Abstract
An ∼4-Mb genomic segment on chromosome 17p11.2, commonly deleted in patients with the Smith-Magenis syndrome (SMS) and duplicated in patients with dup(17)(p11.2p11.2) syndrome, is flanked by large, complex low-copy repeats (LCRs), termed proximal and distal SMS-REP. A third copy, the middle SMS-REP, is located between them. SMS-REPs are believed to mediate nonallelic homologous recombination, resulting in both SMS deletions and reciprocal duplications. To delineate the genomic structure and evolutionary origin of SMS-REPs, we constructed a bacterial artificial chromosome/P1 artifical chromosome contig spanning the entire SMS region, including the SMS-REPs, determined its genomic sequence, and used fluorescence in situ hybridization to study the evolution of SMS-REP in several primate species. Our analysis shows that both the proximal SMS-REP (∼256 kb) and the distal copy (∼176 kb) are located in the same orientation and derived from a progenitor copy, whereas the middle SMS-REP (∼241 kb) is inverted and appears to have been derived from the proximal copy. The SMS-REP LCRs are highly homologous (>98%) and contain at least 14 genes/pseudogenes each. SMS-REPs are not present in mice and were duplicated after the divergence of New World monkeys from pre-monkeys ∼40–65 million years ago. Our findings potentially explain why the vast majority of SMS deletions and dup(17)(p11.2p11.2) occur at proximal and distal SMS-REPs and further support previous observations that higher-order genomic architecture involving LCRs arose recently during primate speciation and may predispose the human genome to both meiotic and mitotic rearrangements.
Smith-Magenis syndrome (SMS) is caused in >90% of cases by a common deletion of an ∼4-Mb gene-rich genomic segment in 17p11.2. Recently, the same chromosome region has been shown to be duplicated in patients with the reciprocal chromosome duplication, dup(17)(p11.2p11.2) (Chen et al. 1997; Potocki et al. 2000). Physical mapping studies have shown that the SMS-common-deletion interval is flanked by large (∼200 kb), highly homologous, low-copy repeat (LCR) gene clusters termed SMS-REPs (Chen et al. 1997). The proximal and distal SMS-REPs likely act as substrates for nonallelic homologous recombination (NAHR; also known as unequal crossing-over), resulting in both common deletions and reciprocal duplications of the same chromosome segment. A third copy, the middle SMS-REP, has been mapped in 17p11.2 between the proximal and distal SMS-REPs (Chen et al. 1997).
The chromosome 17p11.2 genomic region is involved in several other rearrangements. Isodicentric chromosomes idic(17)(p11), with breakpoints mapping within or just adjacent to the SMS critical region, have been identified in various hematological malignancies, including chronic myeloid leukemia, and in solid tumors, such as childhood primitive neuroectodermal tumors (Fioretos et al. 1999; Scheurlen et al. 1999). Chromosome amplifications within 17p11.2 have been described in patients with osteosarcoma and leiomyosarcoma (Tarkkanen et al. 1995; Otaño-Joos et al. 2000). In medulloblastomas, an aberrant hypermethylation of the major breakpoint cluster region in 17p11.2 has been proposed to be an additional genomic feature responsible for the chromosomal fragility (Frühwald et al. 2001). These data, together with identified somatic mosaicism for SMS deletions (Zori et al. 1993; Juyal et al. 1996), indicate that the presence of unique genome architecture features, including highly homologous SMS-REPs, makes the chromosome 17p11.2 a highly unstable region in the human genome, prone to both meiotic and mitotic rearrangements.
To delineate the genomic structure and evolution of the SMS-REPs, we constructed and sequenced a complete bacterial artificial chromosome/P1 artifical chromosome (BAC/PAC) contig. Based on genomic sequence analysis, we elucidated the size and the orientation of each SMS-REP, the extent of homology among the SMS-REPs, genes within the SMS-REPs, and here provide a model for the evolution of SMS-REPs. Sequence-based structural analysis of the SMS-REPs is essential to determine the molecular mechanism of chromosome rearrangements in SMS, as well as other genomic disorders (Lupski 1998). These studies add to a growing body of evidence that implicate genome architecture in DNA rearrangements responsible for genomic disorders (Lupski 1998; Mazzarella and Schlessinger 1998; Ji et al. 2000; Shaffer and Lupski 2000; Emanuel and Shaikh 2001; Stankiewicz and Lupski 2002).
RESULTS
Aligning BAC Genomic Clones to Specific SMS-REPs by cis-Morphism Analysis and DNA Fingerprinting
The identification of genomic clones specific to an individual SMS-REP was possible due to a limited number of sequence differences among them, which we called cis-morphisms. Cis-morphisms refer to sequence variation among nonallelic, highly similar sequence copies on the same chromosome. This term has been proposed to distinguish from polymorphisms resulting from variation between sequences on different chromosome homologs (alleles; Boerkoel et al. 1999). We identified SMS-REP-specific cis-morphisms using Southern blotting with genomic DNA and BAC clones as targets and compared the hybridization band patterns with those of established SMS-REP-specific yeast artificial chromosome (YAC) clones (912D7 and 567A2 for distal SMS-REP; 907E8 and 126H9 for proximal SMS-REP; 951G11 for middle SMS-REP) that were anchored with flanking unique sequences (Chen et al. 1997).
We documented previously that SMS-REPs contain at least four genes or pseudogenes (CLP, TRE, SRP, and KER, a keratin gene cluster) by sequence skimming of SMS-REP-specific cosmid clones (Chen et al. 1997). By utilizing probes from these genes in Southern analysis of genomic DNA and YAC clones anchored by flanking unique sequences, we have detected cis-morphisms that serve as unique identifiers to each specific SMS-REP. Multiple restriction enzyme digestions of large-insert genomic clones were utilized and enabled several cis-morphisms to be identified (Fig. 1; Table 1). Using CLP, TRE, and SRP sequences as probes on a BAC genomic library we identified >50 SMS-REP-like BAC clones. The CLP probe was a 1.1-kb HindIII fragment of the cDNA clone 41G7A (Chen et al. 1997), and TRE and SRP probes were amplified by 3′ TRE and SRP primers, respectively. Cross-hybridizing fragments were then analyzed for SMS-REP specific cis-morphisms to uniquely identify to which SMS-REP (proximal, middle, or distal) each BAC clone mapped. Independently, all SMS-REP cross-hybridizing BAC clones were subjected to DNA-fingerprinting analysis. The restriction patterns of the majority of these clones were highly similar, although minor differences allowed the BACs to be grouped into contigs. Fingerprint analysis was combined with hybridization analyses to select minimal clone sets for genomic sequencing.
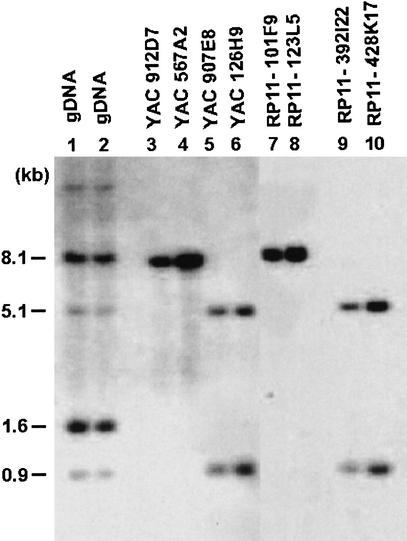
Figure 1.
