Abstract
The mechanisms regulating the synthesis of mRNA, cRNA, and viral genomic RNA (vRNA) by the influenza A virus RNA-dependent RNA polymerase are not fully understood. Early results suggested that the RNA polymerase “switched” from a transcriptase to a replicase during the viral life cycle in response to the expression of viral proteins. However, recently an alternative model suggesting that replication of influenza virus is regulated by stabilization of the replicative intermediates was proposed. According to this model, the virion-associated polymerase is capable of synthesizing both mRNA and cRNA. We now demonstrate that virion-derived viral ribonucleoproteins (vvRNPs) synthesize both mRNA and cRNA in vitro in the absence of non-virion-associated RNA polymerase or nucleoproteins. The authenticity of the in vitro-transcribed mRNA and cRNA was confirmed by terminal sequence analysis. The addition of non-virion-associated polymerase or NP had no effect on vvRNP activity. De novo synthesis of cRNA was found to be more sensitive than the capped primer-dependent synthesis of mRNA to the concentration of ATP, CTP, and GTP. We conclude that vvRNPs intrinsically possess both transcriptase and replicase activities and that there is no switch in the synthesis of mRNA to cRNA during the influenza virus life cycle.
The influenza A virus genome comprises eight segments of negative-sense RNA, encoding at least 11 different viral proteins (5). These include the polymerase subunits PB1, PB2, and PA, which form a heterotrimeric complex and, together with nucleoprotein (NP), associate with the viral genomic RNA (vRNA) to form viral ribonucleoproteins (vRNPs). The polymerase complex initiates both transcription and replication from the vRNA promoter, which is comprised of the 5′-terminal 13 nucleotides and the 3′-terminal 12 nucleotides of each gene segment (reviewed in reference 11). These 5′ and 3′ ends, which are conserved between all eight segments, display partial and inverted complementarity and have been proposed to take on a corkscrew structure by the formation of a duplex region and 5′- and 3′-terminal hairpin loops (9, 10, 23, 24).
Transcription of the genomic RNA (vRNA) template into mRNA is primed by capped RNA fragments which are cleaved from host cell mRNAs by an endonuclease activity associated with the polymerase complex (30). As a consequence, viral mRNAs contain a cap structure and 9 to 15 heterologous nucleotides at their 5′ ends. mRNA synthesis terminates at a sequence of five to seven uridines located 15 to 17 nucleotides from the 5′ end of vRNA templates, followed by the addition of a poly(A) tail (16, 36). In contrast, replication entails the synthesis of cRNA, which is a full-length copy of vRNA. It is neither capped nor polyadenylated and functions as a template for vRNA synthesis. Both cRNA and vRNA have a triphosphorylated nucleotide at their 5′ terminus (17), which implies that the initiation of their synthesis occurs without a primer.
The control of transcription and replication of viral RNA in vivo is not well understood. The viral life cycle comprises an early transcriptive phase followed later by a predominantly replicative phase. Treatment of infected cells with cycloheximide, an inhibitor of protein synthesis, prevents the switch from the transcriptive to the replicative phase, suggesting that replication requires de novo protein synthesis (1, 16, 43). In vitro, purified vRNPs have been found to be capable of synthesizing mRNA (30) but not cRNA, except in the context of infected cell extracts containing soluble viral proteins (3, 41). Viral nucleoprotein (NP), which is associated with viral RNA, was identified as a prime candidate for a switching molecule based on several temperature-sensitive NP mutants defective in replication and RNA binding and biochemical studies that suggested that NP is required for the synthesis of cRNA by preventing premature termination (3, 25, 38). It was proposed that interactions of NP with the polymerase (4, 26) or with the promoter element of the template RNA (21) may alter the mode of transcriptional initiation. PB2 and PA have similarly been implicated in the switch from studies of mutations affecting viral replication but not transcription (15, 18). Various cellular factors that are purported to possibly play a role have also been identified (27, 28, 40). However, the mechanism by which expression of any of these proteins may achieve a switch in activity remained unknown.
We recently demonstrated, by preexpression of viral polymerase and NP, that the virion-associated polymerase synthesizes both mRNA and cRNA during primary transcription in vivo (44). In addition, we found that detection (or “rescue”) of cRNA depended primarily on the preexpression of polymerase and that mutations inhibiting the RNA binding activity of the polymerase inhibited the rescue of cRNA. Therefore, we hypothesized that there is no switch between the synthesis of mRNA and cRNA per se. Rather, the expression of polymerase and NP is required to bind nascent cRNA to protect it from degradation by host cell nucleases. Here, additional evidence in support of this hypothesis is provided. It is demonstrated that preexpressed polymerase and NP bind nascent cRNA synthesized during primary transcription in vivo to form cRNPs. Furthermore, vRNPs isolated from virions are shown to synthesize both mRNA and cRNA in vitro in the absence of any non-virion-associated proteins.
MATERIALS AND METHODS
Strains and culture conditions.
Influenza A/WSN/33 virus stocks were prepared and titrated in MDBK cell monolayers maintained in minimal essential medium with 10% fetal calf serum and antibiotics at 37°C with 5% CO2. All in vivo assays were carried out using human 293T cells under similar culture conditions.
Plasmids.
pcDNA-PB1-D445A/D446A (44), pcDNA-PB2, pcDNA-PA, pcDNA-PA-His6, pcDNA-NP (12), and pcDNA-PA-(His6)TAP (22; abbreviated here as pcDNA-PA-tap for convenience) have been described previously. pcDNA-tap-NP, which expresses NP with an N-terminal TAP tag (33), was provided by Tao Deng.
cRNA rescue and copurification with his-tagged PA.
Transfections of 293T cells in 35-mm dishes and viral infections at a multiplicity of infection of 5 in the presence of 100 μg/ml cycloheximide were carried out as described previously (44). At 3 h postinfection, the cells were washed twice in 1 ml ice-cold phosphate-buffered saline, resuspended in 0.4 ml lysis buffer (50 mM Tris-HCl pH 8.0, 200 mM NaCl, 25% glycerol, 0.5% Igepal CA-630 [Sigma], 1 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 complete mini-EDTA-free protease inhibitor cocktail tablet [Roche] per 10 ml), and incubated on ice for 30 min. All subsequent steps were performed at 4°C. The cell lysate was centrifuged at 16,000 × g for 10 min. The supernatant was transferred into a sealed 1-ml Mobicol (MoBiTec) tube with a 90-μm filter, containing Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN) from a 100-μl-volume suspension prewashed twice in wash buffer (50 mM Tris-HCl, pH 8.0, 200 mM NaCl, 10% glycerol, 1 mM β-mercaptoethanol, 0.1 mM PMSF, and 5 mM imidazole). Imidazole was then added to 5 mM. After gentle mixing for 3 h, the flowthrough was collected by centrifugation at 400 × g for 10 s. The Ni-NTA agarose pellet was washed four times with 0.5 ml wash buffer. RNA was extracted from the flowthrough (unbound fraction) and the Ni-NTA agarose (bound fraction) with 1 ml and 0.25 ml Tri reagent (Ambion), respectively. RNA in the bound fraction was precipitated in the presence of 20 μg glycogen as carrier. One-twentieth of each of the RNA samples was analyzed by primer extension.
