tested in PBL. For PBL assays (A, C, and D), PBL were infected with a standardized input of virus in the presence of various concentrations of inhibitor. Supernatant p24 levels were determined after 7 days. Data points represent the means of triplicate wells within the same assay, and the bars represent the standard error of the mean. PBL assays were repeated between two and five times; mean IC50 are given in Table 6. For the PhenoSense HIV Entry assay (B), luciferase activity was measured to quantify infection of U87CD4+CCR5+ cells by pseudoviruses derived from the Env-recombinant NL4-3 clones in the presence of various concentrations of inhibitor.
Maraviroc-resistant variants retain full susceptibility to other CCR5 antagonists and HIV-1 entry inhibitors.
Pseudotyped viruses derived from maraviroc-resistant CC1/85 and RU570 produced full dose-response curves (reaching 100% inhibition, in contrast to the plateaus seen with maraviroc) in PhenoSense HIV Entry assays using the CCR5 antagonist SCH-C and the fusion inhibitor enfuvirtide (Fig. 2). No differences in the IC50 of SCH-C or enfuvirtide were observed between pre- and post-maraviroc passage viruses for either CC1/85 or RU570 (Table 2). Full dose-response curves were also obtained for all the Env-recombinant NL4-3 clones in PBL assays using aplaviroc and enfuvirtide (Fig. 3). No resistance to either of these inhibitors was observed; rather, there was a slight decrease (approximately twofold) in the susceptibility of MVCres compared with MVCsens (Table 6).
Maraviroc resistance-associated mutations may compromise viral replication in vitro.
In order to understand whether maraviroc resistance was associated with lower infectivity for the CC1/85 and RU570 variants, the output from the PhenoSense HIV entry assay was examined. The RLU values in the PhenoSense HIV Entry assay were lower with maraviroc-resistant RU570 than with start virus (21,037 RLU versus 1,403,070 RLU, n = 2). Also, the eight Env clones derived from maraviroc-resistant RU570 with the three-amino-acid QAI deletion in the V3 crown and a plateau of <90% in maximum percent inhibition had lower RLU readings than the three Env clones without the deletion (3,995 to 15,983 RLU versus 906,269 to 1,823,566 RLU, respectively), further supporting the impact of the maraviroc-resistant genotype on the infectivity of the virus. In contrast, maraviroc-resistant CC1/85 yielded RLU readings that were comparable to those of the start virus (1,049,311 RLU versus 633,142 RLU, n = 2). Consistent with this, the RLU output of the Env clones that gave plateaus (44,453 to 654,546 RLU, n = 8) was not lower than the RLU output of those that did not (4,262 to 464,973 RLU, n = 4).
To further investigate the impact of maraviroc resistance-associated mutations on in vitro replication, we passaged control and maraviroc-resistant CC1/85 and RU570 for 20 weeks in the absence of maraviroc. Individual Env clones were characterized by DNA sequencing of the V3 loop (Table 4) and phenotypic susceptibility assays. Passage of maraviroc-resistant RU570 resulted in the selection of virus without the three-amino-acid QAI deletion. All 12 clones tested were completely susceptible to maraviroc, as indicated by a complete dose-response curve with no shift in IC50 compared to the control virus (data not shown). As discussed above, the maraviroc-resistant RU570 virus stock that was used to initiate the reversion experiment contained a minority of clones that lacked the QAI deletion (Table 4). Therefore, it is likely that this minority variant regained the replicative advantage during the maraviroc-free passage. The 12 RU570 revertant clones also each had a 10-amino-acid deletion in the V1 region that was not present in the starting culture or the maraviroc-resistant variants, indicating that further tissue culture adaptation of this virus likely occurred during serial passage in the absence of drug.