An example of a cis-morphism among three SMS-REPs. Southern blotting with CLP cDNA probe on HindIII-digested large-insert genomic clones revealed an 8.1-kb band for the distal SMS-REP (lanes 3 and 4 for yeast artificial chromosomes [YACs]; lanes 7 and 8 for bacterial artificial chromosomes [BACs]) and 5.1 kb and 0.9 kb for the proximal SMS-REP (lanes 5 and 6 for YACs; lanes 9 and 10 for BACs). Hybridization to total genomic DNA (gDNA in lanes 1 and 2) reveals additional cross-hybridizing bands, which map to different genomic locations.
Table 1.
Cis-Morphisms Between SMS-REPs and the Related Structures. (Sizes in kb)
| Probe | Restriction enzymes | Distal SMS-REP | Middle SMS-REP | Proximal SMS-REP | Other sites |
|---|---|---|---|---|---|
| CLP | Hind III | 8.1 | (−) | 5.1, 0.9 | 1.6, 1.4 |
| TRE | Hind III | 14 | 8.5, 5.3 | 14 | 7 |
| XbaI | 11, 7 | 11, 7 | 11, 4.3 | 15, 9 | |
| PstI | 8, 2.4 | 8, 4.1 | 8, 2.4 | 7 | |
| SRP | PvuII | 2.3 | 2.3 | 0.8 | 14 |
| RsaI | 1.3 | 2.2 | 2.2 | 4.5, 4.8 | |
| TaqI | 4.1 | 4.5 | 4.1 |
Underlined numbers represent the bands unique to each SMS-REP.
Generation of the BAC Contigs Spanning the SMS-REPs
We collected BAC clones identified by hybridization screening of the RPCI-11 library and by computational methods using BLAST searches. Fifty-eight clones were assigned to individual SMS-REPs by cis-morphisms and DNA fingerprinting, and we constructed BAC contigs spanning the three individual SMS-REPs (Fig. 2). A minimal-tiling path of assigned BAC genomic clones, selected using the combined results of hybridization and fingerprinting analyses, was chosen for sequence analysis.
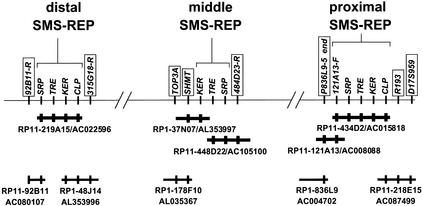
Figure 2.
Bacterial artificial chromosome/P1 artificial chromosome (BAC/PAC) contig map spanning three SMS-REPs. Thick bold lines represent minimal-tiling-path, large-insert clones utilized for genomic finished sequence of SMS-REPs. Clones are designated with their clone name and their GenBank accession number. Markers in boxes represent SMS-REP-flanking sequences used to determine SMS-REP orientations. Final orientation was determined by the construction of a complete BAC/PAC contig spanning the common deletion (Bi et al. 2002).
The mapping of the genomic BAC clones to the SMS-REP region in 17p11.2 was further confirmed by fluorescence in situ hybridization (FISH). Metaphase FISH analysis using the SMS-REP BAC clones displayed hybridization to the 17p11.2 region, whereas interphase FISH analysis showed three hybridization signals for each chromosome (one per chromatid for each of the three SMS-REP copies in this G2 interphase nucleus; Fig. 3). Although FISH analysis can identify SMS-REP cross-hybridizing genomic sequences and distinguish three copies in 17p11.2, it cannot determine from which specific individual SMS-REP the signal arises. Therefore, cell lines from patients with selected 17p11.2 deletions were examined by FISH to further confirm BAC localization to the SMS-common-deletion region.
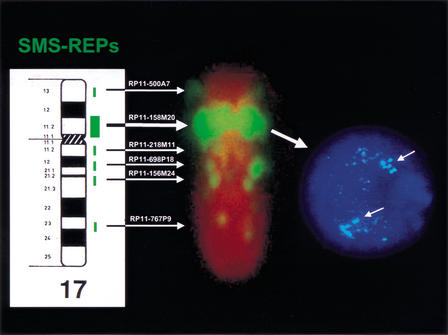
Figure 3.
Human metaphase chromosome 17 and G2 interphase nucleus after fluorescence in situ hybridization (FISH) with SMS-REP-specific bacterial artificial chromosome (BAC) RP11–158M20. To the left is a human-chromosome-17 ideogram with the positions of hybridization signals (green) shown next to specific cytological bands. White horizontal arrows show specific locations of FISH signals with BACs containing the genomic segments listed above the arrows. To the right of the figure is a G2 interphase FISH analysis (the G2 phase of the nucleus was determined without an internal control probe). Note the three copies of SMS-REP (arrows in interphase nucleus) and SMS-REP-like sequences.
Analysis of the Structures and the Sequences of Three SMS-REPs in 17p11.2
Sequencing of the genomic clones in the minimal-tiling paths was performed at the Whitehead Institute/MIT Center for Genome Research. Sequences of two additional PAC clones, RP1–48J14/AL353996 and RP1–37N07/AL353997, were downloaded from the NCBI web site (http://www.ncbi.nlm.nih.gov/). We thus analyzed genomic sequence spanning each individual SMS-REP. Based on the sequence information, a structural map of the SMS-REPs was constructed (Fig. 4) and analyzed for size and orientation of each SMS-REP, the extent of homology among the SMS-REPs, and gene content within the SMS-REPs.
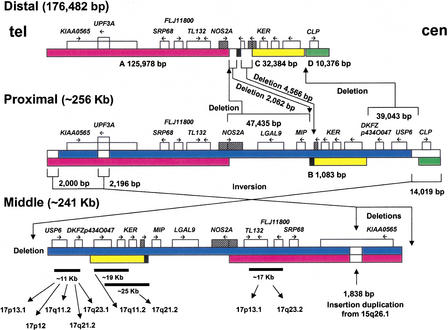
Figure 4.
Sequence-based genomic structure of the SMS-REPs. There are four regions of sequence identity >95% between the proximal and the distal SMS-REPs (A, B, C, and D). The A (red), B (black), and C (yellow) sequence blocks have >98% identity between distal and proximal REPs; the D regions (green) show >95% identity. Blue represents the regions of homology between proximal and middle SMS-REPs. The proximal copy is the largest and is localized in the same orientations as the distal copy. The middle SMS-REP shows almost the same sequence and structure as the proximal copy except for two terminal deletions, an UPF3A gene interstitial deletion and a small (∼2 kb) insertional duplication. However, it is inverted with respect to proximal and distal SMS-REPs. SMS-REP-specific CLP, TRE, and SRP cis-morphisms (Table 1) were confirmed by DNA sequencing. Fourteen genes/pseudogenes were found and are summarized in Table 2. The two additional KER copies in distal SMS-REP represent repeated fragments of the KER pseudogenes, the accession numbers of which are given in Table 2. Crosshatched areas (NOS2A in the proximal and KER [M22927] in the distal) denote two genes spanning the high homology and nonhomology area between the distal and proximal, which suggest a three-step event for the hypothetical model of the evolution of the SMS-REPs (see text). At the bottom, the chromosome 17 distribution of fragments of SMS-REP, which constitutes a chromosome 17 low-copy repeat, LCR17, is shown. The above data were obtained through BLAST analysis of sequence database.
