Abstract
Cells infected with herpes simplex virus type 1 (HSV-1) were conventionally embedded or freeze substituted after high-pressure freezing and stained with uranyl acetate. Electron tomograms of capsids attached to or undergoing envelopment at the inner nuclear membrane (INM), capsids within cytoplasmic vesicles near the nuclear membrane, and extracellular virions revealed the following phenomena. (i) Nucleocapsids undergoing envelopment at the INM, or B capsids abutting the INM, were connected to thickened patches of the INM by fibers 8 to 19 nm in length and ≤5 nm in width. The fibers contacted both fivefold symmetrical vertices (pentons) and sixfold symmetrical faces (hexons) of the nucleocapsid, although relative to the respective frequencies of these subunits in the capsid, fibers engaged pentons more frequently than hexons. (ii) Fibers of similar dimensions bridged the virion envelope and surface of the nucleocapsid in perinuclear virions. (iii) The tegument of perinuclear virions was considerably less dense than that of extracellular virions; connecting fibers were observed in the former case but not in the latter. (iv) The prominent external spikes emanating from the envelope of extracellular virions were absent from perinuclear virions. (v) The virion envelope of perinuclear virions appeared denser and thicker than that of extracellular virions. (vi) Vesicles near, but apparently distinct from, the nuclear membrane in single sections were derived from extensions of the perinuclear space as seen in the electron tomograms. These observations suggest very different mechanisms of tegumentation and envelopment in extracellular compared with perinuclear virions and are consistent with application of the final tegument to unenveloped nucleocapsids in a compartment(s) distinct from the perinuclear space.
Electron microscopic studies have shown that cells infected with herpesviruses contain four types of capsids. Procapsids, believed to represent the progenitor of other capsid types, contain a porous outer shell that is roughly spherical, surrounding an inner protein shell or scaffold (23, 33). Type A capsids consist mostly of a 125-nm-diameter protein shell that comprises 12 vertices of fivefold symmetry (termed pentons) and 20 identical planar facets (53). Collectively, the planar facets are composed of 150 sixfold symmetrical subunits or hexons. Type B capsids have an outer protein shell that is indistinguishable from that of A capsids but also contain an inner, roughly spherical protein shell (19, 52). Type C capsids, or nucleocapsids, also contain an identical outer shell but lack the inner shell, which is replaced with the densely packed linear double-stranded DNA genome (5, 49).
In a process unique to herpesviruses, nucleocapsids bud through the inner nuclear membrane (INM) of infected cells into the perinuclear space, a compartment that is continuous with the lumen of the endoplasmic reticulum (ER), to become virions. The envelope of these nascent virions is therefore derived from the INM. In alphaherpesviruses, nucleocapsids are enveloped preferentially over A or B capsids (40). In one model of virion egress, the virion envelope derived from the INM fuses with the outer nuclear membrane (ONM), dumping the nucleocapsid into the cytoplasm, where it subsequently undergoes a second envelopment step(s) in the Golgi apparatus, in the trans-Golgi apparatus network, or in vesicles derived from these compartments (6, 31, 45). Others have proposed that some nucleocapsids pass through expanded nuclear pores, receiving tegument prior to or at the time of budding into other organelles such as the ER or Golgi apparatus (28, 48). (For purposes of discussion this will be termed the expanded nuclear pore model.) In both of these cases, the final virion envelope is (in the deenvelopment model) or can be (in the expanded nuclear pore model) derived from organelles other than the INM. Alternatively, it has been proposed that one or more enveloped virions bud from the ONM, forming transport vesicles. Fusion of the outer membrane of the transport vesicle with the Golgi apparatus or other membranes would deliver enveloped virions into the Golgi apparatus lumen for glycoprotein processing or, upon fusion with the plasma membrane, would deliver virions within the vesicle to the extracellular space (26). Thus, in this model, the original virion envelope derived from the INM is retained throughout virion egress.
The space between the nucleocapsid surface and the inner surface of the virion envelope is termed the tegument. The tegument of mature extracellular virions contains approximately 20 different proteins of varying stoichiometry, and electron tomography has shown that the tegument is arranged asymmetrically around the nucleocapsid as a series of concentric rings (22). Attachments of the innermost layer of mature tegument to capsid pentons have been observed, suggesting that they serve to anchor the tegument layer to the nucleocapsid (51). The means of attachment of the tegument to the virion envelope is uncertain, although interactions between some tegument proteins and some glycoproteins have been detected (8, 12, 17, 21).
The structure of the nucleocapsid has been determined to a resolution of 0.85 nm from electron cryomicroscopic studies, and this structure presumably reflects that of nucleocapsids within perinuclear virions (52). Other features of perinuclear virions, however, are poorly characterized. Transmission electron microscopic studies have indicated that perinuclear virions are morphologically distinct from extracellular particles. In particular, extracellular virions contain prominent spikes protruding from the virion envelope and a very densely stained tegument layer. Compared to their counterparts in extracellular virions, the external spikes of perinuclear virions are much less prominent, the tegument appears much less dense, and the virion envelope often appears denser (20). It is logical to propose that the tegument of perinuclear virions is relatively less dense because it contains less protein than its counterpart in extracellular virions, but this difference has not been well defined. In any event, these observations suggest that tegument proteins are applied to an unenveloped nucleocapsid, thus supporting either the expanded nuclear pore or deenvelopment model of virion egress.
