Abstract
Alphaviruses are widely distributed throughout the world. During the last few thousand years, the New World viruses, including Venezuelan equine encephalitis virus (VEEV) and eastern equine encephalitis virus (EEEV), evolved separately from those of the Old World, i.e., Sindbis virus (SINV) and Semliki Forest virus (SFV). Nevertheless, the results of our study indicate that both groups have developed the same characteristic: their replication efficiently interferes with cellular transcription and the cell response to virus replication. Transcriptional shutoff caused by at least two of the Old World alphaviruses, SINV and SFV, which belong to different serological complexes, depends on nsP2, but not on the capsid protein, functioning. Our data suggest that the New World alphaviruses VEEV and EEEV developed an alternative mechanism of transcription inhibition that is mainly determined by their capsid protein, but not by the nsP2. The ability of the VEEV capsid to inhibit cellular transcription appears to be controlled by the amino-terminal fragment of the protein, but not by its protease activity or by the positively charged RNA-binding domain. These data provide new insights into alphavirus evolution and present a plausible explanation for the particular recombination events that led to the formation of western equine encephalitis virus (WEEV) from SINV- and EEEV-like ancestors. The recombination allowed WEEV to acquire capsid protein functioning in transcription inhibition from EEEV-like virus. Identification of the new functions in the New World alphavirus-derived capsids opens an opportunity for developing new, safer alphavirus-based gene expression systems and designing new types of attenuated vaccine strains of VEEV and EEEV.
The Alphavirus genus of the Togaviridae family contains a number of important human and animal pathogens (43). Nearly 30 species of the genus are distributed throughout the world and, based on sequencing data, are grouped into eight complexes (48). According to one hypothesis, alphaviruses originated either in the New or Old World and then were distributed to the opposite hemisphere by migratory birds (49). Another hypothesis states that the alphaviruses were transmitted from the New World to the Old World in events occurring separately for those viruses that evolved into Sindbis virus (SINV) and Semliki Forest virus (SFV) complexes (23, 43). Therefore, during the last few thousand years, the New World viruses, including Venezuelan equine encephalitis (VEEV) and eastern equine encephalitis (EEEV) viruses, evolved separately from those of the Old World, i.e., SINV and SFV. It is reasonable to expect that the viruses not only modified their envelope glycoproteins and replicative machinery for replication in particular species of hosts and vectors but may also have evolved different mechanisms of modifying the cellular environment and interfering with the development of an antiviral response. This antiviral reaction is based on the activation of viral stress-inducible genes (41) leading to establishment of an antiviral state in the infected cells and the release of cytokines, making the uninfected cells resistant to successive rounds of infection. To date, more and more DNA and RNA viruses are known to use one or a few encoded proteins to inhibit the development of a cellular response (1-4, 19, 26, 30, 39, 47). These proteins either interfere with the activation of specific genes or, as is the case with picorna- and bunyaviruses (3, 21, 51), inhibit cellular transcription or interfere with mRNA transport from the nucleus.
The alphavirus genome is an 11.5-kb, single-stranded RNA of positive polarity that encodes only a few proteins and contains a 5′ methylguanylate cap and a 3′ polyadenylate tail (42). The genome is directly translated into the viral nonstructural proteins nsP1 to nsP4, which are encoded by the 5′ two-thirds of the genome. Together with host factors, these proteins form the replicative enzyme complex that functions in the replication of the viral genome and transcription of the subgenomic RNA (43). The latter RNA (26S RNA) is synthesized from the promoter located on the minus-strand RNA replicative intermediate and translated into the structural proteins that, together with genomic RNA, form the viral particles (38). The structural proteins are dispensable for RNA replication, and, for some alphaviruses, the RNAs lacking structural genes (replicons) replicate as efficiently as viral genomes.
Previously, SINV nsP2 was found to play a critical role in the downregulation of cellular RNA polymerase I- and II-dependent transcription, and this transcriptional shutoff was hypothesized to be an efficient means of inhibiting the antiviral response developing during SINV replication (13, 14, 16). The point mutations in the carboxy-terminal domain of SINV nsP2 or in the cleavage site between nsP2 and nsP3 make this protein incapable of downregulating transcription but have only a minor effect on viral RNA replication (13, 16). The expression of wild-type (wt) SINV nsP2 from different vectors alone is sufficient to cause transcriptional shutoff and, ultimately, cell death (14).
Compared to SINV, replication of the VEEV- and EEEV-derived replicons appears to be less cytopathic (35). The latter replicons readily establish persistent replication in cells of vertebrate origin that have defects in alpha/beta interferon (IFN-α/β) production or signaling (35). This is an indication that replication of these virus-specific RNAs does not downregulate cellular transcription and translation to levels incompatible with cell survival (28, 35). These data suggest that the New World alphaviruses appear to use a mechanism of interference with cell response, which differs from that described for the Old World alphavirus SINV. These viruses either employ a more specific means of inhibiting transcription of virus-induced stress response genes and/or they use other proteins to function in a mode similar to that described for SINV nsP2 (14). Therefore, it was logical to expect that viral structural proteins, deleted from viral genomes in the course of designing the replicons, might have functions in addition to that of forming the infectious virions.
Alphavirus structural proteins include the capsid, the glycoproteins E2 and E1, and the short proteins E3 and 6K that serve as signaling peptides for the following glycoproteins. After cleavage by furine protease, E3 is released from the cells (29), while E2, E1, and 6K are associated with the membrane-containing cellular compartments (43). Therefore, functioning of these proteins in the regulation of cellular gene expression appears unlikely. The capsid protein is required for packaging viral RNA into viral particles, as it forms an icosahedral nucleocapsid that becomes surrounded by a lipid envelope (containing glycoprotein spikes) during budding through the cellular membrane. Thus, the capsid is distributed in the cellular cytoplasm during virus replication and appears to be a possible candidate for the regulation of cellular functions.
In this study, we compared the mechanisms of the transcriptional shutoffs caused by two New World (VEEV and EEEV) and two Old World (SFV and SINV) alphaviruses. We found that the viruses within these groups employ different (capsid- or nsP2-dependent) mechanisms of transcription inhibition. These data not only provide new insights into alphavirus evolution but also present a plausible explanation for the particular recombination events that lead to the formation of the western equine encephalitis virus (WEEV) from SINV- and EEEV-like ancestors (18, 50). That recombination pattern was essential for WEEV to acquire not only the nonstructural proteins but also capsid protein, functioning in transcription inhibition, from an EEEV-like virus.
MATERIALS AND METHODS
Cell cultures.
BHK-21 cells were kindly provided by Sondra Schlesinger (Washington University, St. Louis, MO). NIH 3T3 cells were obtained from the American Type Culture Collection (Manassas, Va.). Both cell lines were maintained at 37°C in alpha minimum essential medium (α-MEM) supplemented with 10% fetal bovine serum (FBS) and vitamins.
Plasmid constructs.