The proximal SMS-REP is the largest, ∼256 kb (probably 255,808 bp; np 50,751 in RP11–121A13/AC008088 and np 4,010 in RP11–434D2/AC015818), and contains 14 genes/pseudogenes. Based on flanking markers P836L9–5‘ and R193 (Fig. 2), the distal–proximal orientation of proximal SMS-REP is from SRP to CLP (Figs. 2, 4). The distal SMS-REP is 176,482 bp (np 10,612–187,093 in RP11–219A15/AC022596). Distal SMS-REP-flanking markers 92B11-R and 315G18-R showed that the distal SMS-REP is in the same SRP–CLP orientation. Therefore, proximal and distal SMS-REP represent direct repeats within 17p11.2. The markers SHMT and 484D23-R (Fig. 2) flanking middle SMS-REP (241 kb; probably 240,806 bp, np 42,370 in RP11–448D22/AC105100, and np 53,872 in RP1–37N07) showed that it is inverted with respect to proximal and distal SMS-REPs.
There are four regions of high homology between the proximal and the distal SMS-REPs (A, B, C, and D regions in Fig. 4). The sum of these high-sequence-similarity regions is ∼170 kb (169,905 bp), and the homology is greater than 98%, with the exception of the D region (>95%). The largest conserved segment (region A in Fig. 4) is ∼126 kb in size. Two large sequence blocks (between A and B, and C and D) in the proximal SMS-REP are absent in the distal SMS-REP. Two smaller blocks, flanking areas of the B region in the distal SMS-REP, are absent in the proximal SMS-REP.
At least 14 genes/pseudogenes were identified in SMS-REPs and are summarized in Table 2 and Figure 4. Two potential genes in the SMS-REPs include KIAA0565 and FLJ11800. The remaining genes appear to represent pseudogenes or pseudogene-like structures (UPF3A, LGALS9, SRP68, TL-132, NOS2A, MIP, KER cluster, USP6 [also known as TRE], CLP, and several expressed sequence tags: BG108241, AIG53154, AW977532, AL138090, AI913336, BF223454, AW469705, AL136790, AA405442). Importantly, sequence analysis confirmed the presence of all four genes/pseudogenes (SRP, KER, TRE, and CLP) previously identified within SMS-REPs (Chen et al. 1997). Interestingly, many (7 of 14) of the genes/pseudogenes also map to the long arm of chromosome 17 (Table 2). Some parts of the gene sequences are repeated (e.g., a part of TL-132 sequence; np 4173–4326 of AJ012755 is duplicated and inverted). As predicted by previous hybridization studies, no CLP sequence was found in the middle SMS-REP, and the distal SMS-REP is devoid of two genomic fragments containing LGALS9, MIP, and USP6 (Fig. 4). In addition, BLAST analyses identified that an ∼2.3-kb UPF3A gene, present in proximal and distal SMS-REPs, is absent in the middle SMS-REP. Interestingly, adjacent (∼400 bp) to this middle SMS-REP-specific deletion, a 1838-bp insertional duplication from chromosome 15q26.1 was identified (Fig. 4). BLAST analysis revealed that the overall similarity among SMS-REP-homologous segments is 98% identical, indicating that SMS-REPs can likely act as substrates for NAHR, resulting in both deletions and reciprocal duplications of the same chromosome segment.
Table 2.
Genes/Pseudogenes Found in the SMS-REPs
| Name | GenBank accession no. | Size (bp) | Distal SMS-REP (np) | Middle SMS-REP (np) | Proximal SMS-REP (np) | Chromosomal sites of possible real genes | |
|---|---|---|---|---|---|---|---|
| Possible genes | KIAA0565 | AB011137 | 5140 | 1–5140 | In finishing | 1–5140 | 17p11.2 |
| FLJ11800 | AK021862 | 2281 | 1–2281 | 1–2281 | 1–2165 | 17p11.2 | |
| Pseudogenes | LGALS9 | AB006782 | 1725 | (−) | 4–1725 | 4–1725 | 11 |
| UPF3A | AF318575 | 2293 | 155–2269 | (−) | 158–2274 | 13 or 18? | |
| SRP68 | AF195951 | 1860 | 1119–1372 | 1120–1372 | 1119–1372 | 17? | |
| TL132 | AJ012755 | 4413 | 1–4413 (4173–4326)a | 1–4413 (4173–4326)a | 1–4413 (4173–4326)a | 17? | |
| NOS2A | U20141 | 3946 | 2590–2795 | 2590–3946 | 2590–3946 | 17q11.2-q12 | |
| MIP | U72395 | 3015 | (−) | 1–3007 | 1–3007 | ||
| KER | M22928 | 3126 | 1–3126b | (−) | (−) | 17? | |
| KER | M22927 | 1939 | 1–1939b | 1–1382b | 1–1382b | ||
| KER | M37818 | 9697 | 1–9696b | 1–9696b | 1–9696b | ||
| DKFZp434O047 | AL117394 | 3118 | (−) | 1–3015 | 1–3015 | 17q11.2 | |
| USP6 (TRE) | X63596 | 4426 | (−) | 1–245, 1735–4419 | 1–245, 1735–4419 | 17q11 | |
| CLP | L54057 | 1824 | 77–1806 | (−) | 78–1795 | 16q24 |
Repeated in reverse direction.
Some parts repeated.
Nucleotide position (np) denotes the portion of the gene sequence in each SMS-REP.
Fragments of SMS-REPs Are Also Located Outside the SMS Region
DNA-fingerprinting analysis of CLP cross-hybridizing BAC genomic clones indicated several distinct contigs, two of which were mapped to proximal and distal SMS-REPs by cis-morphism analysis. To map other CLP genes/pseudogenes in the human genome, we used the clones from the unique BAC contigs that did not map to proximal or distal SMS-REPs as probes in FISH mapping studies. The majority of these BACs mapped to both chromosome 17p11.2 and 16qter, indicating the presence of a CLP gene on chromosome 16q24.
FISH analysis using SMS-REP BAC clones (distal SMS-REP RP11–219A15/AC022596, middle SMS-REP RP11–158M20/AC023401, and proximal SMS-REP RP11–434D2/AC015818) as probes revealed strong hybridization signals on metaphase chromosome 17p11.2 and three strong signals on the interphase chromosomes. However, SMS-REP-specific BACs also showed weaker hybridization signals in interphase analysis and metaphase spreads; these map to chromosomes 17p13.1, 17p12, 17q11.2, 17q12, 17q21.2, and 17q23.2 (Fig. 3).
Computational analysis also supported the existence of other SMS-REP-like sequences on chromosome 17. In concordance with FISH results, BLAST analysis revealed that ∼11–30-kb fragments of SMS-REPs (Fig. 4) are localized on 17p13.1 (∼28 kb; RP11–500A7/AC008058; RP11–333E1/AC087500; CTD-2013F15/AC074339), 17p12 (∼11 kb; RP11–640I15/AC005324), 17q11.2 (∼30 kb; RP11–218M11/AC011840; RP11–271K11/AC005562), 17q12 (∼11 kb; RP11–698P18/AC067923; RP1191J4/AC003976), 17q21.2 (∼25 kb; RP11–156A24/AC019349), and 17q23.2 (∼28 kb; RP11–767P9/AC023352; RP11–178C3/AC005702).
