Abstract
To determine the demographic history of West Nile virus (WNV) in North America, we employed a coalescent method to envelope coding region data sets for the NY99 and WN02 genotypes. Although the observed genetic diversities in both genotypes were of approximately the same age, the mean rate of epidemiological growth of the WN02 population was approximately three times that of the NY99 population, a finding compatible with the recent dominance of the former genotype. However, there has also been a marked decrease in the recent growth rate of WN02, suggesting that WNV has reached its peak prevalence in North America.
The introduction of exotic agents into naïve ecosystems presents an ongoing challenge to public health, conservation, and biodefense. West Nile virus (WNV; Flavivirus; Flaviviridae) is a single-stranded, positive-sense RNA virus maintained in an enzootic cycle between Culex mosquitoes and birds. Most mammals, notably humans and horses, are dead-end hosts. Infection in vertebrates is usually mild or unapparent, although disease symptoms ranging from mild febrile illness to fatal encephalitis may occur. WNV first appeared in North America in August 1999 in the New York City area, where it resulted in an outbreak of encephalitis in human, avian, and equine communities. Since that time, more than 19,000 human cases have been documented in the United States (9). The first virus strain associated with the North American outbreak, designated NY99 due to its detection in New York in 1999, was most closely related to WNV strains isolated from Israel (8). In 2002, a second U.S. genotype—referred to as WN02 since it was first recognized as a significant entity in 2002—emerged, and although it is closely related to NY99, it belongs to a distinct phylogenetic lineage that seems to have displaced that of its predecessor (1, 3). WN02 has consequently been referred to as the North American genotype of WNV (1). Given the serious health consequences posed by introduced pathogens such as WNV, it is important to determine their epidemiological dynamics as they adapt to a naïve environment and to predict their future impact. To achieve this goal, we performed a Bayesian coalescent analysis of the recent spread of WNV in North America.
Nucleotide sequence data on American WNV isolates were provided in this study or downloaded from GenBank. Sequences generated for this study were obtained from naturally infected birds, mainly American crows (Corvus brachyrhynchos), collected by the New York state WNV surveillance program. Kidney tissue from dead birds was tested for the presence of WNV RNA by quantitative, real-time (TaqMan) reverse transcriptase PCR according to standard methods (7). A total of 39 WNV-positive tissue samples from 2004 and 2005 were selected (Table 1). The complete WNV envelope (E) coding sequence was amplified by reverse transcriptase PCR as three overlapping fragments. Reaction products were electrophoretically separated on a 2% agarose gel, and sequencing was conducted in both directions using a total of nine forward and nine reverse primers (sequences are available upon request) with an ABI 3700 DNA analyzer (Applied Biosystems, Foster City, CA). Raw sequence data were assembled and edited using the software package from DNAStar, Inc. (Madison, WI). A minimum of twofold redundancy was required for sequence data to be considered complete.
TABLE 1.
Isolates of WNV newly sequenced for this study
| Collection date | Strain designation | Sourcea | County of collection (NY) | GenBank accession no. |
|---|---|---|---|---|
| 11 May 2004 | 04000525 | American crow | Columbia | DQ823130 |
| 20 May 2004 | 04000630 | American crow | Cattaraugus | DQ823112 |
| 4 June 2004 | 04000729 | American crow | Suffolk | DQ823113 |
| 18 June 2004 | 04000920 | American crow | Albany | DQ823116 |
| 14 July 2004 | 04001397 | American crow | New York | DQ823114 |
| 14 July 2004 | 04001515 | American crow | Herkimer | DQ823117 |
| 19 July 2004 | 04001462 | American crow | Chautauqua | DQ823115 |
| 24 July 2004 | 04001812 | American crow | Niagara | DQ823118 |