SINV, VEEV, and EEEV replicon-encoding plasmids (SINrep/Pac, VEErep/Pac, and EEErep/Pac) having a single subgenomic promoter that drives the expression of puromycin acetyltransferase (Pac) were described elsewhere (35). The SFrep/Pac replicon had essentially the same design and was developed using a pSFV1 replicon-encoding plasmid (Gibco). Plasmids encoding replicons of Venezuelan equine encephalitis virus (VEErepL) pVEErepL/GFP/Pac and pVEErepL/nsP2SIN/Pac were described elsewhere (14). The plasmid with replicon VEErepL/nsP2SF/Pac had essentially the same design as the previously developed VEErepL/nsP2SIN/Pac (14). SFV nsP2 was fused in frame with a ubiquitin (Ubi) sequence by using standard cloning techniques and cloned under control of the subgenomic promoter. VEErepL replicons expressing capsid proteins of different alphaviruses, VEErepL/CVEE/Pac, VEErepL/CEEE/Pac, VEErepL/CSIN/Pac, and VEErepL/CSF/Pac, had a common design, in which capsid-coding sequences of different alphaviruses were cloned under control of the subgenomic promoter and VEEV 26S RNA 5′ untranslated region (5′UTR). The chimeric VEE/SINV genome was described elsewhere (35). The genome of the second chimeric virus, EEE/SINV, had a very similar design, in which the sequences coding the 5′UTR of SINV 26S RNA and all of the structural genes were cloned into the EEEV genome to replace the corresponding sequences. The 3′UTR-encoding sequence in the chimeric genome was left EEEV specific. All of the constructs that we used are presented in the corresponding figures. Sequences of all of the recombinant plasmids can be provided upon request.
RNA transcriptions.
Plasmids were purified by centrifugation in CsCl gradients. Before being subjected to a transcription reaction, plasmids were linearized using the MluI or NotI restriction sites located downstream of the poly(A) sequence of VEEV replicons. RNAs were synthesized by SP6 RNA polymerase in the presence of a cap analog under previously described conditions (37). The yield and integrity of transcripts were analyzed by gel electrophoresis under nondenaturing conditions. The RNA concentration was measured on a FluorChem imager (Alpha Innotech), and transcription reactions were used for electroporation without additional purification.
Analysis of cytotoxicity of constructs.
BHK-21 cells were electroporated under previously described conditions (25). In all of the experiments, 5 μg of the in vitro-synthesized RNAs was used for electroporation of 5 × 106 cells. Next, the aliquots of the cells were seeded into six-well Costar plates to analyze cell proliferation and viability. Puromycin (Pur) selection (10 μg/ml) was performed between 6 and 48 h posttransfection. Then, cells were incubated in puromycin-free medium, and viable cells were counted at the times indicated in the figures. In parallel, different dilutions of the electroporated cells were seeded into 100-mm tissue culture dishes. At 6 h posttransfection, puromycin was added to the medium to a concentration of 10 μg/ml. Colonies of Purr cells were stained with crystal violet at days 4 to 9 posttransfection, depending on their growth rates. The results are presented in the figures in CFU per μg of RNA used for transfection.
Identification of adaptive mutations in capsid-coding genes.
Total RNA was isolated from the Purr cells by using TRizol according to the procedure recommended by the manufacturer (Invitrogen). Fragments encoding capsid genes were amplified using standard reverse transcription-PCR and the primers specific to the N-terminal fragment of the Pac gene and the C-terminal fragment of nsP4-coding sequence. Sequencing was performed on the gel-purified fragment by using the same primers.
Analysis of cellular transcription.
BHK-21 cells were electroporated with 5 μg of the in vitro-synthesized RNAs, and one-sixth of the cells were seeded into 35-mm culture dishes. At 6 h posttransfection, puromycin was added to the medium to a concentration of 10 μg/ml. At the indicated times postelectroporation, the cellular RNAs were labeled for the time periods indicated in the figure legends in complete α-MEM supplemented with 10% FBS and 20 μCi/ml [3H]uridine without the addition of actinomycin D (ActD). RNA isolation and analysis by agarose gel electrophoresis were performed as previously described (5). For quantitative analysis, the RNA bands were excised from the 2,5-diphenyloxazole-impregnated gels, and the radioactivity was measured by liquid scintillation counting.
In other experiments, we used an advantage of low levels of VEErepL replication, which leads to the presence of very low levels of 3H-labeled virus-specific RNAs in the samples of poly(A)+ and total RNAs isolated from the replicon-containing cells labeled in the absence of ActD. Thus, to assess the total RNA synthesis, the RNA samples on the Whatman 3MM paper were washed with cold 10% trichloroacetic acid, and radioactivity was measured by liquid scintillation counting and normalized based on the number of viable cells determined by the above-described tests. To measure poly(A)+ RNA synthesis, the latter RNA fraction was isolated from the samples of total RNA derived from the cells metabolically labeled with [3H]uridine using oligo(dT) magnetic beads and the procedure recommended by the manufacturer (Ambion). Radioactivity in the eluted samples was determined by liquid scintillation counting and normalized based on the number of viable cells.
Infectious center assay.
One microgram of in vitro-synthesized, full-length RNA transcripts of viral genomes was used per electroporation mixture. Tenfold dilutions of electroporated BHK-21 cells were seeded in six-well Costar plates containing subconfluent naïve cells. After 1 h of incubation at 37°C in a 5% CO2 incubator, cells were overlaid with 2 ml of MEM-containing 0.5% Ultra-Pure agarose supplemented with 3% FBS. Plaques were stained with crystal violet after 2 days incubation at 37°C.
Analysis of protein synthesis.
BHK-21 cells were transfected with replicons or infected with viruses. At the times indicated in the corresponding figure legends, the cells were washed three times with phosphate-buffered saline and then incubated for 1 h at 37°C in 0.8 ml of Dulbecco's modified Eagle's medium lacking methionine and supplemented with 0.1% FBS and 20 μCi/ml of [35S]methionine. After incubation, cells were scraped into the medium, pelleted by centrifugation, and dissolved in 300 μl of standard protein loading buffer. Equal amounts of proteins were loaded onto sodium dodecyl sulfate-10% polyacrylamide gels. After electrophoresis, gels were dried and autoradiographed.
Viral replication analysis.
BHK-21 and NIH 3T3 cells were seeded at a concentration of 5 × 105 cells/35-mm dish. After 4 h of incubation at 37°C, monolayers were infected at a multiplicity of infection (MOI) of 10 PFU/cell for 1 h, washed three times with phosphate-buffered saline, and overlaid with 1 ml of complete medium. At the indicated times postinfection, media were replaced by fresh media and virus titers in the culture fluids were determined by a plaque assay on BHK-21 cells as previously described (22).
RESULTS
The New World and the Old World alphavirus replicons strongly differ in cytotoxicity.
