Abstract
The assembly of bacteriophage PRD1 proceeds via formation of empty procapsids containing an internal lipid membrane, into which the linear double-stranded DNA genome is subsequently packaged. The packaging ATPase P9 and other putative packaging proteins have been shown to be located at a unique vertex of the PRD1 capsid. Here, we describe the isolation and characterization of a suppressor-sensitive PRD1 mutant deficient in the unique vertex protein P6. Protein P6 was found to be an essential part of the PRD1 packaging machinery; its absence leads to greatly reduced packaging efficiency. Lack of P6 was not found to affect particle assembly, because in the P6-deficient mutant infection, wild-type (wt) amounts of particles were produced, although most were empty. P6 was determined not to be a specificity factor, as the few filled particles seen in the P6-deficient infection contained only PRD1-specific DNA. The presence of P6 was not necessary for retention of DNA in the capsid once packaging had occurred, and P6-deficient DNA-containing particles were found to be stable and infectious, albeit not as infectious as wt PRD1 virions. A packaging model for bacteriophage PRD1, based on previous results and those obtained in this study, is presented.
The encapsidation of viral double-stranded DNA (dsDNA) genomes in a protective spherical protein capsid presents a unique set of challenges. First, the genomes of icosahedral dsDNA viruses are enormous in length compared to the inner diameter and volume of the capsids they are to occupy. Packaging DNA to such a high density is energetically extremely unfavorable. Second, the DNA needs to be topologically organized so that when a suitable host cell is encountered, the genome can efficiently be translocated into the cell and a new round of infection can be initiated. Specificity is an additional challenge, because energy and material should not be wasted for nonproductive packaging of host DNA into the viral capsids.
Icosahedral dsDNA viruses have developed ingenious machinery to perform such a complicated task. Encapsidation of the dsDNA genomes proceeds via formation of empty procapsids, devoid of DNA, that subsequently are packaged with DNA by a specific enzyme, called the terminase or packaging ATPase (for recent reviews, see references 24 and 46). This process requires a large amount of energy, usually provided in the form of ATP. It has been estimated that, for example, in the case of bacteriophage phi29, an average of 1 molecule of ATP per each 2 bp of DNA packaged is needed (38). Packaging occurs through a specific vertex of the capsid, which contains a ring-like portal structure, through which the DNA is threaded into the particle. However, packaging machineries are not quite as simple as this; many additional proteins, and in some cases even RNAs, are involved, with the details varying from one virus to another.
The problem with studying these intricate apparatuses is their asymmetric location in the virion, which makes conventional structure determination methods that are based on icosahedral averaging less useful. Several asymmetric cryoelectron microscopic (cryo-EM) reconstructions of bacteriophages epsilon15 and P22, revealing the structures of their packaging machineries within the context of the whole virion in more detail, have been published only very recently (25, 47, 49).
Despite little or no sequence similarity, all portal proteins for which structures have been solved display similar folds and form multimeric ring- or cone-like structures (2, 26, 32, 37, 70, 85, 92). Portals that have been studied as isolated proteins have been 11- to 14-mers, but all portals analyzed within the virion have been composed of 12 subunits of the portal protein (2, 12, 22, 25, 26, 28, 30, 37, 47-49, 54, 64, 69, 70, 85, 92, 94). The presence of a dodecameric portal at the fivefold vertex creates a symmetry mismatch, which has been proposed to facilitate rotation of the portal during DNA packaging (42). Portal proteins not only provide a platform for binding terminases (24, 51) but may also play a more active role in the packaging process, for example, by stimulating terminase activity (20, 66) and by sensing the amount of DNA packaged and triggering the cleavage of concatemeric DNA molecules (23, 49, 69, 90). Portals also have been proposed to play an important role in the initiation and early stages of capsid assembly (29, 63).
Viruses that have concatemeric genomes need terminase enzymes that both translocate and cut the DNA. Terminases are hetero-oligomers of small and large subunits. The small terminase subunit is responsible for recognition and binding of the viral DNA and is necessary for activation of the ATP hydrolysis and/or cleavage activities of the large terminase subunit. When concatemeric DNA is packaged, the cleavage can occur either at a specific sequence, as for bacteriophage lambda cos sites (31), or sequence independently, via a head-full packaging mechanism, as for phages SPP1 and P22 (45, 93). After packaging and cleavage, the terminase-DNA complex is released from the packaged virion and the complex proceeds to bind and package the next procapsid.
In viruses with unit-length genomes, no cleavage is necessary and a simple packaging ATPase is sufficient, as exemplified by the packaging ATPase gp16 of bacteriophage phi29 (38). The phi29 genome is replicated by a protein-primed mechanism, similar to that of adenovirus and of bacteriophage PRD1 (80). Instead of relying on a small terminase subunit for binding DNA, the specificity of packaging in phi29 most probably is achieved through the binding of ATPase gp16 to the covalently attached terminal protein, gp3, at the end of the linear unit-length genome (38).
In addition to terminases, or packaging ATPases, and portal proteins, several other packaging-related proteins in different viruses have been discovered. Some of these proteins are probably part of the actual packaging machinery. For example, lambda gpFI may act by promoting binding of the DNA-terminase complex to the prohead (13, 60, 61). Other proteins may act by sealing or “plugging” the packaged capsids and preventing release of the packaged DNA; examples include the head completion proteins gp15 and gp16 that bind to the portal of DNA-filled SPP1 virions (14, 54, 70), bacteriophage lambda gpW and gpFII (62, 74) and, possibly, the bacteriophage P22 head completion proteins gp4, gp10, and gp26 (87). Some proteins, such as the herpes simplex virus type 1 proteins UL17 and UL25 (91), are more general in function and act by stabilizing the mature packaged virions by binding elsewhere on the capsid, outside the actual packaging machinery. In some cases, host proteins also may be involved in the packaging process: in bacteriophage lambda, integration host factor is proposed to regulate the packaging machinery by binding and bending phage DNA, together with lambda small terminase subunit gpNu1 (55, 71).
Bacteriophage PRD1, the type virus of the family Tectiviridae, differs from the above-described examples of bacteriophages and herpes simplex virus type 1 in that it has a lipid membrane underneath its icosahedral pseudo-T=25 protein capsid (4, 19, 52, 68). PRD1 has a ∼15,000-bp linear dsDNA genome with inverted terminal repeats and covalently attached 5′-terminal proteins (5, 6, 10, 84). At infection, the internal membrane of PRD1 is used to inject the DNA into the host cell and thus represents an alternative method of DNA delivery compared to the tails of the Caudovirales phages (3, 34, 35, 52). A more detailed description of the PRD1 virion architecture can be found in references 1 and 27, which describe the virion structure, solved to high resolution and with the internal membrane visible in the electron density map.
Similar to that of other icosahedral dsDNA phages, assembly of PRD1 has been shown to proceed via the formation of empty procapsids (58). However, in PRD1, the empty procapsids are not formed by assembly of the coat protein on top of a protein scaffold but rather by using a membrane vesicle coated with PRD1 protein P10 as a scaffold (78). As a consequence, scaffolding proteins that would otherwise need removing are not present when packaging is initiated. An expansion of the protein capsid in relation to packaging is not seen for PRD1. Instead, the internal membrane in packaged particles makes closer contact with the inner side of the capsid and adopts a more angular shape, as seen by comparing cryo-EM structures of filled and empty particles (19, 82, 83).
In tailed phages, the portal vertex also is the site of tail attachment. Despite having no tail, PRD1 has been found by immunoelectron microscopy and a combination of genetic and biochemical methods to have its DNA packaging proteins located to one vertex, distinct from the other 11 vertices containing the spike complex (33, 89). This unique packaging vertex contains, at least, minor capsid protein P6, packaging ATPase P9 and two small membrane proteins, P20 and P22. The absence of either P9 or P20 leads to the production of particles devoid of DNA, but in P22− mutants, a small number of packaged particles can be detected in infected cells, where the majority of the particles are empty (57, 89). P9− mutant particles are similar in protein composition to wt procapsids in that they lack only P9 and DNA (57, 89). P20- or P22-deficient particles lack both P6 and P9 (89). Interestingly, instead of recycling of the packaging ATPase, as occurs with the terminase in many other phages, PRD1 P9 stays attached to the mature virion after packaging (58). Recently, an in vitro system was developed for the analysis of PRD1 packaging in more detail (88).
Here, we report the first PRD1 mutant defective in the unique vertex protein P6. An analysis of the effects of the absence of P6 in PRD1 infection and the role of P6 in DNA packaging is described, and a model for PRD1 packaging is proposed.
MATERIALS AND METHODS
Bacterial and phage strains.
Table 1 lists the bacterial strains and viruses used in this study. Wild-type (wt) PRD1 and mutant PRD1-1, carrying a lacZα insert in its genome, were propagated on the nonsuppressor Salmonella enterica strain DS88, and PRD1 amber mutants sus1 and sus621 were propagated on suppressor strain PSA (pLM2).