| 26 July 2004 | 04001893 | American crow | Suffolk | DQ823119 |
| 28 July 2004 | 04001923 | American crow | Jefferson | DQ823120 |
| 29 July 2004 | 04001932 | American crow | Ontario | DQ823121 |
| 12 Aug. 2004 | 04002395 | American crow | Genesee | DQ823122 |
| 20 Aug. 2004 | 04002509 | American crow | Richmond | DQ823123 |
| 25 Aug. 2004 | 04002534 | American crow | Monroe | DQ823124 |
| 15 Sept. 2004 | 04002702 | American crow | Chautauqua | DQ823125 |
| 15 Sept. 2004 | 04002903 | American crow | Ulster | DQ823129 |
| 19 Sept. 2004 | 04002793 | American crow | Oswego | DQ823127 |
| 20 Sept. 2004 | 04002772 | American crow | Nassau | DQ823126 |
| 29 Sept. 2004 | 04002848 | American crow | Queens | DQ823128 |
| 10 Feb. 2005 | 05000918 | American crow | Dutchess | DQ823132 |
| 27 July 2005 | 05001729 | Blue jay | Bronx | DQ823131 |
| 2 Aug. 2005 | 05001782 | Northern mockingbird | Kings | DQ823134 |
| 16 Aug. 2005 | 05001900 | American crow | Lewis | DQ823133 |
| 16 Aug. 2005 | 05001902 | American crow | Nassau | DQ823135 |
| 18 Aug. 2005 | 05001938 | American crow | Onondaga | DQ823136 |
| 18 Aug. 2005 | 05001949 | American crow | Suffolk | DQ823137 |
| 22 Aug. 2005 | 05001962 | American crow | Monroe | DQ823138 |
| 22 Aug. 2005 | 05001970 | American crow | Queens | DQ823140 |
| 23 Aug. 2005 | 05001967 | American crow | Niagara | DQ823139 |
| 25 Aug. 2005 | 05002031 | American crow | Chautauqua | DQ823141 |
| 28 Aug. 2005 | 05002118 | Blue jay | Rockland | DQ823144 |
| 3 Sept. 2005 | 05002079 | American crow | Albany | DQ823142 |
| 6 Sept. 2005 | 05002170 | American crow | Erie | DQ823143 |
| 7 Sept. 2005 | 05002274 | American crow | Broome | DQ823146 |
| 11 Sept. 2005 | 05002374 | House sparrow | Queens | DQ823147 |
| 13 Sept. 2005 | 05002232 | Blue jay | Ontario | DQ823145 |
| 3 Oct. 2005 | 05002412 | American crow | Nassau | DQ823148 |
| 11 Oct. 2005 | 05002553 | American crow | Onondaga | DQ823149 |
| 19 Oct. 2005 | 05002688 | American crow | Rockland | DQ823150 |
American crow, Corvus brachyrhynchos; blue jay, Cyanocitta cristata; northern mockingbird, Mimus polyglottos; house sparrow, Passer domesticus.
To conduct our coalescent analysis, we compared the E coding region sequences from 46 and 110 NY99 and WN02 isolates, respectively, from samples obtained between 1999 and 2005. Approximately 70% of sequences came from samples from avian species. Rates of nucleotide substitution and population growth, as well as times of origin, were estimated using a Bayesian Markov chain Monte Carlo method (MCMC) (program BEAST; http://evolve.zoo.ox.ac.uk/beast/) (2). Four models of demographic history were compared—constant population size and exponential, logistic, and expansion population growth—as well as a Bayesian skyline plot which provides a piecewise graphical depiction of demographic history, and both strict and relaxed (uncorrelated exponential) molecular clocks. Akaike's information criterion was used to determine the best-fit model, with uncertainty in parameter estimates reflected in the 95% highest-probability-density (HPD) values. All MCMC chains were run for a sufficient number of generations to ensure convergence and assessed using the Tracer program (http://evolve.zoo.ox.ac.uk/software.html?id=tracer). The epidemic doubling time (λ) was calculated using the following equation: λ = ln (2)/r, where r is the population growth rate estimated by BEAST. All estimates utilized the HYK85 model of nucleotide substitution.
Mean rates of evolutionary change estimated under the best-fit relaxed molecular clock model were similar for NY99 and WN02, at approximately 3 × 10−4 nucleotide substitutions per site per year (Table 2). These rates are similar to those observed for other RNA viruses, including members of the Flaviviridae (4, 6). At these rates, the mean ages of the sampled genetic diversities (most recent common ancestors) in NY99 and WN02 were 8 and 6 years, respectively. Although these ages are compatible with epidemiological records, they suggest that the WN02 genotype arose some years before it was first detected in 2001.
TABLE 2.
