Abstract
RNA silencing is a potent means of antiviral defense in plants and animals. A hallmark of this defense response is the production of 21- to 24-nucleotide viral small RNAs via mechanisms that remain to be fully understood. Many viruses encode suppressors of RNA silencing, and some viral RNAs function directly as silencing suppressors as counterdefense. The occurrence of viroid-specific small RNAs in infected plants suggests that viroids can trigger RNA silencing in a host, raising the question of how these noncoding and unencapsidated RNAs survive cellular RNA-silencing systems. We address this question by characterizing the production of small RNAs of Potato spindle tuber viroid (srPSTVds) and investigating how PSTVd responds to RNA silencing. Our molecular and biochemical studies provide evidence that srPSTVds were derived mostly from the secondary structure of viroid RNAs. Replication of PSTVd was resistant to RNA silencing, although the srPSTVds were biologically active in guiding RNA-induced silencing complex (RISC)-mediated cleavage, as shown with a sensor system. Further analyses showed that without possessing or triggering silencing suppressor activities, the PSTVd secondary structure played a critical role in resistance to RISC-mediated cleavage. These findings support the hypothesis that some infectious RNAs may have evolved specific secondary structures as an effective means to evade RNA silencing in addition to encoding silencing suppressor activities. Our results should have important implications in further studies on RNA-based mechanisms of host-pathogen interactions and the biological constraints that shape the evolution of infectious RNA structures.
A major focus of current biology is to understand how a pathogen has evolved mechanisms to achieve a balance among several interrelated activities that are crucial to establish a full infection: evading or suppressing host defense, minimizing destructive interference of host metabolism, and maximizing utilization of host factors to support replication and systemic spread. A full understanding of these mechanisms is not only necessary to build a foundation for developing technologies to combat pathogen diseases but also can provide fundamental mechanistic insights into the regulation of basic cellular processes.
Recent studies have discovered small RNA-mediated gene silencing as a powerful antiviral mechanism in plants and animals (6, 22, 25, 47, 49, 50, 72, 77, 84, 87-89, 98). Furthermore, small RNA-mediated gene silencing plays essential roles in regulating a wide variety of growth and development processes (4-6, 11, 13, 17, 23, 28, 42). A key mediator of RNA silencing is several classes of 21- to 24-nucleotide (nt) small RNAs. MicroRNAs (miRNAs) are produced by cleavage of hairpin RNA precursors encoded by the genome of an organism. Short interfering RNAs (siRNAs) are generated by cleavage of double-stranded RNAs (dsRNAs) that may originate from several sources, including cellular genomes, viral replication intermediates, aberrant cellular RNAs, overexpressed transgenes, and transposons. RNase III dicer or dicer-like (DCL) proteins and their associated factors cleave precursor RNAs to produce duplex miRNAs or siRNAs. One of the strands is incorporated into the RNA-induced silencing complex (RISC) to guide sequence-specific cleavage of a target RNA or inhibition of translation or into an RNA-induced initiator of transcriptional silencing complex to guide DNA methylation (1, 6, 14, 19, 26, 39, 42, 58, 85).
It is generally thought that the dsRNAs that form during the replication of plant viral genomic RNAs serve as the substrates of DCL cleavage, and the resulting small RNAs then guide RISC cleavage of viral RNAs. However, critical analyses suggest that the mechanisms are much more complex, and alternative possibilities must be considered (25). The predominant plus-strand polarity and genomic map locations suggest that small RNAs are derived mostly from structured regions of genomic RNAs of several tested positive-strand RNA viruses (59) and the structured region of defective interfering (DI) RNAs (82). It remains to be demonstrated that the structured viral RNAs are direct substrates for DCL activities. Furthermore, it remains to be established that the proposed viral RNA structures indeed exist in vivo, a basic requirement for them to be bona fide substrates for the biogenesis of small RNAs during infection.
As a counterdefense to host gene silencing, many plant and animal viruses studied to date encode silencing suppressors that interfere with distinct steps of RNA silencing pathways (9, 12, 22, 48-50, 69, 72, 79). There is evidence that some satellite RNAs (94) and DI RNAs (82) are less susceptible to RNA silencing. The specific mechanism remains unclear, as it could be attributed to subcellular localization or special RNA structural features (6). The finding that the human adenovirus virus-associated RNAs can function as silencing suppressors by interfering with dicer and RISC activities (3, 53) raises the question of whether some plant viral RNA structures may possess novel silencing suppressor activities. Thus, the specific roles of infectious RNA structures in counterdefense against host silencing remain to be further investigated.
Viroid infection presents a simple system in which to address issues of RNA-based pathogen-host interactions that impact pathogen survival and replication. Without encoding proteins and encapsidation, the small (250- to 400-nt), circular, and noncoding viroid RNAs replicate to high levels in a host cell, move from cell to cell and from organ to organ to establish systemic infection, and can cause devastating diseases (21, 29-31, 83). Therefore, these infectious RNAs are exposed to almost every conceivable means of cellular surveillance, detection, and destruction. Indeed, RNA gel blots revealed small viroid-specific RNAs of both positive and negative polarities that are characteristic of RNA silencing in infected plants, suggesting that viroid RNAs can trigger RNA silencing (40, 45, 56, 57, 61, 74).
How the viroid small RNAs are produced remains poorly understood. Landry and Perreault (44) showed that a hairpin structure of Peach latent mosaic viroid (PLMVd) could be a substrate for dicer-like cleavage in wheat germ extract. Because PLMVd replicates in the chloroplasts that are not known yet to possess RNA silencing machinery, how small RNAs are derived from this viroid in vivo remains an intriguing question to be addressed. The presence of viroid small RNAs suggested activation of host silencing against viroid replication. In general, however, the accumulation of viroid small RNAs does not lead to elimination or even reduced accumulation of PSTVd genomic RNAs. In fact, higher accumulation levels of small RNAs can be associated with higher accumulation of viroid genomic RNAs (40). The secondary structure of a viroid could play a role in resistance to silencing (94), but the role of subcellular localization or novel silencing suppressor activities cannot be excluded.
Thus, the biogenesis and function of viroid and viral small RNAs, as well as how viroid and viral RNAs respond to host RNA silencing, remain to be fully understood. To provide further insights into these issues, we use infection of the 359-nt Potato spindle tuber viroid (PSTVd) as a model system. The secondary structure of PSTVd, the type species of the family Pospiviroidae, consists of five broad domains: (i) central region, (ii) pathogenicity domain, (iii) variable domain, (iv) left-terminal domain, and (v) right-terminal domain (Fig. 1A [43]). Importantly, there is evidence to suggest that the secondary structure of PSTVd exists in vivo (95, 97, 103), making it a compelling model to address the question of whether infectious RNA structures could be substrates for small RNA biogenesis in vivo.
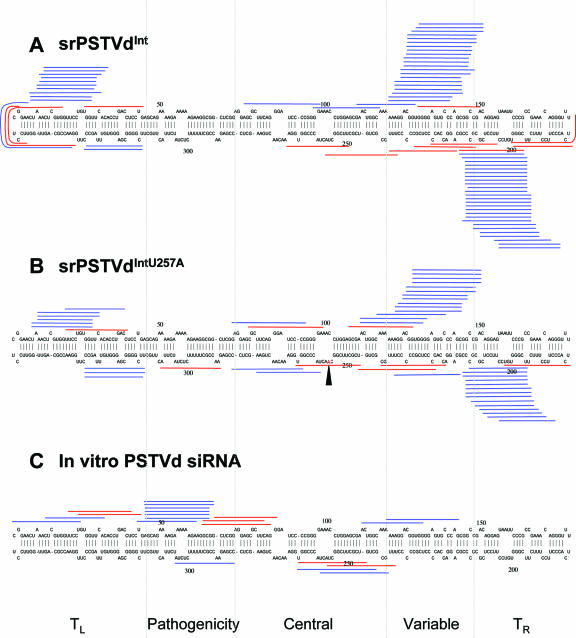
FIG. 1.
Genomic map locations of srPSTVds and PSTVd siRNAs. The single-nucleotide difference between PSTVdInt (A) and PSTVdIntU257A (B) is indicated by the arrowhead. The patterns of PSTVd siRNAs produced in vitro are shown in panel C. Blue and red bars represent plus and minus strands, respectively. Dashed lines delineate the boundaries among the five structural domains (see the text for details). TL, left-terminal domain; TR, right-terminal domain.
PSTVd replicates in the nucleus by utilizing the host DNA-dependent RNA polymerase II (75). During asymmetric rolling-circle replication (10), the unit-length plus circular strands serve as initial templates for the synthesis of concatemeric linear minus strands. Such minus strands then serve as the replication intermediates to direct synthesis of concatemeric, linear plus strands, which are finally cleaved into monomers and circularized. While the minus strands are anchored in the nucleoplasm, the plus strands traffic into the nucleolus, presumably for processing (65).
This study addresses several basic questions concerning the biogenesis and function of viroid small RNAs as well as viroid responses to RNA silencing. (i) Are the viroid small RNAs derived from structured viroid RNAs in vivo? (ii) Are they biologically active in RNA silencing? (iii) How do viroid RNAs evade host RNA silencing-based defense? Here we present molecular and biochemical evidence that small RNAs of PSTVd (srPSTVds) are predominantly produced from the secondary structure of PSTVd RNAs. Our analyses with a reporter system demonstrate that srPSTVds are biologically active guide RNAs in RISC-mediated silencing pathways. Finally, we show that PSTVd replication is resistant to RNA silencing. This resistance can be largely attributed to the resistance of its secondary structure to RISC-mediated cleavage rather than an RNA-based silencing suppressor activity or subcellular localization. We discuss the implications of our findings in studying RNA-based plant-pathogen interactions.
MATERIALS AND METHODS
Plasmids.
Plasmid pRZ6-2, in which the cDNA of PSTVdInt (33) is flanked by ribozymes, was constructed by Hu et al. (38). cDNA of PSTVdInt in pRZ6-2 was replaced with cDNA of other PSTVd strains, PSTVdMild (76), PSTVdIntU257A (105), and PSTVdRG1 (34), giving rise to pRZ:Mild, pRZ:IntU257A, and pRZ:RG1, respectively. The above plasmids (except pRZ:IntU257A) were kindly provided by Robert Owens (USDA Agriculture Research Service, Beltsville, MD) and were used to produce inoculum by in vitro transcription. Plasmid pInter(−) was described by Qi and Ding (67) and used as a transcription template to produce riboprobe specific for plus-PSTVd.