TABLE 1.
Bacterial strains and bacteriophages used in this study
| Bacterial strain or bacteriophage | Descriptiona | Relevant properties | Reference |
|---|---|---|---|
| Bacterial strains | |||
| S. enterica LT2 serovar Typhimurium | |||
| DS88 | SL5676 ΔH2 H1-i::Tn10 (Tcs) (pLM2) | Nonsuppressor host | 8 |
| PSA (pLM2) | supE | Suppressor host for sus621 | 59 |
| DB7154 (pLM2) | DB700 leuA141(Am) hisC527(Am) supD10 | Suppressor host used for isolation of sus621 | 95 |
| E. coli K-12 | |||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi1 relA1 | Cloning host | 40, 81 |
| HMS174 | recA1 hsdR Rfr | Cloning host | 21 |
| HMS174 (DE3) | recA1 hsdR Rfr | Expression strain for PRD1 P6 | 21 |
| HMS174 (pcI857) (pTPH19) | recA1 hsdR Rfr | Expression strain for PRD1 P19 | 73, 77 |
| HB101 | supE44 hsdS20(rB−mB−) recA13 ara14 proA2 lacY1 galK2 rpsL20 xyl5 mtl1 | Cloning host | 16, 17 |
| Bacteriophage PRD1 | |||
| wt | Wild type | 68 | |
| sus1 | Amber mutation in gene IX, two missense mutations in gene VI | Production of particle extract for packaging reaction | 57, 89 |
| PRD1-1 | lacZα insertion (nt 6309) | Production of DNA for packaging reaction | 9 |
| sus621 | Amber mutation in gene VI, two missense mutations in gene VI | This study |
Tcs, tetracycline sensitive; Rfr, rifampin resistant. Nucleotide number refers to the PRD1 genome sequence (GenBank accession no. AY848689).
To obtain the gene VI amber mutant sus621, N-methyl-N′-nitrosoguanidine (NTG) was used to induce mutations in the PRD1 genome, as described previously (57). Amber mutants were isolated by plating them on PSA or DB7154 suppressor strains and then by comparing plating efficiencies on suppressor and nonsuppressor strains. A suppressor-sensitive mutant with defects in gene VI was identified by an in vivo complementation assay using two different plasmid constructions (pNS21 and pNS62) carrying the PRD1 gene VI, performed as described previously (10, 56).
Purification and analysis of virus particles.
wt and mutant virus particles were produced as follows: DS88 cells were grown at 37°C in Luria-Bertani (LB) medium to a cell density of 1 × 109 CFU/ml and infected at a multiplicity of infection of 6 and 8 for the wt and the mutants, respectively. After lysis of the cells, phage particles were purified from the lysate by precipitation with polyethylene glycol 6000 and NaCl, followed by 5 to 20% rate zonal sucrose gradient centrifugation, as previously described (8). The resulting preparation was called “1× purified” virus. When better separation of empty and filled virus particles was required, the 1× purified material was further purified by 20 to 70% equilibrium sucrose gradient centrifugation (Beckman SW41 or Sorvall TH641 rotor, 32,000 rpm, 19 h, 15°C; or Sorvall AH629 rotor, 24,000 rpm, 20 h, 15°C). This preparation was designated “2× purified.” Both 1× and 2× purified particles were collected by differential centrifugation (Beckmann rotor Ti50, 33,000 rpm, 3 h, 5°C; Sorvall rotor T865, 33,000 rpm, 3 h; or Sorvall rotor T647.5, 32,000 rpm, 2.5 h, 5°C, depending on the amount of virus). The 2× purified Sus621 and wt virus particles were plated on suppressor and nonsuppressor hosts to analyze their infectivity. Samples of infected cells were taken at 50 and 60 min postinfection (p.i.), concentrated 10-fold in 20 mM Tris-HCl buffer (pH 7.2), disrupted by sonication, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. Band intensities in SDS-PAGE were quantified by TINA 2.0 software (Raytest), and amounts of P6 and P9 were calculated by comparing their band intensities to those of P2 and P5, with known stoichiometries.
To study the accessibility of the viral membrane within the mutant Sus621 particles, 1× purified particles were treated with 1% SDS for 15 min at 20°C, and the resulting membrane-free particles were separated from residual membrane-containing particles by rate zonal centrifugation in 5 to 20% sucrose gradients (Sorvall rotor TH641, 24,000 rpm, 75 min, 15°C), essentially as described in reference 53.
DNA techniques.
DNA manipulations were performed using standard molecular biology techniques (81). Plasmids used in this study are listed in Table 2. PRD1 gene VI was amplified by PCR from the PRD1 genome using primers specific to the corresponding region in the PRD1 genome (nucleotides [nt] 6784 to 7284, with and without the preceding SD sequence), and the resulting fragments were inserted between the EcoRI and HindIII sites of plasmid pSU18, resulting in constructs pNS21 and pNS22, respectively. Gene VI was cut out from plasmid pNS22 (EcoRI and HindIII) and ligated into plasmid pJJ2, and the resulting construct was named pNS62. The nucleotide sequence of the gene VI insert in pNS62 was verified by DNA sequencing. Sequencing was performed with an automated sequencer (at the DNA Synthesis and Sequencing Laboratory, Institute of Biotechnology, University of Helsinki) and was used to verify the location of the amber mutation in gene VI in the genome of mutant sus621. DNA from sus621 plaques as templates (43) and primers specific to the PRD1 genome were used to create overlapping PCR fragments covering nt 6532 to 8560 and nt 9850 to 10160 of the PRD1 genome (containing genes VI, X, IX, XX, and XXII and open reading frame i, encoding proteins involved in DNA packaging) (89). The PCR fragments then were used as templates for sequencing.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pSU18 | Cloning vector, p15A replicon, Plac, Cmr | 11 |
| pNS21 | PRD1 gene VI with preceding Shine-Dalgarno sequence (nt 6769 to 7284) cloned into pSU18 | This study |
| pNS22 | PRD1 gene VI (nt 6784 to 7284) cloned into pSU18 | This study |
| pJJ2 | Expression vector, Apr | 65 |
| pNS62 | PRD1 gene VI (nt 6784 to 7284) cloned into pJJ2 | This study |
| pJB15 | Encodes the PRD1 receptor | 41 |
| pLM2 | Encodes the PRD1 receptor | 59 |
| pcI857 | Encodes thermosensitive repressor for heat induced expression of proteins under the λ PL promoter | 77 |
| pPLH101 | Expression vector with λ PL promoter | 50 |
| pTPH19 | PRD1 gene XIX cloned into pPLH101 | 73 |
Cmr, chloramphenicol resistance; Apr, ampicillin resistance. Nucleotide numbers refer to the PRD1 genome coordinates (AY848689).
For use in the in vitro packaging system, DNA with the terminal protein P8 attached was extracted from 1× purified PRD1-1 virions by disruption with SDS, two extractions with phenol, and three ether extractions, followed by ethanol precipitation, essentially as described previously (56). For analysis of the DNA content of Sus621 virions, DNA from both 1× and 2× purified Sus621 and wt PRD1 particles was purified essentially as described above but with the addition of protease K during the particle disruption step to digest the genome terminal proteins and with two additional phenol extraction steps to obtain higher-purity DNA. During virus purification, DNase I treatment was performed with the particles prior to the application of the preparations onto sucrose gradients, in order to distinguish between packaged DNA and any residual DNA attached to the outside of the particles.
Nucleic acid extracted from Sus621 particles, together with wt PRD1 DNA as a control, was treated alternatively with DraI restriction enzyme, DNase, RNase, or protease and analyzed, together with untreated samples, by agarose gel electrophoresis and ethidium bromide staining.
DNA packaging.
In vitro DNA packaging was performed essentially as described previously (88). Purified PRD1-1 DNA containing a lacZα insert was mixed with cell extracts containing packaging ATPase P9 and empty virus particles (either from infection with packaging ATPase gene IX defective mutant sus1 or the gene VI mutant sus621), in the presence of polyethylene glycol 8000, salts, and ATP. After incubation for 45 min at 37°C, dilutions of the reaction mix were plated on S. enterica strains PSA and DS88 and Escherichia coli DH5α (pJB15), in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). In the case of P6− Sus621 particles, 0.1 to 20 μl of HMS174 (DE3) pNS62 cell extract containing recombinant P6 also was added to the reaction. Plaques obtained on DS88 or PSA were transferred to DH5α (pJB15) lawns for the production of blue plaques by α-complementation, thus confirming the presence of in vitro packaged PRD1-1 DNA containing the lacZα insert. Since the sus621 particle extract gave rise to a higher background than the sus1 extract, the identity of the background plaques was verified by sequencing of DNA.