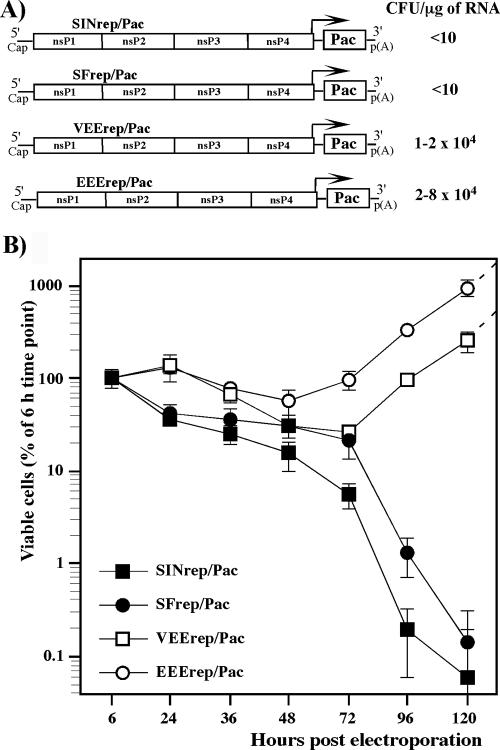
To examine the cytotoxic effect of the replication of different Old World and New World alphavirus-derived replicons, we constructed the VEEV, EEEV, SFV, and SINV genome-based VEErep/Pac, EEErep/Pac, SFrep/Pac, and SINrep/Pac replicons, which all had the same design. The replicons encoded virus-specific nsPs, 5′ and 3′ cis-acting RNA elements required for replication, and the subgenomic promoter driving the transcription of the subgenomic RNA, in which viral structural genes were replaced by a selectable marker, the Pac gene (Fig. 1). BHK-21 cells were transfected with the same amount of in vitro-synthesized replicon RNAs and then incubated in the presence of puromycin to evaluate the effect of replicons on cell growth and their ability to establish persistent replication and to form Purr foci. We assessed both the number of viable cells at different times posttransfection and Purr focus formation per μg of transfected RNA (see Materials and Methods for details). Electroporation delivered replicons into BHK-21 cells very efficiently, and essentially 100% of the cells were resistant to puromycin for at least 24 h. Then, cell growth and cell death depended on the origin of the replicons used (Fig. 1A and B). SINV- and SFV-specific replicons were cytotoxic, and, thus, very few cells survived and formed Purr foci (<10 cells/μg of RNA). The low efficiency of focus formation indicated that replicons in these surviving cells most likely had accumulated adaptive mutations in the nsPs that reduced their cytotoxicity and made them capable of persistent replication. VEEV and EEEV genome-based replicons, in contrast, readily established persistent replication in more than 20% and 90% of transfected cells, respectively. The efficiency of Purr focus formation approached 2 × 104 and 8 × 104 CFU/μg of transfected, in vitro-synthesized RNA, suggesting that the persistent replication of VEErep/Pac and EEErep/Pac in BHK-21 cells did not require adaptive mutations in the nsPs. The efficient growth of Purr cells suggested that changes in cell physiology were not strong, if there were any at all.
FIG. 1.
Analysis of cytotoxicity of the alphavirus replicons. (A) Schematic representation of the New World and the Old World alphavirus replicons. Arrows indicate positions of the subgenomic promoters. Pac indicates the puromycin acetyltransferase gene. RNA transfections and puromycin selection was performed as described in Materials and Methods. Purr colonies were stained with crystal violet, and the results are presented in CFU per μg of RNA used for transfection. (B) Analysis of cell growth and cell death at different times posttransfection. Equal numbers of cells were seeded into six-well Costar plates. Puromycin selection was performed between 6 and 48 h posttransfection and then cells were incubated in puromycin-free medium. The number of viable cells was counted at the indicated time points. The data were normalized based on the number of viable adherent cells determined at 6 h posttransfection. Error bars indicate variations between parallel samples.
These data implied that in spite of limited levels of identity in the nsPs (<70%), some biological attributes of SFV replicons (their cytopathogenicity and the inability to establish persistent replication) were similar to those exhibited by SINV-based constructs. Thus, the cytotoxicity appears to be a common feature of the Old World alphavirus-derived replicons. The New World alphavirus-based constructs, VEErep and EEErep, had a different common feature: they demonstrated a less cytopathic phenotype and were capable of establishing persistent replication.
The Old World alphaviruses use nsP2 to inhibit cellular transcription.
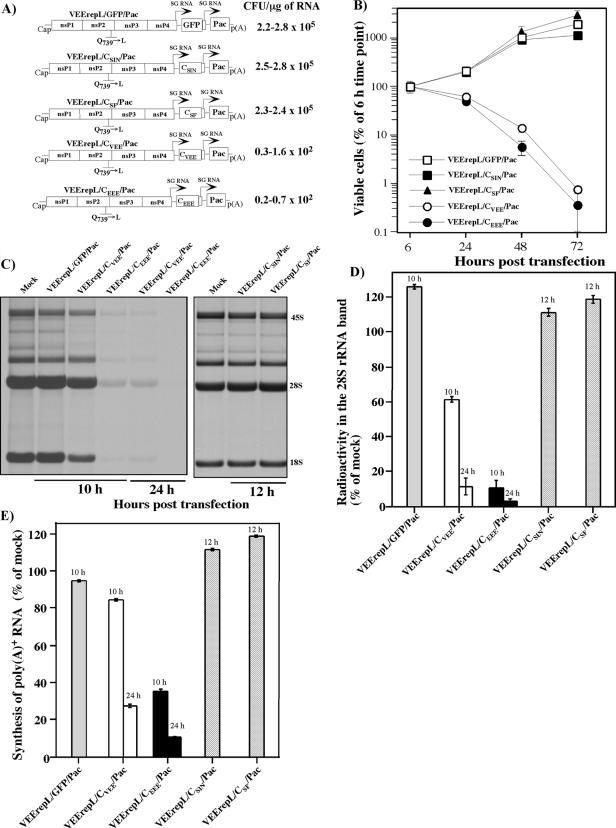
Our previous study indicated that SINV nsP2 is a key regulator of virus-host cell interactions and is directly involved in the inhibition of cellular transcription. To further understand whether these were unique characteristics of SINV nsP2 or common attributes of different Old World viruses, we compared the effects of SFV and SINV nsP2 expression on cell growth and transcription of cellular RNAs. Both nsP2s were expressed from a VEErepL replicon having a Q739→L mutation in its own nsP2 (Fig. 2A). As we demonstrated in the previous work, this replicon had the least effect on cell physiology because of a mutation in the nsP2 gene. It reduced RNA replication by almost 10-fold and made the replicon incapable of downregulating cellular transcription and translation and causing death of BHK-21 cells (14, 35). Most importantly, VEErepL/nsP2SIN/Pac and VEErepL/nsP2SF/Pac did not overexpress heterologous nsP2 (reference 14 and data not shown) but produced it at the levels normally found in virus-infected cells. Such expression made the data biologically relevant. VEErepL/nsP2SF/Pac and VEErepL/nsP2SIN/Pac cassettes contained two subgenomic promoters. One of them drove the expression of SINV and SFV nsP2 (Fig. 2A), fused in frame with the ubiquitin gene (Ubi). The use of Ubi was essential for production of nsP2 with the natural amino-terminal amino acid alanine. The second promoter controlled the expression of the Pac gene, which allowed us to use puromycin selection to eliminate a very low (<5%), but detectable, background of untransfected Purs cells (see Materials and Methods for details). The control replicon, VEErepL/GFP/Pac, encoded green fluorescent protein (GFP) instead of heterologous nsP2 (Fig. 2A).
FIG. 2.