Passage of maraviroc-resistant CC1/85 resulted in loss of the mutation at position 316 in the V3 loop, with retention of I323V. It is possible that variants that lacked the mutation at position 316 existed within the maraviroc-resistant CC1/85 stock and were able to outcompete the maraviroc-resistant virus in the absence of inhibitor. Alternatively, the revertant may have arisen by back mutation of position 316 during the drug-free passage; this is supported by the fact that all 12 Env clones sequenced from the maraviroc-resistant stock had a mutation at this position (Table 4). The revertant virus gave a plateau in maximal inhibition in both the PBL and PhenoSense assays, which is consistent with the phenotype displayed by the MVCres(AAV) SDM clone (Table 6) and indicates that this revertant retained some level of reduced susceptibility to maraviroc compared to the start virus.
The retention of I323V in the CC1/85 revertant implies that a virus carrying this particular mutation is able to replicate efficiently in the absence of maraviroc selection pressure. A single Env clone (of 36 sequenced) was identified in the CC1/85 start culture as having a valine at position 323 (Table 5). However, a virus carrying this mutation was only selected during serial passage in the presence of maraviroc, as the drug-free control virus retained the isoleucine at this position (Table 4). Therefore, it is likely that other amino acid changes carried by the maraviroc-resistant and revertant viruses contribute to the stability of the I323V mutation when these variants are passaged in the absence of maraviroc selective pressure.
The impact of the individual V3 loop mutations in CC1/85 on the in vitro replication capacity of the Env-recombinant NL4-3 clones was assessed by infecting three separate batches of PBL with virus stocks diluted to contain a standardized input according to RT activity. Viral replication was assessed by measuring p24 after 7 days of culture. The p24 levels obtained for MVCsens were between 1,098 and 1,697 ng/ml; in contrast, lower p24 levels between 42 and 96 ng/ml were observed in the MVCres and associated SDM cultures. When a blocked analysis of variance was applied to these data, paired comparisons between MVCsens and each of the other viruses gave P values of less than 0.001. Since these lower p24 levels were seen with all the SDM cultures, including MVCres(AAI), these results indicate that regions outside the V3 loop play a role in determining fitness in CC1/85. Maraviroc-resistant CC1/85, and hence all of the MVCres-derived SDM cultures, carried three non-V3 loop mutations in gp120 (Tables 5 and 6) and an additional eight changes in gp41.
MVCres can use maraviroc-bound receptors to infect target cells.
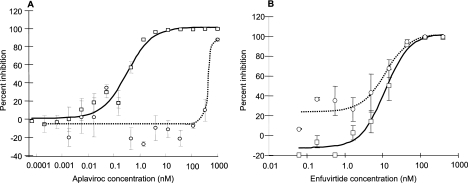
We hypothesized that the lack of inhibition of MVCres by high concentrations of maraviroc in PBL and the plateaus in maximum inhibition by maraviroc observed in the PhenoSense HIV Entry assay occur because MVCres is able to bind to CCR5 in its normal conformation and in the maraviroc-bound conformation. Aplaviroc and maraviroc bind to similar regions of CCR5 (M. Westby et al., Abstr. 12th Conf. Retrovir. Opportunistic Infect., abstr. 96, 2005), but the MVCres clone retains full susceptibility to aplaviroc in PBL. Therefore, we tested our hypothesis by assessing the effect of adding a high concentration of maraviroc to an aplaviroc susceptibility assay in PBL. The addition of 400 nM maraviroc ablated the susceptibility of MVCres to aplaviroc at concentrations of up to 100 nM (Fig. 4). At aplaviroc concentrations of >400 nM, the virus was inhibited, indicating competition between the two agents for binding to CCR5. In contrast, the addition of 400 nM maraviroc had no significant effect on the susceptibility of MVCres to enfuvirtide, which targets viral protein gp41 and would not compete with maraviroc for binding to CCR5 (Fig. 4). Results similar to those obtained with enfuvirtide were obtained with the non-nucleoside RT inhibitor efavirenz (data not shown).
FIG. 4.
Susceptibility of MVCres Env-recombinant NL4-3 clone to aplaviroc (A) and enfuvirtide (B) in the absence (open squares, solid line) or presence (open circles, dotted line) of 400 nM maraviroc in PBL.