The Sus621 particle extract was prepared as previously described for the packaging ATPase-deficient Sus1 particle extract (88): sus621- or sus1-infected cells were collected at 40 min p.i., concentrated 100-fold in 20 mM Tris-HCl buffer (pH 7.2), and disrupted by sonication. For the production of recombinant protein P6 extract, strain HMS174 (DE3) pNS62 was grown at 28°C (to a cell density of approximately 107 CFU/ml) in LB medium in the presence of ampicillin (100 μg/ml). Protein expression was induced by the addition of IPTG to a final concentration of 0.1 mM. Cells were grown overnight at 28°C, collected by centrifugation, and resuspended (100-fold concentrated) in 20 mM Tris-HCl buffer (pH 7.2). The cells were disrupted by two to three passages through a French pressure cell, and the supernatant was cleared by ultracentrifugation (Beckmann rotor Ti50, 33,000 rpm, 2 h, 5°C).
Electron microscopy.
For thin-section electron microscopy, DS88 cells were grown in LB medium to a density of 1 × 109 CFU/ml and infected with wt or mutant virus using a multiplicity of infection of 6 or 8, respectively, and samples were collected at 20, 30, 40, 50, and 60 min p.i. in the case of sus621 and at 30 and 50 min in the case of wt PRD1. The cells were fixed with 3% glutaraldehyde (vol/vol) in 20 mM potassium phosphate buffer (pH 7.2) for ≥20 min at room temperature, collected by centrifugation (3,220 × g, 10 min, 5°C), washed twice with 20 mM potassium phosphate buffer (pH 7.2), and prepared for thin-section transmission electron microscopy as described previously (7).
Both purified and SDS-treated virus particles were analyzed by negative staining and electron microscopy. Samples were absorbed onto carbon-coated grids and stained with 1% (wt/vol) sodium phosphotungstic acid (pH 6). Electron micrographs were taken with a JEOL 1200 CX microscope operating at 60 kV (Electron Microscopy Unit, Institute of Biotechnology, University of Helsinki).
Protein purification.
For the purification of protein P6, the cleared HMS174 (DE3) pNS62 supernatant (described above for in vitro packaging) was subjected to an initial precut with 22% (vol/vol) ammonium sulfate, and the precipitated material was removed by centrifugation. Material was further precipitated by adding ammonium sulfate to a concentration of 26% (vol/vol). The precipitate was collected and resuspended in 20 mM piperazine (pH 5.5). The resuspended material was filtered (0.22-μm filter) and applied to an anion exchange column (Pharmacia Q SP column, 1 ml). The column was washed with 20 mM piperazine followed by 20 mM piperazine with 200 mM NaCl, and P6 was eluted with 20 mM piperazine with 300 mM NaCl. The P6-containing fractions were pooled, and the buffer was changed to 20 mM Tris-HCl (pH 7.2) using a 20-kDa cutoff Centrex UF-2 device (Schleicher & Schuell). This material was filtered and applied to a Hi-Load Superdex 200 gel filtration column (Pharmacia). P6 was collected as a sharp peak, and the material was determined by SDS-PAGE to be homogeneous.
Immunological methods.
Polyclonal serum against protein P6 was raised by immunizing a rabbit three times with 350 μg of purified protein P6 at three-week intervals. Freund's complete adjuvant was used in the primary immunization, and Freund's incomplete adjuvant was used in the subsequent immunizations. The serum was collected two weeks after the last immunization, and the specificity of the serum was determined by Western blotting.
Analytical methods.
The protein concentration of purified virus preparations was determined by the Coomassie brilliant blue method using bovine serum albumin as a standard (18). SDS-PAGE was performed as previously described (67). Western blotting was performed by transferring the proteins from SDS-polyacrylamide gels (15% and 17% acrylamide) onto polyvinylidene difluoride membranes (Millipore). Monoclonal antibodies 6T58, 7A5, 7N41 (41), 11A401 (33), and 16A201 (41, 44) against PRD1 proteins P6, P7/14, P7/14, P11, and P16, respectively, and polyclonal antisera against proteins P6, P2 (34), P5 (41), P31 (79), P9, and P22 (89) were used as primary antibodies. Peroxidase-labeled horse anti-mouse immunoglobulin G (IgG) antibodies (Vector), or horseradish peroxidase-conjugated swine anti-rabbit IgGs (Dako), in the case of monoclonal antibodies or polyclonal antisera, respectively, were used as secondary antibodies. Samples were visualized by using SuperSignal West Pico chemiluminescent substrate (Pierce).
For N-terminal amino acid sequencing, viral proteins were separated using SDS-PAGE (16% acrylamide) and stained by Coomassie brilliant blue. A protein band of the mutant Sus621 preparation corresponding to the size of P19 (∼10.5 kDa) but not present in the wt virion preparation was cut out of the gel and subjected to N-terminal amino acid sequencing (at the Protein Chemistry Research Group and Core Facility, Institute of Biotechnology, University of Helsinki) to determine its identity. For use as a size standard, PRD1 protein P19 was expressed from strain HMS174 (pcI857) (pTPH19) as described previously (73), with strain HMS174 (pcI857) (pPLH101) as a negative control.
RESULTS
Isolation of a mutant phage deficient in unique vertex protein P6.
Mutations were induced by NTG, and amber mutants were identified by plating the phage on suppressor and nonsuppressor strains. The phage mutants were screened for defects in gene VI by an in vivo complementation assay where the recombinant gene VI product, P6, was provided in trans.
A single mutant that could be complemented by P6 expression in trans was found and subsequently named sus621. The titers obtained with sus621 on suppressor host PSA were comparable to the titers obtained with wt virus (1011 to 1012 PFU/ml), and a clear difference (104- to 105-fold) between the titers obtained with nonsuppressor and suppressor hosts could be observed. Titers obtained for the complementation analysis and with suppressor and nonsuppressor hosts are shown in Table 3.
TABLE 3.
Titers of PRD1 gene VI amber mutant sus621 on different hosts
| Strain | Description | Titer (PFU/ml)
|
|
|---|---|---|---|
| sus621 (VI−) | PRD1 wt | ||
| E. coli | |||
| HMS174 (DE3) (pLM2) (pNS62) | Gene VI in pJJ2 | 1.2 × 1011 | 8.0 × 1011 |
| HMS174 (DE3) (pLM2) (pJJ2) | Negative control | 1.9 × 105 | 9.0 × 1011 |
| S. enterica | |||
| PSA | Suppressor host | 3.3 × 1011 | |
| DS88 | Nonsuppressor host | 1.2 × 107a | 1.8 × 1012 |
Plaques were of various sizes.
DNA sequencing of sus621 revealed a C→T transition at nt 7000, resulting in an amber stop codon in gene VI. This should give rise to a 72-amino-acid fragment instead of the 166-amino-acid full-sized protein P6, although no additional fragments could be detected in the particles by SDS-PAGE and Western blotting, suggesting that the fragment was either degraded or not assembled into the particle. Two additional mutations were discovered in the sus621 genome: a C→T mutation at nt 6872 and a C→T mutation at nt 9877, causing Ala→Val changes in P6 and small membrane protein P22, respectively. However, since P6 alone (provided from plasmid pNS62) was enough to fully complement the defects in sus621 (Table 3), the conservative amino acid change in P22 seemed to be irrelevant, and the possibility of any other mutations in the sus621 genome affecting the infectivity also was ruled out.
To confirm that no P6 was expressed during sus621 infection, nonsuppressor DS88 cells were infected with the mutant virus, and the cells were collected at 40 and 60 min postinfection (p.i.). wt PRD1 was used as a control. Cells were disrupted by sonication, and the preparation was analyzed by SDS-PAGE and Western blotting. No P6 production was seen in cells infected with sus621 in samples taken at either 40 min or 60 min p.i., whereas in wt infection, P6 could be detected at both time points (data not shown).
The P6− mutant assembles but has a defect in genome packaging.
