Abstract
The replication of eukaryotic positive-strand RNA virus genomes occurs in the membrane-bound RNA replication complexes. Previously, we found that the extract of evacuolated tobacco BY-2 protoplasts (BYL) is capable of supporting the translation and subsequent replication of the genomic RNAs of plant positive-strand RNA viruses, including Tomato mosaic virus (ToMV). Here, to dissect the process that precedes the formation of ToMV RNA replication complexes, we prepared membrane-depleted BYL (mdBYL), in which the membranes were removed by centrifugation. In mdBYL, ToMV RNA was translated to produce the 130-kDa and 180-kDa replication proteins, but the synthesis of any ToMV-related RNAs did not occur. When BYL membranes were added back to the ToMV RNA-translated mdBYL after the termination of translation with puromycin, ToMV RNA was replicated. Using a replication-competent ToMV derivative that encodes the FLAG-tagged 180-kDa replication protein, it was shown by affinity purification that a complex that contained the 130-kDa and 180-kDa proteins and ToMV genomic RNA was formed after translation in mdBYL. When the complex was mixed with BYL membranes, ToMV RNA was replicated, which suggests that this ribonucleoprotein complex is an intermediate of ToMV RNA replication complex formation. We have named this ribonucleoprotein complex the “pre-membrane-targeting complex.” Our data suggest that the formation of the pre-membrane-targeting complex is coupled with the translation of ToMV RNA, while posttranslationally added exogenous 180-kDa protein and replication templates can contribute to replication and can be replicated, respectively. Based on these results, we discuss the mechanisms of ToMV RNA replication complex formation.
Most plant viruses and many animal viruses, which include viruses of agricultural, clinical, and veterinary importance, are positive-strand RNA viruses that have single-stranded, messenger-sense RNA genomes in virions and which replicate via negative-strand RNA [(−)RNA]. The genomes of positive-strand RNA [(+)RNA] viruses each encode one or more proteins that are involved in viral RNA replication. These replication proteins are synthesized by translation of the genomic RNAs after infection. The replication proteins recruit the genomic RNA to the cytoplasmic faces of the organellar membranes specific for each virus and synthesize (−)RNA, which is sequestered in a membranous compartment that cannot be accessed by the cytoplasmic macromolecules (23, 30). Then, large amounts of (+)RNAs (genomic and, for certain viruses, subgenomic RNAs) are synthesized using the (−)RNA as a template and are released into the cytoplasm (2, 30). In this report, we designate this membrane-bound complex that harbors (−)RNA and is capable of producing (+)RNA the viral RNA replication complex. For a number of positive-strand RNA viruses, it has been demonstrated that replication complexes reside in membranous invaginations called spherules or in other membranous structures formed after virus infection (30, 32).
For Brome mosaic virus (BMV), one of the most well-characterized positive-strand RNA viruses, it was demonstrated that the viral helicase-like 1a replication protein recruits replication template RNA and the viral 2a polymerase protein to the cytoplasmic surface of the endoplasmic reticulum membranes to form the replication complex (4, 30). Genetic and other analyses using the yeast Saccharomyces cerevisiae revealed that template recruitment by BMV 1a requires the host Lsm1p-7p/Pat1p/Dhh1p complex, which is involved in deadenylation-dependent mRNA decapping (6, 21). The initiation of BMV negative-strand RNA synthesis requires a cochaperone, Ydj1p (33), and depends on membrane lipid composition controlled by the Δ9 fatty acid desaturase Ole1p (18). For tombusviruses, other well-characterized positive-strand RNA viruses, it was shown that the p33 replication protein interacts with the p92 polymerase protein and replication template RNA (27-29). It was suggested that a complex containing p33, p92, and template RNA is targeted to the peroxisomal membranes, where the replication complexes are formed (27). The initiation of tombusvirus (−)RNA synthesis requires the molecular chaperone Hsp70 (31). Even for these viruses, however, details of replication complex formation have not been revealed.
Tomato mosaic virus (ToMV) is a positive-strand RNA virus that belongs to the genus Tobamovirus, which also includes Tobacco mosaic virus (TMV). The genome of ToMV is a nonsegmented RNA of 6,384 nucleotides with a 7-methylguanosine cap at the 5′ terminus and a tRNA-like structure at the 3′ terminus. The ToMV genome encodes at least four proteins, of 130 kDa (130K protein), 180 kDa (180K protein), 30 kDa, and 17 kDa (coat protein). The 180K protein is synthesized by suppression of the termination codon of the 130K protein open reading frame (ORF) (3, 14, 24). Among these four ToMV-coded proteins, only the 180K protein is necessary for viral RNA replication; the remaining three proteins are dispensable for replication. However, replacement of the termination codon of the 130K protein ORF with a tyrosine or phenylalanine codon, which results in the inhibition of 130K protein production, reduces the efficiency of viral RNA replication. Thus, balanced synthesis of the 130K and 180K proteins is necessary for efficient RNA replication (3, 11, 12, 14, 20). In keeping with the involvement of the 130K and 180K proteins in RNA replication, these proteins contain the methyltransferase-like and helicase-like domains, as well as the C-terminal, 180K protein-specific region that harbors the polymerase-like domain (2, 16). The ToMV 130K and 180K proteins do not have stretches of amino acid sequence that are predicted to serve as membrane-spanning regions. Nevertheless, in ToMV-infected cells, the activity of ToMV-related RNA synthesis is exclusively associated with membranes, and a fraction of the 130K and 180K protein pool is associated with membranes (9, 23). Several lines of observation suggest that the 130K and 180K replication proteins acquire the ability to synthesize ToMV-related RNA only after multiple interactions with host membranes and proteins, with ToMV RNA, and with themselves (7, 8, 22, 23, 25, 26, 34-37).