Primer extension analysis.
RNA was mixed with 0.1 pmol each of two 32P-labeled DNA primers in 5 μl and denatured by heating at 95°C for 5 min. The mixture was cooled on ice and transferred to 45°C for 2 min prior to the addition of 5 μl of a 2× transcription mix (2× first-strand buffer [Invitrogen], 20 mM dithiothreitol [DTT], 1 mM deoxynucleoside triphosphate mix, 50 U SuperScript II RNase H− reverse transcriptase [Invitrogen]) prewarmed at 45°C for 1 min. The reaction was stopped after 1.5 h by the addition of 8 μl 90% formamide, 10 mM EDTA, heated at 95°C for 3 min, and analyzed on 6% polyacrylamide gels containing 7 M urea in Tris-borate-EDTA buffer. Transcription products were visualized by autoradiography. The NA gene-specific primers were 5′-TGGACTAGTGGGAGCATCAT-3′ (to detect negative-sense RNA) and 5′-TCCAGTATGGTTTTGATTTCCG-3′ (to detect positive-sense RNA). The NS gene-specific primers were 5′-TGATTGAAGAAGTGAGACACAG-3′ and 5′-CGCTCCACTATTTGCTTTCC-3′, respectively.
Terminal sequencing of viral RNAs.
The 5′ and 3′ termini of influenza virus RNA were sequenced according to the method described by Szymkowiak et al. (42). Briefly, RNA isolated by Tri reagent extraction and isopropanol precipitation was treated with tobacco acid pyrophosphatase (Epicenter) in the provided reaction buffer for 1 h at 37°C. Following reextraction with Tri reagent, the RNA was circularized by intramolecular ligation of the 5′-phosphoryl and 3′-hydroxyl ends using T4 RNA ligase (Epicenter) in the provided reaction buffer with 1 mM ATP for 1 h at 37°C. A reverse transcription reaction was performed as described above using 2.5 pmol of either the unlabeled cRNA- or vRNA-specific primer (as specified above). The first-strand cDNA was amplified by PCR using Taq polymerase (Promega) with both the cRNA- and vRNA-specific primers (as specified above). Following agarose gel electrophoresis, amplicons of the expected size were cut out, extracted by Qiaquick gel extraction (QIAGEN), and cloned by TOPO TA cloning (Invitrogen) according to the manufacturers' instructions. Colonies were screened for plasmids with the expected-size inserts by PCR, and minipreps of overnight cultures were then sequenced.
Virion vRNP preparations.
Viral cores were prepared essentially as described previously (37). Briefly, five 175-cm2 flasks of 90% confluent MDBK cells were infected with influenza A/WSN/33 virus at a multiplicity of infection of 0.01 and incubated for 48 h at 37°C. The infected cell medium was collected, and the cell debris was pelleted by centrifugation at 2,000 × g for 30 min at 4°C. The supernatant was further clarified by centrifugation in SW28 tubes at 10,000 rpm for 30 min at 4°C and then loaded onto precooled 30% sucrose cushions in SW28 tubes. Virions were pelleted by centrifugation at 25,000 rpm for 1.5 h at 4°C and lysed by incubation in a disruption buffer (100 mM Tris-HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 3% Triton X-100, 5% glycerol, freshly diluted 10 mg/ml lysolecithin, and 1.5 mM DTT) at 30°C for 30 min. The viral lysate was fractionated by centrifugation on a discontinuous glycerol gradient (1 ml each 70%, 50%, 40%, and 33%) in 50 mM Tris-HCl (pH 7.5) and 150 mM NaCl. The gradients were centrifuged at 45,000 rpm in a Beckman SW55Ti rotor for 4 h at 4°C. Fractions of approximately 300 μl were collected dropwise from the bottom of the tube. Proteins were analyzed by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) and silver stained. Three fractions enriched with RNP cores were pooled and used for in vitro transcription reactions.
In vitro vvRNP polymerase reactions.
Two microliters of vvRNP (approximately 0.5 μg influenza virus core protein) was normally incubated at 30°C overnight (for maximum sensitivity) in the presence of 1 mM ATP, 0.5 mM of each CTP, GTP, and UTP, 5 mM MgCl2, 1 mM DTT, and 1 U RNAsin (Promega), with globin mRNA (Sigma) or 0.5 mM ApG as indicated, in a 4-μl reaction. RNA was extracted with Tri reagent and isopropanol precipitated with 20 μg glycogen carrier before analysis by NA or NS gene-specific primer extension.
Preparation of partially purified recombinant influenza virus RNA polymerase and NP.
pcDNA plasmids expressing an active-site mutant of PB1 (PB1-D445A/D446A), wild-type PB2, C-terminally tap-tagged PA, or N-terminally tap-tagged NP or empty plasmids were transfected into 293T cells in suspension using Lipofectamine 2000 (Invitrogen). After 48 h at 37°C, the cells were lysed in buffer containing 50 mM HEPES, pH 8.0, 200 mM NaCl, 50% glycerol, 0.5% Igepal CA-630 (Sigma), 1 mM β-mercaptoethanol, 0.1 mM PMSF, and 1 complete mini-EDTA-free protease inhibitor cocktail tablet (Roche) per 10 ml. The tap-tagged trimeric polymerase and/or NP were affinity purified on immunoglobulin G-Sepharose and washed with a buffer containing 10 mM HEPES, pH 8.0, 150 mM NaCl, 10% glycerol, 0.1% Igepal CA-630 (Sigma), and 0.1 mM PMSF. The partially purified proteins were released by cleavage with tobacco etch virus (TEV) protease in wash buffer containing 1 mM dithiothreitol and were stored at −20°C in 35% glycerol.
RESULTS
Preexpressed viral polymerase and NP binds full-length cRNA synthesized by the infecting virion during primary transcription.