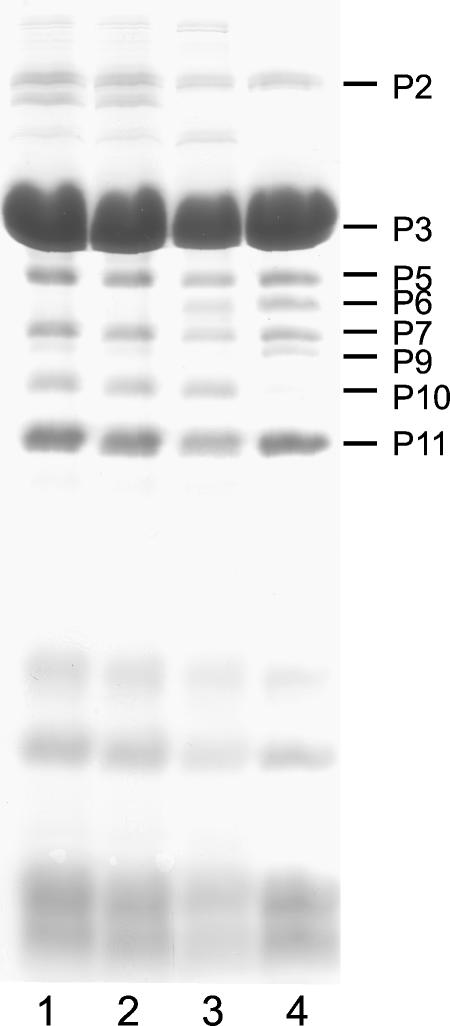
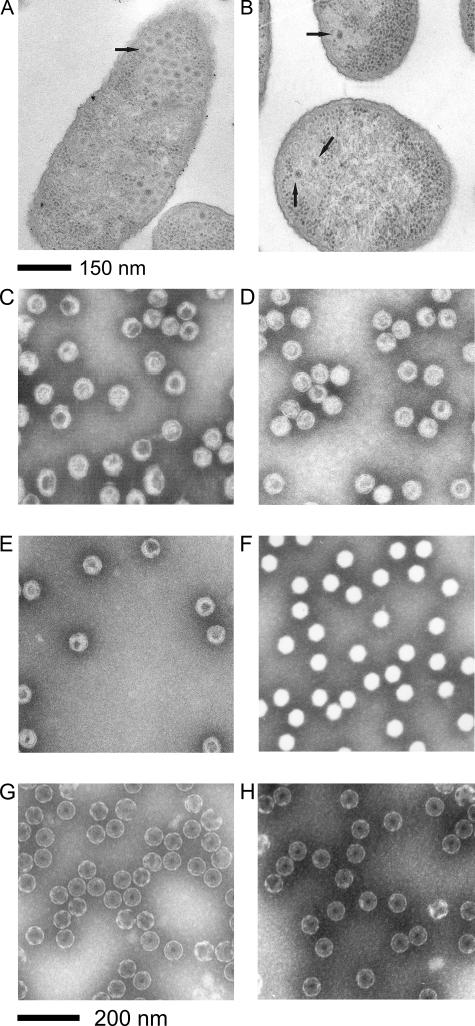
Sus621 and wt PRD1 were grown in DS88 cells and purified by rate zonal centrifugation in 5 to 20% sucrose gradients. Purified virus-like particles were analyzed by SDS-PAGE (Fig. 1). Samples of both infected cells and purified virus-like particles were analyzed by electron microscopy (Fig. 2).
FIG. 1.
Analysis of the protein content of 2× purified virus particles by SDS-PAGE and Coomassie brilliant blue staining. Lane 1, empty Sus621; lane 2, DNA-filled Sus621; lane 3, empty wt; and lane 4. DNA-filled wt particles. Since both 1× and 2× purified preparations produced similar patterns, only 2× particles are shown here.
FIG. 2.
Effect of P6 deficiency on particle assembly. (A and B) Sections of S. enterica DS88 cells infected with (A) wt PRD1 (50 min p.i.) and (B) mutant sus621 (60 min p.i.). Examples of filled particles are depicted with arrows. (C to H) Negatively stained purified virus particles. (C) Empty Sus621 particles, (D) filled Sus621 particles, (E) empty Sus1 particles, (F) filled wt particles, (G) SDS-treated empty Sus621 particles, and (H) SDS-treated empty Sus1 particles.
The properties of growth and lysis of sus621-infected cells were identical to those seen with wt infection. Sus621 was found to produce numbers of virus-like particles approximately equal to those produced with wt infection. However, whereas in wt infection, light-scattering zones that correspond to 20% empty and 80% DNA-filled wt PRD1 particles are formed, with Sus621, only approximately 5 to 10% of the particles sedimented in a manner similar to that of filled wt particles, with the rest behaving in a manner similar to that of empty wt particles. In addition, the zones produced by the mutant virus were not as sharp and easy to separate as zones formed by wt PRD1, and an additional step of equilibrium centrifugation was performed to further separate the empty and filled particles.
Purified virus particles were analyzed by SDS-PAGE (shown in Fig. 1) and Western blotting (data not shown) with all available antibodies against PRD1 structural proteins. The only structural protein found to be missing from the Sus621 particles was P6 (roughly estimated to be 40 copies/virion in wt PRD1; see Materials and Methods), which was confirmed by a polyclonal antiserum and a monoclonal antibody, 6T58, against P6. Both empty and filled Sus621 particles were found to contain packaging ATPase P9, which in wt PRD1 was seen only in DNA-filled particles but never in empty procapsids (89). The amounts of P9 in the DNA-filled and empty Sus621 particles were estimated by SDS-PAGE and Coomassie brilliant blue staining to be similar to one another but less (≤50%) than the amount of P9 in filled wt particles (estimated to be approximately 6 copies/virion) (Fig. 1). Western blotting of samples from infected cells showed no reduction in the amount of P9 produced by sus621 (data not shown), suggesting that the reduced amount of P9 in Sus621 was not due to lower levels of protein expression. DNase I treatment of Sus621 particles did not have any effect on the amount of P9, indicating that P9 was assembled properly into particles and was not loosely attached to the outside of the particles via partially packaged DNA (data not shown).
An additional band not present in wt virions, corresponding to a protein of approximately 10 kDa, was seen in SDS-PAGE analysis of Sus621 empty particles when gels were overloaded. The identity of this band was confirmed by N-terminal amino acid sequencing to be PRD1 single-stranded DNA binding protein P19, which is a nonstructural protein and not seen in empty or filled wt PRD1 particles (57, 73).
Similar to the results of sucrose gradient analysis, only a few DNA-filled particles were seen in thin-sections of sus621-infected cells (Fig. 2A and B). This is in contrast to results seen for wt infection, where very few empty particles were seen, and filled particles accumulated in the cell until lysis occurred. The presence of equivalent amounts of empty particles in both cells and purified virus preparations suggested that the filled particles are stable and the empty particles are not the result of DNA leaking out of packaged particles. However, compared to other PRD1 mutants producing empty particles, such as the packaging ATPase-deficient mutant sus1 (57), the empty Sus621 particles were not as clearly detected due to the poor contrast with the surrounding cellular material.
When Sus621 particles were analyzed by negative staining and EM, they were found to resemble wt virions in both size and shape (Fig. 2C to F). However, DNA-filled Sus621 particles were much more susceptible to stain penetration than wt DNA-filled particles and, based on the electron micrographs alone, it was difficult to distinguish between empty and DNA-filled Sus621 particles. This result was unlike that previously observed in the case of wt PRD1 and in the packaging ATPase-deficient empty Sus1 particles, where the difference between empty and filled particles was clear (Fig. 2E and F).
SDS treatment can be used to remove the PRD1 internal membrane from empty packaging ATPase-deficient Sus1 mutant particles but not from DNA-filled wt particles (53). SDS treatment was used to analyze whether the interior of the empty Sus621 particles is accessible or possibly protected by presence of the packaging ATPase P9. Empty Sus621 particles were found to behave in a manner identical to that of Sus1 particles and to lose their internal membranes when treated with 1% SDS (Fig. 2G and H), suggesting that the presence of P9 did not protect the inside of the virion when P6 was absent.
The packaged DNA in P6-deficient particles is PRD1 specific, and the packaged particles are infectious.
The DNA in the filled Sus621 particles was shown to be approximately 15 kb in length (which is the length of the PRD1 genome) and to form a DraI restriction enzyme digestion pattern identical to that of wt PRD1 DNA (Fig. 3), suggesting that it was PRD1 specific. DNase I treatment during purification of the particles did not affect the amount of PRD1-specific DNA obtained, which suggested that the sus621 DNA was properly packaged.
FIG. 3.
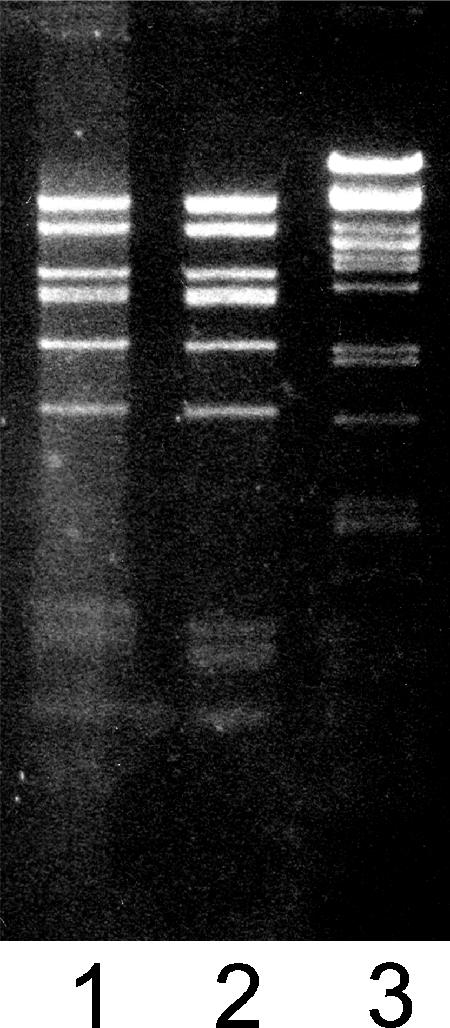
Restriction enzyme analysis of DNA extracted from filled Sus621 and wt PRD1 particles. Lane 1, DraI-digested sus621 DNA; lane 2, DraI-digested wt PRD1 DNA; and lane 3, DNA standard, PstI-digested bacteriophage λ DNA.
