Abstract
Full replication of adeno-associated virus type 5 (AAV5) is sustained by adenovirus type 5 (Ad5) helper functions E1a, E1b, E2a, E4Orf6, and virus-associated (VA) RNA; however, their combined net enhancement of AAV5 replication was comprised of both positive and negative individual effects. Although Ad5 E4Orf6 was required for AAV5 genomic DNA replication, it also functioned together with E1b to degrade de novo-expressed, preassembled AAV5 capsid proteins and Rep52 in a proteosome-dependent manner. VA RNA enhanced accumulation of AAV5 protein, overcoming the degradative effects of E4Orf6, and was thus required to restore adequate amounts of AAV5 proteins necessary to achieve efficient virus production.
Efficient replication of adeno-associated viruses (AAVs) requires helper functions that can be supplied by larger DNA viruses such as adenovirus (Ad) or herpes simplex virus (2). There are five adenovirus type 5 (Ad5) functions required to support both AAV2 replication and production of recombinant AAV (E1a, E1b, E2a, E4Orf6, and virus-associated [VA] RNA), and the roles these factors play during both Ad and AAV infection have been extensively characterized (2, 3, 24, 25). Ad5 E2a provides single-stranded DNA (ssDNA) binding activity presumably required during the strand-displacement phase of AAV2 terminal repeat (TR)-mediated genome replication (20). E4Orf6 has been shown to be essential for second strand synthesis, both in vitro and in vivo (7), and likely plays other roles during AAV2 replication as well (22, 24, 26). VA RNA has been suggested to enhance AAV gene expression at the posttranscriptional level, most likely at the level of protein translation (22, 26). The role of VA RNA in the translation of Ad5 proteins during Ad5 infection is well characterized (12, 13). Ad5 E1a and E1b have been reported to perform a number of essential roles during AAV gene expression, including promoter activation (3, 11, 15, 28); however, other than the E1A's well-defined participation in the regulation of expression of AAV2 P5, the role that adenovirus plays in other aspects of AAV2 gene expression is as yet only partially understood (3, 25).
Adeno-associated virus type 5 (AAV5), which was first isolated from a penile condylomatous lesion, is the most divergent of the AAV serotypes, sharing only 64% overall nucleotide identity with the prototype AAV2 (1, 8). While the basic transcription profile of AAV5 is similar to that of AAV2, there are also significant differences (17). In contrast to AAV2, RNAs generated from both the AAV5 P7 and P19 promoters are efficiently polyadenylated at a site lying within the intron in the center of the genome, and because these RNAs are not spliced, Rep78 and Rep52 are the only Rep proteins detected during AAV5 infection (17). Furthermore, unlike AAV2, neither the AAV5 Rep protein nor additional adenovirus gene products are required to achieve efficient AAV5 promoter activity and pre-mRNA splicing following transfection of an AAV5 rep/cap plasmid clone lacking the inverted terminal repeats (ITRs) into 293 cells (17, 28).
The relatively independent expression of AAV5 compared to AAV2 led us to consider whether AAV5 might have fewer Ad5 requirements for its replication. In this report, we show that full replication of AAV5 required the same five Ad5 gene products as did replication of AAV2. However, closer analysis of the role of the individual Ad5 gene products showed that their combined net enhancement of AAV5 replication was comprised of both positive and negative effects. Specifically, although Ad5 E4Orf6 was required for AAV5 genome replication, it also functioned together with E1b, to degrade de novo-expressed, preassembled AAV5 capsid proteins and Rep52 in a proteasome-dependent manner. VA RNA enhanced accumulation of AAV5 protein, overcoming the degradative effects of E4Orf6, and thus was required to restore adequate amounts of AAV5 proteins necessary for efficient virus production.
MATERIALS AND METHODS
Plasmid constructions.
Ad5 helper expression constructs cytomegalovirus-driven E2a (CMV E2a), CMV E4Orf6, and VA RNA plasmids containing both VAI and VAII RNA, pucE1a and pucE1b (4, 14), were kind gifts from Tom Shenk (Princeton University). P19/P41-Rep52/Cap was constructed by deleting nucleotides (nt) 0 to 310 (NotI-SwaI) from the AAV5 RepCap clone previously described. CMV P19Rep and CMV P41Cap were constructed by inserting AAV5 nt 918 to 2251 and 1891 to 4381, respectively, into pcDNA3 by standard PCR techniques. The construction of P41Cap and the RNase protection probe PGEM3Z-RP has been described previously (16).
Viruses.
AAV5 virus was a gift from Ursula Bantel-Schaal (DKFZ, Heidelberg, Germany).
Analysis of intracellular and extracellular AAV viral replicating DNA.