Because perinuclear virions are difficult to separate from isolated cytoplasmic and extracellular virions, few studies have been undertaken to characterize the biochemical composition of perinuclear virions. By immunogold analyses, it has been determined that perinuclear virions and the INM of infected cells contain at least the integral membrane glycoproteins B, C, D, and M (gB, gC, gD, and gM, respectively), a myristylated and palmitoylated protein encoded by UL11, VP16 (at least when fused to green fluorescent protein), and a protein complex containing the integral membrane protein pUL34 and its interaction partner pUL31 (1, 2, 9, 24, 24, 32, 39, 47). Interestingly, the UL31 and UL34 proteins are largely absent from extracellular virions, suggesting that the initial virion envelope derived from the INM is replaced during virion egress (18, 39).
Under the hypothesis that such information would provide insight into the assembly pathway of herpesviruses and the envelopment reaction in particular, the major goal of this study was to generate a three-dimensional structure of nascent virions undergoing envelopment at the INM.
MATERIALS AND METHODS
At 16.5 h postinfection, herpes simplex virus type 1 (HSV-1) [strain HSV-1(F)]-infected Vero cells were rinsed with serum-free Dulbecco modified Eagle medium and then scraped into cold serum-free Dulbecco modified Eagle medium plus 150 mM sucrose and pelleted at 250 × g for 5 min. As much supernatant as possible was removed without disturbing the pellet. From this point on, the cells were kept on ice.
Conventional fixation.
Cells were fixed with 4% paraformaldehyde (Electron Microscopy Sciences) and 0.25% glutaraldehyde (Electron Microscopy Sciences) in 0.1 M NaP buffer, pH 7.4, for 30 minutes at room temperature and then 90 minutes at 4°C. Cells were washed three times for 5 min each with 0.1 M sodium phosphate buffer and then dehydrated with a graduated series of ethanol concentrations (10%, 30%, 50%, 70%, and 100%) at 4°C and then −20°C. This was followed by stepwise infiltration with Epon 812 resin (Electron Microscopy Sciences catalogue no. 14381) over the course of 48 h at −20°C. Samples were aliquoted into gel capsules, and the resin was polymerized by exposure to UV light.
High-pressure freezing/freeze substitution.
A cell slurry was made by resuspending pelleted infected cells in a small amount of medium. Microdialysis tubing (200-μm diameter; Spectrum Labs) was filled with the cell slurry by capillary action and immediately submerged in 1-hexadecene. The tubing was cut with a scalpel to fit Staehelin freezing hats (Ted Pella). Each hat was filled with four to six pieces of tubing. The remaining empty space was filled with 1-hexadecene. Samples were frozen with liquid nitrogen under high pressure using a Bal-Tec HPM010 machine. Samples were then freeze substituted in 0.5% glutaraldehyde with 0.1% tannic acid in acetone at −90°C for 24 h. The acetone was replaced three times at −90°C for 30 min each time. Substitution was continued with 1% OsO4 made from crystal and 0.1% uranyl acetate in acetone at −90°C for 24 h. The temperature was allowed to rise to −20°C overnight. The acetone was replaced in three sequential steps with Epon 812/araldite, first at −20°C, then at 4°C, and finally at room temperature. Each replacement step took 1 h.
Samples were put into 50/50 uncatalyzed Epon-Araldite-acetone at room temperature overnight and were then changed to 100% uncatalyzed Epon-Araldite for 24 h. Samples were then placed into catalyzed Epon-Araldite for 8 h and then changed into fresh catalyzed Epon-Araldite and cured at 60°C for 48 h.
Electron tomography and three-dimensional reconstruction.
Sections were cut at a 220-nm thickness and collected on Formvar-carbon-coated 200-mesh copper grids. Grids were poststained with 2% uranyl acetate for 20 min and Reynolds lead citrate for 5 min. Dual-axis tomographic tilt series were carried out with an angular range of 120° and a tilt interval of 2°. Tilt images were recorded with a JEOL JEM-4000FX operated at 400 kV in zero-loss mode using a Gatan GIF 2002 imaging energy filter. The image pixel size relative to the specimen was 1 nm. The total electron dose for a dual-axis tilt series was about 10,000 e−/nm2.
Tilt images were aligned, and tomographic reconstructions were computed using SPIDER (16, 36). The limits to resolution in the reconstructions, set by data collection geometry and specimen size, were approximately 5 to 6 nm in the section plane and 7 to 8 nm normal to the section (15). Tomograms were examined slice by slice using WEB (34) and ImageJ (http://rsb.info.nih.gov/nih-image), and 1.0-nm-thick slices were selected for display. Surface-rendered models were made using Amira. The three-dimensional models shown came from a single representative tomogram.