Previously, we found that extracts of evacuolated tobacco BY-2 protoplasts (BYL) were able to translate ToMV RNA to produce the 130K and 180K proteins, and when ribonucleoside triphosphates (rNTPs) were added to the mixture after translation, the ToMV (-)RNAs and (+)RNAs were synthesized in a pattern similar to that observed in vivo (15). BYL contains membranes on which the ToMV RNA replication complex is formed. In the present study, we examined membrane requirements for ToMV RNA replication in BYL and identified a ribonucleoprotein intermediate of ToMV RNA replication complex formation by following the translation of ToMV RNA in membrane-depleted BYL.
MATERIALS AND METHODS
RNA templates for in vitro translation and replication.
ToMV RNA was prepared from purified virions by phenol extraction and alcohol precipitation. The other template RNAs used in this study were synthesized by in vitro transcription using the AmpliCap T7 (for TLW3, TL180SF, TL180, TL130, and D1 RNA) or SP6 (for Rluc RNA) High Yield Message Maker kit (Epicentre Technologies, Madison, WI) in the presence of the cap analog m7G[5′]ppp[5′]G.
TLW3 RNA was synthesized from SmaI-linearized pTLW3(Sma). The pTLW3(Sma) plasmid was constructed by deleting the 3′-terminal A residue of the ToMV RNA sequence and the following 17-nucleotide polylinker sequence of the wild-type, full-length ToMV cDNA clone pTLW3 (17). The nucleotide sequence of pTLW3(Sma) around the 3′-terminal region of ToMV RNA is as follows: 5′-GGCCCGGGAATTC-3′ (ToMV RNA-derived nucleotides are shown in bold letters, the C residue that corresponds to the second nucleotide from the 3′ terminus of ToMV RNA is underlined, and the SmaI recognition sequence is italicized). Capped in vitro transcript synthesized from SmaI-linearized pTLW3(Sma) was infectious for plants and showed similar or slightly higher efficiency of in vitro replication in BYL, compared to the corresponding transcript from MluI-linearlized pTLW3.
TL180SF RNA was synthesized from SmaI-linearized pTL180SF, a derivative of pTLW3(Sma) that encodes a FLAG-tagged 180K protein. The nucleotide sequence between the last codon (underlined) of the 180K protein-coding region and the BstEII site (italicized) located in the coat protein coding region is as follows: 5′-TGTGGAGAGCTCGGAGGTGATTATAAGGATGATGATGATAAGAACTGGTCACATCCTCAATTTGAAAAGTGAGGTAACC-3′ (the termination codon for the 180K-FLAG ORF is indicated by bold letters).
TL180 RNA, which expresses the 180K protein but not the 130K protein, was synthesized from MluI-linearized pTL180. The pTL180 plasmid is a derivative of pTLW3, in which the NheI-KpnI fragment that contains the termination codon of the 130K protein ORF has been replaced with the corresponding fragment from pLFR2 (11).
TL130 RNA and D1 RNA are ToMV RNA derivatives that were synthesized from PCR fragments amplified using modified pTLW3 DNA templates and the following primers: 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′, which corresponds to a region upstream of the T7 promoter, and 5′-TGGGCCCCAACCGGGGGTTC-3′, which anneals to the 3′-terminal region of the ToMV RNA sequence (nucleotides 6384 to 6361). TL130 RNA has a deletion of nucleotides 3424 to 5798, and D1 RNA has a deletion of nucleotides 1346 to 5655.
Rluc RNA was synthesized from EcoRI-linearized pMI27 (5).
In vitro translation and RNA-dependent RNA polymerase (RdRP) reaction in BYL.