Analysis of effects of nsP2 expression on cellular transcription and cell viability. (A) Schematic representation of VEE genome-based replicons expressing SINV and SFV nsP2 and analysis of their ability to establish persistent replication and develop Purr foci. Arrows indicate the positions of the subgenomic promoters. Ubi indicates a ubiquitin sequence fused in frame with the SINV and SFV nsP2 genes. (B) Analysis of growth of cells carrying VEEV replicons expressing GFP or SINV- and SFV-derived nsP2. (C) Inhibition of transcription in BHK-21 cells transfected with VEEV replicons expressing SINV or SFV nsP2. Cells were electroporated with 5 μg of in vitro-synthesized RNAs. At 10 and 24 h posttransfection, cellular RNAs were labeled with [3H]uridine in the absence of ActD for 3 h and analyzed by RNA gel electrophoresis under the conditions described in Materials and Methods. (D) For quantitative analysis of transcription inhibition, aliquots of RNA samples used for the gel shown in panel C were washed on Whatman 3MM filters with trichloroacetic acid as described in Materials and Methods, and the radioactivity was measured by liquid scintillation counting. (E) Another aliquot of each sample was used for isolation of the poly(A)+ RNA as described in Materials and Methods, and the radioactivity was measured by liquid scintillation counting. One of three reproducible experiments is presented; error bars indicate variations between parallel samples.
Upon delivery into BHK-21 cells, VEErepL/nsP2SIN/Pac and VEErepL/nsP2SF/Pac exhibited similar, cytopathic phenotypes that were very different from that of the control, VEErepL/GFP/Pac. In multiple experiments, the SINV and SFV nsP2-expressing replicons inhibited cell growth and, by 72 to 96 h posttransfection, killed the cells at similar rates (Fig. 2B). Compared to control VEErepL/GFP/Pac, both replicons were 1,000-fold less efficient in their ability to establish persistent replication and generated close to 200 Purr foci per μg of transfected RNA (compare to ∼2.5 × 105 foci formed by VEErepL/GFP/Pac RNA). We also sequenced SFV nsP2-encoding RNAs in the replicons present in randomly selected Purr colonies and found that cell survival and growth coincided with an accumulation of out-of-frame deletions, which inactivated the activity of nsP2 (data not shown). The cytotoxic effect of both SFV and SINV nsP2 can be explained, at least partially, by their ability to inhibit cellular transcription (Fig. 2C and D), because it occurred earlier than cell death. Both total RNA transcription (represented mainly by the synthesis of ribosomal RNAs) and RNA polymerase II-dependent cellular transcriptions, the poly(A)+ RNA synthesis (Fig. 2E), were strongly affected in the cells containing replicating VEErepL/nsP2SF/Pac and VEErepL/nsP2SIN/Pac. Taken together, the results indicated that very distantly related Old World viruses, SINV and SFV, inhibited cellular transcription using the nsP2-dependent mechanism, and for both viruses this protein appears to function in cytopathic effect (CPE) development.
The New World alphaviruses employ a capsid-dependent mechanism in the inhibition of transcription.
Replication of VEEV- and EEEV-specific replicons does not lead to cell death (Fig. 1) or inhibit cellular transcription (14). Nevertheless, the replication of VEEV and EEEV causes CPE and plaque formation, which suggests the possibility that structural genes have other functions than merely the formation of the viral particles. We were particularly interested in the functioning of the capsid protein, because in contrast to E2 and E1, this protein is distributed in the cytoplasm of infected cells and might interfere with the development of an antiviral response. Note that the ability of SFV and SINV nsP2 to inhibit cellular transcription does not rule out the possibility that structural proteins of these viruses interfere with the antiviral response as well. Therefore, we expressed the capsid protein of the New World and Old World viruses, VEEV, EEEV, SINV, and SFV, using the same noncytopathic VEErepL replicon (Fig. 3A). The constructs had exactly the same design and encoded capsid and Pac genes that were cloned under the control of separate subgenomic promoters. The replicons expressing VEEV and EEEV capsids formed Purr foci very inefficiently (Fig. 3A) and caused the death of essentially all of the cells within 48 to 72 h (Fig. 3B). In contrast, the expression of SINV and SFV capsids did not affect cellular biology. Transfected cells efficiently formed Purr foci and continued to grow at the same rates as those cells carrying control VEErepL/GFP/Pac replicon (Fig. 3A and B).
FIG. 3.
Effects of capsid expression on cellular transcription and cell growth. (A) Schematic representation of VEEV replicons expressing SINV, SFV, VEEV, and EEEV capsids. Arrows indicate positions of the subgenomic promoters. Different dilutions of the electroporated cells were seeded into 100-mm tissue culture dishes. Puromycin selection was performed as described in Materials and Methods. Purr cell colonies were stained with crystal violet at days 4 to 9 posttransfection, depending on their growth rates. The results are presented in CFU per μg of RNA used for transfection. The ranges indicate variations between the experiments. (B) Analysis of growth of cells transfected with VEEV replicons expressing GFP and different capsids. Equal numbers of cells were seeded into six-well Costar plates. Puromycin selection (10 μg/ml) was performed between 6 and 48 h posttransfection. Then, cells were incubated in puromycin-free medium, and viable cells were counted at the indicated times. The data were normalized based on the number of viable adherent cells determined at 6 h posttransfection. (C and D) Analysis of cellular transcription. RNA labeling was performed with [3H]uridine at the indicated times posttransfection for 2 h. RNA samples were analyzed by gel electrophoresis under the conditions described in Materials and Methods (C). For quantitative analysis, the rRNA bands were excised from the 2,5-diphenyloxazole-impregnated gels (C), and the radioactivity was measured by liquid scintillation counting (D). (E) Equal aliquots of each sample were used for isolation of the poly(A)+ RNA as described in Materials and Methods, and the radioactivity was measured by liquid scintillation counting. One of three reproducible experiments is presented; error bars indicate variations between parallel samples.
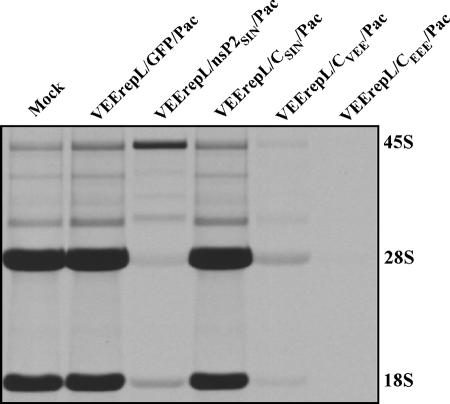
The expression of either VEEV or EEEV capsid led to the development of a phenomenon that was similar to that found in the cells expressing SINV- or SFV-specific nsP2. These capsid proteins were capable of inhibiting the transcription of cellular messenger and ribosomal RNAs (Fig. 3C, D, and E), while the EEEV-derived capsid was reproducibly more efficient than VEEV capsid in causing transcriptional shutoff. Within 6 to 10 h posttransfection with VEErepL/CEEE/Pac replicon, the RNA synthesis was inhibited to the level detected in VEErepL/CVEE/Pac-transfected cells only at 24 h postinfection, despite both capsids being expressed at the same levels (data not shown). Taken together, the data indicated that expression of SINV or SFV nsP2 and VEEV or EEEV capsid caused the same result, namely, development of the transcriptional shutoff. However, these proteins might have different mechanisms of action; in the SINV nsP2-expressing cells, we could reproducibly detect higher levels of 45S rRNA precursor and different patterns of partially processed RNAs than in normal or VEEV and EEEV capsid-expressing cells (Fig. 4). However, this observation needs further experimental support. Moreover, replication of SINV leads to a strong decrease in the cellular mRNA pool, suggesting the possibility of additional active degradation of mRNAs (16), and this does not appear to be the case during VEEV infection. We did not detect the rapid degradation of cellular mRNA templates in the microarray experiments performed on the poly(A)+ RNAs isolated from the cells infected with VEEV TC-83 (data not shown).