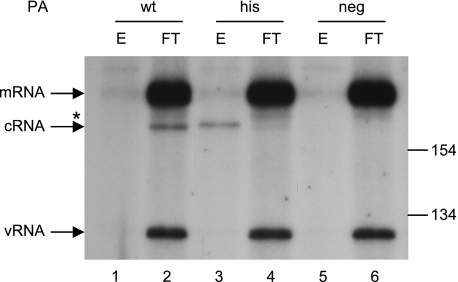
We have previously shown that cRNA can be detected in 293T cells infected with influenza A virus in the presence of cycloheximide if viral polymerase and NP are preexpressed (44). Detection, or rescue, of cRNA was dependent on the competence of the preexpressed viral polymerase to bind viral RNA. This suggested that cRNA was bound to viral polymerase and NP in a cRNP complex. To demonstrate this directly, we have preexpressed viral polymerase and NP as before, but with a his-tagged PA subunit. Following virus infection in the presence of cycloheximide, the PA subunit was immobilized on nickel agarose. Figure 1 shows, by NA gene-specific primer extension, that cRNA copurifies with the immobilized his-tagged polymerase (lane 3), whereas mRNA and the virion-derived vRNA are in the flowthrough fraction (lane 4). The slightly retarded migration of the copurified cRNA-specific band (lane 3) was found in mixing experiments to be an artifact brought about by the differences in primer extension reaction conditions (data not shown). As shown previously (44), cRNA was only detected if trimeric polymerase and NP were preexpressed (compare lanes 1 and 2 with lanes 5 and 6). Copurification of the rescued cRNA was dependent on the preexpressed polymerase possessing a his tag (compare lanes 1 and 2 with lanes 3 and 4). This demonstrates that the rescued cRNA is bound exclusively and completely by the preexpressed polymerase.
FIG. 1.
Influenza virus polymerase and NP bind nascent full-length cRNA synthesized during primary transcription. 293T cells were transfected with plasmids expressing an active-site mutant of PB1 (PB1-D445A/D446A), wild-type PB2 and NP, and either wild-type (wt), his-tagged (his), or no (neg) PA, as indicated. Twelve to 14 h later the cells were infected with influenza A/WSN/33 for 3 h in the presence of cycloheximide. The cell lysates were separated by nickel agarose column chromatography, and the total RNA in the bound (E) and unbound (FT) fractions was isolated and analyzed by NA gene-specific primer extension. The positions of the size markers and the NA vRNA-, cRNA-, and mRNA-specific signals are indicated. The asterisk indicates minor unknown products.
In order to test whether the rescued cRNA represented full-length copies of the genomic RNA, 5′ and 3′ termini of NA and NS gene-specific cRNA isolated by nickel column copurification were amplified by RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) as described by Szymkowiak et al. (42). Sequencing of 20 individual clones clearly demonstrated that all but one of the rescued cRNA transcripts possessed intact 5′ (5′ AGCAAAAGC…)- and 3′ (…UGUUUCUACU 3′)-terminal sequences (Table 1).
TABLE 1.
Number of reverse transcription-PCR clones derived from in vivo-rescued cRNA (NA or NS gene specific) with full-length or shortened terminia
| Terminus | No. of clones tabulated according to no. of terminal nucleotides deleted
|
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| 5′ (NA, NS) | 20 (7, 13) | 0 (0, 0) | 0 (0, 0) | 0 (0, 0) |
| 3′ (NA, NS) | 19 (7, 12) | 1 (0, 1) | 0 (0, 0) | 0 (0, 0) |
The numbers of NA- and NS-specific clones sequenced are indicated separately in parentheses.
Therefore, these data extend our previous findings and support the hypothesis that full-length cRNA is transcribed by the virion polymerase early in infection (44).
Virion vRNPs transcribe both mRNA and cRNA in vitro.
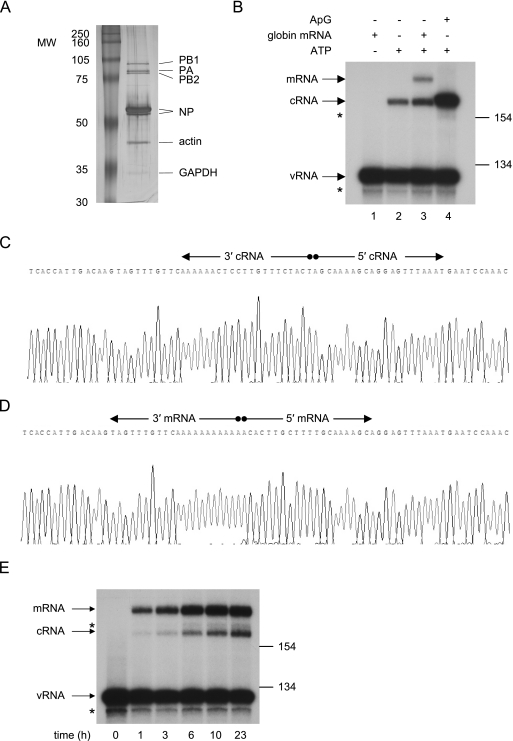
In order to obtain further evidence in support of the hypothesis that there is no switch, per se, between the transcription and replication activities of the influenza A viral polymerase during infection, we sought to develop an in vitro polymerase assay to demonstrate that, in the absence of any added cellular or viral factors, virion vRNPs (vvRNPs) would synthesize both cRNA and mRNA. Influenza A/WSN/33 vvRNPs were purified from virions isolated from infected MDBK cell supernatants and analyzed by SDS-PAGE (Fig. 2A). Besides the viral polymerase and NP, additional protein bands in the vvRNP preparations were identified by liquid chromatography-tandem mass spectrometry and included a presumably differentially phosphorylated (19) or cleaved (45) form of NP, matrix protein (M1; data not shown), actin, and trace amounts of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The presence of any non-vvRNP-associated polymerase or NP in the vvRNP preparations was deemed unlikely. An in vitro polymerase reaction was carried out using the purified vvRNPs in the presence or absence of either an ApG primer or globin mRNA. The viral RNA species were then analyzed by NA gene-specific primer extension (Fig. 2B). The input vRNA is shown in the negative control, which lacked ATP (lane 1). Following incubation in the presence of all four nucleoside triphosphates (NTPs) but in the absence of any primers, a band of the expected length for cRNA was detected (lane 2). If globin mRNA was added to the polymerase reaction, an additional slower migrating band corresponding in length to viral mRNA primed with a capped leader endonucleolytically cleaved from globin mRNA was also detected (lane 3). As expected, the addition of an ApG primer to the polymerase reaction yielded a strong cRNA-specific signal (lane 4). Surprisingly, however, the cRNA-specific band derived from ApG-primed transcription (lane 4) appeared to migrate marginally slower than the cRNA-specific band derived from unprimed transcription (lanes 2 and 3; see below). Nevertheless, these results demonstrate simultaneous primed and unprimed transcription of positive-sense RNA by vvRNPs.
