Abstract
Human immunodeficiency virus type 1 (HIV-1) infection significantly increases the risk of Kaposi's sarcoma (KS) occurrence in individuals infected with Kaposi's sarcoma-associated herpesvirus (KSHV). KSHV infection appears to be necessary but not sufficient for KS development without other cofactors. However, factors that facilitate KSHV to cause KS have not been well defined. Previously, we determined that human herpesvirus 6 was one of the cofactors that activated lytic cycle replication of KSHV. Here, we demonstrate that the Tat protein of HIV-1 is a potentially important factor in the pathogenesis of KS, as determined by production of lytic phase mRNA transcripts and viral proteins in BCBL-1 cells. Mechanistic studies showed ectopic expression of Tat induced the production of human interleukin-6 (huIL-6) and its receptor (huIL-6Ra) and activated STAT3 signaling. Neutralization of huIL-6 or huIL-6R or inhibition of STAT3 signaling enhanced the replication. In addition, IL-4/STAT6 signaling also partially contributed to Tat-induced KSHV replication. These findings suggest that Tat may participate in KS pathogenesis by inducing KSHV replication and increasing KSHV viral load. These data also suggest that JAK/STAT signaling may be of therapeutic value in AIDS-related KS patients.
Kaposi's sarcoma-associated herpesvirus (KSHV, also known as human herpesvirus 8) is a lymphotropic gamma-2 herpesvirus which was originally discovered in 1994 in lesions of AIDS-related KS (13). Today, KSHV has been causally associated with three types of lymphoproliferative diseases: KS, multicentric Castleman disease, and primary effusion lymphoma (PEL, also termed body cavity-based lymphoma, or BCBL) (30). All herpesviruses, including KSHV, share the ability to establish latent infections in their natural host cells. During latent infection, the viral genome persists as an episome, and viral gene expression is highly restricted. Upon chemical induction, KSHV produces varieties of virus-encoded proteins and progeny virions. Regulation of viral replication is critical to disease progression since the tissue deterioration and infection progression is proportionally related to the percentage of virus-infected cells undergoing reactivation. Indeed, studies have shown that KSHV viral load is higher in KS patients than in KSHV-infected individuals without KS, and KSHV viral load also increases during progression of this disease (16, 59). Although KSHV appears to be necessary for development of KS, evidence suggests other factors also play important roles in the pathogenesis of this disease. However, who and how to reactivate latent virus are not well defined.
A couple of agents, such as human herpesvirus 6, human immunodeficiency virus (HIV), and human cytomegalovirus, which are commonly found in immunocompromised individuals have been proposed and considered as cofactors activating KSHV (36, 61, 63). Among them, HIV type 1 (HIV-1) is a potentially important cofactor. Although HIV-1 is clearly not necessary for the development of KS, AIDS-related KS (AIDS-KS) is well recognized as more aggressive, disseminated, and resistant to treatment than other forms of KS disease (8, 21, 58). Previous studies have shown that KS tumor cells are not infected with HIV-1; therefore, it is widely accepted that HIV-1 does not play a directly oncogenic role in AIDS-related KS (17). HIV-1 may function passively through the induction of immunosuppression or more directly in the pathogenesis of this disease. Current evidence strongly supports a role for HIV-1 in the initiation and progression of KS through mechanisms other than immunosuppression. For instance, studies indicated that KS developed almost exclusively in HIV-1-positive, but not HIV-2-positive, patients in Gambia, West Africa, despite essentially equivalent seroprevalence for KSHV and severity of immunosuppression in both groups of patients (5). These findings suggest that other mechanisms, such as production of HIV-1-encoded proteins and induction of cytokine expression, may play a role in KS development. Notably, the Tat protein has long been of particular interest to investigators studying AIDS-KS pathogenesis (6, 19). However, whether Tat can induce KSHV replication is still a highly controversial and arguable subject (24, 28, 42).
Besides the Tat protein, cytokine production induced by HIV-1 may play a role in KS development. For instance, gamma interferon (IFN-γ), oncostatin M, and hepatocyte growth factor/scatter factor (HGF/SF) from HIV-1-infected T cells were found to be partially responsible for KSHV reactivation (43). These cytokines activate Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathways and play important roles in cell growth and proliferation (7, 25, 32). A relationship between the activation of STATs and transcription of viral genes has been shown. For instance, the herpesvirus Saimiri tyrosine kinase-interacting protein Tip-484 activates STAT3 through up-regulation of p56lck, a nonreceptor tyrosine kinase (38). Constitutively activated STATs are also found in human T-cell lymphotropic virus 1-transformed T cells, myeloid leukemia cells, and Epstein-Barr virus (EBV)-related lymphoma cell lines (14, 44). Both ORF50 and latency-associated nuclear antigen of KSHV are able to interact with STAT3, and inhibition of STAT3 signaling can induce apoptosis of KSHV-infected cells (4, 23, 47). These observations collectively indicate that certain viruses modify STATs activities to increase the persistence, replication, or oncogenic potential of the viruses.
In an attempt to better understand the role of Tat in KSHV replication and AIDS-KS pathogenesis, we performed kinetic studies of KSHV replication induced by Tat. We showed that human interleukin-6 (huIL-6) and its receptor down-regulated Tat-induced KSHV replication and inhibition of JAK2/STAT3 signaling partially enhanced the replication. In addition, activation of IL-4/STAT6 signaling partially contributed to Tat-induced KSHV replication. Our data suggest that Tat may activate KSHV lytic cycle replication from latency in part by modulating JAK/STATs pathways. These novel findings are believed to be the first report on the mechanisms of KSHV activation by Tat and shed light on the understanding of AIDS-KS pathogenesis.
MATERIALS AND METHODS
Cells, plasmids, and transfection.
The BCBL-1 and BC-3 cells, both of which are EBV-negative and KSHV-positive PEL cell lines, were obtained through the AIDS Research and Reference Reagent Program, National Institutes of Health. B95-8 cells, which are a KSHV-negative and EBV-positive marmoset B-cell line, and HEK293, NIH 3T3, and HSB2 (a type of T-cell line) cells were obtained from the American Type Culture Collection (Rockville, MD). BCBL-1, B95-8, and HSB2 cells were maintained in RPMI 1640 containing 10% heat-inactivated fetal bovine serum (FBS), 2 mmol/L l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified, 5% CO2 atmosphere. BC-3 cells were grown in RPMI plus 20% FBS (9). HEK293 and NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium plus 10% FBS. Before transfection, both BCBL-1 and BC-3 cells in this study were incubated in serum-free RPMI 1640 medium for a maximum inducibility of KSHV replication as described previously (40). Briefly, cells were first synchronized at G0 by incubating cells in serum-free RPMI 1640 medium for 24 h and then cultured in RPMI plus 10% FBS for an additional 16 h. After 16 h of culture, cells were then used for the following experiments.
The HIV-1 Tat101 gene was synthesized by multiple rounds of overlapping PCR based on the genome sequence of HIV-1 (GenBank accession number M14310.1) as described previously (2, 50). The Flag M2 epitope was fused in frame to the carboxyl-terminal end of the Tat101 open reading frame, a Kozak sequence (GCCACC) was added to the upstream of initiator codon ATG to enhance the expression of the target gene, and the synthesized sequences were engineered with the cut sites of HindIII restriction enzymes at the 5′ and 3′ ends, respectively, to facilitate cloning. The synthesized gene was cloned into the plasmid pcDNA3.1 (Invitrogen, Inc., Carlsbad, CA) and subsequently verified by DNA sequencing to create recombinant plasmid pTat. The KSHV ORF50 (also known as Rta) luciferase reporter construct (p50-Luc), containing the −661 to −7 promoter fragment of the ORF50 promoter region inserted upstream of the luciferase gene in the pGL3 basic vector (Promega, Madison, WI), was generated as described previously (35). The plasmid pTZIII-CAT expressing the chloramphenicol acetyltransferase (CAT) enzyme under the control of the HIV-1 long terminal repeat (LTR) was the kind gift of M. Rusnati (University of Brescia, Brescia, Italy) (51). The human IL-4 luciferase reporter construct (pIL-4-Luc) was generously provided by M. Li-Weber (German Cancer Research Center, Heidelberg, Germany). In this construct, 280 bp of the presumed promoter region (−269 to +11) of IL-4 was inserted in front of the luciferase gene of the pTATALuc vector (34). The promoter regions of huIL-6 (−1235 to −1) (54) and huIL-6 receptor alpha (−1341 to + 3) were amplified using PCR, DNAs of HSB2 cells as templates, and specific primers with MulI and HindIII restriction enzyme cut sites engineered on the ends to facilitate directional cloning, respectively. The PCR products were cloned into the pGL3 basic vector in the sense orientation (designated as pIL-6-Luc and pIL-6Ra-Luc, respectively). The dominant negative STAT6 construct (pST6-DN), containing amino acid 1 to 661 of STAT6, was kindly provided by K. Zhang (UCLA School of Medicine, Los Angeles, CA) (65). The dominant negative STAT3 construct (pMSCV-STAT3D-EGFP, designated pST6-DN in this study) was kindly provided by D. Link (Washington University School of Medicine, St. Louis, MO) (41). All transfection experiments in this study were performed with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions.
Antibodies and reagents.
Both anti-KSHV ORF K8.1 A/B (clone 4A4; immunoglobulin G1 [IgG1]) and ORF59 (clone 11D1; IgG2b) mouse monoclonal antibodies (MAbs) were obtained from Advanced Biotechnologies Inc. (Columbia, Md.). Anti-STAT1 rabbit polyclonal antibody (PAb), anti-phospho-STAT3 (pTyr705) mouse MAb, and anti-phospho-STAT6 (Tyr641) mouse MAb were obtained from Calbiochem (Darmstadt, Germany). Anti-STAT3 rabbit PAb and horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). Anti-phospho-STAT1 (Ser727) rabbit PAb was obtained from Cell Signaling Technologies (Beverly, MA). Anti-STAT6 rabbit PAb was purchased from Bethyl Laboratories Inc. (Montgomery, TX). Anti-Flag M2 mouse MAb was purchased from Sigma (St. Louis, MO). The neutralizing antibodies, including anti-huIL-6 goat PAb, anti-huIL-6Ra goat PAb, and goat IgG (as a control) were obtained from R&D Systems (Minneapolis, MN). Mouse MAb to β-actin (Boster Technologies, Wuhan, Hubei, China) was used to monitor sample loading. The JAK2 tyrosine kinase inhibitor AG490 was obtained from BIOMOL Research Laboratories Inc. (Plymouth Meeting, PA).
Western blot analysis.
After transfection, cells were harvested and lysed in radioimmunoprecipitation assay buffer containing a phosphatase inhibitor cocktail and protease inhibitors. The protein concentration was determined using a Bradford assay (Bio-Rad Laboratories, Hercules, CA). Sixty to eighty micrograms of protein was loaded onto a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, transferred to an Immobilon P (polyvinylidene difluoride) membrane, and blocked with 5% powdered milk in TBST (50 mM Tris, pH 7.5, 150 mM NaCl, 0.01% Tween 20). The membrane was then incubated with primary antibodies diluted in 2.5% powdered milk in TBST, washed extensively, and incubated with horseradish peroxidase-conjugated species-specific secondary antibodies. Proteins were visualized with enhanced chemiluminescence reagents (Amersham Biosciences, Piscataway, NJ) according to the manufacturer's instructions. Even loading of proteins was confirmed by Ponceau S staining and detection of the housekeeping protein β-actin on each blot. Differences in protein expression were determined by densitometry analysis using Scion Image software (Scion Corporation, Frederick, MD). Western blot experiments were repeated at least three times to confirm results.
CAT assay.
BCBL-1 cells (3 × 106) were seeded in six-well dishes and then cotransfected with 2 μg of pTZIII-CAT and 2 μg of pTat. After cotransfection, cells were cultured for 6, 12, 24, 48, 72, 96, and 120 h. At the end of culture, cells were washed three times with the precooled phosphate-buffered saline (PBS; 2 to 8°C) buffer and extracted, and the amount of CAT protein present in the cell extracts was determined using the CAT enzyme-linked immunosorbent assay (ELISA) kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instructions. Each sample was assayed in duplicate, and the assay was repeated a minimum of three times.
RNA isolation and real-time quantitative PCR.
Total RNA was isolated from cells by using TRIzol reagent (Invitrogen). cDNA was synthesized from the isolated RNA using Taqman Gold reverse transcription reagents (Applied Biosystems, Foster City, CA). Reverse transcription was performed by using oligo(dT) primers at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min. Quantitative PCR was performed in a GeneAmp 7300 sequence detection machine (Applied Biosystems). The Taqman PCR core reagents kit and primers/probes for β-actin (Applied Biosystems) were used. The sequences of KSHV-specific primers and probes were as follows: ORF50 5′ primer, GCG CAA GAT GAC AAG GGT AAG, ORF50 3′ primer, CGA GAG GCC GAC GAA GC, and 6-carboxyfluorescein (FAM)-labeled probe, TTC CAC ACA GGA CCG CCG AAG CT (45); ORF26 5′ primer, AGC CGA AAG GAT TCC ACC AT, ORF26 3′ primer, GCT GCG GCA CGA CCA T, and FAM-labeled probe, TGC TCG AAT CCA ACG GAT TTG ACC TC (35); ORF29 5′ primer, CCC GGA GGA CGG TCC A, ORF29 3′ primer, TGT CCC CGA ATG CTG TTC TTA, and FAM-labeled probe, CTC GCT GAT GTG CGC AAC ATG CT (45). The PCR mixture contained AmpErase uracil N-glycosylase to destroy any previously amplified product as described elsewhere (36). Efficiencies of the β-actin and target gene amplification were shown to be approximately equal using a validation experiment as described by the sequence detection system manufacturer.