DISCUSSION
Maraviroc did not select for CXCR4-using variants following prolonged in vitro serial passage experiments with either laboratory-adapted, CCR5-tropic Ba-L or the majority of the CCR5-tropic primary isolates that were used. This is consistent with published in vitro resistance studies of other CCR5 antagonists (18, 19, 28). CXCR4-using variants did emerge following the serial passage of the SF162 strain. However, the CXCR4-using virus emerged at the same time in the passage control culture, indicating that its appearance was not a result of selective pressure by maraviroc. The selection of CXCR4-using variants of SF162 has been described previously, after its serial passage through cells with low cell surface CCR5 expression (MOLT4 and C8166) (5, 11). In these studies and in our experiments, the change in tropism was associated with amino acid changes in the V3 loop that have previously been associated with CXCR4 usage (arginine and valine on either side of the GPGR motif). Whether this change in tropism involved a de novo switch to CXCR4 use or the selection of preexisting CXCR4-using variants within the SF162 population is not clear. Coreceptor tropism switching has been reported as a mechanism of escape from RANTES analogues in SCID Hu mice using a molecular clone that only required a single amino acid change to expand its coreceptor usage to CXCR4 (20). However, evidence from in vitro studies with a range of CCR5-tropic HIV-1 strains suggests that the molecular pathways to coreceptor switching often involve multiple mutations throughout gp160, with transitional intermediates having poor replication fitness and less efficient use of both CCR5 and CXCR4 (22).
Maraviroc-resistant virus was not selected during prolonged serial passage of laboratory-adapted strain Ba-L or three clade B primary isolates of Brazilian origin, suggesting that maraviroc resistance may not be easy for every virus strain to acquire in vitro. No plateaus in maximum percent inhibition were observed for pre- and postpassage Ba-L, and only small shifts in IC50 were measured, which were within the variability of the PBL assay. Maeda et al. (18) were also unable to select for resistance to aplaviroc after 45 passages of Ba-L in PM1 cells. Subsequent experiments were carried out with clinical isolates in PBL in order to produce conditions more representative of the situation within a patient, where a heterogeneous mixture of closely related virus variants replicates in predominantly activated CD4+ cells. Clinical isolates 92BR017, 92BR018, and 92BR023 grew well in PBL; however, the concentrations of maraviroc in the passage cultures could not be significantly increased. The lack of selection of highly resistant isolates from these viruses suggests that there was a high genetic barrier to maraviroc resistance under the conditions used.
Maraviroc-resistant viruses were generated after serial passage of two primary isolates, CC1/85 and RU570. These maraviroc-resistant viruses were not inhibited by the CXCR4 inhibitor AMD3100, were unable to infect CCR5 Δ32/Δ32 PBL, and were inhibited by the CCR5 antagonist SCH-C, demonstrating their continued reliance on CCR5 and not CXCR4 for entry into cells. Resistant viruses generated by sequential passage in the presence of the CCR5 antagonists SCH-C, AD101, and vicriviroc have also been reported to retain CCR5 tropism (15, 19, 28). Several mechanisms have been proposed to explain resistance to CCR5 antagonists in viruses that continue to rely solely on CCR5 for cell entry. An increase in affinity for CCR5 would be associated with an increase in IC50 as a greater proportion of receptors would have to be occupied by the inhibitor before the virus lost its ability to enter the target cells. Consistent with this hypothesis, Trkola et al. (28) observed an increased ability of AD101 escape mutants to use low levels of CCR5, which was associated with a threefold decrease in susceptibility to AD101. It is also possible that the small increases in IC50 observed following serial passage of Ba-L in the presence of maraviroc are due to increased affinity for the receptor and a greater ability to utilize low receptor numbers for entry. However, this does not explain the levels of resistance observed for CC1/85 and RU570 variants against maraviroc or vicriviroc (19) as characterized by dose responses which remain flat at inhibitor concentrations >1,000-fold higher than the wild-type IC50.