Plasmids pRTL2:smGFP and pRTL2:dsGFP (Johansen and Carrington [41]; gifts from James Carrington, Oregon State University, Corvallis) were used for green fluorescent protein (GFP) expression and silencing experiments in protoplasts. pSP:smGFP (68) was used as template to generate riboprobes for soluble-modified GFP (smGFP). For suppressor experiments in protoplasts, pRTL2-based constructs (71) containing tobacco etch virus (TEV) HC-Pro (a gift from James Carrington), tomato bushy stunt virus (TBSV) P19, or turnip crinkle virus (TCV) CP (Qu et al. [70]; gifts from Feng Qu and Jack Morris, University of Nebraska-Lincoln, Lincoln) were used. Binary plasmid vectors, containing TEV HC-Pro (Wang et al. [93]; a gift from David Bisaro, The Ohio State University, Columbus), TBSV P19 (pBIN61-p19; Voinnet et al. [91]; a gift from David Baulcombe, John Innes Centre, Norwich, United Kingdom), and TCV CP (pRP:TCV CP, Qu et al. [70]; a gift from Feng Qu and Jack Morris) were used for Agrobacterium infiltration experiments.
pH2GW7.0:GFP:Sensor and pH2GW7.0:GFP were used to test silencing function of srPSTVds. The GFP gene was amplified by PCR with a forward primer, cacc-GFP-F (caccATGGTGAGCAAGGGCGAG), and a reverse primer, GFP-Sensor-R (ACCCTTCCTTTCTTCGGGTGTCCTTCCTCGCGCCCttacttgtacagctcg; uppercase letters, nucleotide positions 182 to 216 of PSTVd sequence; lowercase letters, the 3′ end of GFP sequence), giving rise to the GFP:Sensor fusion gene. A control GFP gene was amplified by PCR with a primer set, cacc-GFP-F and GFP-R (TTACTTGTACAGCTCG). The PCR products were cloned into pENTR/D-TOPO (Invitrogen) following the manufacturer's instructions. The GFP:Sensor and GFP genes were transferred to pH2GW7.0 Gateway binary vector (Invitrogen) using the Gateway LR clonase (Invitrogen) by following the manufacturer's instructions.
Potato virus X (PVX) viral vector, pP2C2S (Baulcombe et al. [7]; a gift from David Baulcombe, John Innes Centre, Norwich, United Kingdom), was used to express various GFP:PSTVd fusions in Nicotiana benthamiana protoplasts. The GFP gene was amplified from pEGFP-1 (Clontech) with a forward primer, ClaI-GFP-F (CAGTTCATCGATATGGTGAGCAAGGGCG; the ClaI site is underlined), and the following reverse primers: for GFP:PSTVd full-length and GFP:PSTVd right-half constructs, Right/GFP-R (CCAGGTTTCCCCGGGGATC/TTACTTGTACAGCTC), and for the GFP:PSTVd lower-half construct, Lower/GFP-R (AGAAAGGAAGGGTGA/TTACTTGTACAGCTC). The PSTVd cDNA was amplified from infected tomato plants by reverse transcription-PCR (RT-PCR) with the following primer combinations: for the GFP:PSTVd construct, GFP/Right-F (GAGCTGTACAAGTAA/GATCCCCGGGGAAACCTGG) and PSTVd-Left-R-SalI (CAAGGTCGACCCTGAAGCGCTCCTCC; the SalI site is underlined); for the GFP:PSTVd right-half constructs, GFP/Right-F and PSTVd-Right-R-SalI (CAAGGTCGACGTTTCCACCGGGTAGTAG); for the GFP:PSTVd lower-half construct, GFP/Lower-F (GAGCTGTACAAGTAA/TCACCCTTCCTTTCT) and PSTVd-Low-R-SalI (CAAGGTCGACAGGAACCAACTGCGG). The corresponding GFP genes and PSTVd cDNAs were fused by recombinant PCR (37). The fusion genes were digested by ClaI and SalI and subsequently cloned at ClaI and SalI sites of pP2C2S. After insertion of the GFP:PSTVd fusions into pP2C2S, the genes encoding the coat protein and 25K were sequentially deleted from the vectors, first by digestion with SalI and XhoI (for coat protein, see reference 7) followed by ligation and then by digestion with BstZ171 and ApaI (for 25K) followed by ligation. The resulting plasmids were named pP2C2S:GFP:PSTVd:Δ25KΔCP, pP2C2S:GFP:PSTVd-Right:Δ25KΔCP, and pP2C2S:GFP:PSTVd-Lower:Δ25KΔCP, respectively.
Plant materials and growth conditions.
Tomato (Solanum lycopersicum var. Rutgers) and transgenic N. benthamiana line GFP16c (Ruiz et al. [73]; a gift from David Baulcombe) were grown in a growth chamber controlled at 14-h light (27°C)/10-h dark (24°C) cycles. Arabidopsis thaliana (Col) was grown in a growth chamber controlled at 16-h light (22°C)/8-h dark (18°C) cycles. Cultured cells of N. benthamiana were maintained as described previously (67, 81).
In vitro transcription of viroid RNA and inoculation of plants.
Plasmids containing cDNAs of PSTVd (see above) were linearized by HindIII digestion and used as templates for in vitro transcription with T7 RNA polymerase in Maxiscript (Ambion, Austin, TX) following the manufacturer's instructions. The transcripts were purified with a MEGAClear kit (Ambion) and used to inoculate cotyledons of 9- to 10-day-old tomato plants (10 ng/plant) or the first true leaves of 2-week-old N. benthamiana plants (100 ng/plant). Diethyl pyrocarbonate-H2O was used in mock inoculation.
Total RNA isolation.
Total RNA from PSTVd-infected or mock-inoculated plants (1 g of fresh weight) was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) and total RNA from the protoplasts was isolated using an RNeasy Plant Mini kit (QIAGEN, Valencia, CA) following the manufacturers' instructions.
Enrichment for small RNAs.
Small RNA species were enriched by differential precipitation using polyethylene glycol (PEG; molecular weight, 8,000; Sigma, St. Louis, MO). A 1/10 volume of 50% PEG and 5 M NaCl was added to the solution containing total RNA. The mixture was incubated on ice for 10 min and then centrifuged at 13,000 × g for 5 min at 4°C. The supernatant was transferred to a new tube, and small RNAs were precipitated with 3 volumes of ethanol. The pellet was washed with 70% ethanol and resuspended in nuclease-free H2O. Typically, 1/10 of the original amount was recovered in the enriched fraction.
Cloning and sequencing of small RNAs.
The small RNAs were cloned using the procedures of Elbashir et al. (27) and Lau et al. (46) with some modifications. Ten to 100 μg of enriched small RNAs was separated by 15% polyacrylamide gel electrophoresis (PAGE) with 8 M urea and 0.5× Tris-borate-EDTA (TBE). RNAs of 20 to 30 nt in length were isolated from the gel and ligated to a 3′ adapter oligonucleotide (CUGUAGGCACCAUCAAUidT; 5′-phosphate; idT, 3′-inverted deoxythymidine) purchased from Dharmacon Inc. (Dallas, TX) using T4 RNA ligase (New England BioLabs Inc., Beverly, MA) following the manufacturers' instructions. The ligation product (35 to 50 nt) was gel isolated from 15% PAGE-8 M urea-0.5× TBE. The isolated RNA was then ligated to the 5′ adapter oligonucleotide (atcgtAGGCACCUGAAA; lowercase letters, DNA; uppercase letters, RNA). The ligation product (50 to 70 nt) was gel isolated from 10% PAGE-8 M urea-0.5× TBE. The purified RNA was reverse transcribed with an RT primer (ATTGATGGTGCCTACAG; the BanI site is underlined) and ThermoScript reverse transcriptase (Invitrogen) following the manufacturer's instructions. The reverse transcription products were amplified by PCR with a forward primer (ATCGTAGGCACCTGAAA; the BanI site is underlined) and the RT primer. The PCR products were digested with BanI and concatemerized using T4 DNA ligase (Invitrogen). Concatemers of a size range of 200 to 600 bp were gel isolated from 0.8% agarose gel. The purified DNA was incubated with Taq polymerase (Promega, Madison, WI) under standard conditions at 72°C for 15 min to fill the ends and add an A-overhang and was cloned into the pGEM-T vector (Promega) following the manufacturer's instructions. The clones were sequenced at the Plant-Microbe Genomics Facility, Ohio State University, or at the Samuel R. Noble Foundation Genomics Facility.
Small RNA sequences were extracted from the obtained sequences using a hidden Markov model-based computer program. The program was designed to identify partial adapter sequences (“CTGAAA” for the 5′ adapter and “CTGTAG” for the 3′ adapter) in both sense and antisense orientations, allowing up to one “N” in each adapter sequence. The small RNA sequences between the 5′ and 3′ adapter sequences were extracted. The resulting sequences were filtered for sequence lengths between 15 and 30 nucleotides, converted to sense orientation (i.e., from 5′ to 3′), and BLAST searched (2) to identify small RNAs derived from PSTVd.
In vitro production of PSTVd siRNAs.
dsPSTVd RNA (see below) was incubated with recombinant dicer enzyme (Gene Therapy Systems, San Diego, CA) at 37°C overnight to generate PSTVd siRNAs. The siRNAs were ethanol precipitated and subjected to cloning and sequence analysis (see above).
Probe preparation.
Riboprobes were prepared by in vitro transcription using Maxiscript with T7 polymerase (Ambion) in the presence of [α-32P]UTP (Perkin Elmer, Boston, MA). SpeI-digested pInter(−) (67) and HindIII-digested pSP:smGFP (68) were used as templates to generate riboprobes for PSTVd and GFP mRNA, respectively. After the reaction, unincorporated nucleotides were removed by passing the reaction mixture through a Sephadex column (NucAway spin; Ambion).
Northern hybridization.
For detection of PSTVd genome or GFP mRNA, 1 to 5 μg of total RNA was separated on 5% PAGE with 8 M urea and 0.5× TBE. For detection of srPSTVd and GFP siRNA, 5 to 10 μg of enriched small RNAs (see above) was separated on 15% PAGE-8 M urea-0.5× TBE. After electrophoresis, the RNAs were transferred to Hybond-XL nylon membrane (Amersham Biosciences, Piscataway, NJ) and UV cross-linked. The membranes were hybridized in ULTRAhyb Ultrasensitive Hybridization Buffer (Ambion) overnight, washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) for 15 min, and washed twice in 0.2× SSC-0.1% SDS for 15 min. Hybridization and washing were performed at 65°C and at 37°C for detection of large RNA species and small RNA species, respectively. For all experiments, the washed membranes were exposed to Storage Phosphor Screens (Kodak, Rochester, NY). The screen was scanned by Molecular Imager FX using Quantity One-4.1.1 software (Bio-Rad, Hercules, CA).
Silencing and suppressor experiments in N. benthamiana by Agrobacterium infiltration.
Transgenic N. benthamiana line 16c expressing GFP was used for an Agrobacterium tumefaciens infiltration assay. Ten-day-old plants were inoculated with PSTVd or mock inoculated with H2O. Silencing of the GFP transgene was performed as described previously (90). The first true leaves of 17- to 21-day-old plants (7 to 10 days after PSTVd infection) were infiltrated with an Agrobacterium-carrying dsGFP construct. One to 2 weeks after the infiltration, GFP expression was evaluated using a 100-W, long-wave UV lamp (Blak-Ray Model B 100YP; UV Products, Upland, CA). For the suppressor assays, PSTVd-infected leaves (10 days postinoculation) were coinfiltrated with an Agrobacterium-carrying dsGFP construct and respective suppressor constructs (TCV CP, TBSV P19, and TEV HC-Pro; see above) as described by Johansen and Carrington (41). Five days after infiltration, the leaves were examined with the UV lamp. The Agrobacterium-infected area was harvested and subjected to RNA extraction and RNA gel blot analysis.
Preparation of dsPSTVd, dsGFP, synthetic siPSTVd, and siGFP RNAs.