The cell extract of evacuolated BY-2 protoplasts was prepared as described previously (15), using a modified TR buffer (30 mM HEPES-KOH [pH 7.4], 80 mM potassium acetate, 1.8 mM magnesium acetate, 2 mM dithiothreitol, with one tablet of Complete Mini protease inhibitor cocktail, EDTA free [Roche], added to 10 ml of TR buffer before use) (10). The modified TR buffer was used in place of the original TR buffer throughout the experiments. To prepare membrane-depleted BYL (mdBYL), BYL (200 μl) was subjected to centrifugation at 30,000 × g for 15 min at 4°C in a Beckman TLA 100.3 rotor, and the supernatant (180 μl) was recovered. The pellet was resuspended in the residual 20 μl of supernatant to give the p30BYL membrane suspension. In vitro translation of ToMV-related and other RNAs using BYL was performed at 25°C for 1 h as described previously (10, 15). In vitro translation was also performed under the same conditions using mdBYL in place of BYL. The RdRP reaction was performed by adding 5 μl of 5× R buffer (10, 15) to 20 μl of each test sample and then incubating the mixture at 25°C for 1 h.
Sucrose gradient sedimentation analysis.
A linear sucrose gradient was formed using 1.1 ml each of 15% and 40% (wt/vol) sucrose solutions (in 30 mM HEPES-KOH [pH 7.4], 80 mM potassium acetate, 1.8 mM EDTA, 2 mM dithiothreitol) using Gradient Mate (BioComp Instruments, Inc., Fredericton, New Brunswick, Canada) at an angle of 85° at 20 rpm for 1 min. Two hundred microliters of samples were loaded onto the sucrose gradient and subjected to centrifugation (100,000 × g for 2 h at 4°C) in a Beckman TLS-55 rotor.
Protein and RNA analyses.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) and Western analyses of protein samples were performed as described previously (9). Total RNA samples were purified and analyzed by 8 M urea-2.4% PAGE (15) or Northern blotting and hybridization as described previously (13). To detect ToMV (−)RNA, RNA samples were treated with S1 nuclease and subjected to the RNase protection assay using 32P-labeled P2P RNA as the probe, as described previously (15). S1 nuclease treatment was performed as described previously (15).
RESULTS
Membranes are required for ToMV RNA synthesis.
When ToMV RNA was translated in BYL, the 130K and 180K replication proteins were produced (Fig. 1A, lanes 1 and 2); and when rNTPs were added to the translation mixture and incubated further (hereafter referred to as the RdRP reaction; see Materials and Methods), ToMV RNA replication occurred in a pattern similar to that observed in vivo (Fig. 1B, lanes 1 and 2) (15), which suggests that the ToMV RNA replication complex is formed. To investigate the role of membranes in the formation of the ToMV RNA replication complex in this in vitro system, membranes were removed from BYL by centrifugation at 30,000 × g to generate mdBYL. Translation of ToMV RNA in mdBYL yielded amounts of the 130K and 180K replication proteins similar to those produced in BYL (Fig. 1A, lanes 1 to 4). However, no ToMV-specific RNA synthesis was observed in mdBYL after the RdRP reaction (Fig. 1B, lanes 3 and 4). Accumulation of ToMV (−)RNA was detected in BYL but not in mdBYL (Fig. 1C, lanes 1 to 4).
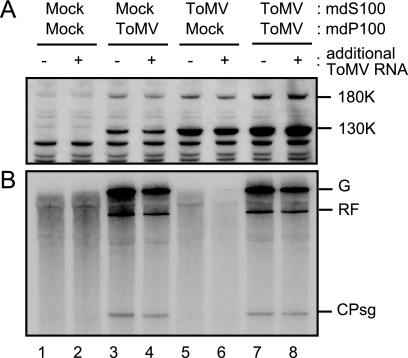
FIG. 1.
ToMV (−)RNA synthesis is dependent upon membranes in the BYL translation-replication system. ToMV RNA (1 μg)-translated (lanes 2, 4, and 6) or mock-translated (lanes 1, 3, and 5) BYL (lanes 1 and 2) or mdBYL (lanes 3 to 6) reaction mixtures (50 μl) were mixed with puromycin (0.2 mM in the resulting mixtures from 10 mM stock solution) and either the P30BYL membrane suspension (lanes 5 and 6) or mdBYL (lanes 1 to 4) (1/20 volume of the resulting mixtures), and incubation was performed at 15°C for 1 h. After this incubation period, an aliquot of each sample was removed for protein analysis for panel A, and the remainder was subjected to the RdRP reaction for panels B and C. (A) Presence of ToMV 130K and 180K replication proteins. Samples were analyzed by the Western blotting method using anti-130K and -180K protein antibody. The positions of the 130K and 180K proteins are indicated to the right. (B) ToMV RdRP activity. The RdRP reaction was carried out in the presence of [α-32P]CTP. RNA products were separated by 8 M urea-2.4% PAGE, and 32P-labeled bands were detected by autoradiography. The positions that correspond to ToMV genomic RNA (G), replicative-form RNA (RF), and the subgenomic RNA for the coat protein (CPsg) are indicated to the right. (C) Accumulation of ToMV (−)RNA. RdRP reactions were performed in the presence of 1 mM of unlabeled CTP. ToMV (−)RNA was detected by the RNase protection method using the 32P-labeled P2P RNA probe (12). Protected RNAs were separated by 8 M urea-3% PAGE and detected by autoradiography.