FIG. 2.
Influenza virion-derived RNPs carry out de novo and globin mRNA-primed transcription in vitro. vRNPs were extracted from virion particles isolated from infected MDBK cell supernatants and purified by glycerol gradient centrifugation. (A) SDS-PAGE analysis of the purified vvRNP pool (approximately 2.5 μg virus core protein), visualized by silver staining. The molecular sizes of the protein markers are indicated in kilodaltons. The identities of the major protein bands in the vvRNP pool (except influenza matrix protein M1, which is not visible on this gel) are indicated. (B) Primer extension analysis of NA gene-specific RNA following in vitro transcription by purified vvRNPs. Transcription was carried out for 14.5 h in the presence of 18 ng/μl globin mRNA (lanes 1 and 3), 0.5 mM ApG (lane 4), or no primer (lane 2). ATP was omitted in the negative control (lane 1). The positions of the size markers (in bases) and the NA vRNA-, cRNA-, and mRNA-specific signals are indicated. The asterisks indicate minor unknown products. (C) A sequence trace of the termini of full-length in vitro-synthesized cRNA (derived from panel B, lane 2), circularized and amplified by RLM-RACE. The ligated 5′- and 3′-terminal sequences of cRNA are indicated. (D) A sequence trace of the termini of in vitro-synthesized mRNA (derived from panel B, lane 3), circularized and amplified by RLM-RACE. The ligated 5′- and 3′-terminal sequences of mRNA are indicated. (E) Primer extension analysis of NA gene-specific RNA following in vitro vvRNP polymerase reactions carried out in the presence of 45 ng/μl globin mRNA for different times, as indicated. The positions of the size markers (in bases) and the NA vRNA-, cRNA-, and mRNA-specific signals are indicated. The asterisks indicate minor unknown products.
Although these results suggested that the synthesis of cRNA occurred by de novo transcription, it was not clear from the primer extension analysis whether the unprimed transcripts represented full-length copies of the virion vRNA. Therefore, the termini of NA (Fig. 2C) and NS (data not shown) gene-specific transcripts synthesized in vitro in the absence of primer were sequenced according to the RLM-RACE method described by Szymkowiak et al. (42). Of the 34 clones sequenced (Table 2), unexpectedly, only 2 had intact 5′ termini (5′ AGCAAAAGC…). Twenty-one of the remaining 32 clones lacked the 5′-terminal A residue, and 10 clones lacked both the 5′-terminal A and G residues. However, all were found to have intact 3′ termini (…UGUUUCUACU 3′), demonstrating that, as expected during de novo transcription (3), antitermination at the polyadenylation site had occurred. However, these data suggested that, surprisingly, initiation of cRNA synthesis in vitro had taken place predominantly at nucleotide 2 of the vRNA template. This explained the small difference in mobility between the cRNA and ApG-primed products observed in Fig. 2B.
TABLE 2.
Number of reverse transcription-PCR clones derived from in vitro-transcribed cRNA (NA or NS gene specific) with full-length or shortened terminia
| Terminus | No. of clones tabulated according to no. of terminal nucleotides deleted
|
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| 5′ (NA, NS) | 2 (1, 1) | 21 (10, 11) | 10 (7, 3) | 1 (0, 1) |
| 3′ (NA, NS) | 34 (18, 16) | 0 (0, 0) | 0 (0, 0) | 0 (0, 0) |
The numbers of NA- and NS-specific clones sequenced are indicated separately in parentheses.
Due to the unexpected nature of these data, the 5′-terminal sequences of the in vitro-transcribed cRNA were confirmed by an independent, direct approach, namely by dideoxynucleotide incorporation during primer extension analysis (data not shown).
A possible explanation for the unexpected finding that cRNA synthesis initiated primarily at residue 2 was that the vRNA template lacked the 3′-terminal residue. Therefore, the termini of the NA- and NS-specific template virion vRNA in two different vvRNP preparations were characterized as before (Table 3). The majority (24 of 35) of the clones sequenced indeed lacked 1 nucleotide at the 3′ terminus, with only 3 of the 35 clones having intact 3′ termini (…GCUUUUGCU 3′). All of the clones had intact 5′ termini (5′ AGUAGAAACA…). As a control, the 5′ and 3′ termini of the NA and NS gene-specific vRNA derived from cells infected with influenza virus in the presence of cycloheximide (Fig. 1, lane 4) were similarly characterized (Table 4). This vRNA is assumed to derive from the infectious virions, as cycloheximide treatment prevents the virus from entering the replicative phase and synthesizing new vRNA. Here, based on the sequencing of 18 clones, all of the vRNA derived from infecting virions also possessed intact 5′-terminal sequences (5′ AGUAGAAACA…). However, only 50% of the vRNA possessed intact 3′-terminal sequences (…GCUUUUGCU 3′), the other 50% lacking the 3′-terminal U residue. We have obtained similar results from the characterization of vRNA extracted directly from the medium of infected cells (data not shown). These results are consistent with the findings of Szymkowiak et al. (42), who reported deletions at the extreme ends of the gene segments, the most common being the first nucleotide from the 3′ end.
TABLE 3.
Number of reverse transcription-PCR clones derived from virion vRNA in viral core extracts (NA or NS gene specific) with full-length or shortened terminia
| Terminus | No. of clones tabulated according to no. of terminal nucleotides deleted
|
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| 5′ (NA, NS) | 35 (27, 8) | 0 (0, 0) | 0 (0, 0) | 0 (0, 0) |
| 3′ (NA, NS) | 3 (0, 3) | 24 (19, 5) | 8 (8, 0) | 0 (0, 0) |
The numbers of NA- and NS-specific clones sequenced are indicated separately in parentheses.
TABLE 4.
Number of reverse transcription-PCR clones derived from virion vRNA in infected cells (NA or NS gene specific) with full-length or shortened terminia
| Terminus | No. of clones tabulated according to no. of terminal nucleotides deleted
|
|||
|---|---|---|---|---|
| 0 | 1 | 2 | 3 | |
| 5′ (NA, NS) | 18 (4, 14) | 0 (0, 0) | 0 (0, 0) | 0 (0, 0) |
| 3′ (NA, NS) | 9 (2, 7) | 9 (2, 7) | 0 (0, 0) | 0 (0, 0) |
The numbers of NA- and NS-specific clones sequenced are indicated separately in parentheses.