RESULTS
HEp-2 cells were infected with HSV-1(F) at 5.0 PFU per cell and at 16.5 h after infection were either Epon embedded and processed conventionally or high-pressure frozen and freeze substituted. In both cases, three-dimensional (tomographic) images were reconstructed from tilt series of recorded images.
Although features in the tegument layer of high-pressure-frozen freeze-substituted samples were similar to those observed in samples processed conventionally, the appearance of DNA within the nucleocapsid varied considerably in samples prepared using the different techniques. High-pressure-frozen, freeze-substituted specimens contained densely stained cores that mostly filled the inner cavity of the nucleocapsid, whereas conventionally fixed material contained considerably more space between the inner surface of the nucleocapsid and the electron opaque core (compare virion in Fig. 6 that was fixed conventionally with virions in Fig. 1, 3, and 5, which were derived from high-pressure-frozen, freeze-substituted material). As a result of these observations and the implication that freeze substitution was less prone to fixation artifacts, features in tomographic reconstructions detailing virion morphology were always verified in freeze-substituted specimens.
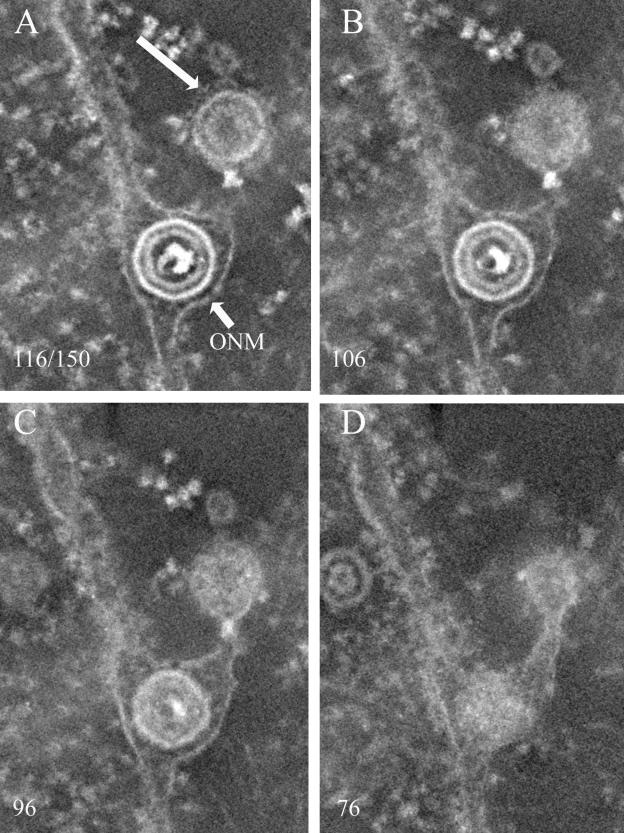
FIG. 6.
Electron tomographic reconstruction of virion in the perinuclear space and putative transport vesicle. Cells were infected with HSV-1(F) and were embedded and stained conventionally. The number of the slice in the reconstruction (of 150 total) is indicated in each image. The membrane of the putative transport vesicle is indicated with a large arrow. The position of the ONM is indicated with a small arrow.
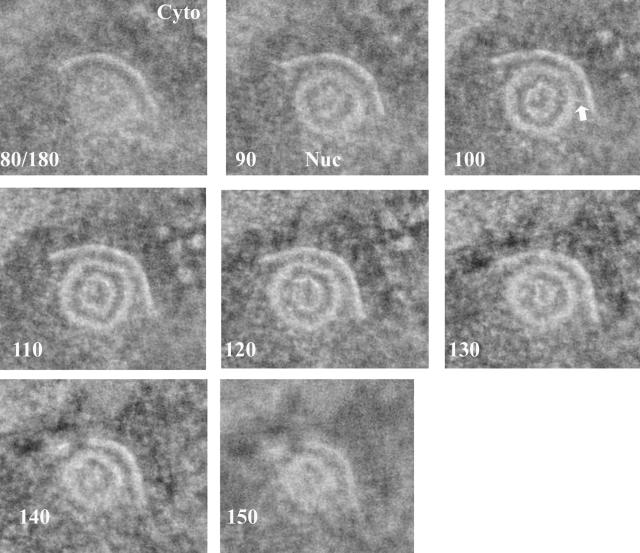
FIG. 1.
One-nanometer-thick slices from a tomographic reconstruction of a type B capsid attached to the INM. Cells infected with HSV-1 were high-pressure frozen and freeze substituted. The slice number of each image (out of a total of 180 slices) is indicated in the lower left of each panel. The cytoplasm is at the top of the image, and the nucleus is at the bottom. An arrow indicates a single bridging rod emanating obliquely from the capsid surface. Cyto, cytoplasm; Nuc, nucleus.