The plus and minus strands of PSTVd RNAs were produced by in vitro transcription (MEGAscript; Ambion) using SpeI-digested pInter(+) and pInter(−) as templates and with T7 polymerase. Sense and antisense smGFP RNAs were produced by in vitro transcription using EcoRI- or HindIII-digested pSP:smGFP as template and with SP6 or T7 RNA polymerase, respectively. Synthetic oligo-RNAs corresponding to nucleotides 194 to 215 of PSTVd with the 5′ end phosphorylated (5′U*UCGGGUGUCCUUCCUCGCGCCTT3′ [plus strand] and 5′G*GCGCGAGGAAGGACACCCGAATT3′ [minus strand]; asterisks denote phosphorylated nucleotides) were custom synthesized at IDT Inc. (Coralville, Iowa). Equal amounts of sense and antisense RNAs were mixed in a buffer (100 mM KC2H3O2, 4 mM MgCl2, 60 mM HEPES-KOH, pH 7.4), boiled for 5 min, and incubated overnight at 37°C to produce dsRNA or siRNA, respectively. The annealed products were purified by ethanol precipitation and dissolved in nuclease-free water. PSTVd 8-28(+), PSTVd 189-210(+), and PSTVd 270-250(−) siRNAs and GFP siRNA (target sequence, 5′-GCA AGC TGA CCC TGA AGT TC 3′) were synthesized by using a Silencer siRNA Construction kit (Ambion) following the manufacturer's instructions.
Protoplast transfection.
Isolation of N. benthamiana cultured cell protoplasts and electroporation were described in detail previously (102). For each electroporation, 1 × 106 protoplasts (in 400 to 500 μl of culture) were transfected with 10 μg of in vitro-transcribed PSTVd RNAs (see above). Cells were collected 3 days after transfection to extract RNA for Northern blot analysis. For RNA silencing assays, dsPSTVd or synthetic PSTVd siRNAs (see above) were cotransfected (see the figures for amounts used). For suppressor assays, 10 μg of the respective suppressor plasmid (pRTL2:suppressor) was cotransfected. For GFP control experiments, 5 μg of pRTL2:smGFP was cotransfected with differing amounts of silencing inducer (siGFP or dsGFP) in the presence or absence of 10 μg of the respective suppressor plasmids. GFP expression was examined under a fluorescence microscope and measured by fluorometer (see below) and RNA gel blot analysis (see above).
Arabidopsis protoplast preparation and PEG-based transfection were performed essentially as described by Sheen (78). Two million cells were transfected with 40 μg of PSTVd RNA. The transfected cells were incubated in 2 ml of Gamborg's B5 media (G5893; Sigma) containing Gamborg vitamins (G1019; Sigma) and 0.4 M of glucose at room temperature (∼20°C) and in darkness for 3 days. The PSTVd replication and srPSTVd accumulation were examined by RNA gel blot analysis as described above.
Purification of Arabidopsis dicer-like protein(s).
The procedure for purification of Arabidopsis dicer-like protein was based on Qi et al. (64) with some modifications. Two grams of Arabidopsis flower buds was homogenized in 2 ml/g of extraction buffer (64). Cell debris was removed by two rounds of centrifugation at 10,000 × g at 4°C for 10 and 20 min, and the supernatant was filtered through a 0.2-μm cellulose membrane. The extract was passed through 20 ml of Bio-Beads SM (catalog no. 152-3920; Bio-Rad) by gravity at 4°C. The extract was then passed through a 1-ml Hi-Trap CM column (Amersham Biosciences). The flowthrough was collected and loaded onto a 1-ml Hi-Trap DEAE column (Amersham Biosciences). Fractionation was performed by fast protein liquid chromatography (Amersham) with 50 to 400 mM NaCl gradient in a total of 5 ml of the extraction buffer (500 μl/fraction). The fractions were assayed for DCL activities as described below.
In vitro processing of PSTVd RNAs.
32P-labeled linear plus PSTVd and single-stranded GFP (ssGFP) in vitro transcripts were prepared as described above. The PSTVd transcripts were run on 5% PAGE with 8 M urea and 0.5× TBE, and the unit-length PSTVd band was isolated from the gel. 32P-labeled mir319 precursor RNA was prepared by in vitro transcription with T7 polymerase (see above) using a cDNA template amplified by RT-PCR from Arabidopsis (Col) total RNA with the primer set T7:PremiR319-F (TTGTAATACGACTCACTATAGGGAATATATATGTAGAGA; the T7 promoter is underlined) and PremiR319-R (GGAATACAAAAGAGAGAGGGA). The transcripts were purified with a MEGAClear kit (Ambion). Substrate RNAs (60,000 cpm each) were incubated in a 10-μl reaction mixture containing 2.5 μl of the extract, 2 μl of 5× reaction buffer (0.5 M NaCl, 5 mM ATP, 1 mM GTP, 6 mM MgCl2, and 2 U of RNAsin RNase Inhibitor), and 5.5 μl of the extraction buffer at 30°C for 2 h. The reaction was stopped by the addition of 10 μl of loading buffer (95% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue, 18 mM EDTA, 0.025% SDS) and 1 μl of phenol and was incubated at 65°C for 15 min. The processing products were separated on a 15% PAGE-8 M urea-0.5× TBE gel. The gel was exposed to a phosphorimager and analyzed as described above.
GFP:Sensor experiment in tomato plants.
Tomato plants were inoculated with PSTVd as described above. Twenty days postinoculation, systemic leaves of PSTVd-infected or noninfected plants were infiltrated with Agrobacterium (strain 1D1249; a gift from Richard Michelmore and Tadeusz Wroblewski, University of California, Davis) carrying pH2GW7.0:GFP:sensor or pH2GW7.0:GFP following the procedure described by Wroblewski et al. (99). Three to 4 days after infiltration, the infiltrated areas of leaves were examined for GFP expression by fluorescence microscopy (see below). The GFP mRNA levels were examined by RT-PCR using Superscript II (Invitrogen) following the manufacturer's instructions. One set of reverse transcription was performed by using GFP-R (see above), followed by PCR with GFP-F (ATGGTGAGCAAGGGCGAG) and GFP-R to amplify cDNA of GFP-specific RNA. The other set of reverse transcription was performed with oligo(dT) primer, followed by PCR with GFP-F to amplify cDNAs of full-length GFP or GFP:Sensor mRNAs, as well as Actin-F (TATTGTGTTGGACTCTGGTG) and Actin-R (ACGGTCAGCAATACCAGGGA) to amplify actin cDNA as an internal control.
Fluorescence microscopy and fluorescence measurement.
GFP expression in plants or transfected protoplasts was examined with an E600 fluorescence microscope (Nikon, Tokyo, Japan). GFP fluorescence was visualized with a filter set consisting of an excitation filter of 450 to 490 nm, a dichroic mirror of 510 nm, and a barrier filter of 520 to 560 nm. Images were captured with a SPOT 2 Slider charge-coupled device camera and the associated software (Diagnostics Instruments, Sterling Heights, MI).
To assess the total GFP fluorescence in protoplasts, 0.2 × 106 cells for each experiment were measured by a Cytofluor 2350 Fluorescence Measurement System with the associated software (Millipore, Billerica, MA), with excitation at 485 nm and emission at 530 nm. The fluorescence values of GFP from control experiments (without dsGFP and without suppressor expression) were arbitrarily set to a value of 1. Fluorescence measurements from all other treatments were shown as relative values.
Silencing assay of PVX:GFP:PSTVd constructs.
PVX constructs, pP2C2S:GFP:PSTVd:Δ25KΔCP, pP2C2S:GFP:PSTVd-Right:Δ25KΔCP, and pP2C2S:GFP:PSTVd-Lower:Δ25KΔCP (see above) were linearized by SpeI and used as templates for capped RNA synthesis using an mMESSAGE mMACHINE with T7 polymerase (Ambion) following the manufacturer's instructions. The transcripts were purified with the MEGAClear kit (Ambion). Three micrograms of transcript was used for N. benthamiana protoplast transfection (see above) in the presence or absence of 3 μg of silencing inducer (dsPSTVd or dsGFP RNAs). Twenty-four hours after transfection, GFP expression was examined by a fluorometer and RNA blot analysis (see above).
RESULTS
srPSTVds were derived predominantly from structured RNAs of PSTVd.
To investigate the characteristics and function of srPSTVds that are produced during infection, we used PSTVd-infected tomato (cv. Rutgers) as our experimental system, because tomato is the classical experimental host of PSTVd for studies on the correlation between viroid structure and function. We analyzed srPSTVds derived from PSTVdInt and PSTVdIntU257A. These two strains, which differ by only one nucleotide (U versus A at genomic position 257 in the central region; Fig. 1A and B), cause intermediate and lethal symptoms, respectively (66). We were interested in determining whether small RNA profiles from strains of different pathogenicities would differ. We isolated and cloned the srPSTVds for sequencing (see Materials and Methods). Sequencing results of approximately 500 clones from five biological replicates are combined and presented.
The sequences of all srPSTVds isolated are presented in Table 1. The plus or minus sign indicates the polarity of an srPSTVd (i.e., the plus sign indicates that an srPSTVd is produced from the plus strand of PSTVd). The numbers indicate the position of an srPSTVd in the PSTVd genome. Several characteristics were notable. First, the srPSTVds were mapped predominantly to the left and right terminal regions as well as the variable domain of the rod-shaped secondary structure (Fig. 1A and B). Very few were mapped to the central region, and even fewer were mapped to the pathogenicity domain for both PSTVd strains. Second, srPSTVds were overwhelmingly plus strands (approximately 90%; Fig. 1A). Only a few minus strands were detected. Interestingly, the plus strands were clustered, whereas the minus strands were not (Fig. 1A and B). Third, the majority of srPSTVds were 21 and 22 nt (Table 1). The above characteristics are not due to cloning artifacts, because the sequence profile of the in vitro-generated PSTVd siRNAs (see Materials and Methods for details) was different from that of srPSTVds (Fig. 1C).