A >19-kb fragment of average >90% homology to a portion of SMS-REP, including the USP6 (TRE) gene, is located within the 17q11.2 BAC clone RP11–271K11/AC005562. This BAC clone also contains ∼85 kb of the neurofibromatosis type 1 LCR, proximal NF1-REP. In addition, we found that the distal NF1-REP on 17q24 (RP11–147L13; AC005332; Dorschner et al. 2000) maps directly adjacent to the breakpoint of the translocation t(1;17) associated with Russell-Silver syndrome (RSS; Dörr et al. 2001).
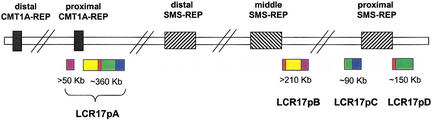
Complex Genome Architecture in Proximal 17p
Analysis of the DNA sequence within proximal 17p, including the SMS common deletion and CMT1A chromosome regions, revealed the presence of several genomic segments of sequence similarity ≥95% to SMS-REP and/or clones that directly flank SMS-REP (Fig. 5). Two LCRs, ∼90-kb LCR17pC and ∼150-kb LCR17pD (also known as, proximal SMS-REP flanking sequences [PSFS]) were found to flank proximal SMS-REP and to be part of at least 410-kb LCR17pA, flanking proximal CMT1A-REP. LCR17pA has also been proposed to be responsible for the evolutionary chromosome translocation 4;19 in gorilla (Stankiewicz et al. 2001). In addition, an at least 210-kb segment LCR17pB within overlapping BAC clones RP11–448D22 and CTD-2145A24, proximally adjacent to the middle SMS-REP, was found to have 98% homology to the other portion of LCR17pA (Fig. 5). Interestingly, the breakpoints of an unusual chromosome deletion in two SMS patients have been identified to map within these LCR17p LCRs (our unpubl. data). The recognition that well-characterized chromosome 17 genomic duplications (SMS, CMT1A, and NF1) are further interspersed with previously unidentified homologous repeated sequences and that these additional LCRs are distributed elsewhere on this and other chromosomes underlines the overall complexity of the chromosome 17 genomic architecture.
Figure 5.
Schematic representation of low-copy repeats (LCRs; >10 kb) in proximal 17p, including SMS and CMT1A chromosome regions. Colored boxes represent positions of highly similar sequence of LCR structures. Note that the at least 410-kb repeat, LCR17pA, flanking the proximal CMT1A-REP has three partial copies: LCR17pB, proximally adjacent to the middle SMS-REP, and LCR17pC and LCR17pD (also described as PSFS; Stankiewicz et al. 2001) flanking the proximal SMS-REP.
Evolutionary Studies
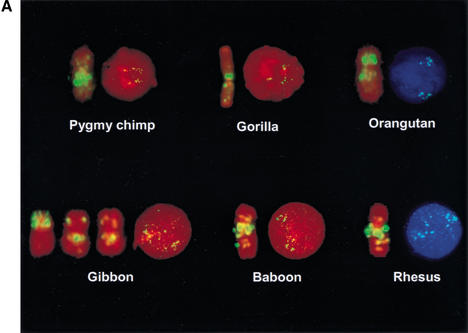
Studies of LCRs involved in several genomic disorders suggest a recent evolutionary origin during primate speciation. In fact, the CMT1A-REP LCR in 17p12 (Pentao et al 1992) is present in human and chimpanzee but not in gorilla, orangutan, and other Old and New World monkeys (Kiyosawa and Chance 1996; Reiter et al. 1997; Boerkoel et al. 1999; Keller et al. 1999). We studied SMS-REP evolution during primate speciation by FISH analysis using human SMS-REP-specific BAC clone probes on different primate cell lines. One BAC from each SMS-REP (distal SMS-REP RP11–219A15/AC022596, middle SMS-REP RP11–158M20/AC023401, and proximal SMS-REP RP11–434D2/AC015818) was used in independent FISH experiments. As expected, the same result was obtained in each of the three separate experiments that used the different SMS-REP probes.
The characteristic fluorescence signal pattern after FISH with SMS-REP-containing BAC clones observed on human metaphase and interphase chromosomes 17 was visible on syntenic metaphase chromosomes of all great apes and Old World monkeys analyzed. The presence of three SMS-REP copies was confirmed by the analysis of the respective interphase nuclei (Fig. 6A). However, presumably due to reduced genomic sequence homology and thus lower hybridization efficiency on the metaphase and interphase chromosomes of New World squirrel monkey, the fluorescent signal after FISH with SMS-REP BAC was of significantly weaker intensity. Therefore it was difficult to determine unequivocally whether all three SMS-REP copies are present in this species.
Figure 6.
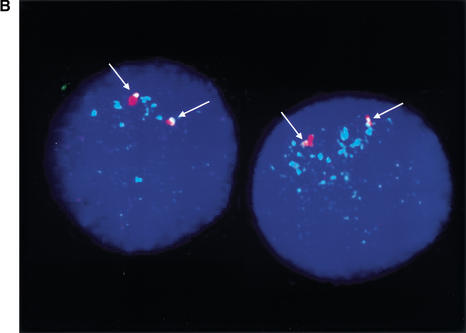
FISH analyses for evolutionary studies. (A) Metaphase and interphase cells of chimpanzee, gorilla, orangutan, gibbon, baboon, and rhesus after hybridization with SMS-REP bacterial artificial chromosome (BAC) RP11–158M20. (B) The interphase nuclei of New World squirrel monkey after cohybridization of differentially labeled SMS-REP-specific BAC (RP11–158M20) and a P1 artificial chromosome clone that directly flanks SMS-REP (RP1–178F10). Note the single yellow foci, indicating the <100-kb physical vicinity of the clones. Similar results were obtained using five other clones directly flanking individual SMS-REPs, showing the presence of three copies of SMS-REPs.
To control for potential misinterpretation of absence of FISH signal secondary to decreased hybridization related to primary sequence divergence, we applied dual-color interphase FISH co-hybridizing SMS-REP BACs with differentially labeled human BAC clones that directly flank SMS-REP. In each of six possible combinations (two flanking unique BAC probes for each three SMS-REPs), nuclei from 50 cells were analyzed. We observed a yellow hybridization signal due to the overlapping of SMS-REP signal (green) and flanking clone signal (red; representative co-FISH in Fig. 6B). These data indicate the presence of all three repeats in squirrel monkey. Therefore, the age of the three SMS-REPs is assessed to be at least ∼40 million years ago (Mya)—the time of divergence of New World monkeys from pre-monkeys (Kumar and Hedges 1998). To further delineate the origin of the SMS-REPs, we investigated evolutionarily older species, including pre-monkey lemur. Only weak unique SMS-region-specific sequence signals were visible. Based on the structure of the individual SMS-REPs and evolutionary investigations using primate cell lines, we propose that an SMS-REP progenitor copy was triplicated after the divergence of New World monkeys and pre-monkeys between ∼40–65 Mya.