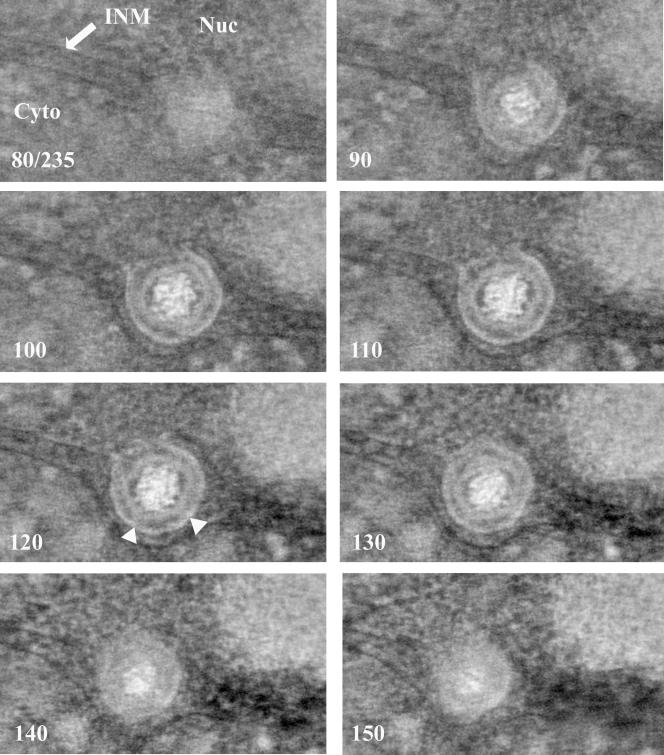
FIG. 3.
One-nanometer-thick slices from a tomographic reconstruction of a nucleocapsid completing envelopment at the INM. The sample was prepared as for Fig. 1. The slice number of each image (out of a total of 235 slices) is indicated in the lower left of each panel. The position of the INM is indicated with an arrow. Triangles in slice 120 indicate two adjacent angular vertices with bridging rods, whereas the relatively planar surface between them is largely devoid of bridging rods. Nuc, nucleus; Cyto, cytoplasm.
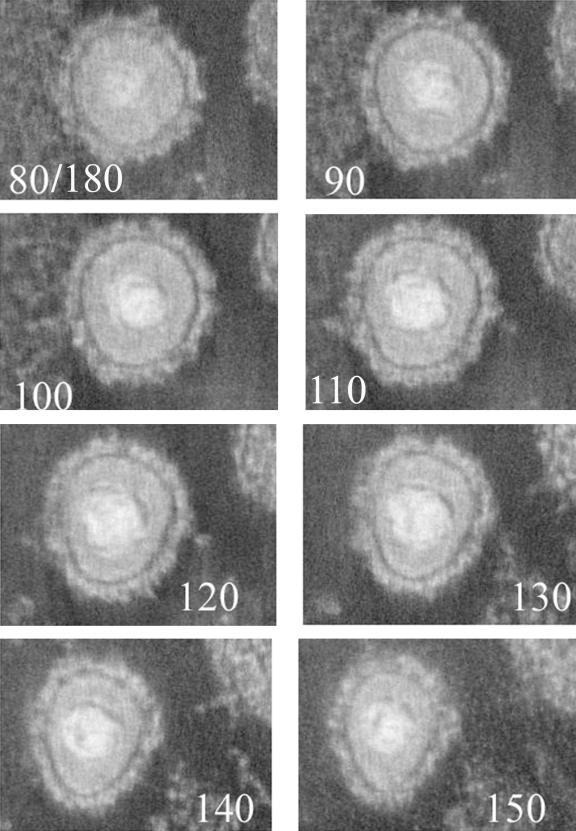
FIG. 5.
One-nanometer-thick slices from a tomographic reconstruction of an extracellular virion. The sample was prepared as described in the legend to Fig. 1. The number of the section (of a total of 180) is indicated at the bottom of each image.
Initial studies focused on capsids abutting the INM. As noted by others viewing single thin sections, capsids both containing and lacking DNA were observed abutting patches of the INM (20, 48). Nucleocapsids (capsids containing DNA) simply abutting the INM or that were partially enveloped at the INM were rare, but fully enveloped nucleocapsids within the perinuclear space were relatively common. B capsids lacking DNA and abutting the INM were also relatively common, but only occasionally were these fully enveloped in the perinuclear space.
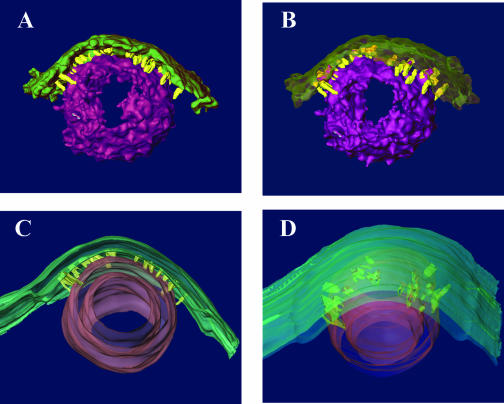
The patches of INM abutting B and C capsids were often markedly more densely stained than the rest of the INM (Fig. 1), although in some preparations this was less apparent, presumably because of variability in staining from section to section. The patches of INM abutting capsids were often indented. These indentations were generally C shaped in single slices taken through an equatorial plane of the capsid (Fig. 1), and in three-dimensional reconstructions, they were generally cup-like, with the densely stained membrane extending over the full area that was adjacent to the capsid. (Fig. 1, and models Fig. 2C and 2D; see also movie S1 of raw data in the supplemental material).