Northern blot analysis.
Twelve micrograms of total RNA was fractionated on a 1% agarose formaldehyde gel and transferred to a nylon membrane (Zetabind; Cuno Inc., Meriden, CT). Even loading of RNA and efficiency of transfer were confirmed by staining of the 18S and 28S bands on the membrane with methylene blue. Membranes were prehybridized with Church's hybridization buffer and probed with [32P]dCTP-labeled probes. Probes were generated by using gel-purified PCR products and a random prime label kit (Roche Applied Science). The membranes were washed with sodium phosphate buffers containing sodium dodecyl sulfate, EDTA, and bovine serum albumin and exposed to Kodak film.
Immunoperoxidase staining.
Cytospin preparations of cultured cells were fixed for 10 min in 50:50 acetone-methanol and air dried. The cells were immunostained to detect two antigens using a highly sensitive avidin-biotin immunoperoxidase technique (Vectastain kit; Vector Laboratories, Burlingame, CA) as previously described (43). The chromogen 3-amino-4-ethylcarbazole was used, producing a positive red reaction. The panel of MAbs used included KSHV ORF K8.1 A/B and KSHV ORF59. Both K8.1 and ORF59 MAbs recognize KSHV lytic cycle proteins and have been previously described (10, 11). To calculate the percentage of positive cells, photographs of at least 10 unique fields were taken of every slide, and the number of positive and negative cells was counted separately by three individuals, including one who was blinded to the results. Immunostaining was performed on samples from three separate experiments.
Luciferase assay.
Typical transfections of cells involved the introduction of 0.4 μg of plasmid effector (pTat or pcDNA) and reporter DNA (p50-Luc, pIL-4-Luc, pIL-6-Luc, or pIL-6Ra-Luc). Cells were treated with 20 ng/ml tetradecanoyl phorbol acetate (TPA) until 24 h posttransfection and then harvested at 48 h posttransfection. Luciferase activity was assayed by using the Promega Bright-N-Glo system. Briefly, cells were collected and resuspended in 150 ml of phosphate-buffered saline. Bright-N-Glo luciferase reagent (150 ml) was added with thorough mixing following the manufacturer's instructions. The sample was incubated for 2 min at room temperature, and the luciferase activity was measured using a Sirius luminometer (Berthold Detection Systems, Pforzheim, Germany). The number of cells present in each sample was also counted to normalize the luciferase activity with the total number of cells (35). All data points were the averages of at least four independent transfections.
RT-PCR.
cDNA was synthesized from isolated RNA using the SuperScript preamplification system for the first-strand cDNA synthesis (Invitrogen) following the manufacturer's instructions. To ensure no DNA contamination of the RNA, which could lead to false positive results, the RNA samples were treated with DNase I (Invitrogen) before reverse transcription. As an additional control, each sample was also subjected to reverse transcription in the absence of reverse transcriptase (RT). Single-stranded cDNA was then amplified using standard PCR techniques as previously described (36). Primers used for analysis in this study are listed in Table 1.
TABLE 1.
Sets of primers used for RT-PCRa
| mRNA | Oligonucleotides | Accession no. | Expected size (bp) | Annealing temp (°C) | Cycles |
|---|---|---|---|---|---|
| GATA-3 | F: 5′-TGT CTG CAG CCA GGA GAG-3′ | NM_002051 | 871 | 57 | 23 |
| R: 5′-ATG CAT CAA ACA ACT GTG GCC A-3′ | |||||
| IL-4 | F: 5′-AAA CTT TGA ACA GCC TCA CAG AGC A-3′ | NM_000589 | 313 | 62 | 36 |
| R: 5′-AGC CTT TCC AAG AAG TTT TCC AAC G-3′ | |||||
| IL-4R | F: 5′-TTC CTA CAG GGC ACG GGT GA-3′ | NM_000418 | 750 | 63 | 29 |
| R: 5′-CAG GGC ATG TGA GCA CTC GTA CTT C-3′ | |||||
| IL-6 | F: 5′-CCC CAT GTG ATT TAC TGT CG-3′ | NM_000600 | 501 | 56 | 27 |
| R: 5′-CAA TCT GAG GTG CCC ATG CTA-3′ | |||||
| IL-6Ra | F: 5′-GCC GCC AGC GGA ACT G-3′ | NM_000565 | 341 | 60 | 28 |
| R: 5′-TCA TAA GGG CTC CGT GGG TC-3′ | |||||
| β-Actin | F: 5′-TGA CGG GGT CAC CCA CAC TGT GCC CAT CTA-3′ | BC016045 | 661 | 56 | 19 |
| R: 5′-CTA GAA GCA TTT GCG GTG GAC GAT GGA GGG-3′ |
The oligonucleotides were selected from the sequences with the indicated accession number. The size of each amplified product, its annealing temperature, and numbers of PCR cycles are indicated. F, forward; R, reverse.
SuperArray analysis.
SuperArray analysis of gene expression was performed according to the manufacturer's directions (GEArray Q series kit, nonradioactive; SuperArray Inc., Bethesda, MD). The total RNA was isolated from cells using the TRIzol reagent. Once isolated, 2.5 μg of mRNA was used as the template for biotin-labeled cDNA probe synthesis. The labeled probes were then hybridized to the GEArray Q series membrane. Two types of gene arrays were used in this report: they were the inflammatory cytokines and receptors pathways and JAK/STAT signaling pathway gene arrays (SuperArray Inc.), each of which contained 96 genes related to relevant signaling. After an overnight incubation in a 60°C incubator, the membranes were washed several times in hybridization tubes. Subsequently, membranes were blocked and subjected to chemiluminescent detection (alkaline phosphatase-conjugated streptavidin; 1:10,000 dilution) with the chemiluminescent substrate for alkaline phosphatase, phenylphosphate-substituted 1,2 dioxetane (CDP-star). The membranes were exposed to X-ray film, the developed X-ray film was scanned to create a raw image file, and this file was analyzed using an image analysis software program (Scanalyze by Michael Eisen). Each GEArray Q series membrane was spotted with negative controls (pUC18 DNA and blanks) and housekeeping genes, such as β-actin, glyceraldehyde-3-phosphate dehydrogenase, cyclophilin A, and ribosomal protein L13a. All raw signal intensities should be corrected for background by subtracting the minimum value to avoid the appearance of negative numbers. All signal intensities should also be normalized to that of a housekeeping gene. The corrected, normalized signals can then be used to estimate the relative abundance of a particular transcript. The results were expressed as the ratio of the normalized spot intensity in the Tat-treated group versus pcDNA vector-treated group. Changes in gene expression were calculated, and two separate experiments were conducted. All changes are reported in Tables 2 and 3, below, and only twofold changes were considered significant.
TABLE 2.
Dynamically regulated host genes encoding inflammatory cytokines and receptors in BCBL-1 cells early during HIV-1 Tat transfection
| Gene function and name | Accession no. | Fold change at time posttransfection
|
|||
|---|---|---|---|---|---|
| 3 h | 6 h | 12 h | 24 h | ||
| Interleukins and receptors | |||||
| IL-10 | NM_000572 | 1.23 | 1.58 | 1.14 | 1.81 |
| IL-10Rα | NM_001558 | 1.58 | 1.59 | 1.57 | 2.94 |
| IL-10Rβ | NM_000628 | 2.66 | 0.73 | 7.17 | NAa |
| IL-11 | NM_000641 | 0.54 | 0.85 | 1.16 | 0.00 |
| IL-11Rα | NM_004512 | 1.33 | 1.08 | 1.40 | 1.72 |
| IL-12A | NM_000882 | 1.26 | 1.08 | 1.18 | 1.89 |
| IL-12B | NM_002187 | 1.33 | 1.08 | 1.35 | 1.95 |
| IL-12Rβ1 | NM_005535 | 1.30 | 1.35 | 1.24 | 1.93 |
| IL-12Rβ2 | NM_001559 | 1.58 | 6.07 | 1.48 | 3.86 |
| IL-13 | NM_002188 | 2.11 | 1.99 | 2.08 | 5.99 |
| IL13Rα1 | NM_001560 | 1.34 | 0.99 | 0.00 | NA |
| IL-13Rα2 | NM_000640 | 1.20 | 1.02 | 2.38 | 0.94 |
| IL-15 | NM_172175 | 1.76 | 1.09 | 2.11 | 7.00 |
| IL-15Rα | NM_002189 | 11.60 | 0.88 | 4.28 | NA |
| IL-16 | NM_004513 | 2.92 | 0.52 | 1.52 | NA |
| IL-17 | NM_002190 | 15.84 | 1.14 | 17.81 | NA |
| IL-17R | NM_014339 | 1.50 | 1.12 | 0.91 | 4.70 |
| IL-18 | NM_001562 | 2.06 | 4.78 | 1.51 | 48.99 |
| IL18R1 | NM_003855 | 1.05 | 1.63 | 1.30 | 1.26 |
| IL-1α | NM_000575 | 1.99 | 0.95 | 0.88 | NA |
| IL-1β | NM_000576 | 1.07 | 1.00 | 1.28 | 1.21 |
| IL-1Rα | NM_000877 | 4.07 | 0.96 | 2.02 | NA |
| IL-1R2 | NM_004633 | 1.66 | 0.81 | 0.73 | NA |
| IL-2 | NM_000586 | 1.49 | 4.24 | 1.46 | 4.05 |
| IL-20 | NM_018724 | 8.68 | 1.06 | 3.41 | NA |
| IL-21 | NM_021803 | 1.28 | 0.96 | 1.07 | 1.77 |
| IL-25 | NM_019107 | 1.21 | 2.38 | 0.90 | 2.00 |
| CD25 | NM_000417 | 1.09 | 2.39 | 1.55 | 2.29 |
| IL-2Rβ | NM_000878 | 1.14 | 1.44 | 1.03 | 1.28 |
| IL-2Rγ | NM_000206 | 1.26 | 1.44 | 1.46 | 1.67 |
| IL-4 | NM_000589 | 5.46 | 5.12 | 3.86 | 6.37 |
| IL-5 | NM_000879 | 1.31 | 1.35 | 1.10 | 2.11 |
| IL-5Rα | NM_000564 | 3.28 | 1.95 | 2.15 | NA |
| IL-6 | NM_000600 | 2.29 | 7.44 | 2.41 | 16.29 |
| IL-6Rα | NM_000565 | 1.50 | 0.97 | 5.38 | 13.38 |
| gp130 | NM_002184 | 1.77 | 0.97 | 1.36 | NA |
| p40 | NM_000590 | 0.94 | 1.08 | 1.03 | 0.88 |
| IL-9Rα | NM_002186 | 1.22 | 1.39 | 1.69 | 1.51 |
| IL-3 | NM_000588 | 1.40 | 1.86 | 2.02 | 2.23 |
| Chemokine receptors | |||||
| CXCR5 | NM_001716 | 3.82 | 0.86 | 37.26 | NA |
| MIP1αR | NM_001295 | 1.74 | 0.98 | 3.13 | NA |
| MCP-1 | NM_000648 | 1.02 | 1.01 | 1.03 | 1.12 |