TABLE 1.
srPSTVds cloned from PSTVd-infected tomato plants
| Genome location | No. of clones | PSTVdInt polarity | PSTVdIntU257A polarity | Length (nt) | Sequence |
|---|---|---|---|---|---|
| 5-26 | 2 | + | 22 | ACUAAACUCGUGGUUCCUGUGG | |
| 6-29 | 1 | + | 24 | CUAAACUCGUGGUUCCUGUGGUUC | |
| 7-27 | 1 | + | 21 | UAAACUCGUGGUUCCUGUGGU | |
| 7-28 | 1 | + | 22 | UAAACUCGUGGUUCCUGUGGUU | |
| 8-29 | 2 | + | 22 | AAACUCGUGGUUUCUGUGGUUC | |
| 9-29 | 1 | + | 21 | AACUCGUGGUUCCUGUGGUUC | |
| 10-30 | 1 | + | 21 | ACUCGUGGUUCCUGUGGUUCA | |
| 10-31 | 1 | + | 22 | ACUCGUGGUUCCUGUGGUUCAC | |
| 77-98 | 1 | + | 22 | AGCGCUUCAGGGAUCCCCGGGG | |
| 97-118 | 1 | + | 22 | GGAAACCUGGAGCGAACUGGCA | |
| 110-130 | 1 | + | 21 | GAACUGGCAAAAAAGGACGGU | |
| 121-141 | 1 | + | 21 | AAAGGACGGUGGGGAGUGCCC | |
| 121-142 | 1 | + | 22 | AAAGGACGGUGGGGAGUGCCCA | |
| 122-144 | 1 | + | 23 | AAGGACGGTGGGGAGTGCCCAGC | |
| 123-144 | 3 | + | 22 | AGGACGGUGGGGAGUGCCCAGC | |
| 125-144 | 1 | + | 20 | GACGGUGGGGAGUGCCCAGC | |
| 125-147 | 1 | + | 23 | GACGGUGGGGAGUGCCCAGCGGC | |
| 125-148 | 1 | + | 24 | GACGGTGGGGAGTGCCCAGCGGCC | |
| 126-145 | 1 | + | 20 | ACGGUGGGGAGUGCCCAGCG | |
| 126-146 | 3 | + | 21 | ACGGUGGGGAGUGCCCAGCGG | |
| 126-147 | 2 | + | 22 | ACGGUGGGGAGUGCCCAGCGGC | |
| 127-147 | 3 | + | 21 | CGGUGGGGAGUGCCCAGCGGC | |
| 127-148 | 3 | + | 22 | CGGUGGGGAGUGCCCAGCGGCC | |
| 129-150 | 1 | + | 22 | GUGGGGAGUGCCCAGCGGCCGA | |
| 130-150 | 1 | + | 21 | UGGGGAGUGCCCAGCGGCCGA | |
| 132-151 | 1 | + | 20 | GGGAGUGCCCAGCGGCCGAC | |
| 183-204 | 2 | + | 22 | CCCUUCCUUUCUUCGGGUGUCC | |
| 186-206 | 1 | + | 21 | UUCCUUUCUUCGGGUGUCCUU | |
| 191-212 | 2 | + | 22 | UUCUUCGGGUGUCCUUCCUCGC | |
| 192-213 | 2 | + | 22 | UCUUCGGGUGUCCUUCCUCGCG | |
| 193-214 | 1 | + | 22 | CUUCGGGUGUCCUUCCUCGCGC | |
| 193-215 | 1 | + | 23 | CUUCGGGUGUCCUUCCUCGCGCC | |
| 194-215 | 6 | + | 22 | UUCGGGUGUCCUUCCUCGCGCC | |
| 195-214 | 1 | + | 20 | UCGGGUGUCCUUCCUCGCGC | |
| 195-215 | 9 | + | 21 | UCGGGUGUCCUUCCUCGCGCC | |
| 196-216 | 2 | + | 21 | CGGGUGUCCUUCCUCGCGCCC | |
| 317-337 | 2 | + | 21 | UCGGGGCGAGGGUGUUUAGCC | |
| 343-5 | 1 | + | 22 | AACCGCAGUUGGUUCCUCGGAA | |
| 2-341 | 1 | − | 21 | CGAAGAACCAACUGCGGUUCC | |
| 16-355 | 1 | − | 21 | CACGAGUUUAGUUCCGAGGAA | |
| 44-24 | 1 | − | 21 | CAGGAGGUCAGGUGUGAACCA | |
| 150-130 | 1 | − | 21 | UCGGCCGCUGGGCACUCCCCA | |
| 199-178 | 1 | − | 22 | CCCGAAGAAAGGAAGGGUGAAA | |
| 200-179 | 1 | − | 22 | ACCCGAAGAAAGGAAGGGUGAA | |
| 226-205 | 1 | − | 22 | GUGGUCCUGCGGGCGCGAGGAA | |
| 233-212 | 1 | − | 22 | GCGAGGGGUGGNCCUGCGGGCG | |
| 239-219 | 1 | − | 21 | AAGGGGGCGAGGGGUGGUCCU | |
| 258-238 | 1 | − | 21 | UAGCCGAAGCGACAGCGCAAA | |
| 270-250 | 1 | − | 21 | UCCACCGGGUAGUAGCCGAAG | |
| 7-27 | 2 | + | 21 | UAAACUCGUGGUUCCUGUGGU | |
| 9-29 | 2 | + | 21 | AACUCGUGGUUCCUGUGGUUC | |
| 10-29 | 1 | + | 20 | ACUCGUGGUUCCUGUGGUUC | |
| 18-38 | 1 | + | 21 | UUCCUGUGGUUCACACCUGAC | |
| 73-94 | 1 | + | 22 | GAGGAGCGCUUCAGGGAUCCCC | |
| 112-133 | 1 | + | 22 | ACUGGCAAAAAAGGACGGUGGG | |
| 113-133 | 1 | + | 21 | CUGGCAAAAAAGGACGGUGGG | |
| 117-139 | 1 | + | 23 | CAAAAAAGGACGGUGGGGAGUGCU | |
| 122-142 | 1 | + | 21 | AAGGACGGUGGGGAGUGCCCA | |
| 123-144 | 2 | + | 22 | AGGACGGUGGGGAGUGCCCAGC | |
| 124-144 | 1 | + | 21 | GGACGGUGGGGAGUGCCCAGC | |
| 126-146 | 2 | + | 21 | ACGGUGGGGAGUGCCCAGCGG | |
| 127-148 | 2 | + | 22 | CGGUGGGGAGUGCCCAGCGGCC | |
| 129-150 | 4 | + | 22 | GUGGGGAGUGCCCAGCGGCCGA | |
| 183-204 | 1 | + | 22 | CCCUUCCUUUCUUCGGGUGUCC | |
| 188-208 | 1 | + | 21 | CCUUUCUUCGGGUGUCCUUCC | |
| 189-210 | 1 | + | 22 | CUUUCUUCGGGUGUCCUUCCUC | |
| 190-211 | 1 | + | 22 | UUUCUUCGGGUGUCCUUCCUCG | |
| 192-212 | 1 | + | 21 | UCUUCGGGUGUCCUUCCUCGC | |
| 192-213 | 1 | + | 22 | UCUUCGGGUGUCCUUCCUCGCG | |
| 193-214 | 1 | + | 22 | CUUCGGGUGUCCUUCCUCGCGC | |
| 194-215 | 2 | + | 22 | UUCGGGUGUCCUUCCUCGCGCC | |
| 195-214 | 1 | + | 20 | UCGGGUGUCCUUCCUCGCGC | |
| 195-215 | 1 | + | 21 | UCGGGUGUCCUUCCUCGCGCC | |
| 195-216 | 2 | + | 22 | UCGGGUGUCCUUCCUCGCGCCC | |
| 216-237 | 1 | + | 22 | CGCAGGACCACCCCUCGCCCCC | |
| 260-280 | 1 | + | 21 | UACCCGGUGGAAACAACUGAA | |
| 264-285 | 1 | + | 22 | CGGUGGAAACAACUGAAGCUCC | |
| 316-337 | 2 | + | 22 | UUCGGGGCGAGGGUGUUUAGCC | |
| 317-337 | 1 | + | 21 | UCGGGGCGAGGGUGUUUAGCC | |
| 39-18 | 1 | − | 22 | GGUCAGGUGUGAACCACAGGAA | |
| 99-79 | 1 | − | 21 | UCCCCGGGGAUCCCUGAAGCG | |
| 128-108 | 1 | − | 21 | CGUCCUUUUUUGCCAGUUCGC | |
| 203-180 | 1 | − | 24 | GACACCCGAAGAAAGGAAGGGUGA | |
| 241-221 | 1 | − | 21 | CAAAGGGGGCGAGGGGUGGUC | |
| 247-226 | 1 | − | 22 | ACAGCGCAAAGGGGGCGAGGGG | |
| 267-246 | 1 | − | 22 | GGUCAGGUGUGAACCACAGGAA | |
| 310-290 | 1 | − | 21 | UAAGAUAGAGAAAAAGCGGUU |
The predominant plus strandedness and mapping to the two terminal regions of the secondary structure of PSTVd genomic RNAs suggest that srPSTVds were derived from the secondary structure of plus-strand RNAs via a pathway distinct from the dsRNA-based siRNA biogenesis. To test this possibility, we examined the effects of several well-characterized viral silencing suppressors, which can interfere with siRNA production from dsRNA precursor, on srPSTVd accumulation. The coat protein of turnip crinkle virus has been shown to selectively inhibit the production of siRNAs (16, 18, 24, 70) but have little effect on the production of miRNAs in plants (16, 24). P19 from tomato bushy stunt virus and P1/HC-Pro from tobacco etch virus were shown to affect siRNA accumulation in some systems (P19 [35, 70, 80] and HC-Pro [24, 35, 52, 54]). Furthermore, P1/HC-Pro can enhance miRNA accumulation (16, 24, 55). To test the effects of these suppressors on srPSTVd accumulation, we used Nicotiana benthamiana for the following reasons: (i) PSTVd actively replicates in N. benthamiana plants (38, 104) as well as in protoplasts of cultured cells (67), and (ii) RNA silencing protocols, including assays of viral suppressor functions, have been well established in N. benthamiana plants (72) and cultured cells (68). Therefore, N. benthamiana provides a convenient experimental system to study RNA silencing and PSTVd biology. We conducted two complementary experiments, as described below.
In the first series of experiments, N. benthamiana plants expressing GFP (line GFP16c [73]) were inoculated with PSTVd. Ten days postinoculation, systemic leaves were infiltrated with Agrobacterium carrying dsGFP and suppressor constructs. It should be noted that srPSTVds were barely detectable at this stage. After 5 more days, RNA was extracted from the infiltrated leaf areas for RNA gel blot analyses. As shown in Fig. 2A, CP and P19 had negligible effects on srPSTVd accumulation, whereas HC-Pro appeared to increase srPSTVd accumulation. In contrast, all suppressors decreased the accumulation of GFP siRNA in the same leaves, with CP being the most effective. Furthermore, the presence of suppressors did not affect the accumulation of genomic PSTVd, whereas it increased the accumulation of GFP mRNA (Fig. 2A). Altogether, these results indicated that the viral suppressors were functional in the infiltrated leaves and that they had different effects on the accumulation of srPSTVd and GFP siRNA in plants.
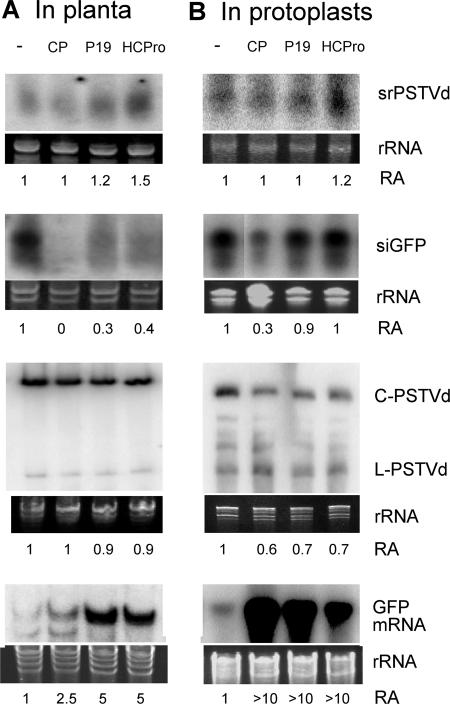
FIG. 2.