293 cells, seeded at 4 × 105 cells/well in six-well dishes, were infected with AAV5 (multiplicity of infection [MOI] of 10) for 1 h at 37 h before transfection with the Lipofectamine reagent and the Plus reagent. Equal amounts of DNA (3 μg per well of a six-well dish) were used to transfect 60 to 80% confluent cells using Lipofectamine and the Plus reagent (Gibco BRL, Gaithersburg, Md.) as previously described (16). At 48 h posttransfection, cells were suspended in spent tissue culture medium and aliquoted equally into three separate centrifuge tubes. One aliquot was used for the analysis of intracellular and extracellular AAV5 DNA, the second aliquot was used for analysis of released virus production, and the third aliquot was used for immunoblot analysis as described below.
For analysis of intracellular and extracellular AAV5 DNA forms, cells were pelleted, resuspended in lysis buffer (2% sodium dodecyl sulfate [SDS], 150 mM NaCl-10 mM Tris-HCl [pH 8], 1 mM EDTA), and incubated at 55°C for 2 h. The samples were sheared by passing them through a syringe fitted with a 25-gauge needle. The spent medium supernatant was adjusted to 2% SDS and 0.15 M NaCl. Proteinase K (0.5 mg/ml) was then added to both the cell lysates and spent medium, and the samples were incubated at 37°C for 12 h. Approximately 104 cell equivalents/lane of cell lysates and spent medium were loaded onto an agarose gel (1%) in TAE buffer (40 mM Tris-HCl [pH 8.5], 50 mM sodium acetate, 10 mM EDTA). After transfer of the DNA to a nitrocellulose membrane, filters were hybridized to a random-primed probe as described previously (17).
For analysis of cell-associated virus production, cells were first separated from the medium by centrifugation (11,000 × g for 3 min), and cell pellets were then resuspended in 10 mM Tris-1 mM EDTA (pH 8.0). After resuspension, cells were subjected to three cycles of freezing and thawing. Debris was centrifuged at 14,000 × g for 1 min, and supernatants were subsequently subjected to slot blot analysis as previously described (17). No virus was detected free in the tissue culture media.
Immunoblot analyses.
Plasmid DNA (6 μg/60-mm-diameter dish) was transfected into 60 to 80% confluent 293 cells using Lipofectamine and Plus reagent (Gibco BRL, Gaithersburg, Md.) as previously described (17). Cell lysates were prepared and subjected to immunoblot analysis as previously described (17). Monoclonal antibody 303.9, raised to an amino-terminally truncated AAV2 Rep78, and anti-Cap antibody (clone B1) were obtained from American Research Products, Inc., Belmont, MA. Antibody to Ad5 E1a (05-599, clone M73) was obtained from Upstate Technologies (Lake Placid, NY), antibody to β-actin (ab-32800-500) was obtained from Abcam (St. Louis, MO), and antibody to 14-3-3 proteins (sc-1020) was obtained from Santa Cruz. Antibodies to Ad5 E2a and E4Orf6 were kind gifts from Tom Shenk (Princeton University), and antibody to Ad5 E1b (9b) was obtained from Arnold Levine (Princeton University).
RNA isolation and RPAs.
Total RNA and cytoplasmic RNA were isolated using guanidine isothiocyanate, and RNase protection assays (RPAs) were performed as previously described (19). Homologous RNase protection probe RP (AAV5 nt 1843 to 2034) was generated from linearized templates by in vitro transcription using SP6 polymerase, as previously described (19). RNA hybridizations for RPAs were done with a substantial probe excess, and RPA signals were quantified with the Molecular Imager FX and Quantity One (version 4.2.2 image software; Bio-Rad, Hercules, CA). Relative molar ratios of individual species of RNAs were determined after adjustment for the number of 32P-labeled uridines in each protected fragment as previously described (17).
CHX chase and proteasomal inhibitor analysis.
293 cells were plated in six-well dishes to 60 to 70% confluence and then transfected with Lipofectamine and the Plus reagent as described above. At 30 h posttransfection, cycloheximide (CHX) (, catalog no. C-7698; Sigma Chemicals, St. Louis, MO) was added to the cells at 100 μg/ml. Control wells that did not receive CHX received dimethyl sulfoxide (DMSO) vehicle instead. Cells were then collected at the time points described in the text and subjected to immunoblot analysis as described above. For proteasome inhibitor experiments, either 10 μM MG132 (catalog no. 474791; Calbiochem), or 5 μM lactacystin (catalog no. 70980; Cayman Chemicals) was applied to cells 40 h posttransfection. Control wells that did not receive MG132 or lactacystin received DMSO vehicle instead. Cells were collected 6 h after addition of the drug and subjected to immunoblot analysis.
Complementation analysis.
293 cells were grown in six-well dishes and infected with AAV5 (MOI of 10). Cells were transfected with equal molar amounts of plasmids as described in the text, using Lipofectamine and Plus reagent. At 48 h posttransfection, cells were collected and divided into two aliquots. One of the aliquots was used for analysis of AAV5 intracellular DNA replicative forms, as described above, and the other aliquot was used for immunoblot analysis, also as described above.