Finally, the termini of the in vitro-transcribed NA mRNA were also determined by RLM-RACE (42) (Fig. 2D). As expected (30, 35), these transcripts possessed 13 5′-terminal residues (plus the cap) derived from β-globin mRNA, followed by NA gene-specific sequences probably transcribed from the third residue of the 3′ terminus of the vRNA template [m7Gpppm6AmC(m)ACUUGCUUUUGCAAAAGC…]; underlined sequence derives from the capped primer. All capped NA gene-specific transcripts characterized (35 clones) were prematurely terminated and/or polyadenylated by reiterative copying at the six U residues 15 nucleotides from the 5′ end of the vRNA template (i.e., no antitermination had occurred), as expected (31). A tails varying in length up to 89 A residues were detected (data not shown). These data confirmed that the in vitro-synthesized cap-primed transcripts were authentic viral mRNA.
The sensitivity of the polymerase assay was examined by measuring the accumulation of NA- and NS-specific mRNA and cRNA signals over time. Both mRNA- and cRNA-specific transcripts were detected after 1 h of incubation by NA gene-specific primer extension analysis, increasing to 24 h (Fig. 2E). Similar profiles were observed for the accumulation of NS gene-specific transcripts (data not shown). The increased ratio of mRNA- to cRNA-specific signals in Fig. 2E compared to Fig. 2B is due to the increased concentration of globin mRNA used in the time course experiment (Fig. 2E). Similarly, preliminary experiments suggest that a minor temporal decrease in the mRNA to cRNA ratio may be related to the fact that transcription, but not replication, requires endonucleolytic cleavage of globin mRNA.
Overall, we conclude that, in vitro, vvRNPs can synthesize full-length cRNA (in the absence of any primers) as well as authentic polyadenylated mRNA (by snatching of capped primers). In addition, these data also suggest, surprisingly, that virion vRNA segments lacking the 3′-terminal U residue can be copied in vitro to cRNA that lacks the corresponding 5′-terminal A residue but is otherwise full length.
Replication and transcription have different NTP concentration requirements.
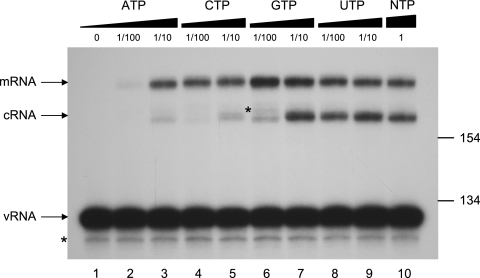
It has previously been found that influenza virus requires a higher ATP concentration for transcriptional initiation than for elongation (20). As transcription entails the elongation of an mRNA-derived capped primer whereas replication requires de novo initiation, we examined whether there was a difference in the requirement of the four different NTPs for the in vitro synthesis of mRNA and cRNA by vvRNPs (Fig. 3). The levels of NA mRNA and cRNA synthesized in vitro at 1 mM ATP and 0.5 mM of each CTP, GTP, and UTP were taken as standard (lane 10). A 1/100 dilution of ATP to 10 μM (lane 2) severely inhibited both mRNA and cRNA levels. However, mRNA transcription was otherwise largely uninhibited by 1/10 or 1/100 dilution of any NTP (lanes 3 to 9). In contrast, cRNA synthesis was strongly inhibited by 1/10 dilution of ATP (100 μM; lane 3) or CTP (50 μM; lane 5) or by 1/100 dilution of GTP (5 μM; lane 6). A 1/10 or 1/100 dilution of UTP (lanes 8 and 9) had little effect on the levels of cRNA synthesized in vitro. Therefore, a relatively higher concentration of ATP, CTP, and GTP is required for de novo replication than for elongation of a capped primer during mRNA transcription. Intriguingly, ATP, GTP, and CTP are the first three nucleotides at the 5′ end of cRNA transcripts, whereas the first UTP residue is at position 13 in NA cRNA (see Discussion).
FIG. 3.
vvRNPs have different nucleotide concentration requirements for in vitro mRNA and cRNA synthesis. In vitro vvRNP transcription reactions were carried out in the presence of 18 ng/μl globin mRNA with varying concentrations of NTPs and analyzed by NA gene-specific primer extension. Lane 10 represents in vitro transcription under standard conditions (1 mM ATP and 0.5 mM of each CTP, GTP, and UTP). The dilution of individual NTPs relative to the standard conditions is indicated. The positions of the size markers (in bases) and the NA vRNA-, cRNA-, and mRNA-specific signals are indicated. The asterisks indicate minor unknown products.
Addition of non-vvRNP-associated polymerase or NP does not affect the transcription or replication activities of vvRNPs.
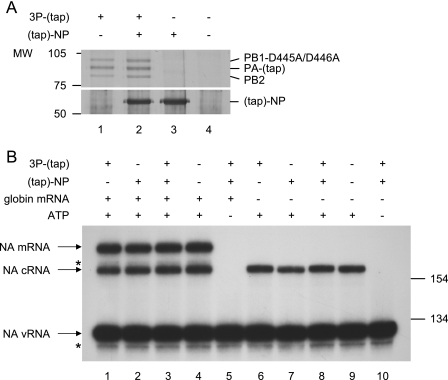
It was long believed that the expression of “free” NP, i.e., NP that is not associated with the virion vRNP, is essential for viral replication (3, 38). In contrast, we have previously shown that the expression of viral polymerase, and to a lesser extent the coexpression of NP, is required for the detection of cRNA synthesis during influenza virus infection of cells (44). In order to examine the effect of viral polymerase and NP on the in vitro activity of vvRNPs, different combinations of an active-site mutant of the trimeric viral polymerase and NP were partially purified from transiently transfected 293T cells by tap tag affinity chromatography. PA-tap and tap-NP have been shown to retain significant replication and transcription activities compared to the wild type both in vivo in transient transfection systems and in vitro (7, unpublished data) and to be compatible with our previously described (44) rescue of cRNA (unpublished data). An active-site mutant of the viral polymerase was used to ensure that the viral transcription and replication signals were derived exclusively from vvRNP-associated polymerase activity. Following purification, the tap tags were removed by TEV cleavage, yielding PA with a 26-amino-acid C-terminal extension and NP with a 4-amino-acid N-terminal extension. Aliquots of each of these partially purified extracts (Fig. 4A, lanes 1 to 4) were added to the in vitro vvRNP polymerase assay in the presence or absence of globin mRNA, and the NA gene-specific transcripts were analyzed by primer extension (Fig. 4B). The addition of partially purified extracts of viral polymerase (lanes 1 and 6), NP (lanes 2 and 7), or polymerase and NP (lanes 3 and 8) produced no noticeable difference in vvRNP transcription and/or replication compared to negative controls (lanes 4 and 9). Therefore, it appears that the addition of non-vvRNP-associated polymerase or NP does not alter the in vitro activity of vvRNP.