DISCUSSION
Chromosome 17 Low-Copy Repeats
Chromosome 17 contains several LCRs. It harbors two Charcot-Marie-Tooth disease type 1A CMT1A-REPs (17p12; Pentao et al. 1992; Reiter et al. 1997), three SMS-REPs (17p11.2; Chen et al. 1997), and three neurofibromatosis type 1 NF1-REPs (17q11.2; Dorschner et al. 2000). These LCRs can serve as substrates for both intra- or interchromosomal NAHR, thus making the respective genomic region unstable and prone to chromosome rearrangements responsible for genomic diseases (for reviews, see Lupski 1998; Ji et al. 2000; Shaffer and Lupski 2000; Emanuel and Shaikh 2001; Stankiewicz and Lupski 2002). Ongoing work is identifying and characterizing additional LCRs, although their contribution to genomic disease is currently unknown.
The proximal SMS-REP spans ∼256 kb and is in the same orientation as the distal SMS-REP, which is shorter (∼176 kb) and devoid of some small repeat fragments. The middle SMS-REP (∼241 kb) is inverted with respect to proximal and distal SMS-REPs. This architecture potentially explains why the common SMS deletions occur between proximal and distal SMS-REPs. The deletions and duplications resulting from NAHR require direct repeats as recombination substrates, whereas inverted repeat recombination substrates may lead to inversions (Lupski 1998).
We have identified recently five SMS patients with smaller-sized deletions who have rearrangements in which their proximal breakpoints map within or adjacent to the middle SMS-REP and the distal breakpoints within or adjacent to the distal SMS-REP (Bi et al. 2002; our unpubl. observations). A candidate genomic structure, which could be potentially responsible for this smaller deletion, is the inverted repeated sequences of 639 bp, including a part of the TL-132 gene (np 4173–4326 of AJ012755). This segment in the distal SMS-REP has the same orientation as that repeated in the middle copy (np 81890–82528 and 69851–70489 in RP11–219A15/AC022596 vs. np 135928–136566 and 147947–148585 in RP11–158M20/AC023401), thus yielding a direct repeat. We also suggest that one of the parents of these 5 SMS patients may be a carrier of a paracentric inversion of the respective segments within 17p11.2, with one chromosome breakpoint involving middle SMS-REP. Similar chromosome polymorphisms involving large genomic regions mediated by olfactory receptor gene cluster LCR on chromosome 8p have been identified recently in 26% and 27% of populations of European and Japanese descent, respectively (Giglio et al. 2001; Matsumoto et al. 2001), in the parents of patients with Williams-Beuren (WBS) syndrome (33%), and in some atypical WBS patients (27%; Osborne et al. 2001).
Evolution of Low-Copy Repeats
Previous hybridization studies suggested the absence of SMS-REP sequences in rodents (Chen et al. 1997; Probst et al. 1999). Here we provide evidence based on FISH analysis of primate cell lines that SMS-REP genomic duplication occurred after the divergence of New World monkeys and pre-monkey lemurs. Evolutionary studies of several different LCRs have shown that they arose recently, apparently during primate speciation (Kiyosawa and Chance 1996; Reiter et al. 1997; Boerkoel et al. 1999; Christian et al. 1999; Keller et al. 1999; Valero et al. 2000; Jenne et al. 2001; Inoue et al. 2001; Shaikh et al. 2001). A high sequence similarity but <100% identity between LCRs appears to be responsible for genomic disorders (Lupski 1998) and suggests that one copy of the LCR is the progenitor and recent genomic duplication gave rise to its additional copies (Eichler 2001).
The two copies of CMT1A-REP have been shown to be absent from rodent species (Pentao et al. 1992). Both CMT1A-REP copies were found in chimpanzee and human, whereas only the distal copy has been identified in gorilla, indicating that it must have been duplicated after the gorilla had diverged from a common human/chimpanzee ancestor (Kiyosawa and Chance 1996; Reiter et al. 1997; Boerkoel et al. 1999; Keller et al. 1999; Inoue et al. 2001). Sequence analysis of CMT1A-REPs revealed that the distal copy is the original and is part of the COX10 gene. The proximal CMT1A-REP represents a duplicated copy of COX10 exon 6 and surrounding intronic sequences (Kiyosawa and Chance 1996; Murakami et al. 1997; Reiter et al. 1997; Boerkoel et al. 1999). Recently, it was shown that the CMT1A-REP insertional/duplicational event divided an ancestral gene, AGIP, into two genes: HREP and CDRT1. These findings illustrate the generation of new genes by DNA rearrangement during mammalian genome evolution (Kennerson et al. 1997; Inoue et al. 2001). Interestingly, sequences flanking proximal CMT1A-REP have been found to be duplicated on gorilla chromosome 19 at the breakpoints of the evolutionary reciprocal translocation t(4;19) in gorilla, equivalent to human t(5;17) (Stankiewicz et al. 2001).
The repeats flanking the Williams syndrome chromosome region were identified in chimpanzee, gorilla, orangutan, and gibbon but were absent in mice, indicating that the duplication event occurred after divergence of rodents and humans (∼80 Mya) but before the diversification of hominoids (∼15–20 Mya; Valero et al. 2000). LCR-sequence comparisons show that block B-mid (common telomeric breakpoint) is probably the ancestral copy. Interestingly, repeat-like sequences were found on human chromosome 7p22 and 7q22 very close to the sites where the breakpoints of evolutionary inversions occurred. It was proposed, therefore, that these segmental duplications had been generated by the evolutionary inversions (DeSilva et al. 1999); however, it is possible these duplicated segments stimulated inversions. The LCR15s, implicated in the pathogenesis of Prader-Willi and Angelman syndromes, were shown to be present in nonhuman primates, including New World monkeys, and only one copy in lemur, suggesting that the LCR duplication may have occurred ∼50 Mya (Christian et al. 1999; Locke et al. 2001). The repeat block LCR15-BP3A was proposed to be an ancestral copy. Analysis of NF1-REPs and LCR22s suggests that their duplication may predate the divergence of the great apes ∼8–9 Mya (Jenne et al. 2001) and New World monkeys ∼40 Mya (Shaikh et al. 2001), respectively.
A Model for the Evolution of the SMS-REPs
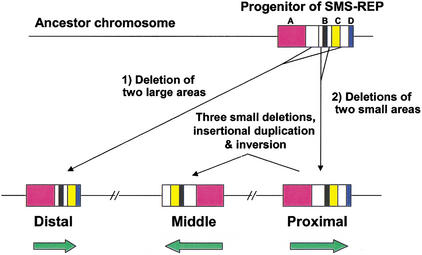
Based on the structural information of three SMS-REPs, we propose a model to explain the evolution of the SMS-REPs. Evidence supporting this model includes the fact that two genes span the border of high- and low-homology areas between the proximal and the distal SMS-REPs, the NOS2A gene in the proximal, and the KER pseudogene (M22927) in the distal SMS-REP (crosshatched area in Fig. 4). A long sequence of the NOS2A gene is located across the border of the A region and its flanking area (A–B in Fig. 4) in the proximal SMS-REP, and a part of the gene in the A–B region is absent in the distal SMS-REP. Also the KER pseudogene (M22927) spans the C region and its flanking area (B–C in Fig. 4) in the distal SMS-REP, and a part of the gene in the B–C region is absent in the proximal SMS-REP. Therefore, only parts of the NOS2A and the KER pseudogene remain in the distal and the proximal SMS-REP, respectively.