| CCR3 | NM_001837 | 3.65 | 1.02 | 4.66 | NA |
| CCR4 | NM_005508 | 1.30 | 1.19 | 1.34 | 1.95 |
| CCR5 | NM_000579 | 1.12 | 1.12 | 1.08 | 1.39 |
| CCR6 | NM_004367 | 1.85 | 1.97 | 1.75 | 22.24 |
| CCR7 | NM_001838 | 3.95 | 1.01 | 3.45 | NA |
| CCR8 | NM_005201 | 1.36 | 1.24 | 2.37 | 1.92 |
| CCR9 | NM_006641 | 1.73 | 1.51 | 1.26 | 58.28 |
| CCXCR1 | NM_005283 | 1.10 | 1.17 | 1.17 | 1.12 |
| CX3CR1 | NM_001337 | 1.97 | 0.95 | 2.30 | 24.79 |
| CXCR4 | NM_003467 | 1.48 | 1.40 | 1.11 | 2.73 |
| Subfamily A (Cys-Cys) | |||||
| I-309/SCYA1 | NM_002981 | 1.00 | 1.45 | 1.14 | 0.79 |
| Eotaxin | NM_002986 | 0.83 | 1.36 | 1.22 | 0.62 |
| MCP-4 | NM_005408 | 0.61 | 1.00 | 1.28 | 0.18 |
| MIP-1δ/MIP-5 | NM_004167 | 2.07 | 2.51 | 3.10 | NA |
| HCC-4 | NM_004590 | 2.04 | 2.55 | 2.48 | NA |
| TARC/SCYA17 | NM_002987 | 1.79 | 1.77 | 2.03 | 5.05 |
| PARC | NM_002988 | 1.56 | 2.46 | 3.63 | 1.47 |
| MCP-3β/SCYA19 | NM_006274 | 1.56 | 3.94 | 2.42 | 1.94 |
| MCP1/SCYA2 | NM_002982 | 0.93 | 1.51 | 1.43 | 0.00 |
| MIP-3α/SCYA20 | NM_004591 | 1.01 | 0.93 | 0.99 | 1.71 |
| SCYA21 | NM_002989 | 1.53 | 1.08 | 1.99 | 3.22 |
| MDC | NM_002990 | 1.40 | 1.00 | 1.64 | 2.40 |
| MPIF-1/SCYA23 | NM_005064 | 1.15 | 0.91 | 1.07 | 1.58 |
| MPIF-2/SCYA24 | NM_002991 | 3.11 | 1.89 | 3.61 | NA |
| TECK | NM_005624 | 1.07 | 1.60 | 1.63 | 0.87 |
| MIP-1α/SCYA3 | NM_002983 | 1.02 | 1.07 | 1.13 | 0.87 |
| MIP-1β | NM_002984 | 1.10 | 1.14 | 1.20 | 1.18 |
| SCYA5/RANTES | NM_002985 | 1.41 | 1.44 | 1.42 | 4.06 |
| MCP-3 | NM_006273 | 11.32 | 0.80 | 2.13 | NA |
| MCP-2 | NM_005623 | 1.72 | 1.65 | 2.11 | 3.79 |
| Subfamily B (Cys-X-Cys) | |||||
| SCYB10/IP-10 | NM_001565 | 1.25 | 1.08 | 1.37 | 1.71 |
| I-TAC/IP9/SCYB11 | NM_005409 | 6.04 | 1.00 | 9.26 | NA |
| SCYB13 | NM_006419 | 1.34 | 1.52 | 1.44 | 1.58 |
| ENA-78/SCYB5 | NM_002994 | 1.09 | 1.65 | 1.43 | 0.91 |
| GCP-2/SCYB6 | NM_002993 | 1.08 | 3.09 | 0.97 | 0.95 |
| SDF1 | NM_000609 | 3.19 | 1.90 | 2.67 | NA |
| Other subfamily members | |||||
| SCYC1 | NM_002995 | 8.51 | 0.98 | NA | NA |
| SCYC2 | NM_003175 | 0.74 | 1.11 | 0.96 | 0.51 |
| SCYD1 | NM_002996 | 1.49 | 0.78 | 0.84 | 2.81 |
| SCYE1 | NM_004757 | 2.50 | 1.99 | 4.28 | NA |
| SDF2 | NM_006923 | 0.92 | 1.13 | 1.02 | 0.80 |
| TGF ligands | |||||
| TGF-α | NM_003236 | 1.32 | 2.46 | 1.22 | 2.28 |
| TGF-β1 | NM_000660 | 1.10 | 0.98 | 0.87 | 1.25 |
| TGF-β2 | NM_003238 | 3.11 | 1.11 | 47.77 | NA |
| TGF-β3 | NM_003239 | 1.20 | 1.07 | 1.31 | 1.41 |
| TNF ligands and receptors | |||||
| TNFB/LT | NM_000595 | 1.33 | 1.26 | 1.25 | 2.07 |
| LT-β | NM_002341 | 2.61 | 5.46 | 1.78 | NA |
| LT-βR | NM_002342 | 75.27 | 1.04 | 32.82 | NA |
| TNFA | NM_000594 | 2.07 | 1.26 | 1.59 | NA |
| TNFR1 | NM_001065 | 1.27 | 0.91 | 1.11 | 2.04 |
| TNFR2/p75 | NM_001066 | 3.22 | 1.45 | 3.22 | NA |
| Other related genes | |||||
| IFN-γ | NM_000619 | 1.46 | 1.21 | 1.05 | 2.64 |
| Leptin | NM_000230 | 1.15 | 0.88 | 1.13 | 1.37 |
| MIF | NM_002415 | 1.03 | 1.93 | 0.96 | 0.76 |
NA, not applicable.
TABLE 3.
Dynamically regulated host genes involved in JAK/STAT signaling pathway in BCBL-1 cells early during HIV-1 Tat transfection
| Gene function and name | Accession no. | Fold change at time posttransfection
|
|||
|---|---|---|---|---|---|
| 3 h | 6 h | 12 h | 24 h | ||
| Jak and Stat proteins | |||||
| JAK1 | NM_002227 | 32.53 | 1.38 | 5.37 | 1.71 |
| JAK2 | NM_004972 | 2.03 | 1.04 | 1.69 | 2.42 |
| JAK3 | NM_000215 | 2.74 | 1.05 | 1.25 | 2.94 |
| Stat1 | NM_007315 | 1.12 | 0.93 | 1.38 | 1.31 |
| ISGF-3 | NM_005419 | 0.87 | 1.00 | 1.24 | 0.87 |
| Stat3 | NM_003150 | 1.28 | 1.04 | 1.99 | 1.38 |
| Stat4 | NM_003151 | 0.93 | 1.61 | 3.27 | 0.88 |
| MGF | NM_003152 | 2.37 | 9.47 | 9.74 | 0.90 |
| Stat5 | NM_012448 | 2.30 | 10.72 | 4.40 | 0.89 |
| Stat6 | NM_003153 | 1.52 | 1.22 | 1.98 | 1.37 |
| Tyk2 | NM_003331 | 0.91 | 6.44 | 2.54 | 0.87 |
| Receptors that bind and activate Jak proteins | |||||
| FMS/CD115 | NM_005211 | 2.13 | 1.05 | 1.90 | 2.54 |
| IL-3Rβ | NM_000395 | 5.91 | 1.38 | 2.65 | 0.94 |
| EGFR | NM_005228 | 1.33 | 1.14 | 0.99 | 1.57 |
| EPOR | NM_000121 | 1.60 | 11.21 | 0.92 | 0.89 |
| IFN-αR1 | NM_000629 | 1.85 | 17.50 | 0.70 | 0.89 |
| IFN-αR2 | NM_000874 | 3.29 | 14.30 | 0.69 | 0.90 |
| IFN-γR-1 | NM_000416 | 18.48 | 1.36 | 2.45 | 1.26 |
| IFN-γR-2 | NM_005534 | 14.58 | 1.60 | 1.17 | 1.23 |
| IL-2Rr | NM_000206 | 10.74 | 1.18 | 3.17 | 2.36 |
| gp130 | NM_002184 | 2.40 | 4.75 | 0.59 | 0.89 |
| IL-10Rα | NM_001558 | 12.18 | 1.29 | 0.78 | 1.17 |
| IL-10Rβ | NM_000628 | 6.89 | 1.34 | 1.02 | 2.09 |
| IL-20Rα | NM_014432 | 4.59 | 3.62 | 0.86 | 1.14 |
| IL-22R | NM_021258 | 1.99 | 11.13 | 1.01 | 0.90 |
| TPOR | NM_005373 | 1.39 | 2.71 | 1.76 | 0.89 |
| Nuclear factors and coactivators that interact with Stat proteins | |||||
| C/EBPβ | NM_005194 | 2.07 | 1.24 | 2.18 | 0.89 |
| CBP | NM_004380 | 10.61 | 1.28 | 1.23 | 1.50 |
| CRKL | NM_005207 | 2.42 | 2.14 | 1.09 | 0.90 |
| HMGIY | NM_145904 | 1.62 | 2.01 | 0.68 | 1.52 |
| p48/IRF9 | NM_006084 | 1.34 | 0.99 | 0.91 | 1.58 |
| v-jun | NM_002228 | 1.20 | 1.00 | 0.98 | 1.29 |
| Smad1 | NM_005900 | 1.94 | 2.05 | 0.36 | 0.88 |
| Smad2 | NM_005901 | 2.56 | 3.38 | 0.74 | 1.71 |
| Smad3 | NM_005902 | 2.32 | 1.34 | 0.96 | 1.56 |
| Smad4/DPC4 | NM_005359 | 0.60 | 4.71 | 20.11 | 0.88 |
| Smad5 | NM_005903 | 1.24 | 0.98 | 3.82 | 0.88 |
| Smad6 | NM_005585 | 2.14 | 1.02 | 1.22 | 3.08 |
| Smad7 | NM_005904 | 2.82 | 1.19 | 1.34 | 2.11 |
| Smad9 | NM_005905 | 0.92 | 1.61 | 1.05 | 0.87 |
| CDC46 | NM_006739 | 1.71 | 1.12 | 0.85 | 1.88 |
| NCOA1 | NM_003743 | 0.56 | 0.77 | 0.72 | 0.87 |
| KBF1 | NM_003998 | 0.97 | 2.27 | 1.11 | 1.20 |
| NMI | NM_004688 | 1.46 | 1.29 | 1.03 | 1.16 |
| SH2B | NM_015503 | 1.10 | 7.24 | 2.41 | 0.87 |
| Sp1 | XM_028606 | 2.74 | 1.65 | 7.49 | 1.60 |
| PU.1 | NM_003120 | 1.29 | 7.88 | 3.46 | 0.88 |
| c-src | NM_005417 | 1.11 | 1.08 | 1.78 | 1.27 |
| STAM | NM_003473 | 0.67 | 1.97 | 2.10 | 0.87 |
| STUB1 | NM_005861 | 1.54 | 1.21 | 8.84 | 1.16 |
| USF1 | NM_007122 | 0.61 | 1.87 | 3.46 | 0.86 |
| YY1 | NM_003403 | 2.85 | 1.52 | 1.31 | 1.03 |
| Genes induced by Stat proteins | |||||
| Induced by Stat1 | |||||
| MIG | NM_002416 | 1.94 | 1.23 | 1.10 | 2.05 |
| INDO | NM_002164 | 1.24 | 0.96 | 0.83 | 1.28 |
| IRF-1 | NM_002198 | 2.10 | 2.63 | 1.44 | 8.28 |
| CIITA | NM_000246 | 1.04 | 1.36 | 1.04 | 0.88 |
| NOS | NM_000625 | 0.62 | 1.35 | 1.45 | 0.78 |
| Induced by Stat3 | |||||
| A2M | NM_000014 | 5.42 | 1.21 | 3.73 | N/A |
| Bcl-x | NM_138578 | 1.71 | 1.09 | 2.86 | 1.77 |
| P21/Waf1/CIP1 | NM_000389 | 11.88 | 2.53 | 1.46 | 1.03 |
| CRP | NM_000567 | 1.01 | 1.02 | 0.86 | 1.04 |
| IRF-1 | NM_002198 | 2.10 | 2.63 | 1.44 | 8.28 |
| Jun-B | NM_002229 | 1.17 | 0.95 | 0.95 | 1.20 |
| Transin | NM_002422 | 2.38 | 1.13 | 2.42 | 1.91 |
| c-Myc | NM_002467 | 0.83 | 13.17 | 1.10 | 0.87 |
| SSI-1/Cish1 | NM_003745 | 0.87 | 0.99 | 0.78 | 0.90 |
| Fas/Apo-1/CD95 | NM_000043 | 0.71 | 14.87 | 4.15 | 0.88 |
| Induced by Stat4 | |||||
| CD23 | NM_002002 | 2.22 | 1.58 | 0.97 | 1.57 |
| FcGR1 | NM_000566 | 2.92 | 1.07 | 1.36 | 2.15 |
| IFN-γ | NM_000619 | 1.01 | 4.33 | 6.99 | 0.88 |
| IRF-1 | NM_002198 | 2.10 | 2.63 | 1.44 | 8.28 |
| c-Myc | NM_002467 | 0.83 | 13.17 | 1.10 | 0.87 |
| Induced by Stat5 | |||||
| Cyclin D1 | NM_053056 | 18.44 | 1.79 | 1.68 | 1.05 |
| P21/Waf1/CIP1 | NM_000389 | 11.88 | 2.53 | 1.46 | 1.03 |
| β-Casein | NM_001891 | 8.55 | 1.59 | 1.35 | 0.95 |
| CD25 | NM_000417 | 0.82 | 2.10 | 4.52 | 0.87 |
| IRF-1 | NM_002198 | 2.10 | 2.63 | 1.44 | 8.28 |
| OSM | NM_020530 | 1.83 | 1.00 | 1.83 | 1.67 |
| Pim-1 | NM_002648 | 0.91 | 1.04 | 0.73 | 0.93 |
| Induced by Stat6 | |||||
| FcER1 | NM_002001 | 1.35 | 1.28 | 1.01 | 1.48 |
| GATA3 | NM_002051 | 23.14 | 1.29 | 1.63 | 1.15 |
| IgHG3 | AI950854 | 37.18 | 1.39 | 1.14 | 1.27 |
| IL-4 | NM_000589 | 5.52 | 5.09 | 3.96 | 6.25 |
| IL-4Rα | NM_000418 | 2.12 | 2.20 | 2.01 | 8.82 |
| c-Maf | NM_005360 | 2.00 | 1.10 | 0.74 | 1.52 |
| Induced by Stat1/Stat1/p48 | |||||
| GBP1 | NM_002053 | 2.39 | 9.17 | 1.54 | 0.90 |
| Induced by Stat1/Stat2/p48 (ISGF3) | |||||
| IFI616 | NM_002038 | 29.77 | 1.21 | 3.87 | 1.38 |
| ISG15 | NM_005101 | 1.31 | 1.03 | 1.02 | 1.50 |
| INDO | NM_002164 | 1.24 | 0.96 | 0.83 | 1.28 |
| OAS1 | NM_002534 | 1.29 | 0.98 | 2.10 | 1.63 |
| Negative regulators of the Jak/Stat pathway | |||||
| PIASX-β | NM_004671 | 1.17 | 1.02 | 2.61 | 1.39 |
| DDXBP1 | NM_016166 | 3.59 | 1.08 | 1.93 | 2.46 |
| PIAS3 | NM_006099 | 0.85 | 1.13 | 1.21 | 0.88 |
| PIASy | NM_015897 | 0.78 | 0.91 | 1.12 | 0.72 |
| PTP-1β | NM_002827 | 0.96 | 0.94 | 0.90 | 0.95 |
| PTPNS1 | NM_080792 | 0.38 | 1.58 | 1.14 | 0.84 |
| Cd45 | NM_002838 | 2.63 | 1.39 | 12.78 | 1.76 |
| SSI-1/Cish1 | NM_003745 | 0.87 | 0.99 | 0.78 | 0.90 |
| StatI2 | NM_003877 | 1.13 | 1.03 | 1.20 | 1.31 |
| SSI-3 | NM_003955 | 1.76 | 1.15 | 1.09 | 1.29 |
| SOCS6/CIS4 | NM_004232 | 1.45 | 1.06 | 0.92 | 1.33 |
| SOCS5 | NM_144949 | 0.74 | 4.17 | 1.74 | 0.88 |
| SOCS4 | NM_080867 | 0.96 | 0.98 | 1.89 | 0.97 |
ELISA.