The infectivity of the P6− mutant particles was analyzed by plating 2× purified virus preparations on PSA and DS88 hosts. The protein concentration of the virus preparations was measured, and the specific activities (PFU per mg of purified virus) were compared. Empty Sus621 and wt particles had approximately similar specific infectivities (1 × 1010 and 5 × 109 PFU/mg of protein, respectively). Filled Sus621 particles produced 10- to 20-fold more PFU (1 × 1011 PFU/mg protein) than empty Sus621 particles, indicating that the filled particles were infectious. However, this was significantly less than the specific infectivity of the filled wt particles (producing up to 1 × 1013 PFU/mg of protein). The stability of the purified virus preparations was evaluated by determining the titers of the viruses once per week for a four-week period. No decreases in the sus621- or wt-specific infectivities were observed over a four-week period, indicating that once packaging has occurred, the Sus621 particles were as stable as wt particles and there was no DNA leakage.
In vitro packaging of Sus621 is very inefficient.
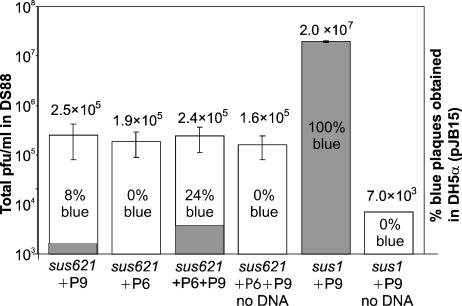
Sus621 particles were tested for their ability to package DNA in vitro. In vitro DNA packaging was performed essentially as described previously for packaging procapsids from a packaging ATPase-deficient mutant (88). Sus621 particles were provided as an extract, and the missing P6 protein was provided as a recombinant protein extract. Packaging ATPase P9 extract also was provided, as in the standard assay, although P9 was expressed in wt amounts in sus621 infection.
In the PRD1 packaging assay, procapsids are filled in vitro with mutant PRD1 DNA containing a lacZα insert which, when plated on a suitable host, gave rise to blue plaques (88). The addition of only P6, but not P9, to the Sus621 packaging reaction did not give rise to any blue plaques. However, when packaging ATPase P9 was added, regardless of whether protein P6 was present, some blue plaques could be detected, but their frequency was less than 25%, compared to the background of white plaques. The titer of in vitro packaged particles obtained with Sus621 extract was, at best, approximately 3% of that obtained with an equal amount of DNA in the packaging system using packaging ATPase-deficient empty Sus1 particles. Not surprisingly, the Sus621 particle extract gave rise to a much higher background of white plaques (10- to 20-fold more) than the packaging ATPase P9-deficient Sus1 particle extract. Results of the in vitro packaging assay are presented in Fig. 4.
FIG. 4.
In vitro packaging of Sus621 particles. Columns indicate the total amount of PFU/ml of packaging reaction obtained in host DS88. This is shown on a logarithmic scale (depicted by the left y axis), and the mean values for each reaction are shown above each column. The gray portion of each column represents the relative frequency of blue plaques (linear scale). In vitro packaging of Sus621 particles was achieved only with the addition of P9 (columns “Sus621+P6+P9” and “Sus621+P9”) but not with the addition of P6 alone (column “Sus621+P6”). The standard in vitro packaging assay with P9-deficient Sus1 particles and P9 extract (Sus1+P9) is shown as a positive control. Negative control reactions for both Sus621 and Sus1 particles, containing all other components except the lacZα-containing DNA, are marked “no DNA.”
DISCUSSION
The DNA packaging apparatuses of tailed dsDNA phages and herpesviruses are located at one vertex of their icosahedral capsids. The location of P6 at the unique packaging vertex of PRD1, and the fact that both XX− and XXII− packaging mutants (deficient in small membrane proteins P20 and P22, respectively) lack P6, already suggested that P6 could play a role in DNA packaging. We have isolated a PRD1 mutant, sus621, which is deficient in P6, to show that the lack of P6 leads to serious deficiencies in DNA packaging.
In viruses, genes coding for proteins functioning together are often located close to each other in the genome. Gene VI, coding for P6, is located in front of gene IX, which codes for packaging ATPase P9. However, gene VI overlaps another open reading frame, gene X, coding for the nonstructural membrane scaffold protein P10. This overlap is conserved in the genomes of many tectiviruses, including the gram-positive bacterium-infecting virus Bam35 (76). Protein P10 is a nonstructural protein that is essential in the formation of virus-specific lipid vesicles, on top of which the PRD1 capsid proteins are assembled (78). Therefore, it was suspected that P6 might play a role in the formation of the procapsid, possibly in initiating capsid assembly or controlling some aspect of vesicle formation. The process of capsid assembly, however, did not seem to be hampered in any detectable way by the lack of P6. Normally sized and shaped particles were found to be produced in wt amounts in sus621 infection.
However, the subsequent stages of maturation were gravely impaired in the P6-deficient mutant, and most of the particles that were produced were empty. Surprisingly, the destruction of packaging activity was not total, and approximately 5% of the particles were able to develop to mature, DNA-containing virions, with the rest being empty.
In wt PRD1 infection, a small percentage (10 to 20%) of empty particles is produced in addition to DNA-filled virions. If lysis is delayed, increased amounts of these empty particles are packaged with DNA. These wt procapsids, similar to packaging ATPase-deficient mutant Sus1 particles, contain all structural proteins of the mature virion except packaging ATPase P9 (89). Neither empty nor filled Sus621 particles were found to lack any structural proteins except P6, but the amount of packaging ATPase P9 in filled particles was reduced compared to that in wt virions. Surprisingly, empty Sus621 particles and filled Sus621 particles contained similar amounts of P9, although P9 is not a part of empty wt procapsids. In addition, a small amount of DNA-binding protein P19 was found attached to empty Sus621 particles. Since no P19 was observed in filled or empty wt particles, we assume that P19 is normally associated with unpackaged DNA and is removed during the packaging process. Thus, P6-deficient, P19-containing empty Sus621 particles could be the products of unsuccessful packaging events.
The packaging mechanisms of other icosahedral dsDNA viruses are known to include proteins that seal the particles after the DNA has been packaged. The possibility of P6 alone being a “plug,” possibly going through a conformational change after packaging and thus closing the particles, could be ruled out by the fact that filled Sus621 particles were found to be stable and infectious. Furthermore, the total amounts of purified particles obtained were similar for sus621 and wt infections, and the ratios of packaged and empty particles were similar between both cell sections and purified particles of sus621. If DNA leakage or capsid stability had been a problem, relatively more packaged particles would have been seen in cell sections than was the case for purified particles, or the relative amount of packaged particles seen in sections of infected cells taken at different time points would have, after an initial increase, decreased during the course of infection. However, this was not the case, and we conclude that P6 is not a packaging completion protein. Since DNA packaging does occur in vivo in sus621 infection, though not efficiently, it seems clear that P6 is not a portal protein either, because lack of a portal most probably would inhibit packaging completely.
It was suggested that the packaging ATPase P9, which stays bound to the PRD1 particles after packaging the DNA, could act as a plug (89). While this may still be the case, it is clear that despite the presence of P9, the internal membrane of the empty P6− particles was susceptible to SDS treatment, unlike results for wt virions. Similarly, the lack of contrast in electron micrographs, in both cell sections and empty and filled negatively stained particles, suggested that material may be exchanged between the exterior and interior of the P6− particles. This could mean that even though DNA does not leak out from packaged P6-deficient particles, both P6 and P9 are needed to tightly seal the particles and to prevent smaller molecules from accessing the particle interior.
It appears that the P9-containing empty P6− particles are products of unsuccessful packaging attempts, where P9 (and some P19) is left attached to the particle but the DNA has failed to be packaged properly. Although large numbers of empty particles were present, only very little packaging could be achieved in vitro, and then could be done only when additional P9 was provided. The interpretation of these results is that the empty particles may consist of two populations: empty, P9-containing “dead-end” particles, for which packaging has been attempted but has failed and which cannot be packaged in vitro, and empty, P9-less procapsids that can be packaged when more P9 is provided to the in vitro system. The latter particles would be equivalent, except for the lack of P6, to normal procapsids.