To explain these phenomena, we adopted the following three-step-event hypothesis (Fig. 7). Our data indicate that in the first step, ∼40–65 Mya, a progenitor SMS-REP must have arisen in an ancient chromosome. Its structure was almost the same as the proximal SMS-REP of today but included flanking sequences on each side of the B region, as is the case of the distal SMS-REP. The distal SMS-REP resulted from the deletions of two large areas between the A and B regions and the C and D regions. Secondly, deletion of both flanking sequences of the B region in the progenitor resulted in the proximal SMS-REP. Finally, two terminal deletions (2 kb and ∼14 kb) and one interstitial deletion (∼2 kb) associated with interchromosomal insertional duplication (∼2 kb), along with an inversion, generated the middle SMS-REP (Fig. 4).
Figure 7.
Hypothetical model for evolution of SMS-REPs. Our data indicate that in the first step, a progenitor SMS-REP must have arisen in an ancient chromosome. Its structure was almost the same as the proximal SMS-REP at the present time, but included the B region flanking sequences on both sides similar to the distal SMS-REP. The distal SMS-REP resulted from the deletions of two large areas between the A and B, and C and D regions. Secondly, deletion of both flanking sequences of the B region in the progenitor resulted in the proximal SMS-REP. Finally, two terminal deletions and one interstial deletion involving the UPF3A gene accompanied by interchromosomal insertional duplication together with an inversion generated the middle SMS-REP.
Based on the structure of the identified LCRs flanking the proximal SMS-REP and proximal CMT1A-REP and adjacent to the middle SMS-REP, we suggest that before the origin of SMS-REPs, an at least 410 kb chromosome segment flanking the proximal CMT1A-REP (LRC17pA) must have undergone partial segmental duplication in an ancestral chromosome syntenic to the proximal HSA 17p, thus further supporting that segmental duplications have accompanied karyotype evolution in primates.
Conclusion
Our molecular analysis of the SMS-REP LCRs reveals a complex structure consisting of at least 14 genes/pseudogenes. There is remarkable preservation of sequence homology, >160 kb of ∼98% identity, providing a sizable substrate for homologous recombination. Portions of the SMS-REP are present on 17q, constituting a chromosome 17 low-copy repeat (LCR17) similar to what has been identified on chromosome 22 (Dunham et al. 1999), whereas some copies of its gene constituents appear to map to other chromosomes. Like other LCRs that mediate DNA rearrangements responsible for genomic disorders, SMS-REP appears to have evolved during primate speciation. Further analyses of proximal chromosome 17p suggest even more complex genome architecture. Why and how large homology segments are preserved in the human genome is not immediately obvious. Nevertheless, in-depth analyses of such LCRs is essential for further determining the higher order architecture of the human genome and to predict regions of susceptibility for rearrangements leading to chromosome imbalance.
METHODS
PCR
PCR was performed with extracted BAC DNA using each of the following primer sets: CLP (TCTGTAAACTGTCTGAGTGCAGAG/CGTCTGCACC-ACACAATCAAAAGG), 3′TRE (ACAGGTAGCACAATCTACTAA/TTCTGTGTTT-ACTTGTATGAGG), SRP (GGAAGCACTTGCTGTCATCC/GCCCAGGCCAAATGG-CCCTGG), KER (CTCTGCTCTGACCCTCTA/AGCCCTGATCCTTGGGGTCCAG), SHMT (TGGACGCACATTTGTCCTAC/CAGGGACCTGCAGAACTGAC), R193 (GGCAGCTCAGGGTGAGCTCTTC/TATTGGCCTTAAATGCATCTCA), RP11–92B11 reverse end (GGCTGAATGTTTTCCCACAT/AAGGAGATGAAAGGCAGCAA), RP11–315G18 reverse end (CTCCACCGAAAAGCCTACAG/TGCCCTGGAGTTACA-AGATG), TOP3 (TGGTTGC TGTTAGCAGAGGA/CCTTGCATTACACCGTCCTT), RP11–484D23 reverse end (AAGTCTCTGGAGCTCTCATTCA/CCCAGGCACACTA-AACCATT), RP5–836L9 5′ end (ACCTCAGAGGCTACCTCACG/CCAAAGACAG CTATCCACTGC), RP11–121A13 forward end (GGTTGTCTGGGCTTGGTAGA/TG AGTGCCAGCTAAGTGCTC), and D17S959 (TCAGATTGAACTCTCGGTAT/GCTG AC ACAGGCAATG).
PCR reactions were performed on a final volume of 25 μL, using 0.7 U of Ampli-Taq polymerase in 200 μM each of dNTPs; 1× GeneAmp PCR buffer (10 mM Tris-HCl at pH8.3 and 25°C; 50 mM KCL; 1.5 mM MgCl2; 0.001% [w/v] gelatin) and 10 pmoles of each primer. PCR conditions were 95°C for 5 min; 35 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min, followed by 72°C for 7 min. The products were visualized on 2% agarose gels.
BAC Library Screening
We screened segments 1 and 2 (11.8×) of RPCI-11 BAC library (BACPAC Resource Center, Children’s Hospital Oakland Research Institute; www.chori.org/bacpac) using CLP, SRP, and SHMT gene probes. CLP probes were cDNA probe (1.1 kb HindIII fragment of the cDNA clone 41G7A), overgo (Cai et al. 1998) oligonucleotide CLP1 (CGGCGATGGCCACCAAGA/GAAGCCTCTTTGTCGATCTTGGTG), and overgo CLP2 (CCAAAGCGACAAGATCGT/CGGGCCTCTCTCCCTAACGATCTT); SRP probe was the PCR products amplified by SRP primer sets; SHMT probe was overgo (GAAGACTACACAGGGCCTTCCTCA/AAGCGACAGGCTTAAGTGAGGAAG). Hybridizations were performed according to the previously described methods (www.chori.org/bacpac; Cai et al. 1998)
BAC end sequences of the assigned clones were obtained from TIGR (http://www.tigr.org). We also identified several candidate clones from BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST) using the BAC end sequences. We checked the extent of the assigned clones by PCR for STSs and BAC ends.
BAC Sequencing and Sequence Analysis
BAC and PAC clone sequencing was performed after generation of m13 and/or plasmid subclone libraries. Individual clones were either sequenced to fourfold coverage (draft quality) and then further sequenced to greater than eightfold coverage or sequenced immediately to greater than eightfold coverage. After greater than eightfold coverage and assembly, directed finishing techniques were applied to determine the sequences of the remaining gaps. All clones were sequenced and assembled independent of each other and then analyzed to confirm their placement in the tiling paths and determine their degree of overlap with neighboring clones. Specific sequencing and finishing procedures were performed as summarized (International Human Genome Sequencing Consortium 2001).
Regional assemblies for each SMS-REP were constructed following BLAST searches against the high-throughput and the nonredundant sequence database (http://www.ncbi.nlm.nih.gov/blast/) and assembled using the Sequencher software (Gene Codes). Genes and transcripts were identified by a database search of the genomic sequences using the BLAST nonredundant database (Altschul et al. 1997).