Production of huIL-6 and huIL-6Ra was measured in pcDNA- and pTat-transfected BCBL-1 cells in a time course by using ELISA kits (DIACONE Research, Fleming, Besancon, France). Undiluted tissue culture supernatants were used as recommended by the supplier. Each sample was assayed in duplicate, and a minimum of three assays was performed.
RESULTS
Overexpression of Tat in PEL cell lines induces KSHV lytic cycle replication.
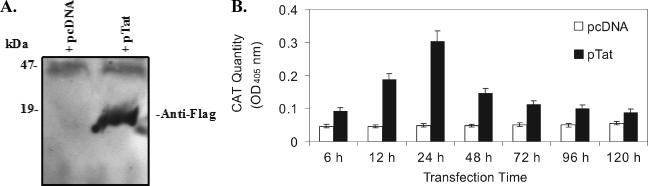
To evaluate whether the Tat protein can affect lytic cycle replication of KSHV, we first examined the activity of the Tat protein expressed in BCBL-1 cells. Western blot analysis showed that Tat protein was readily detected in BCBL-1 cells that were transiently transfected with pTat at 48 h (Fig. 1A). When BCBL-1 cells were cotransfected with pTat and pTZIII-CAT, in which the HIV LTR controls the expression of CAT (Tat protein binds specifically to a stem loop structure at the 5′ end of the viral RNA known as TAR [Tat activation region], resulting in a 10- to 100-fold increase in HIV-1 mRNA production [56]), CAT activity increased up to sixfold. Analysis of data from three independent experiments demonstrated that, on average, CAT expression increased 2.0-fold ± 0.22-fold (mean ± standard deviation) at 6 h, 4.1-fold ± 0.43-fold at 12 h, 6.2-fold ± 0.59-fold at 24 h, 3.1-fold ± 0.35-fold at 48 h, 2.2-fold ± 0.25-fold at 72 h, 2.0-fold ± 0.21-fold at 96 h, and 1.6-fold ± 0.19-fold at 120 h in BCBL-1 cells cotransfected with pTZIII-CAT compared to pcDNA vector at the same time point (Fig. 2B). These results indicate that Tat protein can be expressed and is functionally active in BCBL-1 cells.
FIG. 1.
Expression of HIV-1 Tat and its functional activity in BCBL-1 cells. (A) Tat protein expressed in BCBL-1 cells following transfection with the plasmid pTat. Whole-cell extracts of protein isolated from BCBL-1 cells transfected with the plasmids pcDNA (negative control; + pcDNA) or pTat (+pTat) for 48 h were transferred to an Immobilon P (polyvinylidene difluoride) membrane, and expression of Tat protein was detected by Western blotting with anti-Flag antibody. (B) ELISA for CAT in BCBL-1 cells cotransfected with pTat and pTZIII-CAT. CAT protein expression in BCBL-1 cells cotransfected with pcDNA and pTZIII-CAT (pcDNA) or pTat and pTZIII-CAT (pTat) for 6 to 120 h was quantitated by ELISA. Results presented were from three independent experiments performed in triplicate.
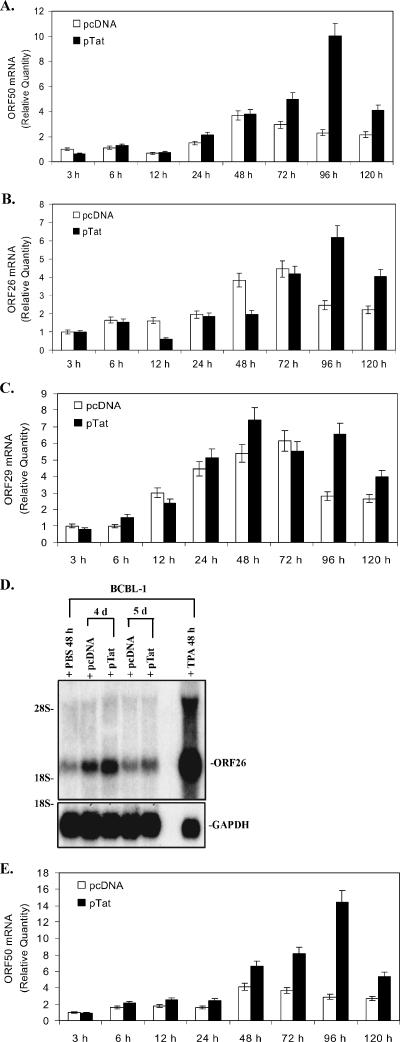
FIG. 2.
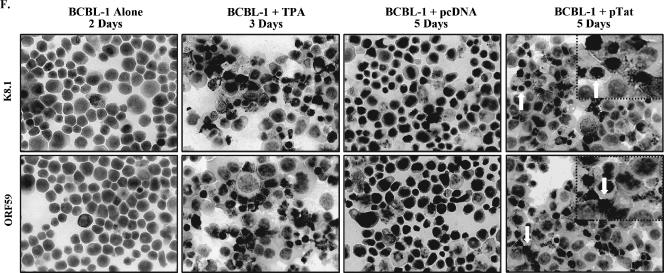
Expression of KSHV lytic cycle RNA and protein in PEL cell lines transfected with Tat. (A) ORF50 mRNA expressed in BCBL-1 cells following transfection with pTat. ORF50 mRNA expression in BCBL-1 cells transfected with pcDNA or pTat for 3, 6, 12, 24, 48, 72, 96, and 120 h was quantitated by real-time quantitative PCR. Relative quantities of ORF50 expression are represented on the y axis. Results shown were from five independent experiments performed in triplicate. (B) ORF26 mRNA expressed in BCBL-1 cells following transfection with pTat. ORF26 mRNA expression in BCBL-1 cells transfected with pcDNA or pTat for 3, 6, 12, 24, 48, 72, 96, and 120 h was quantitated by real-time quantitative PCR. Relative quantities of ORF26 expression are represented on the y axis. Results shown are from five independent experiments performed in triplicate. (C) ORF29 mRNA expressed in BCBL-1 cells following transfection with pTat. ORF29 mRNA expression in BCBL-1 cells transfected with pcDNA or pTat for 3, 6, 12, 24, 48, 72, 96, and 120 h was quantitated by real-time quantitative PCR. Relative quantities of ORF29 expression are represented on the y axis. Results shown are from five independent experiments performed in triplicate. (D) Northern blot analysis for ORF26 mRNA expressed in BCBL-1 cells following transfection with pTat. Total RNA isolated from BCBL-1 cells treated with PBS (negative control) and TPA (positive control) for 48 h, from BCBL-1 cells transfected with pcDNA for 4 and 5 days (pcDNA 4 d and 5 d, respectively), and from BCBL-1 cells transfected with pTat for 4 and 5 days (pTat 4 d and 5 d, respectively) were transferred to a nylon membrane, and ORF26 mRNA expression was detected by Northern blotting. The same membrane was stripped and reprobed for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to demonstrate equal loading of the RNA. Results shown are a representative experiment of at least two independent experiments with similar results. (E) ORF50 mRNA expressed in BC-3 cells following transfection with pTat. ORF50 mRNA in BC-3 cells transfected with pcDNA or pTat for 3, 6, 12, 24, 48, 72, 96, and 120 h was quantitated by real-time quantitative PCR. Relative quantities of ORF50 expression are represented on the y axis. Results shown are from five independent experiments performed in triplicate. (F) Immunohistochemical staining of BCBL-1 cells transfected with pTat (original magnification, × 60). Expression levels of KSHV lytic proteins ORF K8.1 (top panel) and ORF59 (bottom panel) in BCBL-1 cells (negative control; BCBL-1 alone), TPA-treated BCBL-1 cells (positive control; BCBL-1 + TPA), BCBL-1 cells transfected with pcDNA (BCBL-1 + pcDNA), and BCBL-1 cells transfected with pTat (BCBL-1 + pTat) were detected by immunohistochemistry with ORF K8.1 and ORF59 monoclonal antibodies. The lighter-shaded reaction product signifies positive detection of the specified antigen. Arrows highlight examples of positive cells.
To determine whether Tat can activate KSHV replication, BCBL-1 cells were transfected with pTat or pcDNA. The mRNAs of KSHV ORF50 (the molecular switch gene of KSHV from latency to lytic replication) (60), ORF26 (viral minor capsid protein, expressed only during lytic KSHV replication) (52), and ORF29 (viral packaging protein) (52) were analyzed by real-time quantitative PCR. We found that, at up to 72 h, ORF50, ORF26, or ORF29 mRNA did not significantly increase in Tat-transfected BCBL-1 cells. However, a remarkable increase in the peak of ORF50, ORF26, and ORF29 mRNA in Tat-transfected BCBL-1 cells was observed at 96 h (Fig. 2A, B, and C). Analysis of data from five independent experiments between pcDNA- and Tat-transfected cells demonstrated that, on average, ORF50 expression increased 0.63-fold ± 0.05-fold at 3 h, 1.15-fold ± 0.12-fold at 6 h, 1.12-fold ± 0.11-fold at 12 h, 1.44-fold ± 0.13-fold at 24 h, 1.02-fold ± 0.09-fold at 48 h, 1.69-fold ± 0.17-fold at 72 h, 4.33-fold ± 0.49-fold at 96 h (P < 0.05), and 1.9-fold ± 0.26-fold at 120 h (P < 0.05) compared to pcDNA-transfected BCBL-1 cells (Fig. 2A). Meanwhile, ORF26 expression increased 0.99 ± 0.07-fold at 3 h, 0.93 ± 0.1-fold at 6 h, 0.39 ± 0.04-fold at 12 h, 0.94 ± 0.08-fold at 24 h, 0.51 ± 0.04-fold at 48 h, 0.94 ± 0.08-fold at 72 h, 2.52 ± 0.31-fold at 96 h (P < 0.05), and 1.82 ± 0.26-fold at 120 h (P < 0.05), respectively, compared to pcDNA-transfected BCBL-1 cells (Fig. 2B). Moreover, ORF29 expression increased 0.79 ± 0.08-fold at 3 h, 1.57 ± 0.13-fold at 6 h, 0.79 ± 0.06-fold at 12 h, 1.16 ± 0.12-fold at 24 h, 1.37 ± 0.16-fold at 48 h, 0.9 ± 0.08-fold at 72 h, 2.33 ± 0.3-fold at 96 h (P < 0.05), and 1.5 ± 0.22-fold at 120 h, respectively, compared to pcDNA-transfected BCBL-1 cells (Fig. 2C). A statistical analysis of the levels of mRNA in Tat-transfected cells at the time points also showed that ORF50 expression in Tat-transfected BCBL-1 cells increased 2.01-fold ± 0.19-fold at 6 h, 1.17-fold ± 0.13-fold at 12 h, 3.37-fold ± 0.41-fold at 24 h, 6.0-fold ± 0.71-fold at 48 h, 7.86-fold ± 0.89-fold at 72 h, 15.78-fold ± 1.87-fold at 96 h, and 6.45-fold ± 0.59-fold at 120 h compared to pTat-transfected BCBL-1 cells at 3 h (Fig. 2A). ORF26 expression increased 1.57-fold ± 0.17-fold at 6 h, 0.63-fold ± 0.05-fold at 12 h, 1.86-fold ± 0.2-fold at 24 h, 1.98-fold ± 0.21-fold at 48 h, 4.22-fold ± 0.51-fold at 72 h, 6.25-fold ± 0.78-fold at 96 h, and 4.06-fold ± 0.51-fold at 120 h compared to pTat-transfected BCBL-1 cells at 3 h (Fig. 2B). Furthermore, ORF29 expression in Tat-transfected BCBL-1 cells increased 1.92-fold ± 0.18-fold at 6 h, 3.0-fold ± 0.29-fold at 12 h, 6.51-fold ± 0.71-fold at 24 h, 9.37-fold ± 1.1-fold at 48 h, 7.0-fold ± 0.68-fold at 72 h, 8.3-fold ± 0.79-fold at 96 h, and 5.04-fold ± 0.56-fold at 120 h compared to pTat-transfected BCBL-1 cells at 3 h (Fig. 2C). To better visually evaluate the level of KSHV replication, Northern blot analysis for ORF26 mRNA was performed. After transfection of BCBL-1 cells with pTat, the expression of ORF26 mRNA was increased at both 96 and 120 h (Fig. 2D). To examine whether Tat-induced KSHV lytic replication is cell type specific, another KSHV latently infected cell line, BC-3, was used. After Tat transfection, BC-3 cells showed a similar ORF50 mRNA expression pattern to that of BCBL-1 (Fig. 2E). These data suggest that Tat may activate KSHV lytic-phase RNA; this lytic cycle replication is not restricted in BCBL-1 cells, and may be a common mechanism that reactivates KSHV in KS.