A plausible explanation for the resistance that we observed with CC1/85 and RU570 in our study is that the virus has acquired mutations which enable it to enter cells by using CCR5 that is bound to maraviroc. This hypothesis could account for the plateaus in maximal inhibition that we observed when maraviroc was titrated against maraviroc-resistant viruses in PBL and in the PhenoSense assay. Since these plateaus were also obtained with Env recombinant clones in the PhenoSense assay (and with the replication-competent Env recombinant NL4-3 SDM in both assay formats), they cannot be attributed to mixtures of susceptible and resistant viruses. G-protein-coupled receptors such as CCR5 are known to constantly alter their conformation within the cell membrane (2). Natural ligands act by binding to the receptor and stabilizing a particular conformation. It has been hypothesized that CCR5 antagonists stabilize conformations of the receptor which are not recognized by HIV-1 gp120. Mutations in gp120 that allow it to recognize these conformations, for example, by binding to residues in the CCR5 molecule that are relatively unchanged between different conformations, could lead to maraviroc resistance. The level of the maximum inhibition plateau observed in a drug susceptibility assay would depend on the relative affinity of the virus for inhibitor-bound versus free CCR5. The plateau would get lower as the affinity of the virus for inhibitor-bound CCR5 approached its affinity for free CCR5 (i.e., its level of resistance increases). Thus, the Env recombinant clone derived from maraviroc-resistant CC1/85 (MVCres), carrying two V3 loop amino acid mutations, produced a lower plateau than the clones with single mutations in the V3 loop [MVCres(TAI) and MVCres(AAV)], suggesting that the second V3 mutation further increased its affinity for the bound receptor. The absence of a dose-response curve for MVCres in PBL can be described as a plateau at 0%, which is consistent with the recognition of free and bound receptors with equal affinity in this culture system.
Plateaus in maximal percent inhibition as phenotypic markers of maraviroc resistance were qualitatively consistent between the two assay systems used in this study. Although plateaus in the PhenoSense HIV Entry assay were generally higher than those observed in PBL assays, the rank order of plateau heights was the same in both assay systems. Quantitative differences between phenotypic assays are commonly seen with other classes of antiretrovirals, leading to variation in biologically and clinically relevant cutoffs (in IC50) assigned to individual drugs in different resistance tests (10, 21, 24). Differences in the way in which assays are configured (such as a single round versus multiple rounds of infection and the multiplicity of infection and endpoint used) and intrinsic target cell differences (such as surface receptor density) all contribute to variations in cutoffs. Similar assay-specific factors also undoubtedly influence the plateau heights when the PBL and PhenoSense HIV Entry assays used in this study are compared. In a separate study, plateaus in maximal percent inhibition of <90% could reproducibly be identified in the PhenoSense HIV Entry assay as being associated with the maraviroc-resistant viruses (M. Mosley et al., Abstr. 13th Conf. Retrovir. Opportunistic Infect., abstr. 598, 2006). It remains to be seen whether this is a clinically relevant cutoff for resistance to maraviroc in patients.
Corroborating evidence to support the recognition of CCR5 inhibitor-bound receptor comes from our experiment assessing the effect of adding maraviroc to an aplaviroc susceptibility assay with maraviroc-resistant CC1/85 clone MVCres. Aplaviroc binds to a region of CCR5 similar to that bound by maraviroc (Westby et al., Abstr. 12th Conf. Retrovir. Opportunistic Infect.), but the fact that MVCres retains susceptibility to aplaviroc, yielding a full dose-response curve, indicates that it is not able to use aplaviroc-bound CCR5 to infect cells. The addition of 400 nM maraviroc to the susceptibility assay resulted in a completely flat inhibition curve for concentrations of up to 100 nM aplaviroc. At these concentrations, maraviroc is able to effectively compete with aplaviroc for binding to all the available CCR5 molecules and the maraviroc-resistant virus is thus able to infect cells by using the maraviroc-bound receptors. When the concentration of aplaviroc rises above that of maraviroc, aplaviroc can compete with maraviroc for binding to CCR5 and consequently inhibition of the MVCres clone is possible. Plateaus in dose-response curves have been reported for viruses isolated from patients treated with anti-CD4 monoclonal antibody TNX-355 (T. Duensing et al., Abstr. 13th Conf. Retrovir. Opportunistic Infect., abstr. 158LB, 2006) and with the CCR5 antagonists aplaviroc (M. Kitrinos et al., Abstr. 15th Int. HIV Drug Resist. Workshop, abstr. 21, 2006) and vicriviroc (R. Landovitz et al., Abstr. 15th Int. HIV Drug Resist. Workshop, abstr. 18, 2006), indicating that the recognition of an altered conformation of cell surface receptor may be a shared mechanism of escape from noncompetitive entry inhibitors.