RESULTS
Ad5 VA RNA is required for the production of AAV5 progeny single-strand DNA.
To begin our assessment of the roles of individual Ad5 proteins in AAV5 replication, we initially evaluated expression of the Ad5 E2a and E4Orf6 proteins and the VAI RNA, individually and in various combinations, from their endogenous promoters. We found that under these conditions, variable levels of E2a and E4Orf6 were produced, depending upon the combinations used. However, this problem was overcome by expressing E2a and E4Orf6 from CMV promoters, and consistent levels of expression, independent of cotransfected partners, were achieved.
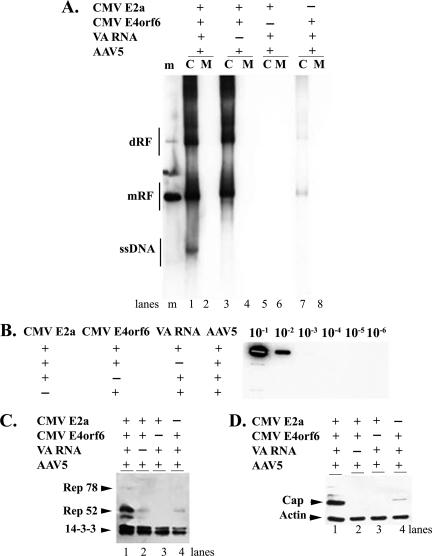
The combination of CMV-expressed Ad5 E2a and E4Orf6 proteins, together with VA RNA expressed from its endogenous promoter, supported full AAV5 replication (Fig. 1A, lanes 1 and 2) and production of cell-associated AAV5 virus (Fig. 1B) in Ad5 E1a and E1b-expressing 293 cells. Assembled virus was not detected free in the media either by the Southern analysis shown in Fig. 1A (lane 2) or in the slot blot analysis shown in Fig. 1B (data not shown), however, cell associated virus generated in response to the full complement of Ad helper products was fully infectious (data not shown.) The levels of both DNA replication forms and infectious virus production in these experiments were similar to those generated following transfection of the widely used pHelper plasmid (data not shown). When E4Orf6 was withheld from these experiments, no replication (Fig. 1A, lanes 5 and 6) or cell-associated virus production (Fig. 1B) was detected. When E2a was withheld, replication was substantially diminished (Fig. 1A, lanes 7 and 8) and cell-associated virus production was below the level of detection (Fig. 1B). Interestingly, when only VA RNA was withheld from these experiments, the AAV5 monomer double-stranded replicative form (mRF) DNA was generated at essentially wild-type levels; however, progeny single-stranded genomic DNA (ssDNA) was undetected (Fig. 1A, lanes 3 and 4). Consistent with this observation, cell-associated virus production was also undetectable in the absence of VA RNA (Fig. 1B). The combination of Ad5 E2A, E4Orf6, and VA RNA could not support replication of AAV5 in HeLa cells which lack E1a and E1b (data not shown). These results suggested that all five Ad products were required for full AAV5 DNA replication and infectious virus production and that, in the absence of VA RNA, there appeared to be a specific block to progeny ssDNA production.
FIG. 1.
(A) Replication of AAV 5 viral DNA in 293 cells in the presence of various combinations of Ad5 gene products. 293 cells were infected with AAV5 at an MOI of 10 and transfected with equal molar quantities of CMV E2a, CMV E4Orf6, and a VAI RNA-expressing clone in the combinations indicated. pBS-SK was used to compensate for the absence of any individual plasmid. Monomer length (mRF), dimer length (dRF), and single-strand progeny (ssDNA) DNA replication forms, either cell associated (C) or released into the media (M), are indicated. Markers (m; lane 1) are a mixture of untreated and denatured 4.6-kb Acc65I fragments from pAV5, which corresponds to the full-length AAV5 genome. (B) Quantification of cell-associated AAV5 generated in 293 cells in the presence of various combinations of Ad5 gene products: Slot blot analysis to detect cell-associated virus production, performed as described in the Materials and Methods, during the same experiment in which replication was assayed in panel A, is shown. The various combinations of Ad5 helper functions are shown on the left, and dilutions of samples are shown to the right. (C and D) Analysis of AAV5 Rep protein (C) and capsid protein (D) production in 293 cells in the presence of various combinations of Ad5 gene products: Immunoblot analysis to determine AAV5 protein expression during the same experiment in which viral replication (A) and virus production (B) was determined as described above, using either antibody against the viral Rep proteins (C) or the viral capsid proteins (D), is shown. The locations of the AAV5 proteins, as well as endogenously expressed 14-3-3 and actin proteins serving as loading controls, are shown.