Although ToMV (−)RNA synthesis was not detectable in mdBYL, it was possible that the replication proteins synthesized in mdBYL were able to form the active RNA replication complex when membranes were supplemented. To test this possibility, we performed translation of ToMV RNA in mdBYL, followed by incubation with puromycin and membranes that had been recovered in the pellet fraction after centrifugation of BYL at 30,000 × g (P30BYL), and then carried out the RdRP reaction. As shown in Fig. 1B and C (lanes 5 and 6), the (−)RNA, as well as the genomic and subgenomic (+)RNAs, was synthesized in patterns similar to and in amounts larger than those observed in the standard reaction using BYL. A similar result was obtained when membranes that were further purified from P30BYL by the flotation method using sucrose gradient centrifugation were added to the ToMV RNA-translated mdBYL (data not shown), which confirms that membranes are essential for the synthesis of (−)RNA and other ToMV-related RNAs.
The P100 fraction of ToMV RNA-translated mdBYL contains components essential for the formation of ToMV RNA replication complex on membranes.
To investigate the molecular basis for the ability to form RNA replication complexes when mixed with P30BYL membranes, as observed for ToMV RNA-translated mdBYL, we first fractionated ToMV RNA-translated mdBYL by centrifugation at 100,000 × g. ToMV 130K and 180K proteins were recovered both in the pellet fraction (mdP100[ToMV]) and in the supernatant fraction (mdS100[ToMV]) (Fig. 2A). When the fractions were incubated with P30BYL membranes, followed by the RdRP reaction, the mdP100[ToMV] fraction but not the mdS100[ToMV] fraction was able to synthesize ToMV-related RNAs (data not shown). To examine whether the inability of the replication proteins in the mdS100[ToMV] fraction to support ToMV RNA replication was caused by the lack of ToMV RNA or other host factors that fractionated only into the mdP100[ToMV] fraction, we mixed the mdS100 and mdP100 fractions either from ToMV RNA-translated mdBYL or mock-translated mdBYL (mdS100[mock] and mdP100[mock] fractions, respectively) in all possible combinations. To these mixtures, we added puromycin with or without additional ToMV RNA and performed the RdRP reaction following incubation with P30BYL membranes. As shown in Fig. 2B, the reaction mixtures that contained the mdP100[ToMV] fraction gave synthesis of ToMV-related RNAs, irrespective of the types of mdS100 or the addition of exogenous ToMV RNA (lanes 3, 4, 7, and 8). These results indicate that the mdP100[ToMV] fraction contains ToMV RNA, which serves as an endogenous template for replication, as well as the replication proteins, which are capable of forming the active RNA replication complex on membranes. In contrast, although the mdS100[ToMV] fraction contained substantial amounts of the replication proteins, this fraction was incapable of synthesizing ToMV-related RNAs even when ToMV RNA, the mdP100[mock] fraction, and P30BYL membranes were supplied (Fig. 2B, lanes 1, 2, 5, and 6).
FIG. 2.
The P100 fraction but not the S100 fraction of ToMV RNA-translated mdBYL is able to synthesize ToMV-related RNAs after the addition of P30BYL membranes. ToMV RNA (4 μg)-translated and mock-translated mdBYL reaction mixtures (200 μl) were centrifuged at 100,000 × g for 30 min at 4°C in a Beckman TLA 100.3 rotor. After centrifugation, 160 μl of supernatants were recovered as the mdS100[ToMV] and mdS100[mock] fractions, and the pellets were resuspended in the residual supernatants (40 μl) to give the mdP100[ToMV] and mdP100[mock] fractions, respectively. The mdS100[ToMV] or mdS100[mock] fraction was mixed with a 1/16 volume of the mdP100[ToMV] or mdP100[mock] fraction in all possible combinations. To each mixture, puromycin (0.2 mM in the resulting mixture) with or without additional ToMV RNA (20 μg/ml in the resulting mixture from 1 mg/ml stock solution) was added, and incubation was performed at 25°C for 30 min. Then, the P30BYL membrane suspension (1/20 volume of the resulting mixture) was added to each reaction mixture, and incubation was performed at 15°C for 1 h. After this incubation period, an aliquot of each sample was removed for protein analysis for panel A, and the remainder was subjected to the RdRP reaction for panel B. (A) Presence of the ToMV 130K and 180K replication proteins. (B) ToMV RdRP activity. The RdRP reaction was performed in the presence of [α-32P]CTP. ToMV replication proteins and RdRP reaction products were analyzed as described in the legend to Fig. 1. For abbreviations, see the legend to Fig. 1.