Genotypic analyses of maraviroc-resistant viruses indicated that changes in the V3 loop play a crucial role in CC1/85 resistance to maraviroc. Considering the importance of the V3 loop in coreceptor recognition and binding (22), it was not surprising that several changes were noted in this region of the resistant viruses, i.e., A316T, A319A/S, and I323V in CC1/85 and a deletion (equivalent to positions 315 to 317 in HXB2) in RU570. These V3 loop changes were different from those seen after passage of similar viruses with other CCR5 antagonists. AD101-resistant CC1/85 has been reported to contain K305R, H308P, A316V, and G321E (15). SCH-C selected for various amino acid substitutions and a two-amino-acid deletion in the subtype G virus JV1083 (J. M. Strizki et al., Abstr. 14th Int. HIV Drug Res. Workshop, abstr. 59, 2005). A search of sequences in the Los Alamos database indicated that the A316T and I323V mutations are extremely rare in clade B viruses—only two of >23,000 were found to contain these substitutions (Los Alamos HIV sequence database, available at http://hiv-web.lanl.gov/content/index, accessed March 2005). None of 366 clade G virus sequences in the database were found to contain deletions in the V3 loop. Analysis of SDM molecular clones derived from maraviroc-resistant CC1/85 showed that the A316T mutation [MVCres(TAI)] had the greatest impact on the maraviroc susceptibility curve, although both A316T and I323V (MVCres) were required for high-level resistance. The fact that A316T was the last V3 loop mutation to appear during the passage experiment suggests that this mutation may be detrimental to the fitness of the virus. This was confirmed by its loss, along with A319S, during in vitro passage of the maraviroc-resistant virus in the absence of maraviroc. The revertant virus still yielded plateaus in maraviroc susceptibility assays (albeit higher than those seen for the parent maraviroc-resistant strain), confirming that the retained I323V V3 loop mutation conferred the ability of the virus to use compound-bound receptor complexes for entry.
Mutations were also seen outside the V3 loop, in the V2, C3, and V4 regions and in gp41, for CC1/85-derived maraviroc-resistant virus and in the V1, C4, V4, and C5 regions for RU570-derived maraviroc-resistant virus. The importance of these changes was not formally investigated in this study; however, the variations in maximum inhibition plateaus observed for different envelope clones derived from the same resistant stock of CC1/85 (sharing the same V3 loop mutations) suggest that regions outside the V3 loop (or the starting sequence of the V3 loop itself) contributed to the resistance phenotype. Also, the maraviroc-sensitive molecular clone MVCsens and the SDM clone MVCres(AAI) yielded maximum inhibition plateaus at <100%, possibly indicating a low level of reduced sensitivity of this virus strain to maraviroc. This suggestion is consistent with similar plateaus in maximal inhibition shown by Kuhmann et al. (15) when measuring the activity of SCH-C against CC1/85 in primary CD4+ T cells.
Mutations outside the V3 loop region could also play a role in viral infectivity. In PBL assays, p24 levels indicated that MVCres(AAI) did not grow as well as the wild-type virus. The mutations in the V2, C3, and V4 regions may help to accommodate the resistance mutations in the V3 loop, while in the absence of the V3 loop mutations they may lead to conformational changes that reduce the infectivity of the virus. The impact of mutations in V2 on the infectivity of CC1/85 was also seen in the control passage, where the SN deletion in this region appeared to enable the virus to grow in PM1 cells, where the CCR5 levels are known to be low. Changes in the V2 loop during in vitro adaptation of CC1/85 to growth in PBL have also been reported by Pugach et al. (23).