FIG. 2.
Models of the tomogram illustrated in Fig. 1. A. Model created by computationally fitting surfaces to densities in the tomogram, representing the capsid surface (purple) and the thickened region of the INM (green). Bridging rods (yellow) were segmented by manual tracing in sequential 1-nm sections. B. Similar to panel A, but the INM is made translucent to better illustrate the attachments of the bridging rods. C. Side view of a model created by manual tracing and surface rendering of the resulting contours. The inner and outer surfaces of the capsid (purple) and the INM (green) are shown, with the bridging rods (yellow) connecting the outer surface of the capsid with the inner surface of the thickened region of the INM. D. Same as panel A, but viewed from the cytoplasmic side to show the orientation of the bridging rods relative to the capsid surface. The inner protein sphere or scaffold does not appear in the model.
Capsids were not observed to directly contact the INM, or they contacted the INM only rarely. Rather, the INM was usually separated from the capsid surface by a consistent distance of approximately 10 nm (Fig. 1). This distance was similar to that maintained between the virion envelope and nucleocapsid of virions located between the INM and ONM (see below). Of interest was the observation that in some sections, thin fibers could be visualized that seemed to bridge the nucleocapsid surface and INM (Fig. 1; see also movie S1 in the supplemental material).
In order to examine the bridging fibers relative to the virion surface in more detail, the fibers in individual slices of a given reconstruction (Fig. 2) and in some cases the capsid surface (Fig. 2A and B) were traced by hand. In both cases, a feature was traced in each slice only if it was apparent in three consecutive slices. The resultant modeled reconstructions revealed the presence of densely stained, rod-like structures bridging the capsid surface and INM. The rods generally projected radially away from the nucleocapsid surface, although many emanated obliquely from the capsid surface, rather than at right angles (an example is indicated by an arrow in Fig. 1). By using the known diameter of the capsid (125 nm), the bridging rods were calculated to be approximately 8 to 19 nm in length with a thickness at or below the resolution of the reconstruction, about 5 nm in the plane of the sections. In most instances, the rod cross sections were elongated by about a factor of 1.3 in the normal direction, a result of the limited range over which the sections were tilted in the electron microscope (±60°). The longest rods were generally located at points within the interface of capsid and INM that were farthest from the center of the capsid. The length of the rods corresponded roughly to the width of the tegumental space located between the nucleocapsid surface and the inner surface of the virion envelope as revealed in tomograms of perinuclear virions (see below). In general, the rods were located only at the interface between the INM and the nucleocapsid surface (i.e., they did not emanate from regions of the nucleocapsid not directly opposed to the INM).
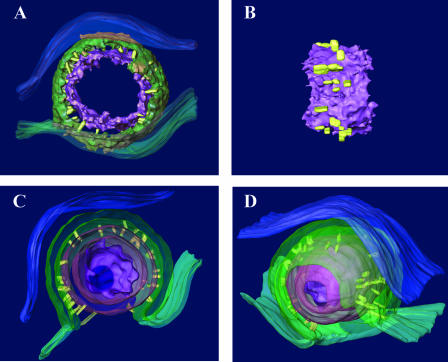
The next set of reconstructions was generated to determine whether features of capsids engaging the INM were reflected in perinuclear virions. Type C capsids within the perinuclear space that were mostly or completely enveloped by the INM in single thick sections were studied by electron tomography. Angular vertices were only occasionally observed in tomographic slices. The spacing of vertices (approximately 60 nm apart) was consistent with that expected of capsid pentons. In single thick sections, the distance between the nucleocapsid surface and inner surface of the virion envelope varied from as little as approximately 4 to 5 nm near the angular portions of the capsid surface to approximately 13 nm opposite relatively flat faces (Fig. 3 and 4; see also movie S2 in the supplemental material).
FIG. 4.
Models of the tomogram shown in Fig. 3. A. Thick slice through the center of the virion. The nucleocapsid shell (purple) and virion envelope (green) are modeled by computationally fitting surfaces to densities in the tomogram, while the ONM (blue), INM (green), and bridging rods (yellow) are modeled by manual tracing. The DNA within the core is not shown. B. Side view of the nucleocapsid surface (purple) and the attached bridging rods (yellow). The horizontal elongation of the rods is an artifact due to the limited tilt range of the tomogram. C and D. Cutaway views of the enveloping nucleocapsid shown as a surface-rendered model after manual tracing. The nucleoplasm is on the bottom and cytoplasm on the top. The near portion of the model has been cut away for illustration. The nucleocapsid (purple), bridging rods (yellow), virion envelope (green), INM (green), and ONM (blue) are shown. As only the surfaces were rendered, the nucleocapsid and nascent virion envelope appear as hollow shells in this cutaway view. Note that the virion envelope is more than twice as thick as the neighboring INM.