The production of AAV5 protein was also analyzed from the same experiments from which AAV5 replication is shown in Fig. 1A. As expected, because full replication of AAV5 was supported, abundant levels of AAV5 structural and nonstructural proteins were produced in 293 cells when all three Ad5 products were cotransfected following AAV5 infection (Fig. 1C and D, lanes 1). (We previously observed that the levels of AAV5 Rep78 were variable during AAV5 infection.) Transfections lacking either Ad5 E2a or E4Orf6 only poorly supported AAV5 replication (Fig. 1A), and thus, as expected, much lower levels of AAV5 proteins were generated following cotransfections lacking either of these proteins (Fig. 1C and D, lanes 4 and 3, respectively). We were surprised, however, to observe that only very low levels of AAV5 protein were generated in cotransfections specifically lacking VA RNA (Fig. 1C and D, lanes 2), since mRF DNA, presumably the transcription template, was produced at high levels (Fig. 1A, lane 3). It has been shown in numerous parvovirus systems that the production of virus capsids and the small nonstructural proteins are required for the production of detectable ssDNA (5, 6, 9, 10, 21, 23, 24), and so we next considered whether different combinations of Ad5 helper functions resulted in different accumulated levels of AAV5 protein.
Ad5 E4Orf6 reduces accumulated levels of AAV5 capsid protein and Rep52, which can be relieved by expression of VA RNA.
We have previously shown that the basal activity of the AAV5 capsid gene promoter P41 is considerably greater in 293 cells than the P40 capsid gene promoter of AAV2 (17, 28). This allowed us to examine the expression of the AAV5 capsid proteins in the presence and absence of combinations of Ad5 helper functions from minimal constructs in the absence of Rep.
The basal level of expression of the minimal P41 expression construct P41Cap is shown in Fig. 2A, panel 1, lane 8. Addition of Ad5 E2a, E4Orf6, and VA RNA together resulted in only a modest change in the level of AAV5 capsid protein expression (Fig. 2A, panel 1, lane 1). Surprisingly, when E4Orf6 was added either alone, or with only E2a, a dramatic decrease in capsid protein accumulation was observed (Fig. 2A, panel 1, compare lanes 6 and 2 to lane 8). Expression of E2a alone resulted in a modest decrease in capsid protein accumulation in this experiment (Fig. 2A, panel 1, lane 5), but in most cases, the effect of E2a alone was negligible (see also Fig. 3, lane 5). Addition of VA RNA alone increased capsid protein expression (Fig. 2A, panel 1, lane 7), and when expressed together with E4Orf6, VA RNA was able to overcome E4Orf6 inhibition of capsid protein accumulation (Fig. 2A, panel 1, lane 4). Expression of various combinations of the Ad5 helper functions did not affect the accumulated levels of either cellular tubulin or actin (Fig. 2A, panels 2 and 3), the endogenous E1a (Fig. 2A, panel 2) or E1b (Fig. 2A, panel 3) proteins expressed in 293 cells, or the ectopically expressed, CMV-driven E2a (Fig. 2A, panel 4) or E4Orf6 proteins (Fig. 2A, panel 5) proteins. Thus, the net accumulated levels of capsid proteins generated in response to the full complement of Ad5 helper functions is the consequence of both positive and negative effects.
FIG. 2.
(A) Accumulation of AAV5 capsid proteins in 293 cells in the presence of various combinations of Ad5 gene products. Immunoblot analysis of protein accumulation in 293 cells following transfection of the P41Cap construct together with various combinations of Ad5 gene products, as indicated, is shown. The complete set of samples was run on five separate gels and after transfer probed with antibodies to either AAV5 capsid proteins and actin (panel 1), E1a and tubulin (panel 2), E1b and actin (panel 3), E2a and actin (panel 4), or E4Orf6 and tubulin (panel 5). (B) Steady-state levels of total and cytoplasmic AAV5 capsid gene RNA generated in 293 cells in the presence of various combinations of Ad5 gene products, from the same experiment shown in panel A. Shown are the results from quantitative RNase protection assays, using the AAV5 RP probe as described in the Materials and Methods, of either total (T) or cytoplasmic (C) RNA generated in 293 cells following transfection of the P41Cap construct together with various combinations of Ad5 gene products, as shown from the same experiment in panel A. Bands protected by P41-generated unspliced and spliced RNAs are indicated on the left. RNase protections using a probe to cellular β-actin served as a loading control. A diagram of the P41Cap construct used for the experiments shown in panels A and B is shown at the bottom, with genetic landmarks and the location of the RP RNase protection probe indicated.
FIG. 3.
Accumulation of AAV5 Rep52 in 293 cells in the presence of various combinations of Ad5 gene products. Immunoblot analysis showing protein accumulation in 293 cells following transfection of the P19/P41-Rep52/Cap construct together with the various combinations of Ad5 gene products as indicated. After transfer, blots were probed together with antibody directed against the AAV Rep proteins and with antibody to cellular 14-3-3 proteins, and these proteins are indicated to the left. A diagram of the P19/P41-Rep52/Cap construct is shown at the bottom with genetic landmarks indicated. The band marked with an asterisk is a stable product generated from the AAV5 Rep open reading frame, which was previously observed (17).