Studies with other CCR5-antagonists have demonstrated that cross-resistance exists between structurally related molecules within this class (AD101, SCH-C, and vicriviroc) (19, 28; Strizki et al., Abstr. 14th Int. HIV Drug Resist. Workshop). Since maraviroc binds to the same transmembrane region of CCR5, it might be expected that a maraviroc-resistant virus would also be cross-resistant to these agents. However, our results show that this is not the case. Maraviroc-resistant virus retained full susceptibility to SCH-C and aplaviroc, suggesting that although the CCR5 binding site for these agents are similar, their impacts on the surface conformation of the receptor (as seen by the virus envelope) are different. Receptor pharmacology studies with CCR5 site-directed mutants coupled with molecular modeling techniques have identified subtle differences in the occupation of the CCR5 transmembrane region by compounds closely related in structure (Westby et al., Abstr. 12th Conf. Retrovir. Opportunistic Infect.). Thus, resistance resulting from recognition of the altered conformation of inhibitor-bound receptor may be agent specific.
In conclusion, our results provide strong evidence to suggest that, in vitro, the envelope proteins of maraviroc-resistant virus are able to recognize and utilize inhibitor-bound CCR5 and indicate that this change involves the ordered accumulation of mutations in the viral envelope, both in the V3 loop and elsewhere within gp120. Although complex, this pathway to resistance presents less of a barrier to escape from maraviroc-associated inhibition than any pathway involving a switch to CXCR4 tropism. An association between the degree of resistance and reduced levels of maximal percent inhibition (rather than an increased IC50) is consistent with the proposed mechanism and requires the development of appropriate interpretation techniques. The relative importance of this mechanism in escape from CCR5 antagonists in treated patients will depend on the dose and the binding efficiency of the inhibitor. An agent with a rapid onset and a short half-life where not all receptors are occupied all the time may select for high-affinity variants. On the other hand, agents whose binding kinetics and long plasma half-life result in 100% receptor occupancy throughout the dosing interval may be more likely to select for viruses that are able to use inhibitor-bound receptor. Resistance is currently being investigated in phase III clinical trials with maraviroc to determine whether this mechanism occurs in vivo.
Acknowledgments
We are grateful to Rolf Kaiser (Cologne University) for provision of CCR5 Δ32/Δ32 PBL, to John Moore for provision of the CC1/85 strain of HIV-1, and to NIBSC for provision of the other viruses used in this study. We also acknowledge Monogram Biosciences Inc. (South San Francisco, CA) for providing genotyping and phenotyping services as specified in Materials and Methods. Finally, we give special thanks to Jeannette Whitcomb and Chris Petropoulos (and colleagues at Monogram Biosciences) and John Moore (Cornell University) for many productive discussions in our joint efforts to better understand plateaus in dose response as phenotypic markers of resistance to CCR5 antagonists.
Footnotes
Published ahead of print on 20 December 2006.