FIG. 4.
RNA synthesis in cells transfected with VEEV replicons expressing GFP, SINV nsP2, or different alphavirus capsids. BHK-21 cells were transfected with replicons expressing different proteins. RNAs were labeled with [3H]uridine in the absence of ActD for 5 h at 24 h posttransfection and analyzed as described in Materials and Methods.
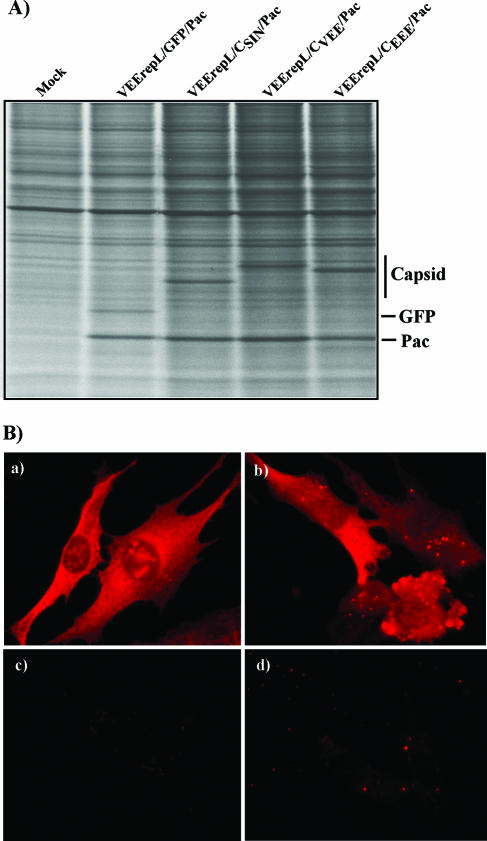
The difference between SINV and VEEV capsid functioning cannot be explained by different levels of their expression and/or intracellular compartmentalization. All of the capsids were synthesized by transfected replicons at very similar rates (Fig. 5A), and at least VEEV and SINV capsids, for which we had specific antibodies, were detected both in the cytoplasm and nucleus of transfected cells (Fig. 5B).
FIG. 5.
Synthesis and distribution of alphavirus capsids in cells transfected with VEEV replicons. (A) Cell were labeled with [35S]methionine at 10 h posttransfection as described in Materials and Methods, and equal amounts of proteins were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis followed by autoradiography. (B) For analysis of capsid distribution, BHK-21 cells were electroporated with in vitro-synthesized RNAs of VEErepL/CSIN/Pac and VEErepL/CVEE/Pac and then, at 20 h posttransfection, stained with mouse anti-SINV (a and c) and anti-VEEV (b and d) antibodies and goat anti-mouse immunoglobulin G-Alexa Fluor 546-labeled secondary antibodies (Molecular Probes). Cells: (a) VEErepL/CSIN/Pac-transfected cells, (b) VEErepL/CVEE/Pac, and (c and d) mock transfection.
The results indicated that the New World alphavirus-derived capsids were capable of downregulating RNA synthesis, as did the nsP2 proteins derived from the Old World alphaviruses. At the same time, the capsids of the Old World alphaviruses were very inefficient in inhibiting transcription, suggesting that these proteins are most likely not involved in regulation of the virus-induced cellular response in the infected cells.
Protease and RNA-binding activity of VEEV capsid is not essential for CPE induction.
It is generally believed that alphavirus capsid functions are limited to processing of the polyprotein precursor, binding to viral RNA, nucleocapsid formation, and interaction with the protein spikes in the viral envelope during virus budding through the cellular membrane. However, as shown above, the capsids in at least two representative members of the New World alphaviruses, VEEV and EEEV, which belong to distantly related complexes, appear to be directly involved in the development of CPE and transcription inhibition. Thus, they may play an additional, critical role(s) in virus replication. We tested if this newly discerned function of VEEV capsid is associated with its RNA-binding, positively charged sequence or with its protease activity. The latter possibility could be the simplest explanation for an inhibitory effect due to cleavage of transcription factors, as previously described for some of the picornaviruses (51).
VEEV capsid genes containing the deletion of amino acids (aa) 81 to 118 in the coding sequence (cluster of positively charged amino acids) and a mutated protease active site (S226→A mutation) were cloned into VEErepL/Pac (Fig. 6A). After transfection of the in vitro-synthesized RNAs, both constructs remained as cytopathic as a replicon expressing wt capsid and killed all of the cells at a similar rate (Fig. 6A and B). Thus, VEEV capsid functioning and its ability to cause cell death and inhibition of cellular transcription appear to be unassociated with protease activity and the RNA-binding domain. An alternative explanation might be that this protein has two activities that have a synergistic effect on CPE development.
FIG. 6.
Analysis of cytotoxicity of VEEV capsid with mutated protease or deleted RNA-binding domain. (A) Schematic representation of VEEV genome-based replicons expressing VEEV capsid containing an S226→A mutation or deletion of aa 81 to 118 and analysis of their abilities to establish persistent replication and develop Purr foci. (B) Survival of cells transfected with the replicons expressing wt capsid or capsid with the indicated mutations. The data were normalized based on the number of viable adherent cells determined at 6 h posttransfection.
To additionally understand the localization of the domain critical for capsid functioning in the inhibition of cellular transcription, we sequenced the capsid gene in the replicons present in the cells transfected with VEErepL/CVEE/Pac and VEErepL/CEEE/Pac and capable of growth in the puromycin-containing medium. A very small number of cells developed Purr foci (Fig. 3A and 6A), indicating that replicons most likely accumulated adaptive mutations, and, based on the previous works (8, 14), we expected mutations in the capsid-coding sequence, in the gene that was responsible for CPE development. The capsid-coding gene in the replicons present in the randomly selected colonies was amplified by reverse transcription-PCR and sequenced. The majority of detected mutations terminated the open reading frame of the CEEEV- and CVEEV-coding sequence (Fig. 7). However, some of the mutations found in the VEEV capsid having the mutated protease were more interesting (variants 8, 9, and 10), because they either changed only one amino acid (K51→E or Q52→P) or a short peptide between aa 57 and 88. The latter mutations suggested that the amino-terminal sequence upstream of the RNA-binding and protease domains is critical for VEEV capsid functions in transcription regulation and/or cytotoxicity. However, the identification of the exact boundaries of this putative functional domain needs more detailed investigation.
FIG. 7.
Accumulation of mutations in VEEV and EEEV capsids encoded by VEErepL replicon. Individual Purr cell colonies that formed after transfection of VEErepL/CVEE/Pac, VEErepL/CEEE/Pac, and VEErepL/CVEEmut/Pac replicons were randomly selected, and the genome fragment encoding capsid was sequenced. Positions of the mutations are indicated. Colony number 2 contained replicons with multiple deletions and/or insertions in the capsid-coding sequence that produced multiple sequences in the indicated fragment during the direct sequencing of the PCR fragment.
Alphaviruses with reduced cytopathogenicity.