Bayesian estimates of population dynamic and evolutionary parameters for North American WNV
| Strain | No. of samples | Range of dates of sample collection | Molecular clock type | Best-fit demographic model | Effective no. of infections (95% HPD) | Substitution ratea (95% HPD) | Mean age (yr) of MRCAb (95% HPD) | Mean rate of population growthc (95% HPD) | Mean epidemic doubling timed (mo) (95% HPD) |
|---|---|---|---|---|---|---|---|---|---|
| WN02 | 110 | 2001-2005 | Relaxed | Logistic growth | 1,508 (97-3,560) | 2.967 × 10−4 (1.493 × 10−4 to 4.451 × 10−4) | 6.070 (4.040-8.588) | 6.015 (1.114-16.382) | 1.370 (0.508-7.467) |
| NY99 | 46 | 1999-2003 | Relaxed | Exponential growth | 18,880 (720-38,110) | 3.663 × 10−4 (0.180 × 10−4 to 6.521 × 10−4) | 7.613 (4.075-15.622) | 1.776 (0.523-2.865) | 4.683 (1.114-15.904) |
Mean number of nucleotide substitutions per site per year.
MRCA, most recent common ancestor.
Mean number of new infections per individual host animal per year.
Time required for the effective number of infections to double in size, calculated using the relation λ = ln (2)/r, where r is the population growth rate.
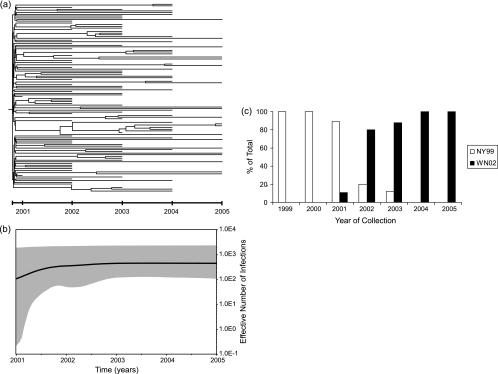
More notable was the contrasting epidemiological dynamics of the NY99 and WN02 genotypes. Whereas a model of exponential population growth was the best-fit model for NY99, as expected given the spread of this genotype in North America, the demographic history of WN02 followed a model of logistic population growth, in which an initially rapid growth phase is followed by a slowdown in the growth rate (Fig. 1). The rapid growth phase is apparent in the bottom-heavy phylogeny for this genotype, where most lineages arose prior to 2002, and corresponds to a mean growth rate of six new infections per individual host animal per year, or an epidemic doubling time of approximately 1 month. In comparison, the mean rate of population growth for NY99 over its sampling period (1999 to 2003) was two infections per host per year, equivalent to an epidemic doubling time of approximately 5 months. The displacement of NY99 by WN02 therefore occurred so rapidly that the decline in the prevalence of NY99 was not apparent in our analysis. These epidemiological dynamics were confirmed with a second analysis of 39 WN02 E gene sequences isolated from 2004 to 2005 for which the exact day of sampling was available (Table 1). Again, a model of logistic population growth was supported, with a mean substitution rate of 3.597 × 10−4 substitutions/site/year (95% HPD, 0.402 × 10−4 to 7.941 × 10−4 substitutions/site/year), an inferred age of 7.714 years (95% HPD, 1.842 to 19.415 years), and an initial growth rate of 10.702 infections year−1 (95% HPD, 0.568 to 33.916 infections year−1). Notably, the period of the highest growth of WN02 (i.e., during its rapid emergence and cocirculation with NY99) coincides with the peak in the number of human cases reported to the U.S. Centers for Disease Control and Prevention in 2002 and 2003 (5).
FIG. 1.
(a) Maximum a posteriori phylogenetic tree of 110 WN02 genotype viruses from samples obtained during the period from 2001 to 2005. For all branches, the times assigned to each tip correspond to the dates of sampling. (b) Bayesian skyline plot for the WN02 genotype. The bold line represents the median estimate of the effective number of infections through time, with the 95% HPD values shown in the shaded area. The effective number of infections, a measure of relative genetic diversity, is given as Neτ, where Ne is the effective population size and τ is the generation time. (c) Relative proportions of the NY99 and WN02 genotypes from 1999 to 2005 among the virus isolates analyzed in this study.