Densely stained rods of approximately 2 to 5 nm in width and 9 to 12 nm in length were seen bridging the outer surface of the nucleocapsid and the inner surface of the virion envelope. As described above, densities corresponding to individual rods that were discerned in at least three consecutive 1.0-nm sections were drawn by hand and included as yellow objects in the models shown in Fig. 4.
The rods were distributed asymmetrically on the surface of the otherwise roughly symmetrical nucleocapsid and projected at various angles from the nucleocapsid surface but generally radially away from the nucleocapsid. Some rods projected obliquely from the nucleocapsid surface within the tegumental space. The rods linked the virion envelope with both angular portions (Fig. 3, indicated by triangles) and reasonably flat faces of the nucleocapsid. Many regions of the capsid surface also lacked obvious bridging rods, and these surfaces were more likely flattened than angular (Fig. 3, region between triangles). Distances between sites of contact of the bridging rods varied between about 4 and 13 nm along the nucleocapsid surface. With the caveat that some bridging rods may lie obliquely in the tegument and therefore might contact sites other than their anchoring points in the nucleocapsid, the presence of bridging rod attachment points at distances closer than 64 nm (theoretical distance between pentons) argues that the bridging rods attach to faces of the capsid (hexons) as well as their vertices (pentons). On the other hand, the observation that some planar surfaces were largely spared of bridging rods argues that pentons are more frequently engaged than hexons.
Portions of the ONM bordering the nascent virion envelope protruded into the cytoplasm, with regions closest to the virion envelope protruding most dramatically (Fig. 3). Interestingly, at no point did the ONM contact the nascent virion envelope. Rather, the space between these membranes was maintained at a minimum distance of approximately 15 nm.
Comparison of perinuclear and extracellular virions.
Three-dimensional reconstructions of extracellular (Fig. 5) and perinuclear (Fig. 3 and 4) virions were compared, and the following were noted. (i) The virion envelope in extracellular virions was studded with rod-like densities that projected radially outward from the virion envelope. These spikes or virion glycoproteins varied in shape and length but were up to about 20 nm long. They were generally narrower in regions closer to the virion envelope and wider at more peripheral regions. Perinuclear virions did not have comparable projections. (ii) The tegument of extracellular virions was much more densely stained than that of perinuclear virions. The bridging rods of perinuclear virions that connected the nucleocapsid surface to the virion envelope were not readily apparent in the tegument of extracellular virions. (iii) The distances and orientations of the nucleocapsid relative to the virion envelope were asymmetric in both perinuclear and extracellular virions. In the electron tomogram of the extracellular virion shown in Fig. 5, for example, some sections in the reconstructions revealed a spherical virion envelope whereas in others the virion envelope was more pear shaped. In others (not shown) the virion envelope contained a bulge of tegument at one pole, much as previously described (22).
Transport vesicles.
Occasionally, single or multiple virions, surrounded by membrane, were noted in the cytoplasm near the nuclear membrane. Such structures have been proposed to represent transport vesicles that are separate from membranous organelles. These vesicles are potentially significant because fusion of the vesicle with the inner surface of the plasma membrane would be expected to deliver virions to the extracellular space, thus providing a simple model of virion egress. We therefore examined whether structures were discontinuous as seen in single sections or were continuous with other organelles such as the ER or nuclear membrane.
Three-dimensional reconstructions of thick sections revealed that, of the putative transport vesicles located near the nuclear membrane that were examined, all were in fact located within extensions of the perinuclear space (Fig. 6). We were unable to substantiate the existence of distinct transport vesicles near the nuclear membrane in these studies and speculate that many of the structures noted previously represent cross sections of nuclear membrane with virions contained within the perinuclear space.
DISCUSSION
Perhaps the most surprising finding in this study was the presence of rods or fibers bridging the surface of the nucleocapsid and the inner surface of the virion envelope. It should be noted that at the present resolution the possibility that these bridging densities represent poorly resolved groups of two or more parallel rods is not precluded. It is tempting to speculate that these rods maintain the space between the virion envelope and nucleocapsid surface, reminiscent of how a canvas tent (the virion envelope) is maintained by a series of poles (the bridging rods), embedded in the ground (the nucleocapsid). The rods that bridge the inner surface of the virion envelope and nucleocapsid surface are more prominent in perinuclear than extracellular virions, presumably because there is less obscuring stain density in the tegument of the former. Alternatively, it is possible that the rods are unique to perinuclear virions derived from envelopment at the inner nuclear membrane and reflect a unique mechanism of virion budding from the nucleus, as opposed to budding into cytoplasmic organelles.