REFERENCES
- 1.Cheng-Mayer, C., and J. A. Levy. 1988. Distinct biological and serological properties of human immunodeficiency viruses from the brain. Ann. Neurol. 23(Suppl.):S58-S61. [DOI] [PubMed] [Google Scholar]
- 2.Christopoulos, A., and T. Kenakin. 2002. G protein-coupled receptor allosterism and complexing. Pharmacol. Rev. 54:323-374. [DOI] [PubMed] [Google Scholar]
- 3.Coakley, E., C. J. Petropoulos, and J. M. Whitcomb. 2005. Assessing chemokine co-receptor usage in HIV. Curr. Opin. Infect. Dis. 18:9-15. [DOI] [PubMed] [Google Scholar]
- 4.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dejucq, N., G. Simmons, and P. R. Clapham. 2000. T-cell line adaptation of human immunodeficiency virus type 1 strain SF162: effects on envelope, vpu and macrophage-tropism. J. Gen. Virol. 81:2899-2904. [DOI] [PubMed] [Google Scholar]
- 6.Dorr, P., M. Westby, S. Dobbs, P. Griffin, B. Irvine, M. Macartney, J. Mori, G. Rickett, C. Smith-Burchnell, C. Napier, R. Webster, D. Amour, D. Price, B. Stammen, A. Wood, and M. Perros. 2005. Maraviroc (UK-427,857): a potent, orally bioavailable and selective small-molecule inhibitor of the chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esté, J. A., K. De Vreese, M. Witvrouw, J. C. Schmit, A. M. Vandamme, J. Anne, J. Desmyter, G. W. Henson, G. Bridger, and E. De Clercq. 1996. Antiviral activity of the bicyclam derivative JM3100 against drug-resistant strains of human immunodeficiency virus type 1. Antiviral Res. 29:297-307. [DOI] [PubMed] [Google Scholar]
- 8.Fätkenheuer, G., A. L. Pozniak, M. A. Johnson, A. Plettenberg, S. Staszewski, A. I. Hoepelman, M. S. Saag, F. D. Goebel, J. K. Rockstroh, B. J. Dezube, T. M. Jenkins, C. Medhurst, J. F. Sullivan, C. Ridgway, S. Abel, I. T. James, M. Youle, and E. van der Ryst. 2005. Efficacy of short-term monotherapy with maraviroc, a new CCR5 antagonist, in patients infected with HIV-1. Nat. Med. 11:1170-1172. [DOI] [PubMed] [Google Scholar]
- 9.Griffin, S. 2003. Structure and organization of HIV, p. 2:1-2:23. In D. D. Richman (ed.), Human immunodeficiency virus. International Medical Press, London, United Kingdom.
- 10.Harrigan, P. R., J. S. Montaner, S. A. Wegner, W. Verbiest, V. Miller, R. Wood, and B. A. Larder. 2001. World-wide variation in HIV-1 phenotypic susceptibility in untreated individuals: biologically relevant values for resistance testing. AIDS 15:1671-1677. [DOI] [PubMed] [Google Scholar]
- 11.Harrowe, G., and C. Cheng-Mayer. 1995. Amino acid substitutions in the V3 loop are responsible for adaptation to growth in transformed T-cell lines of a primary human immunodeficiency virus type 1. Virology 210:490-494. [DOI] [PubMed] [Google Scholar]
- 12.Jacobson, J. M., R. J. Israel, I. Lowy, N. A. Ostrow, L. S. Vassilatos, M. Barish, D. N. Tran, B. M. Sullivan, T. J. Ketas, T. J. O'Neill, K. A. Nagashima, W. Huang, C. J. Petropoulos, J. P. Moore, P. J. Maddon, and W. C. Olson. 2004. Treatment of advanced human immunodeficiency virus type 1 disease with the viral entry inhibitor PRO 542. Antimicrob. Agents Chemother. 48:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 14.Koot, M., R. van Leeuwen, R. E. de Goede, I. P. Keet, S. Danner, J. K. Eeftinck Schattenkerk, P. Reiss, M. Tersmette, J. M. Lange, and H. Schuitemaker. 1999. Conversion rate towards a syncytium-inducing (SI) phenotype during different stages of human immunodeficiency virus type 1 infection and prognostic value of SI phenotype for survival after AIDS diagnosis. J. Infect. Dis. 179:254-258. [DOI] [PubMed] [Google Scholar]
- 15.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuritzkes, D. R., J. Jacobson, W. G. Powderly, E. Godofsky, E. DeJesus, F. Haas, K. A. Reimann, J. L. Larson, P. O. Yarbough, V. Curt, and W. R. Shanahan, Jr. 2004. Antiretroviral activity of the anti-CD4 monoclonal antibody TNX-355 in patients infected with HIV type 1. J. Infect. Dis. 189:286-291. [DOI] [PubMed] [Google Scholar]