Fingerprint Analysis
BAC clones identified following library screens with CLP and SMS-REP markers were fingerprinted and analyzed as described by Marra et al. (1997). Purified BAC DNAs were digested with HindIII, electrophoresed on 1% agarose gels, and stained with SYBR Green I (Roche Diagnostics); then gel images were recorded using a FluorImager SI (Molecular Dynamics). Relative mobility files of observed bands were created using Image (http://www.sanger.ac.uk/Software/Image); then fingerprint data were stored and analyzed using FPC (http://www.sanger.ac.uk/Software/fpc). Clones were grouped into fingerprint contigs as described by Marra et al. (1997), with every internal clone requiring having all of its fragments shared with overlapping clones.
Southern Blotting
Southern hybridization was performed as described by Chen et al. (1997). The target DNAs (human genomic DNA, SMS-REP specific YAC and BAC clones) were cut with various restriction enzymes (HindIII, XbaI, PstI, PvuII, RsaI, TaqI, BglII, XhoI, BamHI, EcoRI KpnI, MspI, and DraI) and hybridized with CLP cDNA probe and the PCR products for TRE and SRP.
Fluorescence In Situ Hybridization
FISH was performed on metaphase and interphase cells of transformed peripheral blood lymphoblasts and skin fibroblasts according to a modified procedure of Shaffer et al. (1997). Briefly, 1 μg of isolated DNA of BAC/PAC clones: RP11–98L14 (proximally adjacent to the proximal SMS-REP), RP11–434D2 (proximal SMS-REP), RP5–836L9 (distally adjacent to the proximal SMS-REP), RP11–28B23 (proximally to the middle SMS-REP), RP11–158M20 (middle SMS-REP), RP1–178F10 (distal to the middle SMS-REP), RP11–416I2 (proximal to the distal SMS-REP), and RP11–219A15 (distal SMS-REP), and RP11–209J20 (distally adjacent to the distal SMS-REP) was labeled by nick-translation reaction using biotin- (Life Technologies-GibcoBRL) or digoxigenin- (Boehringer Mannheim) labeled nucleotides. Biotin was detected with FITC-avidin DCS (fluoresces green; Vector Labs) and digoxigenin was detected with rhodamine-anti-digoxigenin antibodies (fluoresces red; Sigma). Chromosomes were counterstained with DAPI diluted in Vectashield antifade (Vector Labs). Cells were viewed under a Zeiss Axioskop fluorescence microscope equipped with appropriate filter combinations. Monochromatic images were captured and pseudocolored using MacProbe 4.2.2/Power Macintosh G4 system (Perceptive Scientific Instruments, Inc.).
Cell Lines
The human and primate lymphoblastoid cell lines and fibroblasts were grown and harvested using standard methods. The nonhuman-primate-immortalized Epstein-Barr virus stimulated cell lines of lowland gorilla (Gorilla gorilla, CRL 1854), orangutan (Pongo pygmaeus), gibbon (Hylobates lar, TIB-201), New World monkey baboon (Papio hamadryas), Rhesus monkey (Macaca mulatta), and New World squirrel monkey (Saimiri sciureus) were purchased from the American Type Culture Collection (ATCC; http://www.atcc.org/). African green monkey (Cercopithecus aethiops) fibroblasts were also obtained from ATCC. The lemur (Varecia variegates rubber) fibroblasts were obtained from the Center for Reproduction of Endangered Species, Zoological Society of San Diego, (http://www.sandiegozoo.org/conservation/cres_home.html) and the pygmy chimp (Pan paniscus) lymphoblast sample was kindly provided by Dr. D. Nelson from Baylor College of Medicine.
WEB SITE REFERENCES
http://www.atcc.org/; The American Type Culture Collection (ATCC) web site.
http://www.ncbi.nlm.nih.gov/; NCBI web site.
http://www.sanger.ac.uk/Software/Image; Web site for Image software.
http://www.sanger.ac.uk/Software/fpc; The Wellcome Trust Sanger Institute.
http://www.tigr.org; TIGR.
http://www.sandiegozoo.org/conservation/cres_home.html; Center for Reproduction of Endangered Species, Zoological Society of San Diego, San Diego, CA.
www.chori.org/bacpac; Children's Hospital Oakland – BAC-PAC Resources.
Acknowledgments
We appreciate the critical reviews of Drs. B. Bejjani, N. Katsanis, L. Potocki, L. Reiter, and L.G. Shaffer and the excellent technical assistance of M. Withers. We thank Eric Lander for his interest and support. S.-S.P. is supported by Seoul National University Hospital. This study was supported in part by grants from the Muscular Dystrophy Association, the National Institute of Neurological Disorders and Stroke (R01 NS27042), the National Institute of Child Health and Human Development (PO1 HD39420), and the National Human Genome Research Institute (U54 HG02045).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jlupski@bcm.tmc.edu; FAX (713) 798-5073.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.82802.
REFERENCES
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Yan J, Stankiewicz P, Park S-S, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MIM, Inoue K, Lupski JR. Genes in the Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of mouse. Genome Res. 2002;12:713–728. doi: 10.1101/gr.73702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerkoel CF, Inoue K, Reiter LT, Warner LE, Lupski JR. Molecular mechanisms for CMT1A duplication and HNPP deletion. Ann NY Acad Sci. 1999;883:22–35. [PubMed] [Google Scholar]
- Cai WW, Reneker J, Chow C-W, Vaishnav M, Bradley A. An anchored framework BAC map of mouse chromosome 11 assembled using multiplex oligonucleotide hybridization. Genomics. 1998;54:387–397. doi: 10.1006/geno.1998.5620. [DOI] [PubMed] [Google Scholar]
- Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- DeSilva U, Massa H, Trask BJ, Green ED. Comparative mapping of the region of human chromosome 7 deleted in Williams syndrome. Genome Res. 1999;9:428–436. [PMC free article] [PubMed] [Google Scholar]
- Dörr S, Midro AT, Färber C, Giannakudis J, Hansmann I. Construction of a detailed physical and transcript map of the candidate region for Russell-Silver syndrome on chromosome 17q23-q24. Genomics. 2001;71:174–181. doi: 10.1006/geno.2000.6413. [DOI] [PubMed] [Google Scholar]
- Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K. NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum Mol Genet. 2000;9:35–46. doi: 10.1093/hmg/9.1.35. [DOI] [PubMed] [Google Scholar]
- Dunham I, Shimizu N, Roe BA, Chissoe S, Hunt AR, Collins JE, Bruskiewich R, Beare DM, Clamp M, Smink LJ, et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- Eichler EE. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 2001;17:661–669. doi: 10.1016/s0168-9525(01)02492-1. [DOI] [PubMed] [Google Scholar]
- Emanuel BS, Shaikh TH. Segmental duplications: An “expanding” role in genomic rearrangements. Nat Rev Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- Fioretos T, Strömbeck B, Sandberg T, Johansson B, Billström R, Borg Å, Nilsson P-G, Van Den Berghe H, Hagemeijer A, Mitelman F, et al. Isochromosome 17q in blast crisis of chronic myeloid leukemia and in other hematologic malignancies is the result of clustered breakpoints in 17p11 and is not associated with coding TP53 mutations. Blood. 1999;4:225–232. [PubMed] [Google Scholar]