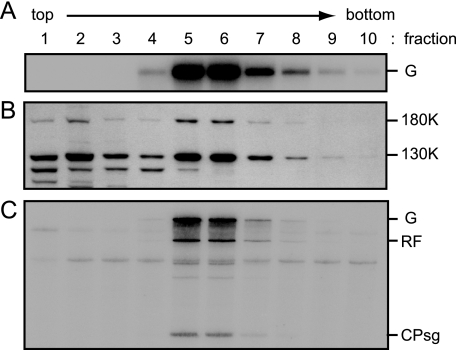
In order to examine more precisely the properties of the replication proteins in ToMV RNA-translated mdBYL, the translation mixture was applied to the top of the 15 to 40% (wt/vol) continuous sucrose density gradient and centrifuged at 100,000 × g for 2 h. After fractionation (fractions 1 to 10, from the top to the bottom of the gradient), each fraction was subjected to Northern and Western analyses to detect ToMV RNA and the replication proteins, respectively. ToMV RNA was fractionated mainly in the fifth and sixth fractions, while the 130K and 180K replication proteins showed fractionation patterns with two peaks at the first-second and fifth-sixth fractions (Fig. 3A and B). The sixth fraction contained the 80S ribosomes (data not shown). In the sucrose gradient centrifugation analysis, the replication proteins in the mdS100[ToMV] fraction were fractionated mainly in the first-second fractions, whereas those in the mdP100[ToMV] fraction were in the fifth-sixth fractions (data not shown). When the fractions were incubated with P30BYL membranes followed by the RdRP reaction, ToMV-related RNA synthesis was observed for the fifth-sixth fractions, which contained both the ToMV replication proteins and ToMV RNA (Fig. 3C).
FIG. 3.
Fractionation of ToMV RNA-translated mdBYL by sedimentation in a sucrose gradient. A ToMV RNA (4 μg)-translated mdBYL reaction mixture (200 μl) was mixed with puromycin (0.2 mM in the resulting mixture) and incubated at 25°C for 10 min. This reaction mixture (200 μl) was loaded onto the 15 to 40% sucrose gradient and subjected to centrifugation. The gradient was manually fractionated into 10 fractions (220 μl per fraction; fractions 1 to 10, from top to bottom of the gradient). RNA and protein samples were prepared from these fractions and analyzed for panels A and B, respectively. In addition, each fraction was mixed with creatine phosphate (30 mM in the resulting mixtures from 1 M stock solution), creatine kinase (0.2 mg/ml in the resulting mixtures from a 10-mg/ml stock solution), ATP (0.75 mM in the resulting mixtures from 37.5 mM stock solution), and the P30BYL membrane suspension (1/20 volume of the resulting mixtures), and incubation was performed at 15°C for 1 h. After the incubation, 20 μl of each mixture was subjected to the RdRP reaction for panel C. (A) Presence of ToMV RNA. ToMV RNA was detected by Northern blot hybridization using a 32P-labeled P1M RNA probe (12). RNAs were denatured with glyoxal and separated in a 1% agarose gel. (B) Presence of ToMV replication proteins. The analysis was performed as described in the legend to Fig. 1. (C) ToMV RdRP activity. The RdRP reaction was performed in the presence of [α-32P]CTP. RdRP reaction products were analyzed as described in the legend to Fig. 1. For abbreviations, see the legend to Fig. 1.
The complex that forms the active ToMV RNA replication complex on membranes contains the ToMV 130K and 180K replication proteins and genomic RNA.
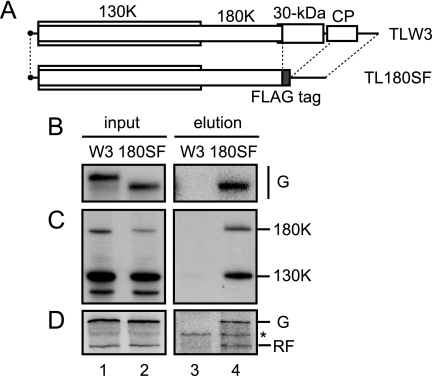
Although the fifth and sixth (approximately 70S) fractions of the sucrose gradient contained both the ToMV replication proteins and ToMV RNA (Fig. 3), it was not clear whether these components were associated with each other in these fractions. To address whether these molecules form a complex, we performed affinity purification of the 180K replication protein using a replication-competent ToMV RNA derivative, TL180SF. This derivative encodes the 180K protein, the C terminus of which is fused to the FLAG tag, while retaining the unmodified 130K protein and carries a deletion in the 30-kDa protein and coat protein coding regions (Fig. 4A). The modification of the 180K protein did not impair the ability of the 180K protein to support ToMV RNA replication in BY-2 cells and in BYL (Fig. 4D, lanes 1 and 2) (23).
FIG. 4.