Viral suppressors do not affect accumulation of srPSTVds or genomic PSTVd. (A) Transgenic N. benthamiana expressing GFP was inoculated with PSTVd. Infected leaves were infiltrated with Agrobacterium carrying the respective suppressors. Five days later, agroinfiltrated areas were collected for RNA gel blot analysis of srPSTVd, siGFP, genomic PSTVd, and GFP mRNA accumulations. (B) N. benthamiana protoplasts were cotransfected with PSTVd and various suppressors. Accumulations of srPSTVd and PSTVd were examined at 3 days postinoculation. Cotransfections of GFP, dsGFP, and suppressors were performed to confirm suppression activities of the suppressors. CP, TCV coat protein; P19, TBSV P19; HCPro, TEV HC-Pro. C-PSTVd, circular genomic PSTVd; L-PSTVd, linear unit-length PSTVd. RA, ratio of RNA accumulation levels obtained from three biological replicates. rRNA bands in PAGE stained with ethidium bromide served as a loading control. C-PSTVd was used for RA assessment.
To corroborate this analysis, we conducted the second series of experiments in which PSTVd inoculum and the plasmids expressing the viral suppressors were introduced into N. benthamiana protoplasts simultaneously by electroporation. As shown in Fig. 2B, CP, P19 and HC-Pro had little effect on srPSTVd accumulation. These observations were similar to those from the experiments with the plants. In control experiments in which silencing of GFP was induced by dsGFP, CP decreased the accumulation of GFP siRNA as expected, whereas P19 and HC-Pro had little effect (Fig. 2B) (68). The dramatic increase in the accumulation of GFP mRNA indicated that the suppressors were functional in this system. The presence of suppressors did not increase the accumulation of genomic PSTVd, similar to the situation in planta (Fig. 2B). Altogether, these data suggest that srPSTVds were unlikely to have been produced via dsRNA-based siRNA pathways and are consistent with the notion that srPSTVds are mostly derived from structured PSTVd RNAs. The observation that none of the tested viral suppressors increased the levels of PSTVd genomic RNA accumulation in both the infected plants and protoplasts also suggested that resistance of PSTVd replication against RNA silencing is intrinsic to the PSTVd RNA (see below).
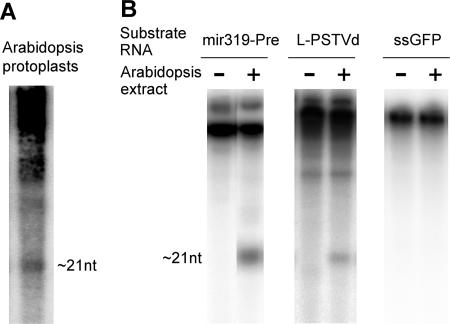
To test further the hypothesis that srPSTVds are derived from structured PSTVd RNAs, we performed biochemical experiments to determine whether structured PSTVd RNAs can indeed be cleaved by DCLs. Partial purification of Arabidopsis DCLs has been reported (64). We observed that PSTVd could replicate in leaf protoplasts of Arabidopsis and that PSTVd-specific small RNAs accumulated in the infected protoplasts (Fig. 3A). Therefore, Arabidopsis possesses enzymatic activities for the biogenesis of srPSTVds. We used chromatography to identify Arabidopsis cell extract fractions that contained DCL activities, as revealed by the processing of mir319 precursor RNA into small RNAs of ∼21 nt (Fig. 3B). The linear, plus-PSTVd RNA, which folds into a rod-like secondary structure similar to that of the circular genomic RNAs (8, 103), was also processed into small RNAs of ∼21 nt in such fractions (Fig. 3B). In contrast, incubation of ssGFP RNAs did not yield any small RNAs or any other types of cleavage products (Fig. 3B). Therefore, production of small RNAs from the substrate RNAs in this system was not attributed to random nuclease activities. These results provide biochemical evidence that the structured PSTVd RNAs can indeed serve as substrates of DCL activities.
FIG. 3.
Processing of structured, linear plus-PSTVd RNAs into srPSTVds by Arabidopsis DCL(s). (A) srPSTVds of ∼21 nt accumulated in PSTVd-transfected Arabidopsis protoplasts. (B) Partially purified Arabidopsis DCLs process miR319 precursor RNAs and PSTVd RNAs into small RNAs of ∼21 nt but not ssGFP RNA. L-PSTVd, linear unit-length PSTVd.
Taken together, the size and genomic map patterns of srPSTVds, the different effects of viral suppressors on the production of srPSTVds and siRNAs, and direct biochemical assays of substrate activities support the hypothesis that the majority of srPSTVds we cloned from the infected plants were derived from the secondary structure of plus-strand PSTVd RNAs.
Replication of PSTVd was resistant to RNA silencing.
The presence of srPSTVds suggested activation of host silencing against PSTVd replication. However, higher accumulation levels of srPSTVds are associated with higher accumulation of viroid genomic RNAs (40). Furthermore, the above experiments showed that viral silencing suppressors did not enhance the accumulation levels of PSTVd genomic RNAs in infected plants and protoplasts. These observations suggest that (i) the level of srPSTVds in a cell is insufficient to trigger silencing against active PSTVd replication, or (ii) replication of PSTVd is resistant to RNA silencing. To address these possibilities, we analyzed PSTVd replication in the presence of PSTVd small RNAs as silencing triggers in the protoplasts of N. benthamiana.
First, we tested whether PSTVd replication would be inhibited in N. benthamiana protoplasts when synthetic srPSTVds were coelectroporated with the PSTVd inoculum to trigger immediate and potent silencing. We initially chose srPSTVd 194-215(+) for this experiment, because it was one of the most abundant srPSTVds. A synthetic duplex of srPSTVd194-215 was synthesized and introduced into N. benthamiana protoplasts. The presence of PSTVd 194-215 siRNAs did not affect PSTVd replication, regardless of the amounts introduced (Fig. 4A). Experiments with three other synthetic PSTVd siRNAs tested [PSTVd 8-28(+), PSTVd 189-210(+), and PSTVd 270-250(−)] produced similar results (Fig. 4B). In contrast, GFP expression was strongly silenced in the presence of GFP siRNAs, indicating effective induction of RNA silencing in this system (Fig. 4C).
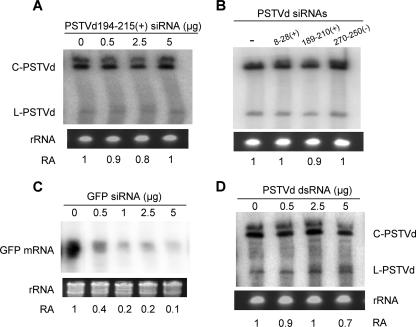
FIG. 4.
Replication of PSTVd is resistant to induced RNA silencing in N. benthamiana protoplasts. (A) PSTVd accumulates at similar levels in the absence or presence of differing amounts of siRNAs. The siRNA sequence corresponds to positions 194 to 215 of the PSTVd genome. (B) PSTVd accumulates at similar levels in the presence of 5 μg of different siRNAs synthesized based on srPSTVds. (C) Accumulation of GFP mRNA decreases with increasing amounts of GFP siRNAs. (D) PSTVd accumulates at similar levels in the absence or presence of differing amounts of dsRNAs. C-PSTVd, circular genomic PSTVd; L-PSTVd, linear unit-length PSTVd. RA, ratio of RNA accumulation levels obtained from three biological replicates. rRNA bands in PAGE stained with ethidium bromide served as a loading control. C-PSTVd was used for RA assessment.
Data from the above experiments showed that srPSTVds originated from different regions of PSTVd failed to trigger silencing to suppress PSTVd replication. To determine whether PSTVd replication was resistant to RNA silencing in general, we investigated the effects of exogenously supplied PSTVd dsRNAs on PSTVd replication in N. benthamiana protoplasts. We have demonstrated that dsRNA is a highly effective silencing inducer in N. benthamiana protoplasts (68; also see Fig. 7). As shown in Fig. 4D, PSTVd replication was not affected in the presence of dsPSTVd. Electroporation of dsPSTVd alone did not result in accumulation of replicating PSTVd (data not shown). Therefore, dsPSTVd did not contribute to PSTVd replication.
FIG. 7.
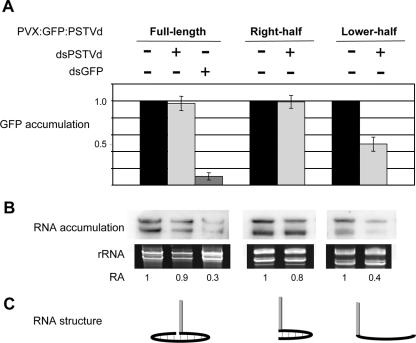
PSTVd secondary structure is resistant to RISC-mediated cleavage. N. benthamiana protoplasts were transfected with PVX vectors to express various GFP:PSTVd fusion constructs (full-length, right-half, and lower-half; see the text for details) in the cytoplasm. Silencing was induced by dsGFP or dsPSTVd as indicated. Shown are the GFP protein levels (A) and RNA levels (B). The two bands in the RNA gel blot represent genomic and subgenomic RNAs of PVX. (C) Schematic view of RNA structures of respective fusion constructs. Gray and black lines represent GFP mRNA and PSTVd RNA, respectively. RA, ratio of RNA accumulation levels obtained from three biological replicates. rRNA bands in PAGE stained with ethidium bromide served as a loading control.
These results indicate that replication of PSTVd is resistant to RNA silencing, which provides one explanation of why the accumulation levels of srPSTVds are not inversely correlated with the levels of PSTVd genomic RNA in infected plants. Without encoding proteins that may suppress silencing and without being encapsidated, how could PSTVd evade cellular RNA silencing? Several possibilities that are not mutually exclusive can be considered. First, srPSTVds are not incorporated into RISCs as functional guide RNAs. Second, PSTVd RNAs have novel silencing suppressor activities in lieu of the protein-based silencing suppressor activities as found in viruses. Third, nuclear localization and/or the PSTVd structure confer resistance to the silencing. These possibilities were tested by the following experiments.
srPSTVds were biologically active in the RISC-mediated cleavage of a target RNA.
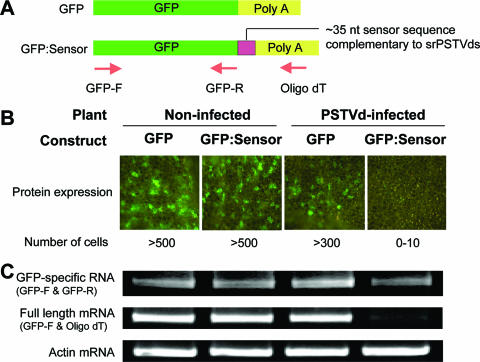
Small RNAs have been detected for many viroids, and their potential roles in RNA silencing are widely discussed. Vogt et al. (86) showed that transgenic reporter genes fused to partial sequences of PSTVd were silenced upon PSTVd infection. However, the mechanistic role(s) of srPSTVds in silencing remains unclear, and there was no evidence yet that srPSTVds were directly involved in silencing. The activity of specific viral small RNAs has not been tested either, leaving uncertainty regarding the specific mechanisms of host silencing against viruses in plants (25). To establish a basis for functional studies of these small RNAs, we tested the silencing activity of srPSTVds in infected tomato by using a GFP reporter system, the strategy used to study plant miRNA-target interactions (62). We created a DNA construct (GFP:Sensor) in which the GFP gene is fused to a sensor sequence with perfect complementarity to the nucleotide positions 182 to 216 of the PSTVd genomic sequence, from which the most abundant srPSTVds were derived (see Fig. 1A and B; see also Table 1). GFP without a sensor sequence was used as a control (Fig. 5A). The GFP:Sensor or GFP constructs were introduced into leaves of noninfected or PSTVd-infected tomato plants by agroinfiltration as described previously (99). Three to 4 days after infiltration, leaf samples were examined under a fluorescence microscope for GFP expression. In noninfected plants, extensive GFP expression was observed in numerous leaf cells (more than 500 cells/leaf) for both constructs. In contrast, in PSTVd-infected plants the expression of GFP:Sensor was nearly abolished, whereas the expression of control GFP was not significantly reduced (Fig. 5B).