The experiments described in the above sections suggested the possibility of designing alphaviruses having reduced levels of cytotoxicity. We hypothesized that recombinant viruses encoding VEEV or EEEV nonstructural genes and SINV structural genes would be less cytopathic than those expressing homologous nonstructural and structural proteins, because they would be incapable of efficiently interfering with cellular transcription. To test this hypothesis, we designed chimeric cDNAs of EEE/SINV genome, encoding homologous EEEV-derived nsP genes, 5′ and 3′ cis-acting RNA elements, and the subgenomic promoter (Fig. 8A). Structural genes and the 5′UTR of 26S RNA were derived from the SINV Toto1101 genome. The recombinant VEE/SINV (Fig. 8A) having the same design and structural genes of SINV strain TE12 came from our previous study (46). The in vitro-synthesized RNAs of both genomes were as infectious as the SINV Toto1101 RNA in the infectious center assay and produced viable viruses. These viruses were still capable of forming plaques in BHK-21 cells under agarose cover containing medium with 2 to 3% FBS. However, in spite of growing to titers higher than 109 PFU/ml, the designed chimeras did not cause CPE in either BHK-21 or NIH 3T3 cells growing in complete medium supplemented with 10% FBS (Fig. 8B, C, and D). In BHK-21 cells, these recombinants developed a persistent infection that did not noticeably interfere with cell growth (Fig. 8C), and the NIH 3T3 cells stopped virus replication and completely cleared the infection by 5 days postinfection (Fig. 8D). The inability to cause CPE could not be explained by low levels of RNA replication or structural protein production, because RNA and SINV structural proteins were synthesized at levels similar to those found in cells infected with nonrecombinant viruses (Fig. 8E).
FIG. 8.
Replication of viruses expressing homologous and heterologous structural and nonstructural proteins. (A) Schematic representation of the viral genomes. SINV-, VEEV-, and EEEV-specific sequences are indicated by white, black, and gray, respectively. (B) BHK-21 and NIH 3T3 cells were infected with the indicated viruses at an MOI of 10 PFU/cell. Cells were stained with crystal violet at day 3 (BHK-21) and day 5 (NIH 3T3) postinfection. (C and D) The media were replaced as described in Materials and Methods at 0, 3, 6, 9, 12 and 22 h postinfection (for all of the viruses), and VEE/SINV- and EEE/SINV-containing samples were also harvested later, at the indicated times. Virus titers were determined as described in Materials and Methods. (E) BHK-21 cells (5 × 105 cells in 35-mm dishes) were infected with SINV Toto1101, VEEV TC-83, VEE/SINV, and EEE/SINV at an MOI of 10 PFU/cell. At 16 h postinfection, proteins were pulse-labeled with [35S]methionine as described in Materials and Methods and analyzed on sodium dodecyl sulfate-10% polyacrylamide gels. The gels were dried and autoradiographed. The E1 proteins of VEE/SINV and EEE/SINV have differing mobilities on the gel because the SINV structural genes were derived from SINV TE12 and SINV Toto1101 strains, respectively, in which E1 differs by two amino acids (27, 37).
Thus, the data indicated that the inability of recombinant alphaviruses to produce proteins affecting cellular transcription, nsP2 or capsid, strongly decreased their ability to cause CPE. Such viruses are capable of either persisting in the BHK-21 cells that have defects in IFN-α/β signaling or are cleared from the NIH 3T3 cells having no defects in the IFN system, as we previously found for SINV variants having adaptive mutations in nsP2 and, thus, incapable of interfering with autocrine activity of IFN-α/β (13).
DISCUSSION
In the infected cells, alphavirus replication appears to initiate two competing processes: viruses redirect cell resources for their efficient replication and, at the same time, they induce a cell response aimed at downregulation of viral replication and activation of cell signaling, making uninfected cells resistant to successive rounds of infection. The balance between these two mechanisms determines the outcome of the infection and the development of viremia required for oral infection of mosquito vectors. As did most, if not all, of the other viruses, alphaviruses developed an efficient means of interference with the cellular response, and our data indicate that at least one of the mechanisms is based on the inhibition of cellular transcription (13, 16). Downregulation of cellular transcription, or pre-mRNA processing, or transport of cellular mRNAs from the nucleus to the cytoplasm appears to be a mechanism used by a variety of viruses. This phenomenon was previously described and intensively investigated for poliovirus and vesicular stomatitis virus infections (51, 53, 54). The nsPs of Rift Valley fever virus was also unambiguously demonstrated to be a critical factor for development of transcriptional shutoff (3, 7), and this phenomenon was found to be an important determinant of Rift Valley fever virus pathogenesis (4). It is reasonable to expect that the number of examples will continue to grow. In our previous studies, we found that SINV nsP2 is a cytotoxic protein and a critical determinant of transcriptional shutoff in infected cells (8, 13, 14, 16), and, based on the similarities between SINV and SFV replication (5, 24), we also expected that the nsP2 protein of the latter virus functions by a similar mode. The results of the present study suggest that this is in fact the case. SFV nsP2 indeed functions in transcription inhibition.
The puzzling moment in our previous studies was when both VEEV- and EEEV-based replicons, the replicons derived from the New World alphaviruses, were observed to be incapable of inhibiting transcription as efficiently as does the SINV-derived replicon (SINrep) (35). In contrast to the latter RNA, replication of VEEV- and EEEV-based constructs does not cause profound changes in cellular biology. Thus, the New World alphaviruses could develop different mechanisms of interference with cellular functions, and this mechanism is mostly independent of nsPs.
The high levels of sequence similarity in the structural and nonstructural genes strongly suggest that alphaviruses have a common ancestor. However, the Old World and the New World alphaviruses have circulated independently in different hemispheres for thousand years or longer and, therefore, developed mechanisms that differ from one another in terms of interference with activation of cellular genes that represent a virus-induced cell response. In both groups, viruses exhibit diversity in structural and nonstructural genes (36). Nevertheless they appear to exhibit common characteristics: (i) VEEV and EEEV (the New World cluster) encode capsid proteins that efficiently inhibit cellular transcription and, as a result, their expression ultimately leads to cell death, and (ii) SINV and SFV (the Old World cluster) nsP2 protein, but not capsid, is capable of performing the same functions, to inhibit transcription and cause CPE. Thus, the expression of these two proteins leads to the development of the same phenomenon that represents an important aspect of virus-host cell interactions. However, the differences in the spectra of rRNA precursors (Fig. 4) suggest that capsid- and nsP2-mediated transcriptional shutoffs might have different mechanisms of development.