To examine whether induction of KSHV lytic cycle RNA by Tat also resulted in induction of lytic cycle proteins, immunostaining of BCBL-1 cells was performed to detect two KSHV lytic cycle proteins (K8.1 and ORF59). After 5 days of transfection of BCBL-1 cells with pTat (BCBL-1 plus pTat), 7.1 ± 0.7% of BCBL-1 cells expressed ORF K8.1 compared to 2.8 ± 0.3% of BCBL-1 cells transfected with pcDNA (BCBL-1 plus pcDNA) and 1.0 ± 0.1% of untreated BCBL-1 cells (P < 0.05) (Fig. 2F, first, third, and fourth panels of the top row). Similarly, 9.8 ± 0.8% of BCBL-1 cells (BCBL-1 plus pTat) expressed ORF59 compared to 3.6 ± 0.4% of BCBL-1 cells transfected with pcDNA (BCBL-1 plus pcDNA) and 1.2 ± 0.2% of untreated BCBL-1 cells (P < 0.05) (Fig. 2F, first, third, and fourth panels of the bottom row). This was consistent with the previous report that the expression of ORF59 occurs earlier and more frequently in the lytic cycle, compared to the expression of ORF K8.1 (66). As positive controls, it was found that 36.3 ± 2.3% and 45.1 ± 2.6% of BCBL-1 cells expressed ORF K8.1 and ORF59, respectively, after treatment with TPA (Fig. 2F, second panel). These results indicate that the induction of KSHV lytic cycle RNA by Tat also results in the induction of lytic cycle proteins.
Tat does not induce KSHV ORF50 promoter activity in PEL cell lines.
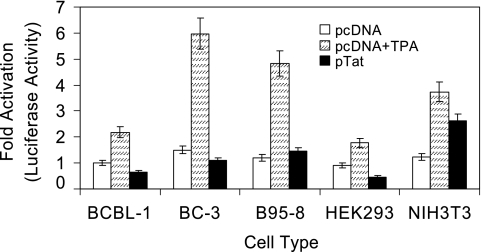
To explore whether Tat induces KSHV lytic cycle replication by direct binding to the ORF50 promoter, we examined the effect of Tat on ORF50 promoter activity in several cell lines. KSHV ORF50 encodes a replication and transcription activator homologous to the EBV Rta, which has been shown to be both necessary and sufficient to activate the KSHV lytic cycle from latency (37, 60). In this assay a 655-bp fragment 5′ to the ORF50 transcriptional start site was used to drive luciferase reporter gene expression (p50-Luc construct). We (35) and Seaman (55) have previously shown that this construct has promoter activity in BCBL-1 cells. Cells cotransfected with p50-Luc and pcDNA showed low baseline levels of luciferase expression (used as a negative control), which was dramatically enhanced (2.19-, 3.98-, 4.02-, 1.98-, and 3.04-fold increases in BCBL-1, BC-3, B95-8, HEK293, and NIH 3T3 cells, respectively) by stimulation with TPA (used as a positive control) (Fig. 3). In contrast, cotransfection of the first four cell types with pTat and p50-Luc did not result in a significant increase in luciferase expression compared to the corresponding control (Fig. 3). This was consistent with the previous report indicating that Tat alone failed to activate the ORF50 promoter (62). Surprisingly, a 2.13-fold increase in luciferase expression was found in NIH 3T3 cells cotransfected with pTat and p50-Luc (Fig. 3), suggesting that induction of ORF50 promoter activity by Tat may be cell type specific. These observations collectively demonstrate that Tat protein is sufficient to induce KSHV lytic cycle replication through an indirect mechanism rather than directly activating ORF50 expression in BCBL-1 cells.
FIG. 3.
Transfection of PEL cell lines with Tat does not promote induction of KSHV ORF50 promoter activity. BCBL-1, BC-3, B95-8, HEK293, and NIH 3T3 cells were cotransfected with p50-Luc and pcDNA (negative control, pcDNA), p50-Luc and pcDNA following treatment with TPA (positive control, pcDNA + TPA), or p50-Luc and pTat (pTat). Luciferase activities were measured as induction. All data points were the averages of four independent experiments performed in triplicate.
Inhibition of HuIL-6 and HuIL-6Ra expression increases Tat-induced KSHV replication.
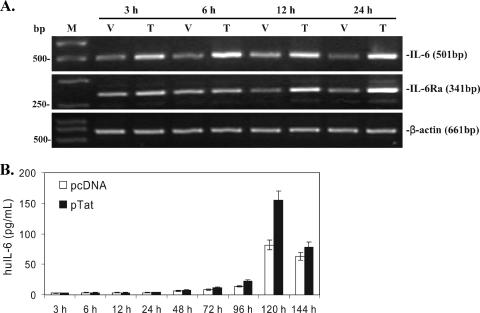
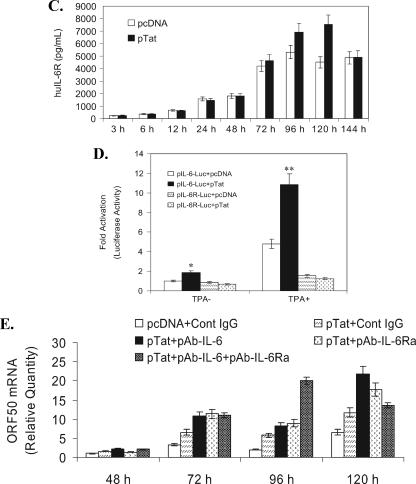
The current facts are that (i) Tat did activate KSHV replication and Tat did not directly activate ORF50 (Fig. 2 and 3) (24, 28, 42, 62); (ii) the induction of KSHV lytic RNA (ORF50, ORF26, and ORF29 mRNA) was delayed (from 3 to 24 h) (Fig. 2); (iii) inflammatory cytokines and receptors induced by HIV-1 play important roles in AIDS-KS pathogenesis by promoting KSHV replication and inducing KS cell malignancy (22, 43, 46). Together, these facts led us to hypothesize that intracellular Tat may first initiate cellular signaling from BCBL-1 cells during the early KSHV replication and subsequently affect viral replication. To test this hypothesis, we first detected gene expressing profile changes affected by Tat expression by using a microarray technique. The Q Series inflammatory cytokines and receptors gene array was used to screen the potential cytokine(s) and receptor(s) involved in this process. As shown in Table 2, mRNAs of many inflammatory cytokine and receptor genes in Tat-transfected BCBL-1 cells were altered to some degree at different time points. Notably, mRNAs of huIL-6 and huIL-6Ra in Tat-transfected BCBL-1 cells consistently increased from 3 to 24 h compared with the corresponding controls (Table 2). RT-PCR, ELISA, and a luciferase reporter assay were then performed to further confirm and extend these results. As expected, both IL-6 and IL-Ra mRNAs increased significantly in Tat-transfected BCBL-1 cells from 3 to 24 h (Fig. 4A), which was consistent with the microarray data (Table 2). The ELISA showed that both huIL-6 and huIL-6Ra produced by Tat-induced BCBL-1 cells could be secreted into the culture after 120 h of transfection (1.92- and 1.73-fold increases of huIL-6 and huIL-6Ra, respectively, compared to that of pcDNA-transfected BCBL-1 cells; P < 0.05) (Fig. 4B and C). Particularly, expression of huIL-6Ra in Tat-transfected BCBL-1 cells reached nanogram levels (Fig. 4C). Scala et al. have previously shown that Tat can activate the IL-6 promoter in MC3 B-lymphoblastoid and HeLa epithelial cells (54). To test whether Tat may directly activate IL-6 and/or IL-6Ra promoters, we performed a luciferase assay. We observed that BCBL-1 cells cotransfected with either pIL-6-Luc and pcDNA or pIL-6Ra-Luc and pcDNA showed baseline levels of luciferase expression with both the absence and presence of TPA (used as a control) (Fig. 4D). Cotransfection of BCBL-1 cells with pIL-6-Luc and pTat resulted in a statistically significant increase in luciferase expression in both the absence and presence of TPA (1.9- and 2.3-fold increases compared to the corresponding control; P < 0.05) (Fig. 4D). However, cotransfection of BCBL-1 cells with pIL-6Ra-Luc and pTat did not lead to any significant increase in luciferase expression in either the absence or presence of TPA compared with the corresponding control (Fig. 4D). These data suggest that in BCBL-1 cells Tat alone is sufficient to induce soluble huIL-6, but not huIL-6Ra.
FIG. 4.
Inhibition of huIL-6 and huIL-6Ra expression increases Tat-induced KSHV replication. (A) RT-PCR analysis of huIL-6 and IL-6Ra mRNA expression in Tat-transfected BCBL-1 cells. HuIL-6 and huIL-6Ra mRNA expression in BCBL-1 cells transfected with pcDNA vector (V) or pTat (T) for 3, 6, 12, and 24 h was detected by RT-PCR. M, DNA molecular marker. β-Actin was used as an internal control to monitor the presence of amplifiable cDNA in all samples. (B) Expression of huIL-6 in BCBL-1 cells transfected for 3 to 144 h with pTat. Supernatants from BCBL-1 cells transfected with pcDNA or pTat for various times were collected for detection of huIL-6 by ELISA. Results are from three independent experiments with duplicates. (C) Expression of huIL-6Ra in BCBL-1 cells transfected for 3 to 144 h with pTat. Supernatants from BCBL-1 cells transfected with pcDNA or pTat for various times were collected for detection of huIL-6Ra by ELISA. Results are from three independent experiments with duplicates. (D) Effects of Tat on huIL-6 and huIL-6Ra promoter activities in BCBL-1 cells. BCBL-1 cells were cotransfected with pIL-6-Luc and pcDNA, pIL-6-Luc and pTat, pIL-6R-Luc and pcDNA, and pIL-6R-Luc and pTat following treatment without TPA or with TPA. Luciferase activities were measured as induction. All data points were the averages of four independent experiments performed in triplicate. * and ** indicate statistically significant increases in luciferase expression in the absence and presence of TPA compared to the corresponding control. (E) Real-time quantitative PCR analysis for ORF50 mRNA expression in a blocking assay with PAb against huIL-6. Real-time quantitative PCR was used to detect relative quantities of ORF50 mRNA in pcDNA-transfected BCBL-1 cells plus control IgG (pcDNA + Cont IgG), pTat-transfected BCBL-1 cells plus control IgG (pTat + Cont IgG), pTat-transfected BCBL-1 cells plus 25 μg/ml of PAb against huIL-6 (pTat + pAb-IL-6), pTat-transfected BCBL-1 cells plus 25 μg/ml of PAb against huIL-6Ra (pTat + pAb-IL-6Ra), and pTat-transfected BCBL-1 cells plus PAbs against huIL-6 and huIL-6Ra in combination (pTat + pAb-IL-6 + pAb-IL-6Ra) for 48, 72, 96, and 120 h. The results from three independent experiments performed in triplicate are shown.