The filled Sus621 particles were found to contain PRD1-specific DNA, but their specific infectivity was two orders of magnitude lower than that of the wt filled particles. It has earlier been shown that a deficiency in membrane protein P22, which also causes the total loss of P6 from particles, has no effect on binding to host cells (58). It is thus assumed that the decrease in infectivity of the P6 mutant sus621 is not due to deficiencies in attachment kinetics.
The poor separation of empty and filled Sus621 particles compared to the wt can probably explain only part of the observed difference in specific infectivity, possibly suggesting an additional role for P6 in DNA delivery. Whether P6 actually plays an active role in the delivery of the PRD1 genome or whether the decreased infectivity of P6− particles is caused by incomplete packaging leading to the DNA not being fully “injection competent” due to, for example, reduced internal capsid pressure or the DNA having an incorrect conformation is not clear. Decreased infectivity could also explain the small amount of in vitro packaging seen, since the assay relies on the in vitro packaged particles being infectious.
When the PRD1 packaging ATPase P9 was used as a starting point for protein database searches (88), almost exactly the same set of viruses, described above, was found as when the coat protein folds and capsid architectures were compared. Thus, a similar packaging mechanism has been envisaged for this group of viruses with internal membranes. Consequently, further information on the packaging of PRD1 may give new insight into the packaging mechanisms of viruses belonging to this lineage.
When comparing PRD1 packaging to packaging in the other tailed bacteriophages or herpesviruses, no obvious counterpart for P6 can be deduced. However, there are certain similarities between P6 and bacteriophage lambda packaging factor gpFI, which is involved in mediating terminase binding to the lambda procapsid and the initial cos cleavage of the DNA (60, 61, 86). Similar to what is seen with gpFI in bacteriophage lambda, lack of PRD1 P6 decreases but does not totally inhibit packaging, and the binding of packaging ATPase P9 to the capsid is altered although, obviously, no cos-type cleavage is necessary for the unit-length PRD1 genome. Although overall sequence similarity is low (∼10%), both gpFI and P6 are very acidic (theoretical pIs, 4.6 and 4.0, respectively), are similar in size, and have similar stretches of glutamate-rich sequences in their N-terminal halves. Interestingly, another glutamate-rich protein, the L4 22-kDa protein, has been shown to be involved in the encapsidation of the adenovirus unit-length DNA (72), although whether adenovirus maturation proceeds via packaging of empty procapsids or by coassembly of DNA and capsid protein is still unresolved.
Similarities between PRD1 and bacteriophage phi29 also exist. The replication systems of PRD1, adenovirus, and phi29 are similar, with all three viruses using a protein-primed mechanism producing unit-length genomes. In phi29, the terminal protein gp3 is essential for packaging and has been suggested to be analogous to the DNA-binding small terminase subunit of phages with concatemeric DNA. Parallels can be drawn between the functions of PRD1 P6 and the packaging RNA molecule of phi29, which is needed for the binding of phi29 ATPase gp16 to the procapsid and for stimulating the ATPase activity of gp16 (36).
Both gpFI of lambda and the packaging RNA of phi29 are transient components of their respective packaging apparatuses (15, 39, 75), whereas PRD1 P6 stays attached to the mature virion. The PRD1 packaging ATPase P9 is also a structural protein, and cleavage of the unit-length PRD1 genome is not necessary. The presence of an internal membrane in PRD1, into which the DNA needs to be translocated, distinguishes the PRD1 packaging mechanism from the packaging systems of other bacteriophages and herpesviruses. No additional stabilizing components are added to the virion after packaging; the ATPase P9 and the genome terminal protein P8 are the only differences in the protein compositions of procapsids and DNA-filled mature particles (89). Stabilization of the packaged particles to prevent leakage could be achieved via a conformational change, for example, in P9 or packaging proteins P20 or P22, triggered by the increased contacts between the capsid and the membrane in the filled virions. The PRD1 terminal protein P8 is highly hydrophobic, which could allow it to interact with the viral membrane in the packaged particle and, thus, to play a role in packaging and in completion of the maturation process.
Based on these findings, previous knowledge of PRD1, and parallels with other viruses, we envisage a model for PRD1 packaging that encompasses several common features of packaging mechanisms in other icosahedral dsDNA bacteriophages but that is clearly a different, unique system. Procapsids containing all other structural components except for P9 and the DNA-P8 complex are first assembled. Packaging probably is initiated by binding of the packaging ATPase P9 to the unit-length genome of PRD1. Whether this occurs through binding of P9 to DNA or the covalently attached terminal protein is still unclear but, based on similarities with phi29, it is probable that the P8 genome terminal protein does play at least some role. The P9-DNA-P8 complex then binds to the empty procapsid. Here, the presence of P6 in the procapsid is crucial, and without it, the affinity of the procapsid to the P8-DNA-P9 complex is lower and the complex may not bind properly, and/or binding does not lead to successful packaging. P6 is connected to the capsid via the small membrane proteins P20 and/or P22 (89). Since the lack of P20 totally abolishes packaging (89) but the lack of P22 still allows for a minute amount of the procapsids to be packaged (58), it can be suggested that P20 is the main candidate for a pore, or membrane-spanning “portal,” through which DNA can be transported into the lipid vesicle inside the capsid. As the membrane vesicle is filled with DNA, contacts between the lipid head groups and the internal part of the major capsid protein are increased, stabilizing the virion. After the PRD1 genome has been packaged, ATPase P9 stays attached to the virion. A conformational change in the packaging proteins or the capsid, or the presence of terminal protein P8, may act to seal the packaging vertex.
Acknowledgments
This investigation was supported by Academy of Finland research grants 1213992 (D.H.B.), 1201964 (J.K.H.B.), and 121 3467 and the Finnish Centre of Excellence Programme (Progamme for Structural Virology, 2002-2005, and Finnish Centre of Excellence in Virus Research, 2006-2011). The Viikki Graduate School of Biosciences is acknowledged for funding N.J.K. and G.Z.
Footnotes
Published ahead of print on 3 January 2007.
REFERENCES
- 1.Abrescia, N. G. A., J. J. B. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. H. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68-74. [DOI] [PubMed] [Google Scholar]
- 2.Agirrezabala, X., J. Martín-Benito, M. Valle, J. M. González, A. Valencia, J. M. Valpuesta, and J. L. Carrascosa. 2005. Structure of the connector of bacteriophage T7 at 8 Å resolution: structural homologies of a basic component of a DNA translocating machinery. J. Mol. Biol. 347:895-902. [DOI] [PubMed] [Google Scholar]
- 3.Bamford, D., and L. Mindich. 1982. Structure of the lipid-containing bacteriophage PRD1: disruption of wild-type and nonsense mutant phage particles with guanidine hydrochloride. J. Virol. 44:1031-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamford, D. H., and H.-W. Ackermann. 2000. Family Tectiviridae, p. 111-116. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, CA.