Affinity purification of the FLAG-tagged 180K replication protein from the TL180SF RNA-translated mdP100 fraction. (A) Schematic representation of TLW3 and TL180SF RNAs. (B to D) TLW3 RNA (12 μg)-translated (W3, lanes 1 and 3) or TL180SF RNA (12 μg)-translated (180SF, lanes 2 and 4) mdBYL reaction mixtures (600 μl each) were centrifuged at 100,000 × g for 30 min at 4°C in a Beckman TLA 100.3 rotor, followed by removal of 480 μl of the supernatants. The pellets were suspended in the residual supernatants (120 μl) (mdP100[TLW3] and mdP100[TL180SF] fractions). One hundred microliters of the mdP100[TLW3] or mdP100[TL180SF] fraction was each mixed with 20 μl of ANTI-FLAG M2 affinity gel (50% suspension; Sigma) and incubated at 4°C for 3 h with occasional shaking. The gels were washed five times with wash buffer (30 mM HEPES-KOH [pH 7.4], 80 mM potassium acetate, 1 mM EDTA) and then eluted in 30 μl of wash buffer that contained 100 μg/ml FLAG peptide (Sigma). The washing and elution steps were performed at 4°C. The mdP100[TLW3] and mdP100[TL180SF] fractions (input; lanes 1 and 2) and the FLAG-purified fractions (elution; lanes 3 and 4) were analyzed for the presence of ToMV RNA for panel B by Northern blotting and hybridization, as described in the legend to Fig. 3, and for the presence of the 130K and 180K proteins for panel C by the Western blotting method, as described in the legend to Fig. 1. For panel D, the mdP100[TLW3] and mdP100[TL180SF] fractions diluted fivefold with wash buffer (input; lanes 1 and 2) and the FLAG-purified fractions (elution; lanes 3 and 4) were each mixed with puromycin (0.2 mM in the resulting mixtures), creatine phosphate (30 mM in the resulting mixtures), creatine kinase (0.2 mg/ml in the resulting mixtures), ATP (0.75 mM in the resulting mixtures), and the P30BYL membrane suspension (1/20 volume of the resulting mixtures), and incubation was performed at 15°C for 1 h. After this incubation period, 20 μl of each mixture was subjected to the RdRP reaction in the presence of [α-32P]CTP. RdRP reaction products were analyzed as described in the legend to Fig. 1. The positions corresponding to the 130K and 180K proteins, genomic RNAs (G), and replicative-form RNAs (RF) of TLW3 and TL180SF are indicated to the right. The asterisk indicates unidentified background signals.
TL180SF RNA was translated in mdBYL, followed by centrifugation at 100,000 × g to give the pellet (mdP100[TL180SF]) fraction. The FLAG-tagged 180K protein in the mdP100[TL180SF] fraction was affinity purified using anti-FLAG immunoglobulin G-conjugated agarose beads. As a control, TLW3 RNA, the wild-type in vitro transcript equivalent to ToMV RNA (we call the in vitro transcript “TLW3” to distinguish it from ToMV RNA isolated from virions) (17) was used. The 180K and 130K proteins were readily detected by Western analysis in the FLAG-purified fraction from the mdP100[TL180SF] fraction, whereas these proteins were barely detectable in the same fraction from the mdP100[TLW3] fraction (Fig. 4C, lanes 3 and 4). Northern analyses revealed that the genomic RNA specifically copurified with the FLAG-tagged 180K protein (Fig. 4B, lanes 3 and 4). ToMV RNA replication was observed when P30BYL membranes were added to the FLAG-purified fraction for TL180SF but not for TLW3 (Fig. 4D, lanes 3 and 4). Taken together, these results suggest that in the mdP100[ToMV] fraction, the 130K and 180K proteins and ToMV RNA form a ribonucleoprotein complex, which is eventually converted into an active RNA replication complex on membranes. We call this ribonucleoprotein complex the “ToMV pre-membrane-targeting complex” (PMTC).
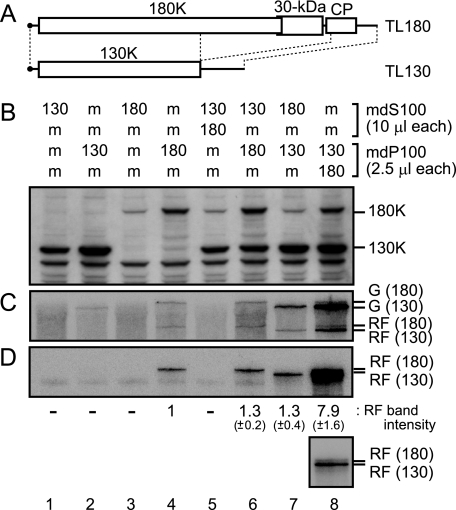
The 180K protein in the mdS100[TL180] fraction participates in replication of TL130 RNA in the mdP100[TL130] fraction.
To gain further insights into the assembly of the PMTC, we constructed the ToMV RNA derivatives TL130 and TL180. TL130 has a large deletion in the read-through region of the 180K protein, 30-kDa protein, and coat protein coding regions and expresses the 130K protein alone (Fig. 5A). TL180 is a derivative of TLW3 that has a point mutation that alters the termination codon of the 130K protein ORF to the tyrosine codon and expresses the 180K protein but not the 130K protein (Fig. 5A). TL180 RNA could replicate by itself, albeit at a lower efficiency than wild-type TLW3 RNA, while 180K protein-deficient ToMV derivatives could not replicate by themselves in BY-2 cells (11, 12; data not shown).