To test the hypothesis that overexpression of huIL-6 and/or huIL-6Ra may modulate Tat-induced KSHV replication, blocking assays of PAbs against huIL-6 and huIL-6Ra were performed. As shown in Fig. 4E, after addition of control IgG to Tat-transfected BCBL-1 cells, ORF50 mRNA was increased 1.98-fold at 72 h, 2.81-fold at 96 h, and 1.78-fold at 120 h, compared to pcDNA-transfected BCBL-1 cells treated with the control IgG, which was almost consistent with the results shown in Fig. 2A, indicating that the control IgG alone has no effect on KSHV replication by Tat. Importantly, after addition of anti-IL-6 PAb to the culture of Tat-transfected BCBL-1 cells, ORF50 mRNA further increased 1.65-fold at 72 h (P < 0.05), 1.43-fold at 96 h, and 1.86-fold at 120 h (P < 0.05), compared to Tat-transfected BCBL-1 cells treated with the control IgG (Fig. 4E). Similarly, ORF50 mRNA also increased 1.73-fold at 72 h (P < 0.05), 1.56-fold at 96 h, and 1.51-fold at 120 h after addition of anti-IL-6Ra PAb to the culture of Tat-transfected BCBL-1 cells (Fig. 4E). Interestingly, a synergism between anti-IL-6 PAb and anti-IL-6Ra PAb was observed to affect ORF50 mRNA expression when anti-IL-6 PAb and anti-IL-6Ra PAb were added in combination to the culture at 96 h (2.43- and 2.23-fold increases compared to anti-IL-6 PAb and anti-IL-6Ra PAb groups, respectively; P < 0.05) (Fig. 4E). These data suggest that huIL-6 and its receptor when induced by Tat protein may, at least in part, modulate the effect of Tat-induced KSHV lytic replication.
Inhibition of the JAK2/STAT3 signaling enhances KSHV replication by Tat.
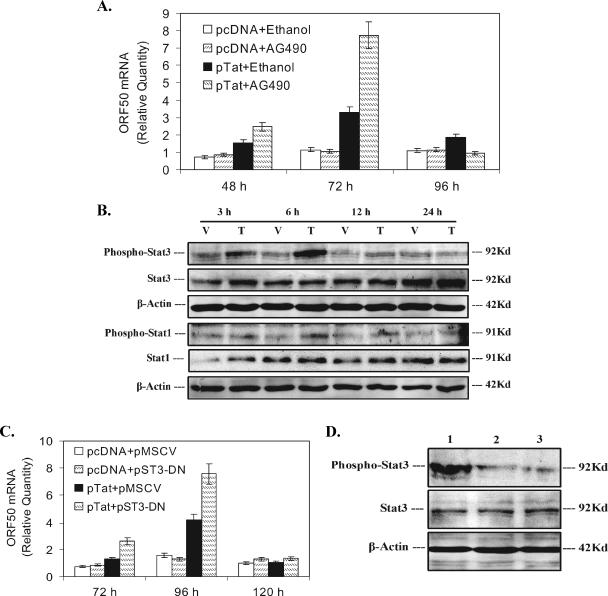
Since huIL-6 and its receptor may down-regulate Tat-induced KSHV lytic replication and signal transduction by IL-6 commonly involves activation of the JAK/STAT pathway (25), we reasoned that activation of JAK/STAT signaling by IL-6 may also be involved in modulating Tat-induced KSHV replication. To address this issue, AG490, a JAK2-specific inhibitor, was added to the cell culture. Real-time quantitative PCR analysis demonstrated ethanol (AG490 was dissolved in the ethanol, which was used as a negative control) alone did not influence Tat-induced KSHV replication; AG490 alone also did not affect KSHV replication in pcDNA-transfected BCBL-1 cells. Interestingly, ORF50 mRNA in Tat-transfected BCBL-1 cells treated with AG490 was increased 1.59-fold at 48 h, 2.43-fold at 72 h, and 0.51-fold at 96 h, compared to Tat-transfected BCBL-1 cells treated with ethanol alone (Fig. 5A). These data indicate that activation of the JAK2 pathway may be, at least in part, involved in down-regulation of Tat-induced KSHV replication. The transcription factor STATs, mainly STAT1 and -3, can be activated by JAK2. To determine whether and which STAT protein is involved in Tat-induced KSHV replication, Western blot analysis on STAT1 and -3 was performed. As shown in Fig. 5B, phosphorylated STAT3 in Tat-transfected BCBL-1 cells was increased 1.56-fold at 3 h, 2.97-fold at 6 h, 1.83-fold at 12 h, and 0.81-fold at 24 h, compared to pcDNA-transfected BCBL-1 cells. We also observed a slight increase of phosphorylated STAT1 in Tat-transfected BCBL-1 cells (Fig. 5B). To examine whether STAT3 plays an important role in KSHV replication by Tat, we inhibited STAT3 by overexpression a dominant negative STAT3. Real-time quantitative PCR indicated that ORF50 mRNA in BCBL-1 cells cotransfected with pTat and pMSCV vector (the pST3-DN construct was cloned in pMSCV vector) was increased 1.72-fold at 72 h, 2.69-fold at 96 h, and 1.43-fold at 120 h, compared to BCBL-1 cells cotransfected with pcDNA and pMSCV (Fig. 5C). Meanwhile, transfection of pST3-DN did not influence KSHV replication in pcDNA-transfected BCBL-1 cells compared to cotransfected BCBL-1 cells with pcDNA and pMSCV (Fig. 5C). Interestingly, ORF50 mRNA in BCBL-1 cells cotransfected with pTat and pST3-DN increased 2.05-fold at 72 h, 1.81-fold at 96 h, and 1.26-fold at 120 h compared to BCBL-1 cells cotransfected with pTat and pMSCV (Fig. 5C). To further confirm that overexpression of dominant negative STAT3 can reduce phosphorylation of STAT3 induced by Tat, Western blot analysis were performed. Although phosphorylated STAT3 in Tat-transfected BCBL-1 cells was increased 2.97-fold at 6 h compared to pcDNA-transfected BCBL-1 cells (Fig. 5B), as shown in Fig. 5D, phosphorylated STAT3 in BCBL-1 cells cotransfected with pTat and pST3-DN was significantly reduced at 6 h and almost reached the same level as that of BCBL-1 cells cotransfected with pcDNA and pMSCV. The experiments also showed that at other various time points expression of phosphorylated STAT3 in BCBL-1 cells cotransfected with pTat and pST3-DN lowered and almost reached the same level as that of BCBL-1 cells cotransfected with pcDNA and pMSCV (data not shown). Together, these data suggest that activation of IL-6-mediated JAK2/STAT3 signaling partially modulates Tat-induced KSHV replication.
FIG. 5.
Inhibition of JAK2/STAT3 signaling by huIL-6 partially enhances Tat-induced KSHV replication. (A) AG490 enhances KSHV replication by Tat. Real-time quantitative PCR was used to detect relative quantities of ORF50 mRNA in pcDNA-transfected BCBL-1 cells plus ethanol, pcDNA-transfected BCBL-1 cells plus AG490, pTat-transfected BCBL-1 cells plus ethanol, and pTat-transfected BCBL-1 cells plus AG490 for 48, 72, and 96 h as indicated. The results from three independent experiments performed in triplicate are shown. (B) Activation STAT1 and -3 in Tat-transfected BCBL-1 cells. BCBL-1 cells were transfected with pcDNA vector (V) or pTat (T) for 3, 6, 12, and 24 h. Lysates were subjected to SDS-PAGE, transferred to a membrane, and then immunoblotted with the indicated anti-phospho antibody. The membrane was stripped and reprobed with the respective antibody or with antiactin to confirm equal amounts of protein in each sample. The results shown are from a representative experiment of at least three independent experiments with similar results. (C) Inhibition of STAT3 activation partially enhances KSHV replication by Tat. Real-time quantitative PCR was employed to detect relative quantities of ORF50 mRNA in BCBL-1 cells cotransfected with pcDNA and pMSCV vector, pcDNA and pST3-DN, pTat and pMSCV vector, or pTat and pST3-DN for 72, 96, and 120 h as indicated. The results from three independent experiments performed in triplicate are shown. (D) Overexpression of dominant negative STAT3 reduces phosphorylation of Tat-induced STAT3. BCBL-1 cells were cotransfected with pTat and pMSCV vector (lane 1), pTat and pST3-DN (lane 2), or pcDNA and pMSCV vector (lane 3) for 6 h. Lysates were subjected to SDS-PAGE, transferred to a membrane, and then immunoblotted with the indicated anti-phospho-STAT3 antibody. The membrane was stripped and reprobed with anti-STAT3 and antiactin antibodies to confirm equal amounts of protein in each sample. The results shown are from a representative experiment of three independent experiments with similar results.
Activation of STAT6 by IL-4 may partially contribute to Tat-induced KSHV replication.
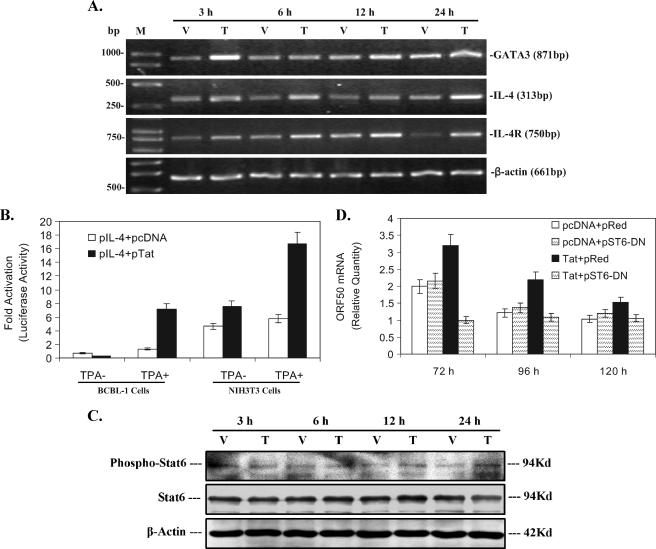
To address whether other JAK and STATs and their signaling were involved in KSHV replication by Tat, the JAK/STAT signal pathway gene array was performed. As shown in Table 3, again we noticed IL-4 and IL-4R mRNAs persistently increased, which was almost consistent with the results shown in Table 2. Meanwhile, both GATA3 and c-Maf, which are upstream genes for IL-4 expression (26, 64), were increased 23.14- and 2-fold at 3 h in Tat-transfected BCBL-1 cells, respectively (Table 3). The data of the RT-PCR analysis further confirmed the microarray results (Fig. 6A). To examine whether Tat can directly induce IL-4 expression, a luciferase reporter assay were performed. We found that cotransfection of BCBL-1 cells with pTat and pIL-4-Luc in the presence of TPA resulted in a significant increase in luciferase expression (5.48-fold increase compared to the corresponding control; P < 0.05) (Fig. 6B). Because a baseline level of IL-4 and/or STAT6 in BCBL-1 cells may induce IL-4 promoter activity, which would complicate the interpretation of the results, we chose NIH 3T3 cells, whose genes for GATA3 and IL-4 are shut off (15), to perform the luciferase assay. It was shown that cotransfection of NIH 3T3 cells with pTat and pIL-4-Luc following treatment with TPA also led to a significant increase in luciferase expression (2.91-fold increase compared to the corresponding control; P < 0.05) (Fig. 6B).
FIG. 6.
Effect of activated STAT6 by IL-4 on KSHV replication by Tat. (A) RT-PCR analysis of GATA3, IL-4, and IL-4R mRNA expression in Tat-transfected BCBL-1 cells. GATA3, IL-4, and IL-4R mRNA expression levels in BCBL-1 cells transfected with pcDNA vector (V) or pTat (T) for 3, 6, 12, and 24 h were detected by RT-PCR. M, DNA molecular marker. β-Actin was used as an internal control to monitor the presence of amplifiable cDNA in all samples. (B) Effect of Tat on IL-4 promoter activity in BCBL-1 and NIH 3T3 cells. BCBL-1 and NIH 3T3 cells were cotransfected with pIL-4-Luc and pcDNA or pIL-4-Luc and pTat following treatment without TPA or with TPA. Luciferase activities were measured as induction. All data points were the averages of four independent experiments performed in triplicate. (C) Activation of STAT6 in Tat-transfected BCBL-1 cells. BCBL-1 cells were transfected with pcDNA vector (V) or pTat (T) for 3, 6, 12, and 24 h. Lysates were subjected to SDS-PAGE, transferred to a membrane, and then immunoblotted with the indicated anti-phospho antibody. The membrane was stripped and reprobed with the respective antibody or with antiactin to confirm equal amounts of protein in each sample. The results shown are from a representative experiment of at least three independent experiments with similar results. (D) Activation of STAT6 partially contributes to KSHV replication by Tat. Real-time quantitative PCR was used to detect relative quantities of ORF50 mRNA in BCBL-1 cells cotransfected with pcDNA and pRed vector, pcDNA and pST6-DN, pTat and pRed vector, or pTat and pST6-DN for 72, 96, and 120 h as indicated. The results from three independent experiments performed in triplicate are shown.