- 5.Bamford, D. H., T. McGraw, G. Mackenzie, and L. Mindich. 1983. Identification of a protein bound to the termini of bacteriophage PRD1 DNA. J. Virol. 47:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bamford, D. H., and L. Mindich. 1984. Characterization of the DNA-protein complex at the termini of the bacteriophage PRD1 genome. J. Virol. 50:309-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bamford, D. H., and L. Mindich. 1980. Electron microscopy of cells infected with nonsense mutants of bacteriophage phi6. Virology 107:222-228. [DOI] [PubMed] [Google Scholar]
- 8.Bamford, J. K. H., and D. H. Bamford. 1990. Capsomer proteins of bacteriophage PRD1, a bacterial virus with a membrane. Virology 177:445-451. [DOI] [PubMed] [Google Scholar]
- 9.Bamford, J. K. H., and D. H. Bamford. 2000. A new mutant class, made by targeted mutagenesis, of phage PRD1 reveals that protein P5 connects the receptor binding protein to vertex. J. Virol. 74:7781-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bamford, J. K. H., A.-L. Hänninen, T. M. Pakula, P. M. Ojala, N. Kalkkinen, M. Frilander, and D. H. Bamford. 1991. Genome organization of membrane-containing bacteriophage PRD1. Virology 183:658-676. [DOI] [PubMed] [Google Scholar]
- 11.Bartolomé, B., Y. Jubete, E. Martínez, and F. de la Cruz. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102:75-78. [DOI] [PubMed] [Google Scholar]
- 12.Bazinet, C., J. Benbasat, J. King, J. M. Carazo, and J. L. Carrascosa. 1988. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry 27:1849-1856. [DOI] [PubMed] [Google Scholar]
- 13.Becker, A., H. Murialdo, H. Lucko, and J. Morell. 1988. Bacteriophage lambda DNA packaging. The product of the FI gene promotes the incorporation of the prohead to the DNA-terminase complex. J. Mol. Biol. 199:597-607. [DOI] [PubMed] [Google Scholar]
- 14.Becker, B., N. de la Fuente, M. Gassel, D. Gunther, P. Tavares, R. Lurz, T. A. Trautner, and J. C. Alonso. 1997. Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol. 268:822-839. [DOI] [PubMed] [Google Scholar]
- 15.Benchimol, S., A. Becker, H. Murialdo, and M. Gold. 1978. The role of the bacteriophage lambda Fl gene product during phage head assembly in vitro. Virology 91:205-221. [DOI] [PubMed] [Google Scholar]
- 16.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 17.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification system of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 18.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 19.Butcher, S. J., D. H. Bamford, and S. D. Fuller. 1995. DNA packaging orders the membrane of bacteriophage PRD1. EMBO J. 14:6078-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camacho, A. G., A. Gual, R. Lurz, P. Tavares, and J. C. Alonso. 2003. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J. Biol. Chem. 278:23251-23259. [DOI] [PubMed] [Google Scholar]
- 21.Campbell, J. L., C. C. Richardson, and F. W. Studier. 1978. Genetic recombination and complementation between bacteriophage T7 and cloned fragments of T7 DNA. Proc. Natl. Acad. Sci. USA 75:2276-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carazo, J. M., L. E. Donate, L. Herranz, J. P. Secilla, and J. L. Carrascosa. 1986. Three-dimensional reconstruction of the connector of bacteriophage phi 29 at 1.8 nm resolution. J. Mol. Biol. 192:853-867. [DOI] [PubMed] [Google Scholar]
- 23.Casjens, S., E. Wyckoff, M. Hayden, L. Sampson, K. Eppler, S. Randall, E. T. Moreno, and S. P. 1992. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J. Mol. Biol. 224:1055-1074. [DOI] [PubMed] [Google Scholar]
- 24.Catalano, C. E. 2005. Viral genome packaging machines: an overview. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structure and mechanism. Landes Bioscience, Austin, TX.
- 25.Chang, J., P. Weigele, J. King, W. Chiu, and W. Jiang. 2006. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14:1073-1082. [DOI] [PubMed] [Google Scholar]
- 26.Cingolani, G., S. D. Moore, P. E. J. Prevelige, and J. E. Johnson. 2002. Preliminary crystallographic analysis of the bacteriophage P22 portal protein. J. Struct. Biol. 139:46-54. [DOI] [PubMed] [Google Scholar]
- 27.Cockburn, J. J. B., N. G. A. Abrescia, J. M. Grimes, G. C. Sutton, J. M. Diprose, J. M. Benevides, G. J. Thomas, J. K. H. Bamford, D. H. Bamford, and D. I. Stuart. 2004. Membrane structure and interactions with protein and DNA in bacteriophage PRD1. Nature 432:122-125. [DOI] [PubMed] [Google Scholar]
- 28.Driedonks, R. A. 1981. The quaternary structure of the T4 gene product 20 oligomer. Prog. Clin. Biol. Res. 64:315-323. [PubMed] [Google Scholar]
- 29.Dröge, A., M. A. Santos, A. C. Stiege, J. C. Alonso, R. Lurz, T. A. Trautner, and P. Tavares. 2000. Shape and DNA packaging activity of bacteriophage SPP1 procapsid: protein components and interactions during assembly. J. Mol. Biol. 296:117-132. [DOI] [PubMed] [Google Scholar]
- 30.Dube, P., P. Tavares, R. Lurz, and M. van Heel. 1993. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 12:1303-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feiss, M., and D. A. Siegele. 1979. Packaging of the bacteriophage lambda chromosome: dependence of cos cleavage on chromosome length. Virology 92:190-200. [DOI] [PubMed] [Google Scholar]
- 32.Fokine, A., P. R. Chipman, P. G. Leiman, V. V. Mesyanzhinov, V. B. Rao, and M. G. Rossmann. 2004. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. USA 101:6003-6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gowen, B., J. K. H. Bamford, D. H. Bamford, and S. D. Fuller. 2003. The tailless icosahedral membrane virus PRD1 localizes the proteins involved in genome packaging and injection at a unique vertex. J. Virol. 77:7863-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grahn, A. M., J. Caldentey, J. K. H. Bamford, and D. H. Bamford. 1999. Stable packaging of phage PRD1 DNA requires adsorption protein P2, which binds to the IncP plasmid-encoded conjugative transfer complex. J. Bacteriol. 181:6689-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grahn, A. M., R. Daugelavičius, and D. H. Bamford. 2002. Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol. Microbiol. 46:1199-1209. [DOI] [PubMed] [Google Scholar]
- 36.Grimes, S., and D. Anderson. 1990. RNA dependence of the bacteriophage phi29 DNA packaging ATPase. J. Mol. Biol. 215:559-566. [DOI] [PubMed] [Google Scholar]
- 37.Guasch, A., J. Pous, B. Ibarra, F. X. Gomis-Ruth, J. M. Valpuesta, N. Sousa, J. L. Carrascosa, and M. Coll. 2002. Detailed architecture of a DNA translocating machine: the high-resolution structure of the bacteriophage φ29 connector particle. J. Mol. Biol. 315:663-676. [DOI] [PubMed] [Google Scholar]
- 38.Guo, P., C. Peterson, and D. Anderson. 1987. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J. Mol. Biol. 197:229-236. [DOI] [PubMed] [Google Scholar]
- 39.Guo, P. X., S. Erickson, and D. Anderson. 1987. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science 236:690-694. [DOI] [PubMed] [Google Scholar]
- 40.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 41.Hänninen, A.-L., D. H. Bamford, and J. K. H. Bamford. 1997. Probing the phage PRD1 specific proteins during infection by monoclonal and polyclonal antibodies. Virology 227:198-206. [DOI] [PubMed] [Google Scholar]
- 42.Hendrix, R. W. 1978. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. USA 75:4779-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huiskonen, J. T., L. Laakkonen, M. Toropainen, M. Sarvas, D. H. Bamford, and J. K. Bamford. 2003. Probing the ability of the coat and vertex protein of the membrane-containing bacteriophage PRD1 to display a meningococcal epitope. Virology 310:267-279. [DOI] [PubMed] [Google Scholar]
- 44.Jaatinen, S. T., S. J. Viitanen, D. H. Bamford, and J. K. H. Bamford. 2004. Integral membrane protein P16 of bacteriophage PRD1 stabilizes the adsorption vertex structure. J. Virol. 78:9790-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson, E. N., D. A. Jackson, and R. J. Deans. 1978. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 118:365-388. [DOI] [PubMed] [Google Scholar]
- 46.Jardine, P. J., and D. L. Anderson. 2005. DNA packaging in double-stranded DNA phages, p. 49-65. In R. Calendar (ed.), The bacteriophages, 2nd ed. Oxford University Press, New York, NY.