Although reliance on viruses drawn largely from birds raises the possibility that our sampling is not representative, the results of the coalescent analyses are highly concordant with epidemiological and epizootiological records, indicating that the approach is robust. In addition, phylogenetic trees of North American WNV show little spatial structure, and there is no evidence for host-dependent evolutionary patterns in WNV. Therefore, sampling bias is unlikely to have had a significant impact on our findings.
We propose that an increased mosquito transmission efficacy of WN02 is most likely responsible for its displacement of NY99. WN02 strains are transmitted by Culex pipiens after approximately two fewer days of extrinsic incubation than NY99, leading to significant increases in the vectorial capacity of WN02- compared to NY99-infected mosquitoes (3). Our data on genotype-specific growth rates and epidemic doubling times support this observation, although future experimental verification may shed additional light on the mechanistic basis for the genotype displacement. Finally, although WN02 has displaced NY99, there is no evidence that the population of this currently dominant genotype is growing. In sum, these results suggest that WNV has reached peak prevalence in North America. Consequently, in the absence of additional fitness increases produced by ongoing WNV evolution, future epidemics in North America are likely to be driven by host and environmental factors.
Nucleotide sequence accession numbers.
The sequence data newly generated here have been deposited in GenBank and assigned the accession numbers DQ823112 to DQ823150.
Acknowledgments
We thank the New York State Department of Health, the New York State Wildlife Pathology Unit, the Wadsworth Center Arbovirus Laboratories, and the Wadsworth Center Molecular Genetics Core facility.
This work was funded in part by National Institutes of Health contract N01-AI25490 and Centers for Disease Control and Prevention contract U50/CCU223671-02.
Footnotes
Published ahead of print on 20 December 2006.
REFERENCES
- 1.Davis, C. T., G. D. Ebel, R. S. Lanciotti, A. C. Brault, H. Guzman, M. Siirin, A. Lambert, R. E. Parsons, D. W. Beasley, R. J. Novak, D. Elizondo-Quiroga, E. N. Green, D. S. Young, L. M. Stark, M. A. Drebot, H. Artsob, R. B. Tesh, L. D. Kramer, and A. D. T. Barrett. 2005. Phylogenetic analysis of North American West Nile virus isolates, 2001-2004: evidence for the emergence of a dominant genotype. Virology 342:252-265. [DOI] [PubMed] [Google Scholar]
- 2.Drummond, A. J., S. Y. W. Ho, M. J. Phillips, and A. Rambaut. 14 March 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ebel, G. D., J. Carricaburu, D. Young, K. A. Bernard, and L. D. Kramer. 2004. Genetic and phenotypic variation of West Nile virus in New York, 2000-2003. Am. J. Trop. Med. Hyg. 71:493-500. [PubMed] [Google Scholar]
- 4.Hanada, K., Y. Suzuki, and T. Gojobori. 2004. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol. Biol. Evol. 21:1074-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes, E. B., N. Komar, R. S. Nasci, S. P. Montgomery, D. R. O'Leary, and G. L. Campbell. 2005. Epidemiology and transmission dynamics of West Nile virus disease. Emerg. Infect. Dis. 11:1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenkins, G. M., A. Rambaut, O. G. Pybus, and E. C. Holmes. 2002. Rates of molecular evolution in RNA viruses: a quantitative phylogenetic analysis. J. Mol. Evol. 54:152-161. [DOI] [PubMed] [Google Scholar]
- 7.Kauffman, E. B., S. A. Jones, A. P. Dupuis II, K. A. Ngo, K. A. Bernard, and L. D. Kramer. 2003. Virus detection protocols for West Nile virus in vertebrate and mosquito specimens. J. Clin. Microbiol. 41:3661-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 9.Monta, T. P., J. Liu, N. Kanesa-Thasan, G. A. Myers, R. Nichols, A. Deary, K. McCarthy, C. Johnson, T. Ermak, S. Shin, J. Arroyo, F. Guirakhoo, J. S. Kennedy, F. A. Ennis, S. Green, and P. Bedford. 2006. A live, attenuated recombinant West Nile virus vaccine. Proc. Natl. Acad. Sci. USA 103:6694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]