Total and cytoplasmic RNA generated by P41Cap in the same experiment shown in Fig. 2A was also analyzed. There was no difference in either the steady-state levels of total spliced RNA or the relative levels of RNA exported to the cytoplasm in response to expression of the various combinations of Ad5 helper functions (Fig. 2B). Thus, both the inhibitory effect of E4Orf6 and the enhancing effect of VA RNA on AAV5 capsid protein accumulation occurred posttranscriptionally at a step subsequent to the export of the capsid protein-encoding mRNAs.
The E4Orf6 protein also prevented accumulation of the AAV5 P19-generated Rep 52 protein in 293 cells. The basal-level expression of Rep52 from the P19 minimal construct P19/P41-Rep52/Cap is shown in Fig. 3, lane 8. As seen for the capsid proteins, coexpression of E4Orf6, either by itself or together with E2a, resulted in a dramatic decrease in accumulated levels of Rep52 (Fig. 3, compare lanes 6 and 2 to lane 8). Expression of VA RNA enhanced Rep52 expression (here only slightly; Fig. 3, lane 7), and when cotransfected with E4Orf6 restored capsid protein expression to the levels seen when the full complement of helper functions were present (Fig. 3, lane 4). Expression of E2a had little effect on expression of Rep52 (Fig. 3, lanes 5 and 2).
These results indicated that while the E4Orf6 protein had a critical role during genomic DNA replication, in 293 cells its expression reduced the accumulated amounts of the AAV5 capsid proteins and Rep52 posttranscriptionally, which could be returned to necessary levels by the enhancing activity of VA RNA. Because of the inherent instability in our hands of the AAV5 Rep78 protein (also see reference 17), we were unable to determine the effect of E4Orf6 on its accumulated level.
E4Orf6 causes the degradation of AAV5 Rep52 and preassembled capsid proteins.
Inhibition by E4Orf6 of accumulation of de novo-expressed AAV5 capsid protein was reversed both by MG132, a proteasome substrate analog which competitively inhibits both the chymotrypsin-like and the peptidylglutamyl-peptide-hydrolyzing (PGPH) activity of the 20S proteasome (Fig. 4A, compare lanes 3 to 4), and lactacystin, which specifically inhibits the chymotrypsin-like, trypsin-like, and PGPH activity of the 20S proteasome by irreversibly modifying all catalytic subunits (Fig. 4B, compare lanes 3 to 4). These results suggest that E4Orf6 functions to bring about the proteasome-dependent degradation of the AAV5 capsid proteins. Consistent with this model, we found that while ectopically expressed AAV5 capsid proteins were very stable in the presence of a cycloheximide block to additional translation (Fig. 4C, compare lanes 1 to 4 to 5 to 8), in the presence of E4Orf6, accumulated levels of AAV5 capsid proteins were still lost very quickly under these conditions (Fig. 4C, lanes 13 to 16).
FIG. 4.
(A) Inhibition of E4Orf6 activity by proteasome inhibitor MG132. 293 cells were transfected with P41Cap (lanes 1 and 2) and P41Cap with E4Orf6 (lanes 3 and 4). At 40 h posttransfection, 10 μM MG132 (lanes 2 and 4) or DMSO (lanes 1 and 3) was added to each of the samples as noted. The cell lysates were collected at 48 h posttransfection and then probed with antibody to AAV5 capsids. (B). Inhibition of E4Orf6 activity by proteasome inhibitor lactacystin. 293 cells were transfected with P41Cap (lanes 1 and 2) and P41 with E4Orf6 (lanes 3 and 4). At 40 h posttransfection, 5 μM lactacystin (lanes 2 and 4) or DMSO (lanes 1 and 3) was added to each of the samples as noted. The cell lysates were collected at 48 h posttransfection and then probed with antibody to AAV5 capsids. (C) E4Orf6 degrades AAV5 capsid proteins in the presence of cycloheximide. 293 cells were transfected with P41Cap and either pBS-SK or with CMV E4Orf6. At 30 h posttransfection, cycloheximide (100 μg/ml) or DMSO was added to each of the samples as noted above. Lysates were then collected at 30 h, 33 h, 36 h, or 39 h posttransfection. The samples then underwent 10% SDS-polyacrylamide gel electrophoresis and were immunoblotted with antibody to AAV5 capsid proteins.
E4Orf6 effects on AAV5 protein accumulation depend on the Ad5 E1b protein.