IL-4 activates STAT6 in T cells and plays multiple roles in regulation of the immune system (27). To determine whether persistent expression of IL-4 can also activate STAT6 in BCBL-1 cells, Western blot analysis was performed. We found that phosphorylated STAT6 in Tat-transfected BCBL-1 cells slightly increased (1.83-fold at 12 h and 2.12-fold at 24 h) (Fig. 6C) compared to the corresponding control. To address the question of whether the slight activation of STAT6 by IL-4 plays a role in KSHV replication by Tat, we inhibited STAT6 by overexpression of a dominant negative STAT6. Real-time quantitative PCR analysis demonstrated that cotransfection of pTat and pST6-DN into BCBL-1 cells significantly lowered ORF50 mRNA expression compared to cotransfection of pTat plus pRed vector (the pST6-DN was cloned in pRed) (Fig. 6D). Meanwhile, transfection of pST6-DN did not influence KSHV replication in pcDNA-transfected BCBL-1 cells compared to cotransfected BCBL-1 cells with pcDNA and pRed vector BCBL-1 cells (Fig. 6D). To further confirm that overexpression of dominant negative STAT6 can reduce phosphorylation of STAT6 induced by Tat, Western blot analysis was performed. It was demonstrated that phosphorylated STAT6 in BCBL-1 cells cotransfected with pTat and pST6-DN was reduced and almost reached the same level as that of BCBL-1 cells cotransfected with pcDNA and pRed for 3 to 24 h (data not shown). Together, these observations suggest that Tat protein may stimulate GATA3 protein in BCBL-1 cells and then activated GATA3 further induces IL-4 expression, resulting in activation of STAT6; activated STAT6 partially contributes to Tat-induced KSHV replication.
DISCUSSION
During the course of HIV-1 infection, normal host cells are consistently exposed to free HIV-1 viral particles and circulating HIV-1-associated proteins. While not directly infected with HIV, exposure of these normal cells to HIV-1-associated proteins can have a profound effect on their gene expression and normal function. Aside from the glycoprotein 120 (gp120), soluble HIV-1 Tat is secreted from infected cells and can bind to both αvβ3 and α5β1 integrins on the surface of target cells (6, 18). This binding occurs through the highly conserved Asp-Gly-Arg (RGD) sequence, which is found in the carboxyl terminus of Tat and may function to induce intracellular signals that ultimately lead to changes in cellular gene expression. In addition, extracellular Tat nonspecifically binds to cells membranes and is internalized (19, 56). Like intracellular Tat, this internalized Tat may directly interact with cellular genes to alter gene expression. In this study, we investigated the kinetic of KSHV replication by intracellular Tat and explored the possible mechanisms by which Tat activates KSHV cycle replication. Our results reveal several novel points in understanding AIDS-KS disease progression.
First, our results provide direct experimental evidence that intracellular Tat does activate KSHV lytic cycle replication from latency. Since Ensoli and colleagues previously demonstrated extracellular Tat could promote the growth and proliferation of KS tumor cells (6, 19), the role of Tat as a cofactor in enhancing KSHV replication in PEL cell lines is always a highly controversial and arguable subject (24, 28, 42, 62). For instance, studies from three groups consistently demonstrated that both intracellular and extracellular Tat could strongly activate KSHV well (24, 28, 42). On the contrary, one group showed an absolutely contradictory observation that either intracellular or extracellular Tat alone failed to induce KSHV replication (62). In this study, we modified the protocol for cell culture by first synchronizing BCBL-1 cells at G0 by 24 h of incubation in serum-free medium and then incubating BCBL-1 cells in normal serum medium for an additional 16 h to get to the S phase of the cell cycle. Tat transfection of BCBL-1 cells at S phase led to a maximum inducibility of KSHV replication. We definitely demonstrated that transfection of PEL cell lines with Tat is able to induce KSHV lytic replication, suggesting that Tat may promote KS progression by reactivating KSHV lytic replication and increasing viral load.
Second, we have provided experimental evidence suggesting that huIL-6 and its receptor produced by Tat-transfected BCBL-1 cells play an important role in modulating KSHV activation. Previous studies have shown that Tat can activate the huIL-6 promoter in MC3 B-lymphoblastoid and HeLa epithelial cells(54). Subsequent studies further indicated that Tat induces the expression of the IL-6 gene by binding to the IL-6 leader RNA and by interacting with CAAT enhancer-binding protein β (NF-IL-6) transcription factors(3). In our study, although we did not find that Tat acted directly on the promoters of huIL-6Ra in BCBL-1 cells, we have provided direct evidence that the Tat protein not only activates huIL-6 and its promoter activity, but also induces IL-6Ra expression, reaching nanogram levels in BCBL-1 cells. With respect to huIL-6 and KSHV activation, this was also an arguable subject. A previous study implied that huIL-6 could induce KSHV lytic replication in both marrow cultures and BCBL-1 cells (1, 57). However, recent reports demonstrated that huIL-6 failed to activate KSHV in BCBL-1 cells (12, 43). On the contrary, it significantly inhibited KSHV replication in experimentally infected human microvascular endothelial cells, the precursor of KS (45). Here we have provided direct experimental evidence that both huIL-6 and its receptor may mediate modulation of KSHV lytic replication in Tat-transfected BCBL-1 cells. Indeed, we also demonstrated that a synergism between anti-IL-6 PAb and anti-IL-6Ra PAb appeared to affect ORF50 mRNA expression when anti-IL-6 PAb and anti-IL-6Ra PAb were added in combination to the culture at 96 h in a neutralization antibody blocking assay. One possible interpretation of this observation is that there was an optimal concentration of antigen-antibody binding between expressed huIL-6, its receptor, and anti-IL-6 PAb and anti-IL-6Ra PAb in the culture system at 96 h. This optimal concentration of antigen-antibody binding can effectively neutralize huIL-6 and its receptor, leading to a maximal inducibility of KSHV replication. On the other hand, the maximal inducibility of KSHV replication at 96 h may also activate the other signals that further facilitate KSHV replication. At this time point, blocking of huIL-6 and its receptor will enhance activation of these signals which, in turn, promote KSHV to be further reactivated, leading to detectable ORF50 mRNA increase. However, whether other cytokines, growth factors, or their soluble receptors produced by or in response to Tat-transfected PEL cell lines may also be involved in this process is still unknown.
Third, we showed that inhibition of JAK2/STAT3 signaling significantly enhances Tat-induced KSHV replication. Commonly, activation of JAK2/STAT3 signaling by IL-6 functions to stimulate cell proliferation, mediate survival signals, and prevent apoptosis. In support of this idea, STAT3 activity has been shown to be modulated by many viral proteins, such as EBV, human T-cell lymphotropic virus 1, and herpesvirus Saimiri, to increase the persistence and oncogenic potential of viruses (14, 38, 44). In this study, we found that inhibition of JAK2 and STAT3 activities was able to enhance Tat-induced KSHV activation. The possible mechanism of this observation is that after introduction with Tat, expressed huIL-6 and its receptor subsequently activate JAK2/STAT3 signaling. On one hand, activated STAT3 signaling directly contributes to malignant progression of BCBL-1 cells by preventing apoptosis, acting through the prosurvival protein survivin (4). On the other hand, phosphorylation of STAT3 interacts with latency-associated nuclear antigen of KSHV, which is critical to the persistence of viral episomes and functions in this capacity by tethering viral episomes to chromosomes during mitosis (47), leading to undetectable ORF50 mRNA. Besides JAK/STAT signaling, huIL-6 also involves activation of mitogen-activated protein kinase and phosphatidylinositol-3-kinase/AKT pathways. More recent studies have indicated that Raf/MEK/ERK signaling modulates TPA-induced reactivation of KSHV latency (20, 25); therefore, our results did not eliminate the possibility that another pathway(s) by huIL-6 may also be involved in this process.
Finally, we demonstrated that activation of IL-4/STAT6 signaling partially contributes to Tat-induced KSHV replication. Usually, IL-4/STAT6 signaling is involved in activation, differentiation, and proliferation of Th2 and B cells. Early studies showed that the Tat protein not only up-regulated IL-4R on Raji cells (a human B-lymphoblastoid cell line) but also doubled the germinal center B-cell differentiation and proliferation induced by CD40 MAb and IL-4, suggesting that Tat might directly affect the normal B-cell differentiation process in HIV-positive patients and favor the occurrence of AIDS-associated B-cell lymphomas (29, 33, 49). A recent study further demonstrated that Tat protein also induced IL-4 release from basophils and mast cells (39). Here we have provided experimental evidence to suggest that while up-regulating IL-4 and IL-4R Tat may induce a marginal activation of STAT6, which in turn contributes to Tat-induced KSHV replication in BCBL-1 cells. Activation of IL-4/STAT6 signal may lead to cell death and thereby effectively inhibit cell growth. However, we did not find changes in the cell number or viability in our experiment (C. Lu, unpublished data). We believe that huIL-6/STAT3 signaling may compensate the effect of IL-4/STAT6 by preventing apoptosis (4). Moreover, cellular and KSHV-derived factors, such as activated NF-κB, nerve growth factor, and viral bcl-2, might serve as a growth factor or antiapoptotic factor (31, 48, 53).
In summary, we have experimentally shown the possible roles of IL-6/STAT3 and IL-4/STAT6 signaling in KSHV replication by Tat. Since Tat can induce multiple signaling pathways and has many functions in AIDS-KS pathogenesis, further studies are needed to better understand whether other cytokines and their signals by Tat are also involved in KSHV replication in AIDS-KS patients.
Acknowledgments
We thank M. Rusnati, M. Li-Weber, K. Zhang, and D. Link for plasmids pTZIII-CAT, pIL-4-Luc, pST6-DN, and pMSCV-STAT3D-EGFP, respectively.
This work was supported by grants from the National Natural Science Foundation of China (30670096 to C.L.), Fok Ying Tung Education Foundation (101038 to C.L.), Program for New Century Excellent Talents in University of China (NCET-05-0506 to C.L.), and the Ministry of Science and Technology of Jiangsu Province (BK2006524 to C.L.).
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Agbalika, F., J. P. Marolleau, and J. C. Brouet. 2000. Interleukin-6 stimulates HHV-8 replication in bone marrow cultures and infected cell lines. Hematol. J. 1:48-52. [DOI] [PubMed] [Google Scholar]
- 2.Aldovini, A., C. Debouck, M. B. Feinberg, M. Rosenberg, S. K. Arya, and F. Wong-Staal. 1986. Synthesis of the complete trans-activation gene product of human T-lymphotropic virus type III in Escherichia coli: demonstration of immunogenicity in vivo and expression in vitro. Proc. Natl. Acad. Sci. USA 83:6672-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambrosino, C., M. R. Ruocco, X. Chen, M. Mallardo, F. Baudi, S. Trematerra, I. Quinto, S. Venuta, and G. Scala. 1997. HIV-1 Tat induces the expression of the interleukin-6 (IL-6) gene by binding to the IL-6 leader RNA and by interacting with CAAT enhancer-binding protein beta (NF-IL-6) transcription factors. J. Biol. Chem. 272:14883-14892. [DOI] [PubMed] [Google Scholar]
- 4.Aoki, Y., G. M. Feldman, and G. Tosato. 2003. Inhibition of STAT3 signaling induces apoptosis and decreases survivin expression in primary effusion lymphoma. Blood 101:1535-1542. [DOI] [PubMed] [Google Scholar]
- 5.Ariyoshi, K., M. Schim van der Loeff, P. Cook, D. Whitby, T. Corrah, S. Jaffar, F. Cham, S. Sabally, D. O'Donovan, R. A. Weiss, T. F. Schulz, and H. Whittle. 1998. Kaposi's sarcoma in the Gambia, West Africa is less frequent in human immunodeficiency virus type 2 than in human immunodeficiency virus type 1 infection despite a high prevalence of human herpesvirus 8. J. Hum. Virol. 1:193-199. [PubMed] [Google Scholar]
- 6.Barillari, G., R. Gendelman, R. C. Gallo, and B. Ensoli. 1993. The Tat protein of human immunodeficiency virus type 1, a growth factor for AIDS Kaposi sarcoma and cytokine-activated vascular cells, induces adhesion of the same cell types by using integrin receptors recognizing the RGD amino acid sequence. Proc. Natl. Acad. Sci. USA 90:7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bromberg, J. 2002. Stat proteins and oncogenesis. J. Clin. Investig. 109:1139-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchbinder, A., and A. E. Friedman-Kien. 1992. Clinical aspects of Kaposi's sarcoma. Curr. Opin. Oncol. 4:867-874. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 10.Chan, S. R., C. Bloomer, and B. Chandran. 1998. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology 240:118-126. [DOI] [PubMed] [Google Scholar]