We speculate that transcriptional shutoff might play a critical role in inhibiting the autocrine effect of IFN-α/β, in making cells incapable of downregulating viral replication in response to released IFN. However, the ability of the alphavirus-specific proteins to cause transcriptional shutoff and CPE development is certainly not the only process that determines viral pathogenesis. Moreover, its importance for virus replication in vivo remains to be determined. Nevertheless, the accumulated evidence suggests these functions play significant roles in virus replication: (i) point mutations in the carboxy-terminal part of nsP2 inactivate the ability of SINV or SFV to interfere with cellular transcription and development of host response (17) and make viruses highly attenuated in vivo, even after intracranialinoculation of young mice (data not shown). SINV and SFV replicons having nsP2-specific mutations are capable of persistent replication (34). (ii) Capsid-specific mutants of VEEV and EEEV have not yet been described; however, the recombinant viruses expressing heterologous SINV capsid (Fig. 8) demonstrate a less cytopathic phenotype that is very similar to those described for SIN nsP2 mutants (13). In accord with their reduced ability to induce CPE, these viruses are attenuated in mice and cause no mortality even after intracranial inoculation (data not shown). (iii) The nonhomologous recombination between two alphavirus-specific RNAs is a very efficient process (15, 40, 52) that readily proceeds in the fragment covering the junction between nonstructural and structural genes and usually leads to duplication of the subgenomic promoter. In contrast, the WEEV genome has been formed by two sophisticated recombination events between EEEV- and SINV-like viruses: one recombination occurred in the E3-coding sequence and the second was in the 3′UTR (18). The resulting virus had a genome encoding EEEV-derived nsPs and capsid, and only the envelope glycoproteins E2 and E1 were SINV specific. Most likely, this recombination strategy was essential not only for more efficient packaging of viral genetic material into nucleocapsids by homologous EEEV capsid, because capsids package genomes of heterologous alphaviruses very efficiently (9, 12, 31), but also for saving the EEEV-specific capsid-coding sequence in the recombinant virus to make its replication capable of interference with cell defense mechanisms. In addition, SINV glycoproteins (in contrast to envelope glycoproteins of other alphaviruses) are capable of making a shell around many heterologous nucleocapsids (I. Frolov, unpublished data). Initially, formation of these virions with heterologous capsids was probably less efficient, but alphaviruses are well known for a rapid accumulation of adaptive mutations. Thus, the cytoplasmic domain of WEEV E2 could readily adjust for better interaction with the nucleocapsid.
Alphavirus nucleocapsid assembly and capsid protein structure have been intensively studied by a number of research groups. The knowledge accumulated to date about the identity of structural elements is summarized in Fig. 9. The C-terminal domain contains a protease activity required for self-cleavage of capsid during the processing of structural polyprotein (6). The N-terminal fragment contains a highly positively charged RNA-binding domain that plays a critical role in the packaging of virus-specific RNA and a helix I that was suggested to play a central role in the assembly of nucleocapsid cores through coiled-coil interactions (33). In addition, the sequences coding the amino-terminal part of the Old World alphavirus capsids contain enhancers that strongly increase the translation of subgenomic RNA under conditions of translational shutoff caused by alphavirus infection (10, 11). The results of this work imply that this is not the entire list of capsid-related functions. The expression of VEEV- and EEEV-derived capsids induces CPE and strongly affects transcription of cellular ribosomal and messenger RNAs. These additional activities appear to be independent of protease functioning and the major RNA-binding domain of VEEV capsid, but they are at least partially determined by the sequence located between helix I and the RNA-binding, positively charged peptide. This peptide demonstrates a high level of conservation among the New World alphaviruses (VEEV, WEEV, and EEEV) and strongly differs from the sequence found downstream of helix I in the capsids of the Old World alphaviruses (Fig. 9). These variations certainly correlate with an inability of SINV and SFV capsids to downregulate transcription, but these structure-function differences need more detailed investigation. The mutations found in aa 51 to 87 of the VEEV capsid had a strong effect on its ability to inhibit transcription and cause CPE (Fig. 7 and 9), and the deletion of aa 81 to 118 had no effect on capsid function (Fig. 6). Therefore, further investigation of the function of the amino-terminal 81 aa represents a good starting point for future studies.
FIG. 9.
Schematic representation of VEEV capsid and sequence alignment with other alphavirus capsids. VEEV, EEEV, SINV, and SFV sequences are derived from references 20, 45, 42, and 44, respectively. All of the mutations identified in the capsids of replicons incapable of causing CPE (see variants 8, 9, and 10 in Fig. 7) are indicated in blue. Helix I sequences are indicated in red. Residues identical to those in the VEEV sequence are indicated by dashes. Stars indicate positions of the deletions introduced for better alignment of the sequences. The arrow indicates the beginning of the deletion made in the capsid of VEErepL/CVEEdel+/Pac mutant.
We will probably never be able to identify the mechanism or mechanisms that were used by the alphaviruses' ancestor for interfering with the development of the host cell response. In the original virus, such a mechanism may have been both capsid and nsP2 dependent or determined by one of the proteins or by none of them at all. Nevertheless, we hypothesize that at least one of the mechanisms is required for efficient circulation of the viruses under natural conditions. Chimeras between VEEV and SINV or between EEEV and SINV expressing no proteins capable of interfering with cellular transcription are less cytopathic and very attenuated. However, it should be noted that the combination of SINV nsP2 and VEEV or EEEV capsids in the reciprocal chimeric viruses, SIN/VEEV and SIN/EEEV, does not make them more pathogenic than parental constructs (31, 32). They cause very efficient CPE but remain attenuated in animal models because of the inefficient functioning of SINV replicative machinery in primary cells and/or less efficient packaging of the RNA into heterologous structural proteins.
In conclusion, we have demonstrated that (i) the Old World alphaviruses SINV and SFV developed the mechanism of interference with cellular transcription that depends on the nsP2, but not on the capsid, functioning. (ii) The New World alphaviruses EEEV and VEEV (and most likely WEEV) developed an alternative mechanism of inhibition of cellular transcription that is mainly determined by capsid protein, but not nsP2, functioning. (iii) Recombinant, chimeric viruses expressing the Old World alphavirus-derived capsid and the New World alphavirus-derived nsP2 are less cytopathic. They persistently replicate in cells with defects in IFN-α/β signaling, and cells without known defects in IFN-α/β production and signaling stop virus replication and clear the infection. (iv) The ability of VEEV capsid to inhibit cellular transcription appears to be determined by the amino-terminal fragment of the protein but not by its protease activity and the positively charged RNA-binding domain.
These new findings open an opportunity for developing new, safer alphavirus-based gene delivery and expression systems and new types of vaccines against VEEV and EEEV infections.
Acknowledgments
We thank Scott Weaver, Shinji Makino, and Mardelle Susman, technical editor, for critical reading and editing of the manuscript.
This work was supported by Public Health Service grant AI050537. S.P. was supported by NIH K08 grant AI059491.
Footnotes
Published ahead of print on 15 November 2006.