The identity of the protein(s) comprising the bridging rods is not known, and it is not known if it corresponds to similar-appearing fibers noted in extracellular virions, speculated to be composed of filamentous actin (22). The viral proteins that are most likely to form the bridging rods would be components of perinuclear virions that have a size sufficient to account for the observed 8- to 19-nm length. The list of virally encoded proteins potentially fulfilling these criteria includes the pUL31/pUL34 protein complex. These proteins are particularly strong candidates because the UL31 and UL34 proteins are essential for envelopment at the inner nuclear membrane in a number of different cell lines and are known to become incorporated into perinuclear virions (7, 29, 38, 41). UL34 encodes a type II integral membrane protein embedded in the INM with a predicted 27,265-Da portion of the 29,789-Da protein located in the nucleoplasm, whereas the UL31 protein with an Mr of 33,951 directly interacts with UL34 and anchors it to the nuclear lamina, possibly by also interacting with lamin A/C (30, 37-39, 43, 44). Another possible candidate is the UL25 protein that has been shown to associate with the external surfaces of purified capsids (27, 34, 46). Although ample B capsids abut the INM in HSV-infected cells, C capsids are preferentially enveloped (40). The presence of many capsids abutting the INM of cells infected with a pseudorabies virus mutant lacking UL25 argues against the possibility that UL25 is required to tether nucleocapsids to the INM (27). On the other hand, the observation that C capsids contain more UL25 than B capsids is consistent with the notion that UL25 protein might be involved in selective envelopment of C capsids (42).
It is also possible that the bridging rods comprise one or more proteins derived from the host cell rather than from the virus. A large number of cellular proteins potentially match the size and INM location of the rods observed in these studies including actin, lamins, and lamin receptors. Actin is a strong candidate given the presence of actin in herpesvirions, the association of F actin with capsids in infected-cell nuclei, and the dependence of active intranuclear movement of capsids on actin (3, 10, 11, 13, 14, 25, 50). Lamins are less likely because they are locally depolymerized to allow access of nucleocapsids to the INM; thus, lamin filaments would not be expected to become incorporated into nascent virions (4, 35, 37, 44). Efforts to identify the components of the bridging rods are under way.
Previously noted differences between perinuclear and extracellular virions fixed in glutaraldehyde and embedded in glycid ether 100 (17) were confirmed in this work using high-pressure freeze-substitution techniques. Specifically, the tegument of perinuclear virions is much less dense overall and the virion envelope is considerably denser. Moreover, projections emanating from the external surface of the extracellular virion envelope are much more prominent, wider in diameter, and longer. Although the precise reasons for these differences are not known, it is logical to presume that the extensive changes in the external spikes are a consequence of more extensive glycosylation in the mature glycoproteins of extracellular virions. The increased stain density of the tegument of extracellular virions also suggests that additional tegument proteins are applied after the initial envelopment step at the INM. These data are therefore most consistent with a model in which the virion envelope initially obtained from the INM is lost in a subsequent fusion event (deenvelopment) and then replaced by budding into a cytoplasmic membrane compartment (reenvelopment), as originally proposed by others (45). These data are also theoretically consistent with the possibility that the tegument is applied to nucleocapsids that pass through expanded nuclear pore complexes into the cytoplasm (28). On the other hand, we were unable to substantiate the expansion of nuclear pores as reported in previous studies, although our search was not exhaustive (28, 48).
One argument supporting the possibility that the original virion envelope is maintained throughout virion egress has been the longstanding observation that perinuclear virions can be observed within cytoplasmic vesicles. A simple model of virion egress suggests that fusion of this transport vesicle with the plasma membrane should deliver the enclosed virion to the extracellular space. In this report, however, we noted that a number of apparent vesicles containing virions were, when followed through a three-dimensional reconstruction, actually irregular extensions of the nuclear membrane, with lumens continuous with the perinuclear space or endoplasmic reticulum. These observations are also consistent with the deenvelopment model of virion egress. On the other hand, we cannot exclude the possibility that distinct vesicles containing nascent virions exist; instead, we would argue that if such transport vesicles do exist, they are more rare than previously supposed. Electron tomograms of the entire nuclear membrane and contiguous ER of infected cells would be useful to confirm the existence of such vesicles.
Supplementary Material
Acknowledgments
We thank V. Mandayam Parthasarathy and the staff of the Cornell Integrated Microscopy Center for useful discussions and help with the freeze substitution.
The studies at Cornell University were supported by NIH grant AI 52341 to J.D.B. The microscopy and image processing work was supported by NIH grant RR01219 (J. Frank, Principal Investigator) through the NCRR Biomedical Research Technology Program.