- 11.Chandran, B., C. Bloomer, S. R. Chan, L. Zhu, E. Goldstein, and R. Horvat. 1998. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology 249:140-149. [DOI] [PubMed] [Google Scholar]
- 12.Chang, J., R. Renne, D. Dittmer, and D. Ganem. 2000. Inflammatory cytokines and the reactivation of Kaposi's sarcoma-associated herpesvirus lytic replication. Virology 266:17-25. [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 14.Chen, H., L. Hutt-Fletcher, L. Cao, and S. D. Hayward. 2003. A positive autoregulatory loop of LMP1 expression and STAT activation in epithelial cells latently infected with Epstein-Barr virus. J. Virol. 77:4139-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, J. X., S. Watanabe, A. Muto, A. Miyajima, T. Yokota, and K. Arai. 1994. Activation of early response genes and cell proliferation by human interleukin-3, granulocyte-macrophage colony-stimulating factor, and interleukin-5 receptors: comparison with human interleukin-4 receptor signaling. J. Allergy Clin. Immunol. 94:605-611. [DOI] [PubMed] [Google Scholar]
- 16.Decker, L. L., P. Shankar, G. Khan, R. B. Freeman, B. J. Dezube, J. Lieberman, and D. A. Thorley-Lawson. 1996. The Kaposi sarcoma-associated herpesvirus (KSHV) is present as an intact latent genome in KS tissue but replicates in the peripheral blood mononuclear cells of KS patients. J. Exp. Med. 184:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delli Bovi, P., E. Donti, D. M. Knowles II, A. Friedman-Kien, P. A. Luciw, D. Dina, R. Dalla-Favera, and C. Basilico. 1986. Presence of chromosomal abnormalities and lack of AIDS retrovirus DNA sequences in AIDS-associated Kaposi's sarcoma. Cancer Res. 46:6333-6338. [PubMed] [Google Scholar]
- 18.Ensoli, B., G. Barillari, S. Z. Salahuddin, R. C. Gallo, and F. Wong-Staal. 1990. Tat protein of HIV-1 stimulates growth of cells derived from Kaposi's sarcoma lesions of AIDS patients. Nature 345:84-86. [DOI] [PubMed] [Google Scholar]
- 19.Ensoli, B., L. Buonaguro, G. Barillari, V. Fiorelli, R. Gendelman, R. A. Morgan, P. Wingfield, and R. C. Gallo. 1993. Release, uptake, and effects of extracellular human immunodeficiency virus type 1 Tat protein on cell growth and viral transactivation. J. Virol. 67:277-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford, P. W., B. A. Bryan, O. F. Dyson, D. A. Weidner, V. Chintalgattu, and S. M. Akula. 2006. Raf/MEK/ERK signalling triggers reactivation of Kaposi's sarcoma-associated herpesvirus latency. J. Gen. Virol. 87:1139-1144. [DOI] [PubMed] [Google Scholar]
- 21.Friedman-Kien, A. E., L. J. Laubenstein, P. Rubinstein, E. Buimovici-Klein, M. Marmor, R. Stahl, I. Spigland, K. S. Kim, and S. Zolla-Pazner. 1982. Disseminated Kaposi's sarcoma in homosexual men. Ann. Intern. Med. 96:693-700. [DOI] [PubMed] [Google Scholar]
- 22.Gallo, R. C. 1998. Some aspects of the pathogenesis of HIV-1-associated Kaposi's sarcoma. J Natl. Cancer Inst. Monogr 1998:55-57. [DOI] [PubMed] [Google Scholar]
- 23.Gwack, Y., S. Hwang, C. Lim, Y. S. Won, C. H. Lee, and J. Choe. 2002. Kaposi's sarcoma-associated herpesvirus open reading frame 50 stimulates the transcriptional activity of STAT3. J. Biol. Chem. 277:6438-6442. [DOI] [PubMed] [Google Scholar]
- 24.Harrington, W., Jr., L. Sieczkowski, C. Sosa, S. Chan-a-Sue, J. P. Cai, L. Cabral, and C. Wood. 1997. Activation of HHV-8 by HIV-1 tat. Lancet 349:774-775. [DOI] [PubMed] [Google Scholar]
- 25.Heinrich, P. C., I. Behrmann, G. Muller-Newen, F. Schaper, and L. Graeve. 1998. Interleukin-6-type cytokine signalling through the gp130/Jak/STAT pathway. Biochem. J. 334:297-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho, I. C., M. R. Hodge, J. W. Rooney, and L. H. Glimcher. 1996. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell 85:973-983. [DOI] [PubMed] [Google Scholar]
- 27.Hou, J., U. Schindler, W. J. Henzel, T. C. Ho, M. Brasseur, and S. L. McKnight. 1994. An interleukin-4-induced transcription factor: IL-4 Stat. Science 265:1701-1706. [DOI] [PubMed] [Google Scholar]
- 28.Huang, L. M., M. F. Chao, M. Y. Chen, H. Shih, Y. P. Chiang, C. Y. Chuang, and C. Y. Lee. 2001. Reciprocal regulatory interaction between human herpesvirus 8 and human immunodeficiency virus type 1. J. Biol. Chem. 276:13427-13432. [DOI] [PubMed] [Google Scholar]
- 29.Husain, S. R., P. Leland, B. B. Aggarwal, and R. K. Puri. 1996. Transcriptional up-regulation of interleukin 4 receptors by human immunodeficiency virus type 1 Tat gene. AIDS Res. Hum. Retrovir. 12:1349-1359. [DOI] [PubMed] [Google Scholar]
- 30.Katano, H., and T. Sata. 2000. Human herpesvirus 8 virology, epidemiology and related diseases. Jpn. J. Infect. Dis. 53:137-155. [PubMed] [Google Scholar]
- 31.Keller, S. A., E. J. Schattner, and E. Cesarman. 2000. Inhibition of NF-κB induces apoptosis of KSHV-infected primary effusion lymphoma cells. Blood 96:2537-2542. [PubMed] [Google Scholar]
- 32.Kotanides, H., M. Moczygemba, M. F. White, and N. C. Reich. 1995. Characterization of the interleukin-4 nuclear activated factor/STAT and its activation independent of the insulin receptor substrate proteins. J. Biol. Chem. 270:19481-19486. [DOI] [PubMed] [Google Scholar]
- 33.Lefevre, E. A., R. Krzysiek, E. P. Loret, P. Galanaud, and Y. Richard. 1999. Cutting edge: HIV-1 Tat protein differentially modulates the B cell response of naive, memory, and germinal center B cells. J. Immunol. 163:1119-1122. [PubMed] [Google Scholar]
- 34.Li-Weber, M., P. Salgame, C. Hu, I. V. Davydov, O. Laur, S. Klevenz, and P. H. Krammer. 1998. Th2-specific protein/DNA interactions at the proximal nuclear factor-AT site contribute to the functional activity of the human IL-4 promoter. J. Immunol. 161:1380-1389. [PubMed] [Google Scholar]
- 35.Lu, C., G. M. Gordon, B. Chandran, B. J. Nickoloff, and K. E. Foreman. 2002. Human herpesvirus 8 reactivation and human immunodeficiency virus type 1 gp120. Arch. Pathol. Lab. Med. 126:941-946. [DOI] [PubMed] [Google Scholar]
- 36.Lu, C., Y. Zeng, Z. Huang, L. Huang, C. Qian, G. Tang, and D. Qin. 2005. Human herpesvirus 6 activates lytic cycle replication of Kaposi's sarcoma-associated herpesvirus. Am. J. Pathol. 166:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 38.Lund, T. C., R. Garcia, M. M. Medveczky, R. Jove, and P. G. Medveczky. 1997. Activation of STAT transcription factors by herpesvirus Saimiri Tip-484 requires p56lck. J. Virol. 71:6677-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marone, G., G. Florio, A. Petraroli, M. Triggiani, and A. de Paulis. 2001. Role of human FɛRI+ cells in HIV-1 infection. Immunol. Rev. 179:128-138. [DOI] [PubMed] [Google Scholar]
- 40.McAllister, S. C., S. G. Hansen, I. Messaoudi, J. Nikolich-Zugich, and A. V. Moses. 2005. Increased efficiency of phorbol ester-induced lytic reactivation of Kaposi's sarcoma-associated herpesvirus during S phase. J. Virol. 79:2626-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLemore, M. L., S. Grewal, F. Liu, A. Archambault, J. Poursine-Laurent, J. Haug, and D. C. Link. 2001. STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity 14:193-204. [DOI] [PubMed] [Google Scholar]
- 42.Merat, R., A. Amara, C. Lebbe, H. de The, P. Morel, and A. Saib. 2002. HIV-1 infection of primary effusion lymphoma cell line triggers Kaposi's sarcoma-associated herpesvirus (KSHV) reactivation. Int. J. Cancer 97:791-795. [DOI] [PubMed] [Google Scholar]
- 43.Mercader, M., B. Taddeo, J. R. Panella, B. Chandran, B. J. Nickoloff, and K. E. Foreman. 2000. Induction of HHV-8 lytic cycle replication by inflammatory cytokines produced by HIV-1-infected T cells. Am. J. Pathol. 156:1961-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Migone, T. S., J. X. Lin, A. Cereseto, J. C. Mulloy, J. J. O'Shea, G. Franchini, and W. J. Leonard. 1995. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science 269:79-81. [DOI] [PubMed] [Google Scholar]
- 45.Milligan, S., M. Robinson, E. O'Donnell, and D. J. Blackbourn. 2004. Inflammatory cytokines inhibit Kaposi's sarcoma-associated herpesvirus lytic gene transcription in in vitro-infected endothelial cells. J. Virol. 78:2591-2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 47.Muromoto, R., K. Okabe, M. Fujimuro, K. Sugiyama, H. Yokosawa, T. Seya, and T. Matsuda. 2006. Physical and functional interactions between STAT3 and Kaposi's sarcoma-associated herpesvirus-encoded LANA. FEBS Lett. 580:93-98. [DOI] [PubMed] [Google Scholar]
- 48.Pica, F., A. Volpi, A. Serafino, M. Fraschetti, O. Franzese, and E. Garaci. 2000. Autocrine nerve growth factor is essential for cell survival and viral maturation in HHV-8-infected primary effusion lymphoma cells. Blood 95:2905-2912. [PubMed] [Google Scholar]
- 49.Puri, R. K., and B. B. Aggarwal. 1992. Human immunodeficiency virus type 1 tat gene up-regulates interleukin 4 receptors on a human B-lymphoblastoid cell line. Cancer Res. 52:3787-3790. [PubMed] [Google Scholar]
- 50.Qian, C., D. Qin, Q. Tang, Y. Zeng, G. Tang, and C. Lu. 2006. Identification of a B-cell antigenic epitope at the N-terminus of SARS-CoV M protein and characterization of monoclonal antibody against the protein. Virus Genes 33:147-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice, A. P., and M. B. Mathews. 1988. Transcriptional but not translational regulation of HIV-1 by the tat gene product. Nature 332:551-553. [DOI] [PubMed] [Google Scholar]
- 52.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sarid, R., T. Sato, R. A. Bohenzky, J. J. Russo, and Y. Chang. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat. Med. 3:293-298. [DOI] [PubMed] [Google Scholar]
- 54.Scala, G., M. R. Ruocco, C. Ambrosino, M. Mallardo, V. Giordano, F. Baldassarre, E. Dragonetti, I. Quinto, and S. Venuta. 1994. The expression of the interleukin 6 gene is induced by the human immunodeficiency virus 1 TAT protein. J. Exp. Med. 179:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seaman, W. T., D. Ye, R. X. Wang, E. E. Hale, M. Weisse, and E. B. Quinlivan. 1999. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology 263:436-449. [DOI] [PubMed] [Google Scholar]
- 56.Selby, M. J., E. S. Bain, P. A. Luciw, and B. M. Peterlin. 1989. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 3:547-558. [DOI] [PubMed] [Google Scholar]
- 57.Song, J., T. Ohkura, M. Sugimoto, Y. Mori, R. Inagi, K. Yamanishi, K. Yoshizaki, and N. Nishimoto. 2002. Human interleukin-6 induces human herpesvirus-8 replication in a body cavity-based lymphoma cell line. J. Med. Virol. 68:404-411. [DOI] [PubMed] [Google Scholar]
- 58.Strathdee, S. A., P. J. Veugelers, and P. S. Moore. 1996. The epidemiology of HIV-associated Kaposi's sarcoma: the unraveling mystery. AIDS 10(Suppl. A):S51-S57. [PubMed] [Google Scholar]
- 59.Sturzl, M., C. Blasig, A. Schreier, F. Neipel, C. Hohenadl, E. Cornali, G. Ascherl, S. Esser, N. H. Brockmeyer, M. Ekman, E. E. Kaaya, E. Tschachler, and P. Biberfeld. 1997. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi's sarcoma. Int. J. Cancer 72:68-71. [DOI] [PubMed] [Google Scholar]
- 60.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Varthakavi, V., P. J. Browning, and P. Spearman. 1999. Human immunodeficiency virus replication in a primary effusion lymphoma cell line stimulates lytic-phase replication of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:10329-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varthakavi, V., R. M. Smith, H. Deng, R. Sun, and P. Spearman. 2002. Human immunodeficiency virus type-1 activates lytic cycle replication of Kaposi's sarcoma-associated herpesvirus through induction of KSHV Rta. Virology 297:270-280. [DOI] [PubMed] [Google Scholar]
- 63.Vieira, J., P. O'Hearn, L. Kimball, B. Chandran, and L. Corey. 2001. Acti-vation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J. Virol. 75:1378-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng, W., and R. A. Flavell. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89:587-596. [DOI] [PubMed] [Google Scholar]
- 65.Zhou, C., A. Saxon, and K. Zhang. 2003. Human activation-induced cytidine deaminase is induced by IL-4 and negatively regulated by CD45: implication of CD45 as a Janus kinase phosphatase in antibody diversification. J. Immunol. 170:1887-1893. [DOI] [PubMed] [Google Scholar]
- 66.Zoeteweij, J. P., S. T. Eyes, J. M. Orenstein, T. Kawamura, L. Wu, B. Chandran, B. Forghani, and A. Blauvelt. 1999. Identification and rapid quantification of early- and late-lytic human herpesvirus 8 infection in single cells by flow cytometric analysis: characterization of antiherpesvirus agents. J. Virol. 73:5894-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]