FIG. 5.
srPSTVds are functional in RISC-mediated cleavage of a target RNA. (A) Schematic view of mRNAs produced from DNA constructs used in this experiment. The GFP:Sensor construct contains an ∼35-nt sensor sequence that is complementary to the most abundant srPSTVds. The arrows denote the primers used for RT-PCR analyses. (B) The DNA constructs were introduced by agroinfiltration into noninfected or PSTVd-infected tomato leaves. Three to 4 days after infiltration, GFP protein expression was examined with a fluorescence microscope. (C) RNA levels were determined by RT-PCR. Three biological replicates gave similar results.
To assess mRNA accumulation, we initially performed RNA gel blot analysis using a GFP-specific probe. Surprisingly, we found little difference in GFP RNA accumulation among the samples (data not shown), raising the possibility of translational inhibition by srPSTVds. To determine whether or not mRNA from the GFP:Sensor construct was cleaved by srPSTVds, we designed primers for RT-PCR to amplify either GFP-specific RNA (GFP-R as a reverse primer) or full-length mRNA containing a sensor sequence [oligo(dT) as a reverse primer; see Fig. 5A for primer positions]. The levels of GFP-specific RNA were similar to those of samples in RNA blot analysis, whereas the level of the full-length mRNA was greatly reduced for the GFP:Sensor construct in the infected leaves (Fig. 5C). The results indicate that the diminished expression of GFP:Sensor protein was due to specific cleavage of mRNA and not due to inhibition of translation. The results also indicate that the expression of reporter genes was not silenced at the transcriptional level, eliminating the possibility of transcriptional silencing caused by PSTVd RNA-mediated DNA methylation of the transgenes (63, 96).
Taken together, srPSTVds produced in the infected plants are incorporated into RISC and are functional in guiding sequence-specific cleavage of a target RNA. Therefore, srPSTVds produced during viroid infection are biologically active in RNA silencing.
PSTVd did not function as an RNA silencing suppressor at the cellular and whole-plant levels.
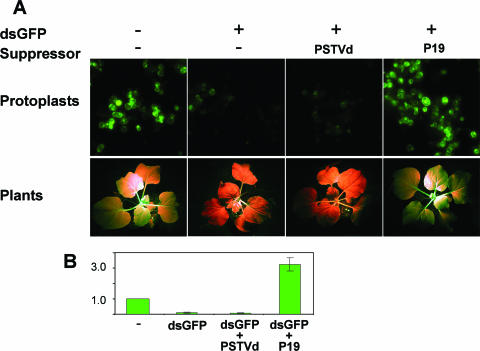
We tested the potential silencing suppressor activities of PSTVd RNAs by two sets of experiments, one in N. benthamiana protoplasts and the other in N. benthamiana plants. To examine potential suppressor activities of PSTVd at the cellular level, we induced RNA silencing against GFP as described above in the protoplasts in the presence of replicating PSTVd. As shown in Fig. 6A, GFP fluorescence was observed in a few cells with dim intensity, indicating that PSTVd did not suppress dsGFP RNA-induced RNA silencing targeted to GFP. This observation was substantiated by a quantitative analysis of total GFP fluorescence (Fig. 6B). Replication of PSTVd in the protoplasts was confirmed by RNA gel blots (data not shown). In control experiments, coexpression of TBSV P19, a well-established silencing suppressor (72), led to efficient suppression of silencing, as demonstrated by the strong GFP fluorescence (Fig. 6).
FIG. 6.
PSTVd does not possess silencing suppressor activities or elicit endogenous silencing suppressor activities. (A) Silencing against GFP was induced in N. benthamiana protoplasts (upper panel) or in transgenic N. benthamiana (lower panel). PSTVd infection did not suppress the induced silencing, whereas cotransfection of TBSV P19 suppressed silencing. (B) Quantitative comparison of GFP expression in the protoplasts with different treatments. The level of GFP expression without any treatment (control) is set to 1.0. The expression levels with different treatments are expressed relative to this control value. Averages for three biological replicates are presented with standard deviations.
In the second set of experiments, we used N. benthamiana plants that transgenically express GFP (line GFP16c [73]) to test PSTVd suppressor activity. When the plants were inoculated with water (mock control) or PSTVd inoculum, GFP fluorescence was unaltered (data not shown). Systemic silencing of GFP expression was observed 1 week after introduction of a dsGFP construct into a leaf via agroinfiltration (Fig. 6A), as reported previously (73). Presence of PSTVd did not suppress dsGFP RNA-induced silencing targeted to GFP expression (Fig. 6A; Table 2). Systemic infection of the plant by PSTVd was confirmed by RNA gel blots (data not shown).
TABLE 2.
Number of silenced or nonsilenced transgenic N. benthamiana plants (GFP16c) in silencing suppression assays
| Treatment | No. silenced (%) | No. nonsilenced (%) |
|---|---|---|
| None | 0 (0) | 12 (100) |
| PSTVd | 0 (0) | 9 (100) |
| dsGFP | 31 (96.9) | 1 (3.1) |
| dsGFP + PSTVd | 32 (91.5) | 3 (8.5) |
| dsGFP + P19 | 1 (12.5) | 7 (87.5) |
Taken together, these results showed that presence of replicating PSTVd did not affect siRNA-mediated silencing targeted to a reporter gene. Therefore, we concluded that PSTVd does not have silencing suppressor activities or elicit endogenous silencing suppressor activities.
PSTVd secondary structure was resistant to RISC-mediated cleavage.
Because srPSTVds are functional guide RNAs in RISC and because PSTVd does not have silencing suppressor activities, resistance of PSTVd replication may be simply attributed to its nuclear localization, especially its localization in the nucleolus (36, 65), shielding the viroid RNA molecules from the RISC machinery that is localized in the cytoplasm. However, RNA silencing at the posttranscriptional level can occur in the nucleus in Caenorhabditis elegans (60), and silencing of nucleolar RNAs has been observed in trypanosomatids (51), although whether these occur in the plant nucleus remains to be determined. Furthermore, the viroid RNA molecules must move from the nucleus into the cytoplasm to establish systemic infection, exposing them to RISC. Therefore, the observed resistance to silencing could be attributed to the secondary structure of PSTVd.
To test these alternative possibilities, we used an engineered Potato virus X (PVX) vector (7) to express GFP:PSTVd fusions in the cytoplasm of N. benthamiana protoplasts. The genes encoding the viral suppressor 25K (92) and coat protein, which may interfere with RNA silencing, were removed from the vector. The GFP gene was fused to three different forms of PSTVd (see Fig. 7C): (i) the full-length PSTVd genomic RNA that will form a rod-shaped secondary structure, (ii) the right-half segment of PSTVd that is predicted to form the secondary structure, and (iii) the lower-half segment of PSTVd that is not predicted to form the secondary structure. If the secondary structure confers resistance to RISC-mediated silencing, the first two forms of PSTVd (full length and right half) are expected to be resistant, whereas the third form (lower half) is not. Because of the cytoplasmic localization of PVX replication, this system allows us to examine simultaneously the contributions of subcellular localization and secondary structure of substrate RNAs to RNA silencing resistance.
The above three constructs were delivered into protoplasts together with dsGFP RNAs or dsPSTVd RNAs as silencing triggers. GFP expression and stability of GFP:PSTVd RNAs were monitored by quantification of GFP fluorescence and RNA gel blot analyses, respectively. As shown in Fig. 7, dsGFP RNAs triggered efficient silencing against GFP:PSTVd, indicating operation of the RNA silencing machinery in this system. In contrast, introduction of dsPSTVd RNAs did not affect protein and RNA accumulation of the GFP:PSTVd full-length and right-half fusions, whereas it reduced that of the lower-half fusion by approximately 50 to 60% (Fig. 7).
These results provide direct experimental evidence that the RNA secondary structure plays a critical role in the protection of PSTVd against RISC-mediated silencing. Nuclear localization appears not to be essential against RISC activity but may provide additional protection.
DISCUSSION
How small RNAs are derived from pathogenic RNAs in plants remains to be fully understood. One of the main issues is the form of substrate RNAs for DCL activities. Although it is generally assumed that small RNAs of RNA viruses are derived from dsRNAs formed during replication, direct experimental evidence remains to be provided. The sequence profiles and strand polarities of small RNAs from several tested RNA viruses suggest that they are produced from structured regions of viral RNAs (59, 82).
A hairpin structure of PLMVd could be a substrate for DCL cleavage in wheat germ extract (44). Whether it also acted as an in vivo substrate for DCLs remains to be determined. We have obtained several lines of in vivo as well as in vitro evidence to suggest that srPSTVds are produced mostly from the secondary structure of PSTVd RNAs. First, sequence profiles showed that srPSTVds are predominantly plus strands and are clustered in the two terminal regions of the PSTVd secondary structure. Second, viral suppressors well demonstrated to interfere with the production of siRNAs from dsRNA substrates did not affect the accumulation of srPSTVds. Lastly, our biochemical assays provide direct evidence that the secondary structure of PSTVd can be processed by DCLs into small RNAs. Importantly, there is evidence suggesting that the secondary structure of PSTVd exists in vivo (95, 97, 103). Thus, there is a solid basis for a structured PSTVd RNA to serve as a DCL substrate in vivo. It should be noted that a previous study showed that the plus-PSTVd RNA was resistant to cleavage by human dicer (15). Whether this is attributed to some differences in the biochemical activities of human and plant dicers or in technical procedures is unknown.
Considering the RNA forms that may exist during the rolling circle replication, the major substrate RNAs could include the circular plus-genomic RNA and the secondary structures in the linear unit-length and concatemeric plus RNAs. The small percentage of minus srPSTVds could arise from dsPSTVd RNAs formed between the plus- and minus-concatemeric RNAs or dsPSTVd RNAs generated via a cellular RNA-directed RNA polymerase. It is also possible that the minus-PSTVd monomers or multimers can form certain structures that are recognized by DCLs, albeit at a relatively low efficiency. The specific DCLs that are responsible for the biogenesis of srPSTVds remain to be elucidated. Based on the sequence profiles of srPSTVds and the known characteristics of Arabidopsis DCLs (32, 64, 100, 101), it is likely that the biogenesis of srPSTVds involves more than one DCL in tomato. Further studies to elucidate the specific DCLs and their associated factors for the biogenesis of srPSTVds will await molecular and functional characterization of all tomato DCLs. Denti et al. (20) reported cytoplasmic localization of srPSTVds based on cell fractionation studies. Whether the observation implies that srPSTVds are produced in the cytoplasm or are produced in the nucleus and then efficiently exported into the cytoplasm remains to be determined.