FIG. 5.
In vitro replication of TL130 and TL180 RNAs. (A) Schematic representation of TL180 and TL130 RNAs. (B to D) TL130 or TL180 RNA (4 μg each)-translated and mock-translated mdBYL reaction mixtures (100 μl each) were centrifuged at 100,000 × g for 30 min at 4°C in a Beckman TLA 100.3 rotor. After centrifugation, 80 μl of each supernatant was recovered as the mdS100[TL130] (130), mdS100[TL180] (180), or mdS100[mock] (m) fraction. The pellets were resuspended in residual supernatant (20 μl each) to give the mdP100[TL130] (130), mdP100[TL180] (180), and mdP100[mock] (m) fractions. These fractions were mixed as indicated in panel B and were incubated at 15°C for 30 min. To each mixture (25 μl), the P30BYL membrane suspension (1/20 volume of the resulting mixture) and puromycin (0.2 mM in the resulting mixtures) were added, and incubation was performed at 15°C for 1 h. After this incubation period, an aliquot of each sample was removed for protein analysis for panel B, and the remainder was subjected to the RdRP reaction for panel C. For panel B, the 130K and 180K replication proteins were detected by the Western blotting method as described in the legend to Fig. 1. For panel C, the RdRP reaction was carried out in the presence of [α-32P]CTP, and the reaction products were analyzed as described in the legend to Fig. 1. For panel D, the 32P-labeled RNA products analyzed in panel C were treated with S1 nuclease. S1 nuclease-resistant RNA was separated by 8 M urea-2.4% PAGE, and the 32P-labeled bands were detected by autoradiography. The intensities of the S1 nuclease-resistant RNA bands relative to that in lane 4 are indicated below panel D. The values are averages and standard deviations for three experiments using different preparations of BYL. In lane 8, the TL180 and TL130 RF bands were too close to be separately quantified, and the quantified areas contained both bands. The bottom inset shows a shorter exposure of lane 8. The positions that correspond to the 130K and 180K proteins, TL130 and TL180 genomic RNAs [G (130) and G (180), respectively], and the TL130 and TL180 replicative-form RNAs [RF (130) and RF (180), respectively] are indicated to the right.
The in vitro transcripts of TL130 and TL180 were separately translated in mdBYL, the reaction mixtures were fractionated into the P100 and S100 fractions (mdP100[TL130], mdS100[TL130], mdP100[TL180], and mdS100[TL180]), and each fraction was subjected to the RdRP reaction following incubation with P30BYL membranes. As expected, only the mdP100[TL180] fraction showed a detectable level of viral RNA synthesis (Fig. 5C and D, lanes 1 to 4). Although a faint band that corresponds to the TL130 genomic RNA was observed in the mdP100[TL130] fraction (Fig. 5C, lane 2), this does not represent replication, since the genome-length double-stranded RNA (replicative form) band was not detected, even after S1 nuclease treatment (Fig. 5D, lane 2). Instead, this band probably represents terminal labeling of the input TL130 RNA by host ATP(CTP):tRNA nucleotidyltransferase (1). We mixed these fractions in all possible heterologous combinations, incubated the mixtures with P30BYL membranes, and then performed the RdRP reaction. When the mdP100[TL130] and mdS100[TL180] fractions were mixed, the replication of TL130 RNA was observed (Fig. 5C and D, lane 7), which suggests that a PMTC-like complex is formed in TL130-translated mdBYL and that the 180K protein in the mdS100[TL180] fraction contributes to the replication of TL130 RNA contained in the PMTC-like complex. When the mdP100[TL130] and mdP100[TL180] fractions were mixed, strong replication of TL130 RNA was observed (Fig. 5C and D, lane 8). The level of TL180 RNA replication was not remarkably altered by the presence of the 130K protein (compare lanes 6 and 8 with lane 4 in Fig. 5C and D), which suggests that the 130K protein does not contribute to the replication of TL180 RNA in trans. In the combination of the mdS100[TL130] and mdS100[TL180] fractions, ToMV-related RNA replication was not detected (Fig. 5C and D, lane 5). This result is consistent with the absence of PMTC (or PMTC-like complexes) in the mdS100 fractions.
PMTC is able to accept exogenous ToMV-related RNAs as replication templates.
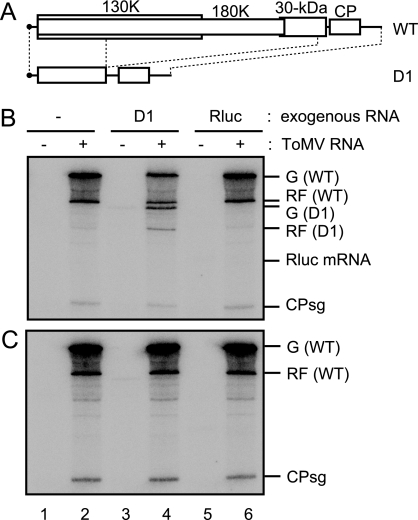
We next examined whether PMTC could accept exogenous RNA as replication templates. As a model replicon, a derivative of ToMV RNA, termed D1 RNA, that carries a deletion between nucleotides 1346 to 5655 was used (Fig. 6A). D1 RNA mimics the well-characterized TMV RNA derivative ΔCla, which does not code for intact replication proteins but can be replicated efficiently with the assistance of wild-type helper TMV RNA in protoplasts (19).