Footnotes
Published ahead of print on 10 January 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., E. G. Wills, J. Pennington, R. J. Jacob, and B. Roizman. 2007. Glycoprotein M of herpes simplex virus type 1 is incorporated into virions during budding at the inner nuclear membrane. J. Virol. 81:800-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjerke, S. L., and R. J. Roller. 2006. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347:261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booy, F. P., W. W. Newcomb, B. L. Trus, J. C. Brown, T. S. Baker, and A. C. Steven. 1991. Liquid crystalline, phage-like packaging of encapsidated DNA in herpes simplex virus. Cell 64:1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopment during egress. J. Virol. 70:4311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi, J. H., C. A. Harley, A. Mukhopadhyay, and D. W. Wilson. 2005. The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J. Gen. Virol. 86:253-261. [DOI] [PubMed] [Google Scholar]
- 9.Crump, C. M., B. Bruun, S. Bell, L. E. Pomeranz, T. Minson, and H. M. Browne. 2004. Alphaherpesvirus glycoprotein M causes the relocalization of plasma membrane proteins. J. Gen. Virol. 85:3517-3527. [DOI] [PubMed] [Google Scholar]
- 10.Davison, A. J., and M. D. Davison. 1995. Identification of structural proteins of channel catfish virus by mass spectrometry. Virology 206:1035-1043. [DOI] [PubMed] [Google Scholar]
- 11.del Rio, T., C. J. DeCoste, and L. W. Enquist. 2005. Actin is a component of the compensation mechanism in pseudorabies virus virions lacking the major tegument protein VP22. J. Virol. 79:8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feierbach, B., S. Piccinotti, M. Bisher, W. Denk, and L. W. Enquist. 2006. Alpha-herpesvirus infection induces the formation of nuclear actin filaments. PLoS Pathog. 2:763-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forest, T., S. Barnard, and J. D. Baines. 2005. Active intranuclear movement of herpesvirus capsids. Nat. Cell Biol. 7:429-431. [DOI] [PubMed] [Google Scholar]
- 15.Frank, J., and M. Radermacher. 1986. Three-dimensional reconstruction of nonperiodic macromolecular assemblies. Adv. Tech. Biol. Electron Microsc. III:1-64. [Google Scholar]
- 16.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, W., and B. Roizman. 1972. Proteins specified by herpes simplex virus. VIII. Characterization and composition of multiple capsid forms of subtypes 1 and 2. J. Virol. 10:1044-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gross, S. T., C. A. Harley, and D. W. Wilson. 2003. The cytoplasmic tail of herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317:1-12. [DOI] [PubMed] [Google Scholar]
- 22.Grunewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302:1396-1398. [DOI] [PubMed] [Google Scholar]
- 23.Heymann, J. B., N. Cheng, W. W. Newcomb, B. L. Trus, J. C. Brown, and A. C. Steven. 2003. Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol. 10:334-341. [DOI] [PubMed] [Google Scholar]
- 24.Jensen, H. L., and B. Norrild. 1998. Herpes simplex virus type 1-infected human embryonic lung cells studied by optimized immunogold cryosection electron microscopy. J. Histochem. Cytochem. 46:487-496. [DOI] [PubMed] [Google Scholar]
- 25.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, D. C., and P. G. Spear. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1101-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp, B. G., H. Granzow, G. M. Keil, and T. C. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leuzinger, H., U. Ziegler, E. M. Schraner, C. Fraefel, D. L. Glauser, I. Heid, M. Ackermann, M. Mueller, and P. Wild. 2005. Herpes simplex virus 1 envelopment follows two diverse pathways. J. Virol. 79:13047-13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, L., M. Tanaka, Y. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 329:68-76. [DOI] [PubMed] [Google Scholar]
- 30.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 31.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naldinho-Souto, R., H. Browne, and T. Minson. 2006. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 80:2582-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newcomb, W. W., F. L. Homa, D. R. Thomsen, F. P. Booy, B. L. Trus, A. C. Steven, J. V. Spencer, and J. C. Brown. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432-446. [DOI] [PubMed] [Google Scholar]
- 34.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 80:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penczek, P., M. Marko, K. Buttle, and J. Frank. 1995. Double-tilt electron tomography. Ultramicroscopy 60:393-410. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina of cells induced by herpes simplex virus type 1 require genes UL31 and UL34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reynolds, A. E., B. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizman, B., and D. Furlong. 1974. The replication of herpesviruses, p. 229-403. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology. Plenum Press, New York, NY.
- 41.Roller, R., Y. Zhou, R. Schnetzer, J. Ferguson, and D. Desalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, Y. Yamauchi, O. Koiwai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 81:2397-2405. [DOI] [PubMed] [Google Scholar]
- 44.Simpson-Holley, M., J. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stackpole, C. W. 1969. Herpes-type virus of the frog renal adenocarcinoma. I. Virus development in tumor transplants maintained at low temperature. J. Virol. 4:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torrisi, M. R., C. Di Lazzaro, A. Pavan, L. Pereira, and G. Campadelli-Fiume. 1992. Herpes simplex virus envelopment and maturation studies by fracture label. J. Virol. 66:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wild, P., M. Engels, C. Senn, K. Tobler, U. Ziegler, E. M. Schraner, E. Loepfe, M. Ackermann, M. Mueller, and P. Walther. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 79:1071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wildy, P., W. C. Russell, and R. W. Horne. 1960. The morphology of herpes virus. Virology 12:204-222. [DOI] [PubMed] [Google Scholar]
- 50.Wong, M. L., and C. H. Chen. 1998. Evidence for the internal location of actin in the pseudorabies virion. Virus Res. 56:191-197. [DOI] [PubMed] [Google Scholar]
- 51.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 A. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
- 53.Zhou, Z. H., B. V. Prasad, J. Jakana, F. J. Rixon, and W. Chiu. 1994. Protein subunit structures in herpes simplex virus A-capsid determined from 400 kV spot-scan electron cryomicroscopy. J. Mol. Biol. 242:456-469. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.