Previous characterizations of E4Orf6 have shown that it can function to promote protein degradation by forming an E3 ligase complex together with the Ad5 E1b protein (18). We chose to test whether E4Orf6 degradation of AAV5 protein required E1b using HeLa cells; however, AAV5 promoters are active only at very low levels in HeLa cells (28). Therefore, we examined the accumulation in HeLa cells of AAV5 capsid proteins expressed from a minimal capsid gene construct driven by the CMV promoter. Consistent with the requirement for E1b, expression of the E4Orf6 protein in HeLa cells did not lead to a decrease in the accumulated level of the AAV5 capsid proteins expressed from the CMV-driven minimal capsid gene product (Fig. 5, compare lane 7 to lane 8). However, when E1b was cotransfected together with E4Orf6, accumulation of AAV5 capsid proteins was dramatically reduced (Fig. 5, lane 3). E1a alone provided no help to E4Orf6 in this regard (Fig. 5, lane 2), while the combination of E1b and E1a affected E4Orf6 function similarly to E1b alone (Fig. 5, lane 1). E1a and E1b in the absence of E4Orf6 had no effect on the accumulation of AAV5 capsid proteins (Fig. 5, lanes 4 to 6). These results demonstrated that, as seen for other substrates, the effect of E4Orf6 on AAV5 capsid protein accumulation required E1b.
FIG. 5.
Accumulation of AAV5 capsid proteins in HeLa cells in the presence of various combinations of Ad5 gene products. The results of immunoblot analysis showing protein accumulation in HeLa cells following transfection of the CMV P41Cap construct together with the various combinations of Ad5 gene products are shown. The complete set of samples was run on four separate gels and after transfer probed with antibodies to either AAV5 capsid proteins and actin (panel 1), E1a and tubulin (panel 2), E1b and actin (panel 3), or E4Orf6 and tubulin (panel 4). The locations of individual proteins are indicated.
The inhibitory effect of E4Orf6 on the production of AAV5 genomic ssDNA could be overcome by overexpression together of Rep52 and the capsid proteins.
Subtraction of VA RNA from the three helper factors exogenously required for AAV5 replication in 293 cells resulted in a lack of production of AAV5 ssDNA in these cells (Fig. 1A, compare lanes 1 and 2 to 3 and 4). If this was specifically due to reduced levels of preassembled capsid proteins and Rep52 as a consequence of their degradation by E4Orf6, we hypothesized that we should be able to regain full replication and ssDNA production in the presence of E4Orf6 following overexpression of these AAV5 proteins. As can be seen in Fig. 6A, in the presence of E2a and E4Orf6 and the absence of VA RNA, all AAV5 replicative DNA forms were generated when Rep52 and capsid proteins were supplied at high levels by CMV-driven expression vectors (Fig. 6A and B, lanes 7), but not when either was supplied alone (Fig. 6A and B, lanes 3 and 5). The levels of ssDNA and AAV5 proteins generated were similar to that generated following cotransfection of all three helper functions (Fig. 6A and B, lanes 1). These results suggest that at least one function of VA RNA in supporting AAV5 replication was to overcome the degradative activity of E4Orf6, thereby increasing the accumulated levels of Rep52 and the capsid proteins to levels necessary for ssDNA production. In these experiments, either full replication or replication of the mRF transcription template led to a substantial increase in AAV5 protein production. When protein was expressed from both the replicating viral transcription template and the expression plasmid, the levels of AAV5 proteins were apparently such that there was some escape from E4Orf6 degradation (Fig. 6B, lanes 1, 3, and 7). This was not observed following ectopic expression of either AAV5 Rep52 or capsid proteins from expression plasmids, either alone or together in the absence of the AAV5 genome (Fig. 6B, lanes 4, 6, and 8). That complementation could be achieved without supplying Rep78 suggested that either E4Orf6 does not fully degrade this protein or only small amounts were necessary for replication.
FIG. 6.
Overexpression of AAV5 Rep52 and capsid proteins allows full replication of AAV5 in the absence of VA RNA. (A) 293 cells were infected with AAV5 at an MOI of 10 and transfected with equal molar quantities of CMV E2a, CMV E4Orf6, a VAI RNA-expressing clone, AAV5 Rep52-expressing CMV P19 Rep, and AAV5 capsid protein expressing CMV P41Cap, in the combinations indicated. pBS-SK was used to compensate for the absence of any individual plasmid. Monomer length (mRF), dimer length (dRF), and single-strand progeny (ssDNA) DNA replication forms are indicated. Markers (m; lane 9) are a mixture of untreated and denatured 4.6-kb Acc65I fragments from pAV5, which corresponds to the full-length AAV5 genome. (B) Accumulation of AAV5 capsid proteins in 293 cells in the presence of various combinations of Ad5 gene products, AAV5 Rep 52, and AAV5 capsid proteins. Shown are the results of immunoblot analysis demonstrating protein accumulation from the same experiment in which AAV5 replication was determined in the presence of various combinations of Ad5 and AAV5 gene products, as shown in panel A. The complete set of samples was run on two separate gels and after transfer probed with antibodies to either the viral Rep proteins and cellular 14-3-3 or the viral capsid proteins and cellular actin. The locations of the AAV5 proteins, as well as endogenously expressed 14-3-3 and actin proteins serving as loading controls, are shown. The band marked with an asterisk is a stable product generated from the AAV5 Rep open reading frame as previously observed (17).