REFERENCES
- 1.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billecocq, A., M. Spiegel, P. Vialat, A. Kohl, F. Weber, M. Bouloy, and O. Haller. 2004. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J. Virol. 78:9798-9806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouloy, M., C. Janzen, P. Vialat, H. Khun, J. Pavlovic, M. Huerre, and O. Haller. 2001. Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J. Virol. 75:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi, H.-K., L. Tong, W. Minor, P. Dumas, U. Boege, M. G. Rossman, and G. Wengler. 1991. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature 354:37-43. [DOI] [PubMed] [Google Scholar]
- 7.Dasgupta, A. 2004. Targeting TFIIH to inhibit host cell transcription by Rift Valley Fever virus. Mol. Cell 13:456-458. [DOI] [PubMed] [Google Scholar]
- 8.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Prágai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frolov, I., E. Frolova, and S. Schlesinger. 1997. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 71:2819-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frolov, I., and S. Schlesinger. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70:1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolov, I., and S. Schlesinger. 1994. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol. 68:8111-8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolova, E., I. Frolov, and S. Schlesinger. 1997. Packaging signals in alphaviruses. J. Virol. 71:248-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and alpha/beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garmashova, N., R. Gorchakov, E. Frolova, and I. Frolov. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geigenmuller-Gnirke, U., B. Weiss, R. Wright, and S. Schlesinger. 1991. Complementation between Sindbis viral RNAs produces infectious particles with a bipartite genome. Proc. Natl. Acad. Sci. USA 88:3253-3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorchakov, R., E. Frolova, and I. Frolov. 2005. Inhibition of transcription and translation in Sindbis virus-infected cells. J. Virol. 79:9397-9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorchakov, R., E. Frolova, B. R. Williams, C. M. Rice, and I. Frolov. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 78:8455-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, C. S., S. Lustig, E. G. Strauss, and J. H. Strauss. 1988. Western equine encephalitis virus is a recombinant virus. Proc. Natl. Acad. Sci. USA 85:5997-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings, S., L. Martinez-Sobrido, A. Garcia-Sastre, F. Weber, and G. Kochs. 2005. Thogoto virus ML protein suppresses IRF3 function. Virology 331:63-72. [DOI] [PubMed] [Google Scholar]
- 20.Kinney, R. M., B. J. B. Johnson, J. B. Welch, K. R. Tsuchiya, and D. W. Trent. 1989. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology 170:19-30. [DOI] [PubMed] [Google Scholar]
- 21.Le May, N., S. Dubaele, L. Proietti De Santis, A. Billecocq, M. Bouloy, and J. M. Egly. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541-550. [DOI] [PubMed] [Google Scholar]
- 22.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levinson, R. S., J. H. Strauss, and E. G. Strauss. 1990. Complete sequence of the genomic RNA of O'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees. Virology 175:110-123. [DOI] [PubMed] [Google Scholar]
- 24.Liljeström, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. BioTechnology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 25.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, W. J., H. B. Chen, X. J. Wang, H. Huang, and A. A. Khromykh. 2004. Analysis of adaptive mutations in Kunjin virus replicon RNA reveals a novel role for the flavivirus nonstructural protein NS2A in inhibition of beta interferon promoter-driven transcription. J. Virol. 78:12225-12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lustig, S., A. Jackson, C. S. Hahn, D. E. Griffin, E. G. Strauss, and J. H. Strauss. 1988. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 62:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mason, P. W., A. V. Shustov, and I. Frolov. 2006. Production and characterization of vaccines based on flaviviruses defective in replication. Virology 351:432-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayne, J. T., C. M. Rice, E. G. Strauss, M. W. Hunkapiller, and J. H. Strauss. 1984. Biochemical studies of the maturation of the small Sindbis virus glycoprotein E3. Virology 134:338-357. [DOI] [PubMed] [Google Scholar]
- 30.Munoz-Jordan, J. L., G. G. Sanchez-Burgos, M. Laurent-Rolle, and A. Garcia-Sastre. 2003. Inhibition of interferon signaling by dengue virus. Proc. Natl. Acad. Sci. USA 100:14333-14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paessler, S., R. Z. Fayzulin, M. Anishchenko, I. P. Greene, S. C. Weaver, and I. Frolov. 2003. Recombinant sindbis/Venezuelan equine encephalitis virus is highly attenuated and immunogenic. J. Virol. 77:9278-9286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paessler, S., H. Ni, O. Petrakova, R. Z. Fayzulin, N. Yun, M. Anishchenko, S. C. Weaver, and I. Frolov. 2006. Replication and clearance of Venezuelan equine encephalitis virus from the brains of animals vaccinated with chimeric SIN/VEE viruses. J. Virol. 80:2784-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perera, R., K. E. Owen, T. L. Tellinghuisen, A. E. Gorbalenya, and R. J. Kuhn. 2001. Alphavirus nucleocapsid protein contains a putative coiled coil alpha-helix important for core assembly. J. Virol. 75:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perri, S., D. A. Driver, J. P. Gardner, S. Sherrill, B. A. Belli, T. W. Dubensky, Jr., and J. M. Polo. 2000. Replicon vectors derived from Sindbis virus and Semliki forest virus that establish persistent replication in host cells. J. Virol. 74:9802-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrakova, O., E. Volkova, R. Gorchakov, S. Paessler, R. M. Kinney, and I. Frolov. 2005. Noncytopathic replication of Venezuelan equine encephalitis virus and eastern equine encephalitis virus replicons in mammalian cells. J. Virol. 79:7597-7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Powers, A. M., A. C. Brault, Y. Shirako, E. G. Strauss, W. Kang, J. H. Strauss, and S. C. Weaver. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75:10118-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice, C. M., and J. H. Strauss. 1981. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc. Natl. Acad. Sci. USA 78:2062-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruggli, N., J. D. Tratschin, M. Schweizer, K. C. McCullough, M. A. Hofmann, and A. Summerfield. 2003. Classical swine fever virus interferes with cellular antiviral defense: evidence for a novel function of Npro. J. Virol. 77:7645-7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlesinger, S., and B. G. Weiss. 1994. Recombination between Sindbis virus RNAs. Arch. Virol. Suppl. 9:213-220. [DOI] [PubMed] [Google Scholar]
- 41.Sen, G. C., and S. N. Sarkar. 2005. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytokine Growth Factor Rev. 16:1-14. [DOI] [PubMed] [Google Scholar]
- 42.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 43.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takkinen, K. 1986. Complete nucleotide sequence of the non-structural protein genes of Semliki Forest virus. Nucleic Acids Res. 14:5667-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volchkov, V. E., V. A. Volchkova, and S. V. Netesov. 1991. Complete nucleotide sequence of the eastern equine encephalomyelitis virus genome. Mol. Genet. Mikrobiol. Virusol. 5:8-15. [PubMed] [Google Scholar]
- 46.Volkova, E., R. Gorchakov, and I. Frolov. 2006. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1-3 synthesis. Virology 344:315-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weaver, S. C., and I. Frolov. 2005. Togaviruses, p. 1010-1024. In B. W. J. Mahy and V. Meulen (ed.), Virology, vol. 2. IRL Press, Salisbury, United Kingdom. [Google Scholar]
- 49.Weaver, S. C., A. Hagenbaugh, L. A. Bellew, S. V. Netesov, V. E. Volchkov, G. J. Chang, D. K. Clarke, L. Gousset, T. W. Scott, D. W. Trent, et al. 1993. A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology 197:375-390. [DOI] [PubMed] [Google Scholar]
- 50.Weaver, S. C., W. Kang, Y. Shirako, T. Rumenapf, E. G. Strauss, and J. H. Strauss. 1997. Recombinational history and molecular evolution of western equine encephalomyelitis complex alphaviruses. J. Virol. 71:613-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weidman, M. K., R. Sharma, S. Raychaudhuri, P. Kundu, W. Tsai, and A. Dasgupta. 2003. The interaction of cytoplasmic RNA viruses with the nucleus. Virus Res. 95:75-85. [DOI] [PubMed] [Google Scholar]
- 52.Weiss, B. G., and S. Schlesinger. 1991. Recombination between Sindbis virus RNAs. J. Virol. 65:4017-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan, H., S. Puckett, and D. S. Lyles. 2001. Inhibition of host transcription by vesicular stomatitis virus involves a novel mechanism that is independent of phosphorylation of TATA- binding protein (TBP) or association of TBP with TBP-associated factor subunits. J. Virol. 75:4453-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuan, H., B. K. Yoza, and D. S. Lyles. 1998. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results from inactivation of TFIID. Virology 251:383-392. [DOI] [PubMed] [Google Scholar]