FIG. 4.
Addition of non-virion-associated viral polymerase and NP has no effect on the in vitro transcription and replication activity of vvRNPs. (A) SDS-PAGE analysis of partially purified extracts of RNA polymerase (3P-tap, comprising PB1-D445A/D446A, PB2, and PA-tap; lane 1), RNA polymerase and NP (lane 2), NP (lane 3), or a mock control (lane 4) derived from 293T cells transiently transfected with the relevant expression plasmids and visualized by silver staining. The positions of PB1-D445A/D446A, PA-tap, PB2, and tap-NP are indicated. The tap tags are shown in parentheses to indicate that the bulk of the tags were removed during purification. The molecular sizes of the protein markers are indicated in kilodaltons. (B) NA gene-specific primer extension analysis of in vitro vvRNP polymerase reactions carried out in the presence or absence of 18 ng/μl globin mRNA and the partially purified extracts shown in panel A. In vitro vvRNP polymerase activity was assayed in the presence of partially purified RNA polymerase derived from lane 1 of panel A (lanes 1 and 6), partially purified NP derived from lane 3 of panel A (lanes 2 and 7), partially purified RNA polymerase and NP derived from lane 2 of panel A (lanes 3, 5, 8, and 10), or partially purified extracts of mock-transfected cells derived from lane 4 of panel A (lanes 4 and 9). Lanes 5 and 10 are negative controls of in vitro vvRNP polymerase activity in the absence of ATP. The positions of the size markers (in bases) and the NA vRNA-, cRNA-, and mRNA-specific signals are indicated. The asterisks indicate minor unknown products.
DISCUSSION
The control of influenza viral transcription and replication has remained a conundrum despite more than two decades of research. Following infection, the virion-derived polymerase appears to initially synthesize exclusively mRNA which, it was thought, must be translated to viral proteins, specifically NP, before replication of the viral genome into cRNA commences (3, 38). However, we have previously demonstrated that, if viral polymerase and NP are preexpressed, both cRNA and mRNA synthesis can be detected early in the viral infection cycle (44). This rescue of cRNA was shown to be dependent on the RNA binding activity of the free viral polymerase. We hypothesized that free viral polymerase and NP binds nascent cRNA, thereby stabilizing and protecting it from degradation. In order to test this hypothesis, we have in this report preexpressed a tagged polymerase and NP to rescue cRNA in vivo and were able to demonstrate, by copurification, that the preexpressed polymerase and NP do indeed bind exclusively to the nascent cRNA (Fig. 1). The bound NA and NS gene-specific cRNA were characterized by sequencing of both the 5′ and 3′ termini and were shown to be authentic full-length copies of vRNA (Table 1). It has previously been shown (29, 39) that influenza virus polymerase binds viral mRNA. However, these experiments were carried out in vitro using synthetic truncated viral mRNA, and it could not be concluded whether binding of authentic viral mRNA by the viral polymerase occurs in vivo. Our present data (Fig. 1) suggest that, if such binding occurs in vivo, it is at undetectable levels. Similarly, our finding that no vRNA copurified with the preexpressed tagged polymerase (Fig. 1) suggests that the virion-derived vRNA remained inaccessible for competitive binding by the preexpressed polymerase. This would imply, perhaps surprisingly (13, 32), that the 5′ end of template vRNA is not released by the virion-derived viral polymerase during transcription or replication. Overall, these data support our stabilization model for the switch from transcription to replication during the viral life cycle (44).
However, it might be argued that preexpression of viral proteins could alter the cellular conditions prior to infection or the virion-derived polymerase upon infection, thereby artifactually inducing the transcription of both mRNA and cRNA. We have, therefore, developed an in vitro assay using virion-derived vRNPs to test the activity of the polymerase in the absence of any added cellular or free non-vvRNP-associated viral factors. This polymerase assay uses primer extension to analyze transcripts synthesized in vitro by the polymerase activity of purified vvRNPs on the intrinsic vRNA templates. The advantage of this approach was that we were not constrained by the need to add a radiolabeled NTP to the transcription reaction and the concentration of all the NTPs could therefore be easily controlled. The in vitro-synthesized transcripts could also be further characterized by sequencing of their termini. In this manner, polyadenylated transcripts initiated from capped primers endonucleolytically cleaved from globin mRNA, and full-length transcripts synthesized in the absence of added primer, were detected (Fig. 2B). Therefore, we conclude that virion-derived polymerase both transcribes (mRNA synthesis) and replicates (cRNA synthesis) the intrinsic negative-sense vRNA templates.
The 3′-terminal residues of vRNA (of all gene segments) are 3′ UCGC/UUUUCGU…, which are replicated to 5′ pppAGCG/AAAAGCA… in cRNA by de novo initiation with ATP. Surprisingly, however, sequencing of the 5′ and 3′ termini revealed that a significant proportion of the template RNA in vRNPs derived from virions, whether obtained by extraction from purified virions (Table 3) or from cycloheximide-treated infected cells (Table 4), lacked the 3′-terminal U residue. The cause of these 3′-terminal deletions in the virion vRNA, also found by Szymkowiak et al. (42), is not clear. Intriguingly, however, our data suggest that, in vitro, vRNA lacking the 3′-terminal U residue can be copied by de novo initiation to give rise to cRNA that lacks the corresponding 5′-terminal A residue but is otherwise full length (Table 2). In contrast, cRNA synthesized by vvRNP in vivo and rescued by tagged polymerase and NP is almost exclusively full length (Table 1). One possible explanation for these apparently contradictory results is that cRNA transcripts lacking the 5′-terminal A residue, if synthesized in vivo, would not be expected to be recognized by free viral polymerase (6) and would, we speculate, probably be degraded by cellular exonucleases (44). On the other hand, full-length cRNA transcripts synthesized from intact vvRNPs in vivo are bound by free viral polymerase and NP to form cRNPs and would thus be selectively protected from degradation by such exonucleases.