- Frühwald MC, O'Dorisio MS, Dai Z, Rush LJ, Krahe R, Smiraglia DJ, Pietsch T, Elsea SH, Plass C. Aberrant hypermethylation of the major breakpoint cluster region in 17p11.2 in medulloblastomas but not supratentorial PNETs. Genes Chromosomes Cancer. 2001;30:38–47. [PubMed] [Google Scholar]
- Giglio S, Broman KW, Matsumoto N, Calvari V, Gimelli G, Neumann T, Ohashi H, Voullaire L, Larizza D, Giorda R, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am J Hum Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Dewar K, Katsanis N, Reiter LT, Lander ES, Devon KL, Wyman DW, Lupski JR, Birren B. The 1.4-Mb CMT1A duplication/HNPP deletion genomic region reveals unique genome architectural features and provides insights into the recent evolution of new genes. Genome Res. 2001;11:1018–1033. doi: 10.1101/gr.180401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Jenne DE, Tinschert S, Reimann H, Lasinger W, Thiel G, Hameister H, Kehrer-Sawatzki H. Molecular characterization and gene content of breakpoint boundaries in patients with neurofibromatosis type 1 with 17q11.2 microdeletions. Am J Hum Genet. 2001;69:516–527. doi: 10.1086/323043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- Juyal RC, Kuwano A, Kondo I, Zara F, Baldini A, Patel PI. Mosaicism for del(17)(p11.2p11.2) underlying the Smith-Magenis syndrome. Am J Med Genet. 1996;66:193–196. doi: 10.1002/(SICI)1096-8628(19961211)66:2<193::AID-AJMG13>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Keller MP, Seifried BA, Chance PF. Molecular evolution of the CMT1A-REP region: A human- and chimpanzee-specific repeat. Mol Biol Evol. 1999;16:1019–1026. doi: 10.1093/oxfordjournals.molbev.a026191. [DOI] [PubMed] [Google Scholar]
- Kennerson ML, Nassif NT, Dawkins JL, DeKroon RM, Yang JG, Nicholson GA. The Charcot-Marie-Tooth binary repeat contains a gene transcribed from the opposite strand of a partially duplicated region of the COX10 gene. Genomics. 1997;46:61–69. doi: 10.1006/geno.1997.5012. [DOI] [PubMed] [Google Scholar]
- Kiyosawa H, Chance PF. Primate origin of the CMT1A-REP repeat and analysis of a putative transposon-associated recombinational hotspot. Hum Mol Genet. 1996;5:745–753. doi: 10.1093/hmg/5.6.745. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Locke DP, Yavor AM, Lehoczky J, Chang J, Dewar K, Zhao S, Nichols RD, Schwartz S, Eichler EE. Structure and evolution of genomic duplication in 15q11-q13. Am J Hum Genet. 2001;69:179. , Suppl. 1, Abstract 17. [Google Scholar]
- Lupski JR. Genomic disorders: Structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. doi: 10.1016/s0168-9525(98)01555-8. [DOI] [PubMed] [Google Scholar]
- Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Harada N, Giglio S, Kuroiwa K, Ledbetter DH, Niikawa N. Inverted low copy repeats and a common 8p23 inversion polymorphism. Am J Hum Genet. 2001;69:789. , Suppl. 1, Abstract 318. [Google Scholar]
- Mazzarella R, Schlessinger D. Pathological consequences of sequence duplications in the human genome. Genome Res. 1998;8:1007–1021. doi: 10.1101/gr.8.10.1007. [DOI] [PubMed] [Google Scholar]
- Murakami T, Reiter LT, Lupski JR. Genomic structure and expression of the human heme A:farnesyltransferase (COX10) gene. Genomics. 1997;42:161–164. doi: 10.1006/geno.1997.4711. [DOI] [PubMed] [Google Scholar]
- Osborne LR, Li M, Pober B, Chitayat D, Bodurtha J, Mandel A, Costa T, Grebe T, Cox S, Tsui L-C, et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaño-Joos M, Mechtersheimer G, Ohl S, Wilgenbus KK, Scheurlen W, Lehnert T, Willeke F, Otto HF, Lichter P, Joos S. Detection of chromosomal imbalances in leiomyosarcoma by comparative genomic hybridization and interphase cytogenetics. Cytogenet Cell Genet. 2000;90:86–92. doi: 10.1159/000015640. [DOI] [PubMed] [Google Scholar]
- Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR. Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet. 1992;2:292–300. doi: 10.1038/ng1292-292. [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen K-S, Park S-S, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, et al. Molecular mechanism for duplication 17p11.2—The homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Probst FJ, Chen K-S, Zhao Q, Wang A, Friedman TB, Lupski JR, Camper SA. A physical map of the mouse shaker-2 region contains many of the genes commonly deleted in Smith-Magenis syndrome (del17p11.2p11.2) Genomics. 1999;55:348–352. doi: 10.1006/geno.1998.5669. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR. The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs. Hum Mol Genet. 1997;6:1595–1603. doi: 10.1093/hmg/6.9.1595. [DOI] [PubMed] [Google Scholar]
- Scheurlen WG, Schwabe GC, Seranski P, Joos S, Harbott J, Metzke S, Döhner H, Poustka A, Wilgenbus K, Haas OA. Mapping of the breakpoints on the short arm of chromosome 17 in neoplasms with an i(17q) Genes Chromosomes Cancer. 1999;25:230–240. doi: 10.1002/(sici)1098-2264(199907)25:3<230::aid-gcc5>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- Shaffer LG, Kennedy GM, Spikes AS, Lupski JR. Diagnosis of CMT1A duplications and HNPP deletions by interphase FISH: Implications for testing in the cytogenetics laboratory. Am J Med Genet. 1997;69:325–331. [PubMed] [Google Scholar]
- Shaikh TH, Kurahashi H, Emanuel BS. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: An update and literature review. Genet Med. 2001;3:6–13. doi: 10.1097/00125817-200101000-00003. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Park S-S, Inoue K, Lupski JR. The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res. 2001;11:1205–1210. doi: 10.1101/gr.181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarkkanen M, Karhu R, Kallioniemi A, Elomaa I, Kivioja AH, Nevalainen J, Böhling T, Karaharju E, Hyytinen E, Knuutila S, et al. Gains and losses of DNA sequences in osteosarcomas by comparative genomic hybridization. Cancer Res. 1995;55:1334–1338. [PubMed] [Google Scholar]
- Valero MC, de Luis O, Cruces J, Perez Jurado LA. Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: The low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s) Genomics. 2000;69:1–13. doi: 10.1006/geno.2000.6312. [DOI] [PubMed] [Google Scholar]
- Zori RT, Lupski JR, Heju Z, Greenberg F, Killian JM, Gray BA, Driscoll DJ, Patel PI, Zackowski JL. Clinical, cytogenetic, and molecular evidence for an infant with Smith-Magenis syndrome born from a mother having a mosaic 17p11.2p12 deletion. Am J Med Genet. 1993;47:504–511. doi: 10.1002/ajmg.1320470414. [DOI] [PubMed] [Google Scholar]