DISCUSSION
In this report, we show that full replication of AAV5 required the same five Ad5 gene products as did replication of AAV2. However, upon closer inspection, we found that while these factors in combination achieved a net enhancement of AAV5 replication, individually they exhibited both positive and negative effects. Ad5 E4Orf6 was required for AAV5 genomic DNA replication; however, it also functioned to degrade AAV5 capsid proteins and Rep52. VA RNA enhanced accumulation of AAV5 RNA, overcoming the degradative effects of E4Orf6, and was thus required to restore adequate amounts of AAV5 proteins necessary for efficient virus production. Our experiments were simplified because, in contrast to AAV2, we were able to analyze AAV5 expression in the absence of Rep. However, to achieve consistent levels of expression of Ad5 proteins in all combinations in our experiments, we found it necessary to express these proteins from CMV-driven expression vectors. Because both 293 cells and our E1b expression vector potentially express the Ad pIX protein, we cannot rule out a role for this protein in our experiments.
The inhibition of accumulation of AAV5 capsid proteins and Rep52 by E4Orf6 was posttranscriptional. Under conditions in which capsid protein and Rep52 were significantly reduced, the levels of fully spliced mRNAs present in the cytoplasm were found to be unaltered. The effect of E4Orf6 was not promiscuous. In experiments in which a decrease in the accumulation of AAV5 protein was observed, there was no detectable effect on accumulated levels of cellular actin, tubulin, 14-3-3 proteins, the endogenous E1a and E1b expressed in 293 cells, or ectopically expressed E2a or E4Orf6 proteins. It is important to note that whether E4Orf6 has an effect on accumulated levels of AAV5 Rep78 is not yet clear, because of the inherently unstable nature of this protein in our hands. However, since complementation for replication could be achieved without added Rep78 (Fig. 6), either E4Orf6 only partially degrades Rep78, or only a small amount of this protein is required to support replication. Whether E4Orf6 affects accumulated levels of AAV2 proteins is currently under investigation.
The effect of E4Orf6 on AAV5 protein levels was prevented by the proteasome inhibitors MG132 and lactacystin. This suggested that E4Orf6 reduced AAV5 protein levels via a pathway dependent upon degradation by the proteasome. Loss of resident pools of AAV5 protein in the presence of E4Orf6 continued in the presence of the translational inhibitor cycloheximide, consistent with a model in which the E4Orf6-dependent decrease in accumulated AAV5 protein was due to degradation. Taken together, these results suggested that E4Orf6 brought about the degradation of AAV5 capsid proteins and likely Rep52.
We have also performed pulse-chase experiments to analyze the degradation of AAV5 protein in the presence of E4Orf6. We have observed that a significant portion of labeled material is chased into a fraction resistant to further degradation (R. Nayak and D. Pintel, unpublished data). While not yet unequivocally confirmed, it seems most likely that these resistant forms are assembled capsids, and hence that E4Orf6 degrades de novo-generated capsid proteins only prior to assembly. Previous studies have shown ubiquitylation of both recombinant AAV2 and AAV5 virions following transduction. Although the role of such ubiquitylation during the entry process is unknown, it was also shown that heat-denatured viral particles were preferential substrates for in vitro ubiquitylation (27).
Previous reports have demonstrated that E4Orf6 can participate in the degradation of cellular targets including p53, and Mre11 by forming as an E3-ubiquitin ligase complex together with the E1b55K protein. We found that the ability of E4 Orf6 to degrade AAV5 protein also required the Ad5 E1b55K protein. Additionally, initial experiments demonstrate that an E4Orf6 mutant deficient in its E3 ligase function was less able to reduce accumulated levels of AAV5 protein (Nayak and Pintel, unpublished). These results suggest that E4Orf6 achieves the reduction of AAV5 protein by forming an E3 ligase complex along with E1b55K. Whether the effect of such an E4Orf6/E1b55K E3 ligase complex is directly on AAV5 proteins themselves or is due to an indirect effect on an intermediary is currently under investigation.
Overexpression of Rep and Cap proteins in the presence of E4Orf6 (and E2a) in the absence of VA RNA could recover the full replication of AAV5 DNA. Presumably, high levels of Rep and Cap proteins overwhelmed the effects of E4Orf6-mediated degradation and allowed viral production to take place. This result supported a model in which at least one required role of VAI RNA during AAV5 infection was to increase the levels of AAV5 Rep and Cap proteins, thereby surmounting the degradative effects of E4Orf6 and thus allowing efficient virus production.
The experiments presented here evaluate the role of Ad5 helper functions supplied in the absence of adenovirus. It remains possible that in our experiments, the expression of E4Orf6 together with E1b55K have an effect not seen during adenovirus infection and that there may be other Ad gene products, not necessarily part of the minimal AAV5 helper contingent, that could counteract the degradative effects of the E4Orf6/E1b55K complex on AAV5 protein.