While the synthesis of nonfunctional cRNA transcripts destined for degradation might be considered “wasteful,” virion vRNAs with a 3′-terminally deleted residue would still serve as templates for the synthesis of fully functional mRNA. Transcription of viral mRNA utilizes a capped primer endonucleolytically cleaved from host mRNA approximately 10 to 13 nucleotides from their 5′ caps, usually after a purine residue (2, 30). It has been shown that the influenza viral polymerase shows some preference for CA-terminated capped fragments as primers for transcriptional initiation, by extension with a G residue directed by the penultimate C nucleotide at the 3′ end of the vRNA template, i.e., m7GpppN(8-11)CAGCG/AAAAGCA… (2, 34). However, initiation with 3′ G-terminated capped primer by incorporation of a C residue, directed by the G residue at position 3 in the vRNA, has also been observed (14). Indeed, as shown here (Fig. 2D) and elsewhere (30, 35), capped fragments endonucleolytically cleaved from globin mRNA probably after the G residue at position 13 serve as suitable primers for the initiation of in vitro mRNA transcription on the third residue from the 3′ end of the vvRNP template, i.e., 5′ m7Gpppm6AmC(m)ACUUGCUUUUGCG/AAAAGCA… . Therefore, it can be envisaged that virion vRNPs lacking the 3′-terminal U residue would presumably be functional templates for mRNA transcription in vivo with a subset of endonucleolytically cleaved capped primers (those with a 3′-terminal G residue) whose elongation is independent of the 3′-terminal U residue of the vRNA template.
Preliminary in vivo experiments studying viral polymerase-driven replication and transcription of RNA polymerase I-derived vRNA or cRNA templates lacking a 3′-terminal U or a 5′-terminal A residue, respectively, support these views. Specifically, deletion of the 5′-terminal U residue rendered cRNA templates inactive, whereas vRNA with a 3′-terminal U residue deletion was still a capable template for transcription but not for replication (data not shown). Interestingly, 3′-terminal U residue deletions of cRNA are known to be corrected by the internal prime and realignment mechanism used by influenza virus polymerase for the initiation of replication on a cRNA promoter (8).
It has previously been reported that influenza virus transcription requires a higher ATP concentration for initiation than for elongation (20), but these observations were based on the incorporation of α-32P-labeled CTP or GTP during vvRNP transcription with an ApG primer rather than by de novo initiation. As described above, replication involves de novo initiation with ATP, whereas initiation of transcription involves extension of a capped primer with either a G or a C residue. We found that at least 10-fold higher concentrations of ATP, CTP, and GTP were required for the synthesis of cRNA than for mRNA transcription (Fig. 3). Our laboratory has previously shown, by incorporation of α-32P-labeled CTP or GTP during de novo replication of a synthetic vRNA template, that the viral polymerase synthesizes a significant number of abortive transcripts, specifically pppApG, pppGpC, and pppApGpC (8). We also found that the detection of longer (15-nucleotide) in vitro-synthesized de novo transcripts by this method requires the use of a radiolabeled nucleotide which is incorporated further downstream (8). Our current data (Fig. 3) provide a reasonable explanation for this observation, namely that the viral polymerase requires a higher concentration of ATP, CTP, and GTP to overcome abortive transcription during de novo initiation of replication than for extension of a capped primer during mRNA transcription.
Early evidence suggested that de novo transcription and antitermination (cRNA synthesis) by the viral polymerase requires NP not associated with vvRNPs (3, 38). We have previously shown that cRNA synthesized in vivo by vvRNPs in the presence of cycloheximide (an inhibitor of protein expression) can be detected if viral polymerase, capable of binding viral RNA, and NP are preexpressed in the cell (44). We hypothesized that these preexpressed proteins were required to protect nascent cRNA from degradation by host cell nucleases, but it could not be ruled out entirely that such preexpressed viral polymerase and NP did not modify the infecting vvRNP to transcribe cRNA. We now report that partially purified viral polymerase and/or NP have no effect on the in vitro transcription and replication activities of purified vvRNPs (Fig. 4B). However, it is not possible to rule out that host protein(s) not present in these partially purified polymerase and/or NP preparations may influence transcription or replication. Nonetheless, this result supports our contention that viral polymerase and NP expressed during an in vivo viral infection are required to protect and replicate nascent cRNA but do not affect the infecting viral polymerase.
In conclusion, we have provided evidence that vvRNPs synthesize both cRNA and mRNA in the absence of any other added proteins, either viral or cellular. cRNA which is synthesized in infected cells will, however, be degraded by cellular pathways unless free viral polymerase and NP is present to bind the nascent transcript, thereby stabilizing it.
Acknowledgments
We thank Ervin Fodor for useful discussions, Julian Robinson for DNA sequencing, and Tao Deng for the expression plasmid pcDNA-tap-NP.
This study was supported by MRC grants G9523972 and G9901312 to G.G.B.
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Barrett, T., A. J. Wolstenholme, and B. W. J. Mahy. 1979. Transcription and replication of influenza virus RNA. Virology 98:211-225. [DOI] [PubMed] [Google Scholar]
- 2.Beaton, A. R., and R. M. Krug. 1981. Selected host cell capped RNA fragments prime influenza viral RNA transcription in vivo. Nucleic Acids Res. 9:4423-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaton, A. R., and R. M. Krug. 1986. Transcription antitermination during influenza viral template RNA-synthesis requires the nucleocapsid protein and the absence of a 5′ capped end. Proc. Natl. Acad. Sci. USA 83:6282-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, S. K., P. L. Boutz, and D. P. Nayak. 1998. Influenza virus nucleoprotein interacts with influenza virus polymerase proteins. J. Virol. 72:5493-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, W., P. A. Calvo, D. Malide, J. Gibbs, U. Schubert, I. Bacik, S. Basta, R. O'Neill, J. Schickli, P. Palese, P. Henklein, J. R. Bennink, and J. W. Yewdell. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 6.Crow, M., T. Deng, M. Addley, and G. G. Brownlee. 2004. Mutational analysis of the influenza virus cRNA promoter and identification of nucleotides critical for replication. J. Virol. 78:6263-6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, T., J. Sharps, E. Fodor, and G. G. Brownlee. 2005. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J. Virol. 79:8669-8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, T., F. T. Vreede, and G. G. Brownlee. 2006. Different de novo initiation strategies are used by influenza virus RNA polymerase on its cRNA and viral RNA promoters during viral RNA replication. J. Virol. 80:2337-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flick, R., and G. Hobom. 1999. Interaction of influenza virus polymerase with viral RNA in the ‘corkscrew’ conformation. J. Gen. Virol. 80:2565-2572. [DOI] [PubMed] [Google Scholar]
- 10.Flick, R., G. Neumann, E. Hoffmann, E. Neumeier, and G. Hobom. 1996. Promoter elements in the influenza vRNA terminal structure. RNA 2:1046-1057. [PMC free article] [PubMed] [Google Scholar]
- 11.Fodor, E., and G. G. Brownlee. 2002. Influenza virus replication, p. 1-29. In C. W. Potter (ed.), Influenza. Elsevier Science, New York, NY.