- 17.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. Reitz, Jr. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeda, K., H. Nakata, Y. Koh, T. Miyakawa, H. Ogata, Y. Takaoka, S. Shibayama, K. Sagawa, D. Fukushima, J. Moravek, Y. Koyanagi, and H. Mitsuya. 2004. Spirodiketopiperazine-based CCR5 inhibitor which preserves CC-chemokine/CCR5 interactions and exerts potent activity against R5 human immunodeficiency virus type 1 in vitro. J. Virol. 78:8654-8662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marozsan, A. J., S. E. Kuhmann, T. Morgan, C. Herrera, E. Rivera-Troche, S. Xu, B. M. Baroudy, J. Strizki, and J. P. Moore. 2005. Generation and properties of a human immunodeficiency virus type 1 isolate resistant to the small molecule CCR5 inhibitor, SCH-417690 (SCH-D). Virology 338:182-199. [DOI] [PubMed] [Google Scholar]
- 20.Mosier, D. E., G. R. Picchio, R. J. Gulizia, R. Sabbe, P. Poignard, L. Picard, R. E. Offord, D. A. Thompson, and J. Wilken. 1999. Highly potent RANTES analogues either prevent CCR5-using human immunodeficiency virus type 1 infection in vivo or rapidly select for CXCR4-using variants. J. Virol. 73:3544-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkin, N. T., N. S. Hellmann, J. M. Whitcomb, L. Kiss, C. Chappey, and C. J. Petropoulos. 2004. Natural variation of drug susceptibility in wild-type human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48:437-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pastore, C., A. Ramos, and D. E. Mosier. 2004. Intrinsic obstacles to human immunodeficiency virus type 1 coreceptor switching. J. Virol. 78:7565-7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugach, P., S. E. Kuhmann, J. Taylor, A. J. Marozsan, A. Snyder, T. Ketas, S. M. Wolinsky, B. T. Korber, and J. P. Moore. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 321:8-22. [DOI] [PubMed] [Google Scholar]
- 24.Ross, L., R. Boulme, R. Fisher, J. Hernandez, A. Florance, J. C. Schmit, and V. Williams. 2005. A direct comparison of drug susceptibility to HIV type 1 from antiretroviral experienced subjects as assessed by the antivirogram and PhenoSense assays and by seven resistance algorithms. AIDS Res. Hum. Retrovir. 21:933-939. [DOI] [PubMed] [Google Scholar]
- 25.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strizki, J. M., S. Xu, N. E. Wagner, L. Wojcik, J. Liu, Y. Hou, M. Endres, A. Palani, S. Shapiro, J. W. Clader, W. J. Greenlee, J. R. Tagat, S. McCombie, K. Cox, A. B. Fawzi, C. C. Chou, C. Pugliese-Sivo, L. Davies, M. E. Moreno, D. D. Ho, A. Trkola, C. A. Stoddart, J. P. Moore, G. R. Reyes, and B. M. Baroudy. 2001. SCH-C (SCH 351125), an orally bioavailable, small molecule antagonist of the chemokine receptor CCR5, is a potent inhibitor of HIV-1 infection in vitro and in vivo. Proc. Natl. Acad. Sci. USA 98:12718-12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westby, M., M. Lewis, J. M. Whitcombe, M. Youle, A. Pozniak, I. James, T. Jenkins, M. Perros, and E. van der Ryst. 2006. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J. Virol. 80:4909-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.WHO Network for HIV Isolation and Characterization. 1994. HIV type 1 variation in World Health Organization-sponsored vaccine evaluation sites: genetic screening, sequence analysis, and preliminary biological characterization of selected viral strains. AIDS Res. Hum. Retrovir. 10:1327-1343. [DOI] [PubMed] [Google Scholar]
- 31.Xu, L., A. Pozniak, A. Wildfire, S. A. Stanfield-Oakley, S. M. Mosier, D. Ratcliffe, J. Workman, A. Joall, R. Myers, E. Smit, P. A. Cane, M. L. Greenberg, and D. Pillay. 2005. Emergence and evolution of enfuvirtide resistance following long-term therapy involves heptad repeat 2 mutations within gp41. Antimicrob. Agents Chemother. 49:1113-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]