Acknowledgments
We thank Lisa Burger for excellent technical assistance and Jianming Qiu for helpful discussion.
This work was supported by PHS grants RO1 AI46458 and RO1 AI56310 from NIAID to D.J.P.
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Bantel-Schaal, U., and H. zur Hausen. 1984. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology 134:52-63. [DOI] [PubMed] [Google Scholar]
- 2.Berns, K. I., and C. Giraud. 1996. Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218:1-23. [DOI] [PubMed] [Google Scholar]
- 3.Bowles, D., J. E. Rabinowitz, and R. J. Samulski. 2006. The genus Dependovirus, p. 15-24. In J. Kerr et al., (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 4.Chang, L.-S., and T. Shenk. 1990. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J. Virol. 64:2103-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi, E.-Y., A. E. Newman, L. Burger, and D. Pintel. 2005. Replication of minute virus of mice DNA is critically dependent on accumulated levels of NS2. J. Virol. 79:12375-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotmore, S. F., and P. Tattersall. 2006. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p. 171-188. In J. Kerr et al. (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 7.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georg-Fries, B., S. Biederlack, J. Wolf, and H. zur Hausen. 1984. Analysis of proteins, helper dependence, and seroepidemiology of a new human parvovirus. Virology 134:64-71. [DOI] [PubMed] [Google Scholar]
- 9.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleinschmidt, J. A., and J. A. King. 2006. Molecular interactions involved in assembling the viral particle and packaging the genome, p. 305-319. In J. Kerr et al. (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 11.Laughlin, C. A., N. Jones, and B. J. Carter. 1982. Effect of deletions in adenovirus early region 1 genes upon replication of adeno-associated virus. J. Virol. 41:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma, Y., and M. B. Mathews. 1996. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J. Virol. 70:5083-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathews, M. B. 1995. Structure, function, and evolution of adenovirus virus-associated RNAs. Curr. Top. Microbiol. Immunol. 199:173-187. [DOI] [PubMed] [Google Scholar]
- 14.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. USA 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrove, J. M., and K. I. Berns. 1980. Adenovirus early region 1b gene function required for rescue of latent adeno-associated virus. Virology 104:502-505. [DOI] [PubMed] [Google Scholar]
- 16.Qiu, J., R. Nayak, and D. J. Pintel. 2004. Alternative polyadenylation of adeno-associated virus type 5 RNA within an internal intron is governed by both a downstream element within the intron 3′ splice acceptor and an element upstream of the P41 initiation site. J. Virol. 78:83-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qiu, J., R. Nayak, G. E. Tullis, and D. J. Pintel. 2002. Characterization of the transcription profile of adeno-associated virus type 5 reveals a number of unique features compared to previously characterized adeno-associated viruses. J. Virol. 76:12435-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 181:22-34. [DOI] [PubMed] [Google Scholar]
- 20.Stracker, T. H., G. D. Cassell, P. Ward, Y.-M. Loo, B. van Breukelen, S. D. Carrington-Lawrence, R. K. Hamatake, P. C. van der Vliet, S. K. Weller, T. Melendy, and M. D. Weitzman. 2004. The Rep protein of adeno-associated virus type 2 interacts with single-stranded DNA-binding proteins that enhance viral replication. J. Virol. 78:441-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tratschin, J.-D., I. L. Miller, and B. J. Carter. 1984. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J. Virol. 51:611-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trempe, J. P., and B. J. Carter. 1988. Regulation of adeno-associated virus gene expression in 293 cells: control of mRNA abundance and translation. J. Virol. 62:68-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tullis, G. E., L. R. Burger, and D. J. Pintel. 1993. The minor capsid protein VP1 of the autonomous parvovirus minute virus of mice is dispensable for encapsidation of progeny single-stranded DNA but is required for infectivity. J. Virol. 67:131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward, P. 2006. Replication of adeno-associated virus DNA, p. 189-212. In J. Kerr et al. (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 25.Weitzman, M. D. 2006. The parvovirus life cycle: an introduction to molecular interactions important for infection, p. 143-156. In J. Kerr et al. (ed.), Parvoviruses. Hodder Arnold, London, United Kingdom.
- 26.West, M. H., J. P. Trempe, J. D. Tratschin, and B. J. Carter. 1987. Gene expression in adeno-associated virus vectors: the effects of chimeric mRNA structure, helper virus, and adenovirus VA1 RNA. Virology 160:38-47. [DOI] [PubMed] [Google Scholar]
- 27.Yan, Z., R. Zak, G. W. Gant Luxton, T. C. Ritchie, U. Bantel-Schaal, and J. F. Engelhardt. 2002. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 76:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye, C., J. Qiu, and D. J. Pintel. 2006. Efficient expression of the adeno-associated virus type 5 p41 capsid gene promoter in 293 cells does not require Rep. J. Virol. 80:6559-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]