Encoding silencing suppressor proteins is a major mechanism that viruses have evolved to counteract host silencing in plants and animals (9, 22, 48, 49, 69, 72). Viroids do not encode any proteins and therefore must have evolved alternative mechanisms to evade host RNA silencing. PSTVd sequences are shown to be resistant to silencing in planta (94), suggesting the importance of PSTVd secondary structure in resistance against silencing. However, other potential mechanisms have not been tested. These include subcellular localization and RNA-based silencing suppressor activity. In this study, we conducted a series of experiments to test these alternative possibilities. We first established that replication of PSTVd in the protoplast is resistant to RNA silencing in the presence of diverse PSTVd-specific silencing triggers. Further analyses showed that PSTVd RNA does not have or does not elicit silencing suppressor activities. Finally, we showed that the PSTVd RNA structure confers resistance to RISC activity in the cytoplasm. The reason for partial silencing (40 to 60%) of GFP fused with the lower half of PSTVd is not known. It is possible that the lower half of PSTVd forms an unknown structure conferring a limited level of resistance. Nonetheless, the data provide clear indications that the secondary structure of PSTVd confers significant protection against RISC activity. Mutational analysis may reveal the specific structural features critical for the resistance. Nuclear localization may provide additional protection during various stages of the viroid replication cycle.
The importance of RNA structure in protection against RNA silencing is unlikely limited to viroids. Secondary structural features of a DI RNA (82) and satellite RNA (94) also may confer resistance, although the contributions of other factors such as subcellular localization in these cases still need to be investigated (6). It will be of great interest to further investigate the role of viral RNA secondary structures in protection against silencing.
The discovery of viroid small RNAs led to the postulate that viroid small RNAs may silence host gene expression to cause symptoms (30, 31, 56, 61, 94). Consistent with this hypothesis, the level of srPSTVd accumulation is positively correlated with symptom severity of a PSTVd strain (40). A correlation between small RNA accumulation and symptom severity was also reported for Avocado sunblotch viroid (56). Furthermore, symptom development is correlated with production of siRNAs in transgenic tomato expressing nonreplicating, dsPSTVd RNAs (94). It has been postulated that small RNA-guided silencing of host genes may be a mechanism of pathogenesis not only for viroid infection but also for viral infection (25). It should be noted, however, that the correlation between viroid small RNA accumulation and symptom expression is not universal among different viroid strains or species (56, 61) as well as among different tissues or developmental stages of a plant (56, 74). Direct evidence that any viroid small RNAs play a role in targeting host gene expression remains outstanding. Our present work has not uncovered srPSTVds derived from the pathogenicity domain. To determine whether viroid small RNAs do play a role in silencing host genes to cause symptoms, in-depth sequencing efforts will be needed to identify all viroid small RNAs. This, however, may be insufficient to deduce their roles until the complete genome sequence information for tomato and other viroid host species becomes available to allow prediction and experimental validation of possible host target genes for any viroid small RNAs.
Could viroid small RNAs have any biological roles for infection? Although the secondary structure of PSTVd confers efficient resistance to RISC cleavage, it is a substrate for DCLs. Therefore, all biological constraints combined have not allowed the evolution of an infectious RNA structure that can escape successfully both DCL and RISC activities. This may mean, alternatively, that the small RNAs derived from DCL cleavage could have a constructive role in infection, considering the fact that viroid RNAs do not encode proteins to facilitate various aspects of infection. For instance, an srPSTVd may inhibit the expression of a host defense-related gene or serve as guide RNAs to direct the transcription or processing of PSTVd RNAs during the rolling circle replication. Addressing these and other possibilities regarding the role of viroid small RNAs will be an exciting challenge for further studies.
Without encoding proteins, a viroid RNA has evolved remarkable structural features that recruit all the necessary cellular factors to accomplish infection as well as to outwit the sophisticated RNA silencing system in a host cell. The results reported here help establish a new foundation to probe the molecular mechanisms of viroid-host interactions with regard to viroid survival, replication, and pathogenicity.
Acknowledgments
We are most grateful to Venkat Gopalan and Lien Lai for their generous help and use of their fast protein liquid chromatography system, for contributing the idea of using Arabidopsis flower buds for DCL activity analyses, and for valuable discussions about experimental designs and results. We are indebted to Yuhong Tang for her generous assistance with the sequencing of srPSTVds. We thank David Baulcombe, Jack Morris, and Feng Qu for cDNA constructs and Kristin Kasschau and James Carrington for protocols of small RNA isolation. We thank David Baulcombe for providing N. benthamiana line GFP16c and Richard Michelmore and Tadeusz Wroblewski for providing the Agrobacterium strain 1D1249. We are indebted to James Carrington, Edward Allen, Detlev Weigel, Shou-Wei Ding, and David Bisaro for many helpful discussions and suggestions. We express our gratitude to Erich Grotewold for his generous support.
This work was supported by grants from the National Science Foundation (IBN-0238412 and IOB-050515745), the United States Department of Agriculture National Research Initiative Competitive Grants (2004-35304-15005), and the Ohio Plant Biotechnology Consortium.
Footnotes
Published ahead of print on 3 January 2007.
REFERENCES
- 1.Allen, E., Z. Xie, A. M. Gustafson, and J. C. Carrington. 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207-221. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersson, M. G., P. C. Haasnoot, N. Xu, S. Berenjian, B. Berkhout, and G. Akusjarvi. 2005. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 79:9556-9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel, B., and D. P. Bartel. 2003. MicroRNAs: at the root of plant development? Plant Physiol. 132:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. [DOI] [PubMed] [Google Scholar]
- 6.Baulcombe, D. 2004. RNA silencing in plants. Nature 431:356-363. [DOI] [PubMed] [Google Scholar]
- 7.Baulcombe, D. C., S. Chapman, and S. Santa Cruz. 1995. Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 7:1045-1053. [DOI] [PubMed] [Google Scholar]
- 8.Baumstark, T., A. R. Schroder, and D. Riesner. 1997. Viroid processing: switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 16:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bisaro, D. M. 2006. Silencing suppression by geminivirus proteins. Virology 344:158-168. [DOI] [PubMed] [Google Scholar]
- 10.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNA's. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 11.Carrington, J. C., and V. Ambros. 2003. Role of microRNAs in plant and animal development. Science 301:336-338. [DOI] [PubMed] [Google Scholar]
- 12.Carrington, J. C., K. D. Kasschau, and L. K. Johansen. 2001. Activation and suppression of RNA silencing by plant viruses. Virology 281:1-5. [DOI] [PubMed] [Google Scholar]
- 13.Carthew, R. W. 2006. Gene regulation by microRNAs. Curr. Opin. Genet. Dev. 16:203-208. [DOI] [PubMed] [Google Scholar]
- 14.Chan, S. W., D. Zilberman, Z. Xie, L. K. Johansen, J. C. Carrington, and S. E. Jacobsen. 2004. RNA silencing genes control de novo DNA methylation. Science 303:1336. [DOI] [PubMed] [Google Scholar]
- 15.Chang, J., P. Provost, and J. M. Taylor. 2003. Resistance of human hepatitis delta virus RNAs to dicer activity. J. Virol. 77:11910-11917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman, E. J., A. I. Prokhnevsky, K. Gopinath, V. V. Dolja, and J. C. Carrington. 2004. Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev. 18:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, X. 2005. MicroRNA biogenesis and function in plants. FEBS Lett. 579:5923-5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deleris, A., J. Gallego-Bartolome, J. Bao, K. D. Kasschau, J. C. Carrington, and O. Voinnet. 2006. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313:68-71. [DOI] [PubMed] [Google Scholar]
- 19.Denli, A. M., and G. J. Hannon. 2003. RNAi: an ever-growing puzzle. Trends Biochem. Sci. 28:196-201. [DOI] [PubMed] [Google Scholar]
- 20.Denti, M. A., A. Boutla, M. Tsagris, and M. Tabler. 2004. Short interfering RNAs specific for potato spindle tuber viroid are found in the cytoplasm but not in the nucleus. Plant J. 37:762-769. [DOI] [PubMed] [Google Scholar]
- 21.Ding, B., and A. Itaya. 2007. Viroid: a useful model for studying the basic principles of infection and RNA biology. Mol. Plant-Microbe Interact. 20:7-20. [DOI] [PubMed] [Google Scholar]
- 22.Ding, S. W., H. Li, R. Lu, F. Li, and W. X. Li. 2004. RNA silencing: a conserved antiviral immunity of plants and animals. Virus Res. 102:109-115. [DOI] [PubMed] [Google Scholar]
- 23.Dugas, D. V., and B. Bartel. 2004. MicroRNA regulation of gene expression in plants. Curr. Opin. Plant Biol. 7:512-520. [DOI] [PubMed] [Google Scholar]
- 24.Dunoyer, P., C. H. Lecellier, E. A. Parizotto, C. Himber, and O. Voinnet. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235-1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Dunoyer, P., and O. Voinnet. 2005. The complex interplay between plant viruses and host RNA-silencing pathways. Curr. Opin. Plant Biol. 8:415-423. [DOI] [PubMed] [Google Scholar]
- 26.Ekwall, K. 2004. The RITS complex-A direct link between small RNA and heterochromatin. Mol. Cell 13:304-305. [DOI] [PubMed] [Google Scholar]
- 27.Elbashir, S. M., W. Lendeckel, and T. Tuschl. 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15:188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finnegan, E. J., and M. A. Matzke. 2003. The small RNA world. J. Cell Sci. 116:4689-4693. [DOI] [PubMed] [Google Scholar]
- 29.Flores, R., J. A. Daros, and C. Hernandez. 2000. Avsunviroidae family: viroids containing hammerhead ribozymes. Adv. Virus Res. 55:271-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flores, R., S. Delgado, M. E. Gas, A. Carbonell, D. Molina, S. Gago, and M. De la Peña. 2004. Viroids: the minimal non-coding RNAs with autonomous replication. FEBS Lett. 567:42-48. [DOI] [PubMed] [Google Scholar]
- 31.Flores, R., C. Hernandez, A. E. Martinez de Alba, J. A. Daros, and F. Di Serio. 2005. Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43:117-139. [DOI] [PubMed] [Google Scholar]
- 32.Gasciolli, V., A. C. Mallory, D. P. Bartel, and H. Vaucheret. 2005. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15:1494-1500. [DOI] [PubMed] [Google Scholar]
- 33.Gross, H. J., H. Domdey, C. Lossow, P. Jank, M. Raba, H. Alberty, and H. L. Sanger. 1978. Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273:203-208. [DOI] [PubMed] [Google Scholar]
- 34.Gruner, R., A. Fels, F. Qu, R. Zimmat, G. Steger, and D. Riesner. 1995. Interdependence of pathogenicity and replicability with potato spindle tuber viroid. Virology 209:60-69. [DOI] [PubMed] [Google Scholar]
- 35.Hamilton, A., O. Voinnet, L. Chappell, and D. Baulcombe. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21:4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harders, J., N. Lukacs, M. Robert-Nicoud, T. M. Jovin, and D. Riesner. 1989. Imaging of viroids in nuclei from tomato leaf tissue by in situ hybridization and confocal laser scanning microscopy. EMBO J. 8:3941-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA.