- 47.Jiang, W., J. Chang, J. Jakana, P. Weigele, J. King, and W. Chiu. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kocsis, E., M. E. Cerritelli, B. L. Trus, N. Cheng, and A. C. Steven. 1995. Improved methods for determination of rotational symmetries in macromolecules. Ultramicroscopy 60:219-228. [DOI] [PubMed] [Google Scholar]
- 49.Lander, G. C., L. Tang, S. R. Casjens, E. B. Gilcrease, P. Prevelige, A. Poliakov, C. S. Potter, B. Carragher, and J. E. Johnson. 2006. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791-1795. [DOI] [PubMed] [Google Scholar]
- 50.Liljeström, P., I. Laamanen, and E. T. Palva. 1988. Structure and expression of the ompB operon, the regulatory locus for the outer membrane porin regulon in Salmonella typhimurium LT-2. J. Mol. Biol. 201:663-673. [DOI] [PubMed] [Google Scholar]
- 51.Lin, H., V. B. Rao, and L. W. Black. 1999. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J. Mol. Biol. 289:249-260. [DOI] [PubMed] [Google Scholar]
- 52.Lundström, K. H., D. H. Bamford, E. T. Palva, and K. Lounatmaa. 1979. Lipid-containing bacteriophage PR4: structure and life cycle. J. Gen. Virol. 43:538-592. [DOI] [PubMed] [Google Scholar]
- 53.Luo, C., S. Butcher, and D. H. Bamford. 1993. Isolation of a phospholipid-free protein shell of bacteriophage PRD1, an Escherichia coli virus with an internal membrane. Virology 194:564-569. [DOI] [PubMed] [Google Scholar]
- 54.Lurz, R., E. V. Orlova, D. Gunther, P. Dube, A. Droge, F. Weise, M. van Heel, and P. Tavares. 2001. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 310:1027-1037. [DOI] [PubMed] [Google Scholar]
- 55.Maluf, N. K., Q. Yang, and C. E. Catalano. 2005. Self-association properties of the bacteriophage lambda terminase holoenzyme: implications for the DNA packaging motor. J. Mol. Biol. 347:523-542. [DOI] [PubMed] [Google Scholar]
- 56.McGraw, T., H. L. Yang, and L. Mindich. 1983. Establishment of a physical and genetic map for bacteriophage PRD1. Mol. Gen. Genet. 190:237-244. [DOI] [PubMed] [Google Scholar]
- 57.Mindich, L., D. Bamford, C. Goldthwaite, M. Laverty, and G. Mackenzie. 1982. Isolation of nonsense mutants of lipid-containing bacteriophage PRD1. J. Virol. 44:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mindich, L., D. Bamford, T. McGraw, and G. Mackenzie. 1982. Assembly of bacteriophage PRD1: particle formation with wild-type and mutant viruses. J. Virol. 44:1021-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mindich, L., J. Cohen, and M. Weisburd. 1976. Isolation of nonsense suppressor mutants in Pseudomonas. J. Bacteriol. 126:177-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murialdo, H., and D. Tzamtzis. 1997. Mutations of the coat protein gene of bacteriophage lambda that overcome the necessity for the Fl gene; the EFi domain. Mol. Microbiol. 24:341-353. [DOI] [PubMed] [Google Scholar]
- 61.Murialdo, H., D. Tzamtzis, M. Berru, W. L. Fife, and A. Becker. 1997. Mutations in the terminase genes of bacteriophage lambda that bypass the necessity for FI. Mol. Microbiol. 24:937-952. [DOI] [PubMed] [Google Scholar]
- 62.Murialdo, H., X. Xing, D. Tzamtzis, A. Haddad, and M. Gold. 2003. The product of the bacteriophage lambda W gene: purification and properties. Biochem. Cell Biol. 81:307-315. [DOI] [PubMed] [Google Scholar]
- 63.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2005. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J. Virol. 79:10540-10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ojala, P. M., J. T. Juuti, and D. H. Bamford. 1993. Protein P4 of double-stranded RNA bacteriophage φ6 is accessible on the nucleocapsid surface: epitope mapping and orientation of the protein. J. Virol. 67:2879-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oliveira, L., A. O. Henriques, and P. Tavares. 2006. Modulation of the viral ATPase activity by the portal protein correlates with DNA packaging efficiency. J. Biol. Chem. 281:21914-21923. [DOI] [PubMed] [Google Scholar]
- 67.Olkkonen, V. M., and D. H. Bamford. 1989. Quantitation of the adsorption and penetration stages of bacteriophage phi6 infection. Virology 171:229-238. [DOI] [PubMed] [Google Scholar]
- 68.Olsen, R. H., J. Siak, and R. H. Gray. 1974. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J. Virol. 14:689-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Orlova, E. V., P. Dube, E. Beckmann, F. Zemlin, R. Lurz, T. A. Trautner, P. Tavares, and M. van Heel. 1999. Structure of the 13-fold symmetric portal protein of bacteriophage SPP1. Nat. Struct. Biol. 6:842-846. [DOI] [PubMed] [Google Scholar]
- 70.Orlova, E. V., B. Gowen, A. Droge, A. Stiege, F. Weise, R. Lurz, M. van Heel, and P. Tavares. 2003. Structure of a viral DNA gatekeeper at 10 Å resolution by cryo-electron microscopy. EMBO J. 22:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ortega, M. E., and C. E. Catalano. 2006. Bacteriophage lambda gpNu1 and Escherichia coli IHF proteins cooperatively bind and bend viral DNA: implications for the assembly of a genome-packaging motor. Biochemistry 45:5180-5189. [DOI] [PubMed] [Google Scholar]
- 72.Ostapchuk, P., M. E. Anderson, S. Chandrasekhar, and P. Hearing. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pakula, T. M., J. Caldentey, C. Gutiérrez, O. V. M., M. Salas, and D. H. Bamford. 1993. Overproduction, purification, and characterization of DNA-binding protein P19 of bacteriophage PRD1. Gene 126:99-104. [DOI] [PubMed] [Google Scholar]
- 74.Perucchetti, R., W. Parris, A. Becker, and M. Gold. 1988. Late stages in bacteriophage lambda head morphogenesis: in vitro studies on the action of the bacteriophage lambda D-gene and W-gene products. Virology 165:103-114. [DOI] [PubMed] [Google Scholar]
- 75.Peterson, C., M. Simon, J. Hodges, P. Mertens, L. Higgins, E. Egelman, and D. Anderson. 2001. Composition and mass of the bacteriophage phi29 prohead and virion. J. Struct. Biol. 135:18-25. [DOI] [PubMed] [Google Scholar]
- 76.Ravantti, J. J., A. Gaidelyte, D. H. Bamford, and J. K. Bamford. 2003. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrates a viral lineage. Virology 313:401-414. [DOI] [PubMed] [Google Scholar]
- 77.Remaut, E., H. Tsao, and W. Fiers. 1983. Improved plasmid vectors with a thermoinducible expression and temperature-regulated runaway replication. Gene 22:103-113. [DOI] [PubMed] [Google Scholar]
- 78.Rydman, P. S., J. K. Bamford, and D. Bamford. 2001. A minor capsid protein P30 is essential for bacteriophage PRD1 capsid assembly. J. Mol. Biol. 313:785-795. [DOI] [PubMed] [Google Scholar]
- 79.Rydman, P. S., J. Caldentey, S. J. Butcher, S. D. Fuller, T. Rutten, and D. H. Bamford. 1999. Bacteriophage PRD1 contains a labile receptor-binding structure at each vertex. J. Mol. Biol. 291:575-587. [DOI] [PubMed] [Google Scholar]
- 80.Salas, M. 1991. Protein-priming of DNA replication. Annu. Rev. Biochem. 60:39-71. [DOI] [PubMed] [Google Scholar]
- 81.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 82.San Martín, C., R. M. Burnett, F. de Haas, R. Heinkel, T. Rutten, S. D. Fuller, S. J. Butcher, and D. H. Bamford. 2001. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure 9:917-930. [DOI] [PubMed] [Google Scholar]
- 83.San Martín, C., J. T. Huiskonen, J. K. Bamford, S. J. Butcher, S. D. Fuller, D. H. Bamford, and R. M. Burnett. 2002. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nat. Struct. Biol. 9:756-763. [DOI] [PubMed] [Google Scholar]
- 84.Savilahti, H., and D. H. Bamford. 1986. Linear DNA replication: inverted terminal repeats of five closely related Escherichia coli bacteriophages. Gene 49:199-205. [DOI] [PubMed] [Google Scholar]
- 85.Simpson, A. A., P. G. Leiman, Y. Tao, Y. He, M. O. Badasso, P. J. Jardine, D. L. Anderson, and M. G. Rossmann. 2001. Structure determination of the head-tail connector of bacteriophage φ29. Acta Crystallogr. D 57:1260-1269. [DOI] [PubMed] [Google Scholar]
- 86.Sippy, J., and M. Feiss. 2004. Initial cos cleavage of bacteriophage lambda concatemers requires proheads and gpFI in vivo. Mol. Microbiol. 52:501-513. [DOI] [PubMed] [Google Scholar]
- 87.Strauss, H., and J. King. 1984. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol. 172:523-543. [DOI] [PubMed] [Google Scholar]
- 88.Strömsten, N. J., D. H. Bamford, and J. K. Bamford. 2005. In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses. J. Mol. Biol. 348:617-629. [DOI] [PubMed] [Google Scholar]
- 89.Strömsten, N. J., D. H. Bamford, and J. K. H. Bamford. 2003. The unique vertex of bacterial virus PRD1 is connected to the viral internal membrane. J. Virol. 77:6314-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tavares, P., M. A. Santos, R. Lurz, G. Morelli, H. de Lencastre, and T. T. A. 1992. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J. Mol. Biol. 225:81-92. [DOI] [PubMed] [Google Scholar]
- 91.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Trus, B. L., N. Cheng, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tye, B. K., and D. Botstein. 1974. P22 morphogenesis. II. Mechanism of DNA encapsulation. J. Supramol. Struct. 2:225-238. [DOI] [PubMed] [Google Scholar]
- 94.Valpuesta, J. M., J. J. Fernandez, J. M. Carazo, and J. L. Carrascosa. 1999. The three-dimensional structure of a DNA translocating machine at 10 Å resolution. Structure 7:289-296. [DOI] [PubMed] [Google Scholar]
- 95.Winston, F., D. Botstein, and J. H. Miller. 1979. Characterization of amber and ochre suppressors in Salmonella typhimurium. J. Bacteriol. 137:433-439. [DOI] [PMC free article] [PubMed] [Google Scholar]