- 12.Fodor, E., M. Crow, L. J. Mingay, T. Deng, J. Sharps, P. Fechter, and G. G. Brownlee. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989-9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1994. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 68:4092-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fodor, E., D. C. Pritlove, and G. G. Brownlee. 1995. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J. Virol. 69:4012-4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gastaminza, P., B. Perales, A. M. Falcon, and J. Ortin. 2003. Mutations in the N-terminal region of influenza virus PB2 protein affect virus RNA replication but not transcription. J. Virol. 77:5098-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay, A. J., B. Lomniczi, A. R. Bellamy, and J. J. Skehel. 1977. Transcription of the influenza virus genome. Virology 83:337-355. [DOI] [PubMed] [Google Scholar]
- 17.Hay, A. J., J. J. Skehel, and J. McCauley. 1982. Characterization of influenza-virus RNA complete transcripts. Virology 116:517-522. [DOI] [PubMed] [Google Scholar]
- 18.Huarte, M., A. Falcon, Y. Nakaya, J. Ortin, A. Garcia-Sastre, and A. Nieto. 2003. Threonine 157 of influenza virus PA polymerase subunit modulates RNA replication in infectious viruses. J. Virol. 77:6007-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kistner, O., K. Muller, and C. Scholtissek. 1989. Differential phosphorylation of the nucleoprotein of influenza A viruses. J. Gen. Virol. 70:2421-2431. [DOI] [PubMed] [Google Scholar]
- 20.Klumpp, K., M. J. Ford, and R. W. H. Ruigrok. 1998. Variation in ATP requirement during influenza virus transcription. J. Gen. Virol. 79:1033-1045. [DOI] [PubMed] [Google Scholar]
- 21.Klumpp, K., R. W. H. Ruigrok, and F. Baudin. 1997. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 16:1248-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung, T. E., and G. G. Brownlee. 2006. A new promoter-binding site in the PB1 subunit of the influenza A virus polymerase. J. Gen. Virol. 87:679-688. [DOI] [PubMed] [Google Scholar]
- 23.Leahy, M. B., H. C. Dobbyn, and G. G. Brownlee. 2001. Hairpin loop structure in the 3′ arm of the influenza A virus virion RNA promoter is required for endonuclease activity. J. Virol. 75:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leahy, M. B., D. C. Pritlove, L. L. Poon, and G. G. Brownlee. 2001. Mutagenic analysis of the 5′ arm of the influenza A virus virion RNA promoter defines the sequence requirements for endonuclease activity. J. Virol. 75:134-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medcalf, L., E. Poole, D. Elton, and P. Digard. 1999. Temperature-sensitive lesions in two influenza A viruses defective for replicative transcription disrupt RNA binding by the nucleoprotein. J. Virol. 73:7349-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mena, I., E. Jambrina, C. Albo, B. Perales, J. Ortin, M. Arrese, D. Vallejo, and A. Portela. 1999. Mutational analysis of influenza A virus nucleoprotein: identification of mutations that affect RNA replication. J. Virol. 73:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momose, F., C. F. Basler, R. E. O'Neill, A. Iwamatsu, P. Palese, and K. Nagata. 2001. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J. Virol. 75:1899-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Momose, F., H. Handa, and K. Nagata. 1996. Identification of host factors that regulate the influenza virus RNA polymerase activity. Biochimie 78:1103-1108. [DOI] [PubMed] [Google Scholar]
- 29.Peng, Q., J. M. Galarza, L. Shi, and D. F. Summers. 1996. Influenza A virus RNA-dependent RNA polymerase cleaves influenza mRNA in vitro. Virus Res. 42:149-158. [DOI] [PubMed] [Google Scholar]
- 30.Plotch, S. J., M. Bouloy, I. Ulmanen, and R. M. Krug. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23:847-858. [DOI] [PubMed] [Google Scholar]
- 31.Poon, L. L. M., D. C. Pritlove, E. Fodor, and G. G. Brownlee. 1999. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J. Virol. 73:3473-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pritlove, D. C., L. L. M. Poon, E. Fodor, J. Sharps, and G. G. Brownlee. 1998. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J. Virol. 72:1280-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 34.Rao, P., W. Yuan, and R. M. Krug. 2003. Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J. 22:1188-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson, H. D., E. Dickson, S. J. Plotch, and R. M. Krug. 1980. Identification of the RNA region transferred from a representative primer, beta-globin mRNA, to influenza mRNA during in vitro transcription. Nucleic Acids Res. 8:925-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson, J. S., M. Schubert, and R. A. Lazzarini. 1981. Polyadenylation sites for influenza virus mRNA. J. Virol. 38:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seong, B. L., and G. G. Brownlee. 1992. A new method for reconstituting influenza polymerase and RNA in vitro: a study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology 186:247-260. [DOI] [PubMed] [Google Scholar]
- 38.Shapiro, G. I., and R. M. Krug. 1988. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J. Virol. 62:2285-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shih, S.-R., and R. M. Krug. 1996. Surprising function of the three influenza viral polymerase proteins: selective protection of viral mRNAs against the cap-snatching reaction catalyzed by the same polymerase proteins. Virology 226:430-435. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu, K., H. Handa, S. Nakada, and K. Nagata. 1994. Regulation of influenza virus RNA polymerase activity by cellular and viral factors. Nucleic Acids Res. 22:5047-5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skorko, R., D. F. Summers, and J. M. Galarza. 1991. Influenza A virus in vitro transcription: roles of NS1 and NP proteins in regulating RNA synthesis. Virology 180:668-677. [DOI] [PubMed] [Google Scholar]
- 42.Szymkowiak, C., W. S. Kwan, Q. Su, T. J. Toner, A. R. Shaw, and R. Youil. 2003. Rapid method for the characterization of 3′ and 5′ UTRs of influenza viruses. J. Virol. Methods 107:15-20. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, J. M., R. Illmensee, S. Litwin, L. Herring, B. Broni, and R. M. Krug. 1977. Use of specific radioactive probes to study transcription and replication of the influenza virus genome. J. Virol. 21:530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vreede, F. T., T. E. Jung, and G. G. Brownlee. 2004. Model suggesting that replication of influenza virus is regulated by stabilization of replicative intermediates. J. Virol. 78:9568-9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhirnov, O. P., T. E. Konakova, W. Garten, and H. Klenk. 1999. Caspase-dependent N-terminal cleavage of influenza virus nucleocapsid protein in infected cells. J. Virol. 73:10158-10163. [DOI] [PMC free article] [PubMed] [Google Scholar]