- 38.Hu, Y., P. A. Feldstein, J. Hammond, R. W. Hammond, P. J. Bottino, and R. A. Owens. 1997. Destabilization of potato spindle tuber viroid by mutations in the left terminal loop. J. Gen. Virol. 78:1199-1206. [DOI] [PubMed] [Google Scholar]
- 39.Hutvagner, G., and P. D. Zamore. 2002. RNAi: nature abhors a double-strand. Curr. Opin. Genet. Dev. 12:225-232. [DOI] [PubMed] [Google Scholar]
- 40.Itaya, A., A. Folimonov, Y. Matsuda, R. S. Nelson, and B. Ding. 2001. Potato spindle tuber viroid as inducer of RNA silencing in infected tomato. Mol. Plant-Microbe Interact. 14:1332-1334. [DOI] [PubMed] [Google Scholar]
- 41.Johansen, L. K., and J. C. Carrington. 2001. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones-Rhoades, M. W., D. P. Bartel, and B. Bartel. 2006. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57:19-53. [DOI] [PubMed] [Google Scholar]
- 43.Keese, P., and R. H. Symons. 1985. Domains in viroids: evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 82:4582-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Landry, P., and J. P. Perreault. 2005. Identification of a peach latent mosaic viroid hairpin able to act as a Dicer-like substrate. J. Virol. 79:6540-6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landry, P., D. Thompson, and J. P. Perreault. 2004. The role of viroids in gene silencing: the model case of Peach latent mosaic viroid. Can. J. Plant Pathol. 26:31-38. [Google Scholar]
- 46.Lau, N. C., L. P. Lim, E. G. Weinstein, and D. P. Bartel. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858-862. [DOI] [PubMed] [Google Scholar]
- 47.Lecellier, C. H., and O. Voinnet. 2004. RNA silencing: no mercy for viruses? Immunol. Rev. 198:285-303. [DOI] [PubMed] [Google Scholar]
- 48.Li, F., and S. W. Ding. 2006. Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 60:503-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li, H. W., and S. W. Ding. 2005. Antiviral silencing in animals. FEBS Lett. 579:5965-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, W. X., and S. W. Ding. 2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 51.Liang, X. H., Q. Liu, and S. Michaeli. 2003. Small nucleolar RNA interference induced by antisense or double-stranded RNA in trypanosomatids. Proc. Natl. Acad. Sci. USA 100:7521-7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Llave, C., K. D. Kasschau, and J. C. Carrington. 2000. Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc. Natl. Acad. Sci. USA 97:13401-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu, S., and B. R. Cullen. 2004. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and microRNA biogenesis. J. Virol. 78:12868-12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mallory, A. C., L. Ely, T. H. Smith, R. Marathe, R. Anandalakshmi, M. Fagard, H. Vaucheret, G. Pruss, L. Bowman, and V. B. Vance. 2001. HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13:571-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mallory, A. C., B. J. Reinhart, D. Bartel, V. B. Vance, and L. H. Bowman. 2002. A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc. Natl. Acad. Sci. USA 99:15228-15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markarian, N., H. W. Li, S. W. Ding, and J. S. Semancik. 2004. RNA silencing as related to viroid induced symptom expression. Arch. Virol. 149:397-406. [DOI] [PubMed] [Google Scholar]
- 57.Martinez de Alba, A. E., R. Flores, and C. Hernandez. 2002. Two chloroplastic viroids induce the accumulation of small RNAs associated with posttranscriptional gene silencing. J. Virol. 76:13094-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matzke, M. A., and J. A. Birchler. 2005. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6:24-35. [DOI] [PubMed] [Google Scholar]
- 59.Molnar, A., T. Csorba, L. Lakatos, E. Varallyay, C. Lacomme, and J. Burgyan. 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 79:7812-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montgomery, M. K., S. Xu, and A. Fire. 1998. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95:15502-15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Papaefthimiou, I., A. Hamilton, M. Denti, D. Baulcombe, M. Tsagris, and M. Tabler. 2001. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 29:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parizotto, E. A., P. Dunoyer, N. Rahm, C. Himber, and O. Voinnet. 2004. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 18:2237-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pelissier, T., S. Thalmeir, D. Kempe, H. L. Sanger, and M. Wassenegger. 1999. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 27:1625-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi, Y., A. M. Denli, and G. J. Hannon. 2005. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19:421-428. [DOI] [PubMed] [Google Scholar]
- 65.Qi, Y., and B. Ding. 2003. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell 15:2566-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Qi, Y., and B. Ding. 2003. Inhibition of cell growth and shoot development by a specific nucleotide sequence in a noncoding viroid RNA. Plant Cell 15:1360-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qi, Y., and B. Ding. 2002. Replication of Potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: the role of specific nucleotides in determining replication levels for host adaptation. Virology 302:445-456. [DOI] [PubMed] [Google Scholar]
- 68.Qi, Y., X. Zhong, A. Itaya, and B. Ding. 2004. Dissecting RNA silencing in protoplasts uncovers novel effects of viral suppressors on the silencing pathway at the cellular level. Nucleic Acids Res. 32:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qu, F., and T. J. Morris. 2005. Suppressors of RNA silencing encoded by plant viruses and their role in viral infections. FEBS Lett. 579:5958-5964. [DOI] [PubMed] [Google Scholar]
- 70.Qu, F., T. Ren, and T. J. Morris. 2003. The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J. Virol. 77:511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Restrepo, M. A., D. D. Freed, and J. C. Carrington. 1990. Nuclear transport of plant potyviral proteins. Plant Cell 2:987-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roth, B. M., G. J. Pruss, and V. B. Vance. 2004. Plant viral suppressors of RNA silencing. Virus Res. 102:97-108. [DOI] [PubMed] [Google Scholar]
- 73.Ruiz, M. T., O. Voinnet, and D. C. Baulcombe. 1998. Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sano, T., and Y. Matsuura. 2004. Accumulation of short interfering RNAs characteristics of RNA silencing precedes recovery of tomato plants from severe symptoms of Potato spindle tuber viroid infection. J. Gen. Plant Pathol. 70:50-53. [Google Scholar]
- 75.Schindler, I. M., and H. P. Mühlbach. 1992. Involvement of nuclear DNA-dependent RNA polymerases in potato spindle tuber viroid replication: a reevaluation. Plant Sci. 84:221-229. [Google Scholar]
- 76.Schnölzer, M., B. Haas, K. Ramm, H. Hofmann, and H. L. Sänger. 1985. Correlation between structure and pathogenicity of potato spindle tuber viroid (PSTV). EMBO J. 4:2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schutz, S., and P. Sarnow. 2006. Interaction of viruses with the mammalian RNA interference pathway. Virology 344:151-157. [DOI] [PubMed] [Google Scholar]
- 78.Sheen, J. 2002. A transient expression assay using Arabidopsis mesophyll protoplasts. http://genetics.mgh.harvard.edu/sheenweb/.
- 79.Silhavy, D., and J. Burgyan. 2004. Effects and side-effects of viral RNA silencing suppressors on short RNAs. Trends Plant Sci. 9:76-83. [DOI] [PubMed] [Google Scholar]
- 80.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tavazza, and J. Burgyan. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sunter, G., and D. M. Bisaro. 2003. Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplasts. Virology 305:452-462. [DOI] [PubMed] [Google Scholar]
- 82.Szittya, G., A. Molnar, D. Silhavy, C. Hornyik, and J. Burgyan. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tabler, M., and M. Tsagris. 2004. Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci. 9:339-348. [DOI] [PubMed] [Google Scholar]
- 84.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants-defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 85.Vazquez, F., V. Gasciolli, P. Crete, and H. Vaucheret. 2004. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14:346-351. [DOI] [PubMed] [Google Scholar]
- 86.Vogt, U., T. Pelissier, A. Putz, F. Razvi, R. Fischer, and M. Wassenegger. 2004. Viroid-induced RNA silencing of GFP-viroid fusion transgenes does not induce extensive spreading of methylation or transitive silencing. Plant J. 38:107-118. [DOI] [PubMed] [Google Scholar]
- 87.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 88.Voinnet, O. 2005. Non-cell autonomous RNA silencing. FEBS Lett. 579:5858-5871. [DOI] [PubMed] [Google Scholar]
- 89.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 90.Voinnet, O., and D. C. Baulcombe. 1997. Systemic signalling in gene silencing. Nature 389:553. [DOI] [PubMed] [Google Scholar]
- 91.Voinnet, O., S. Rivas, P. Mestre, and D. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949-956. [DOI] [PubMed] [Google Scholar]
- 92.Voinnet, O., P. Vain, S. Angell, and D. C. Baulcombe. 1998. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95:177-187. [DOI] [PubMed] [Google Scholar]
- 93.Wang, H., L. Hao, C. Y. Shung, G. Sunter, and D. M. Bisaro. 2004. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15:3020-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang, M. B., X. Y. Bian, L. M. Wu, L. X. Liu, N. A. Smith, D. Isenegger, R. M. Wu, C. Masuta, V. B. Vance, J. M. Watson, A. Rezaian, E. S. Dennis, and P. M. Waterhouse. 2004. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc. Natl. Acad. Sci. USA 101:3275-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang, Y., X. Zhong, A. Itaya, and B. Ding. 2006. Evidence for the existence of the loop E motif of Potato spindle tuber viroid in vivo. J. Virol. 81:2074-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wassenegger, M., S. Heimes, L. Riedel, and H. L. Sanger. 1994. RNA-directed de novo methylation of genomic sequences in plants. Cell 76:567-576. [DOI] [PubMed] [Google Scholar]
- 97.Wassenegger, M., S. Heimes, and H. L. Sanger. 1994. An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J. 13:6172-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waterhouse, P. M., M. B. Wang, and T. Lough. 2001. Gene silencing as an adaptive defence against viruses. Nature 411:834-842. [DOI] [PubMed] [Google Scholar]
- 99.Wroblewski, T., A. Tomczak, and R. Michelmore. 2005. Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol. J. 3:259-273. [DOI] [PubMed] [Google Scholar]
- 100.Xie, Z., E. Allen, A. Wilken, and J. C. Carrington. 2005. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 102:12984-12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoshikawa, M., A. Peragine, M. Y. Park, and R. S. Poethig. 2005. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19:2164-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhong, X., A. Itaya, and B. Ding. 2005. Transfecting protoplasts by electroporation to study viroid replication, p. 16D.14.11-16D.14.11. In R. Coico, T. Kowalik, J. M. Quarles, B. Stevenson, and R. K. Taylor (ed.), Current protocols in microbiology, vol. 1. John Wiley & Sons, Inc., New York, NY. [DOI] [PubMed] [Google Scholar]
- 103.Zhong, X., N. B. Leontis, S. Qian, A. Itaya, Y. Qi, K. Boris-Lawrie, and B. Ding. 2006. Tertiary structural and functional analyses of Loop E motif in viroid RNA reveal its essential role in RNA-templated RNA replication by the nuclear transcription machinery. J. Virol. 80:8566-8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu, Y., L. Green, W. Y.-M. R. Owens, and B. Ding. 2001. Cellular basis of potato spindle tuber viroid systemic movement. Virology 279:69-77. [DOI] [PubMed] [Google Scholar]
- 105.Zhu, Y., Y. Qi, Y. Xun, R. Owens, and B. Ding. 2002. Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130:138-146. [DOI] [PMC free article] [PubMed] [Google Scholar]