FIG. 6.
The ability of PMTC to support the replication of exogenously added defective ToMV RNA. (A) Schematic representation of wild-type ToMV RNA (WT) and D1 RNA. (B and C) A ToMV RNA (4.0 μg)-translated or mock-translated mdBYL reaction mixture (200 μl) was centrifuged at 100,000 × g for 30 min at 4°C in a Beckman TLA 100.3 rotor, followed by the removal of 160 μl of supernatant. The pellets were resuspended in residual supernatant (40 μl) to generate the mdP100[ToMV] and mdP100[mock] fractions. For panel B, the mdP100[ToMV] (lanes 2, 4, and 6) or mdP100[mock] fraction (lanes 1, 3, and 5) (5 μl) was mixed with puromycin (0.24 mM in the resulting mixtures from 1.65 mM stock solution) and D1 RNA (0.15 μg) (lanes 3 and 4) or Rluc RNA (0.08 μg) (lanes 5 and 6) or with puromycin alone (0.24 mM in the resulting mixtures) (lanes 1 and 2) to give a final volume of 7 μl (with H2O), followed by incubation at 25°C for 30 min. To the mixtures, 1.25 μl of P30BYL membrane suspension was added, and incubation was performed at 15°C for 30 min. After this incubation period, each mixture was diluted with TR buffer that contained 0.2 mM puromycin to give a final volume of 25 μl, and the samples (20 μl) were subjected to the RdRP reaction in the presence of [α-32P]CTP. For panel C, the mdP100[ToMV] (lanes 2, 4, and 6) or mdP100[mock] (lanes 1, 3, and 5) fraction (5 μl) was mixed with 1.25 μl of P30BYL membrane suspension and puromycin (0.23 mM in the resulting mixtures from 1.65 mM stock solution), followed by incubation at 15°C for 30 min. Then, D1 RNA (0.15 μg) (lanes 3 and 4), Rluc RNA (0.08 μg) (lanes 5 and 6), or H2O (lanes 1 and 2) was added to give a final volume of 8.25 μl, and the mixture was incubated at 25°C for 30 min. After this incubation period, each mixture was diluted with TR buffer that contained 0.2 mM puromycin to give a final volume of 25 μl, and the samples (20 μl) were subjected to the RdRp reaction in the presence of [α-32P]CTP. The RdRP reaction products were analyzed as described in the legend to Fig. 1. The positions that correspond to the genomic RNA [G (D1)] or replicative-form RNA [RF (D1)] of D1 and Rluc mRNA are indicated to the right. For the other abbreviations, see the legend to Fig. 1.
D1 RNA was incubated with the mdP100[ToMV] fraction that contained puromycin, and then an RdRP reaction was carried out after incubation with P30BYL membranes. As a control, a capped Rluc mRNA carrying an A30 tract was used in place of D1 RNA. As shown in Fig. 6B, single-stranded and double-stranded D1 RNAs were synthesized (lane 4), which was not the case when the mdP100[mock] fraction was used (lane 3) or when D1 RNA was omitted (lane 2). In contrast, Rluc RNA-specific bands were not detected (lane 6). These results suggest that the D1 RNA was specifically recognized by PMTC and was replicated. The replication of D1 RNA was not observed when the mixture of mdS100[ToMV] and mdP100[mock] fractions was used (data not shown), which confirms that the replication proteins in the mdS100[ToMV] fraction lack the ability to replicate ToMV-related RNAs. Finally, we examined whether the order of the addition of D1 RNA and P30BYL membranes affected the efficiency of D1 RNA replication. As shown in Fig. 6C, the replication of D1 RNA was not detected when the mdP100[ToMV] fraction was incubated with P30BYL membranes prior to the addition of D1 RNA (lane 4). This result is consistent with the observation that the components of replication complexes of positive-strand RNA viruses are sequestered in membranous compartments that are inaccessible to cytoplasmic macromolecules (23, 30).
DISCUSSION
The purpose of this study was to dissect the process of ToMV RNA replication complex formation in vitro. Initially, we showed that the initiation of ToMV (−)RNA synthesis requires membranes (Fig. 1). We then showed that a ribonucleoprotein complex named PMTC, which contains the genomic RNA and the 130K and 180K replication proteins, is formed in ToMV RNA-translated mdBYL (Fig. 4). The PMTC had an approximate sedimentation coefficient of 70S (Fig. 3) and was fractionated into the P100 (mdP100[ToMV]) fraction (Fig. 2). While PMTC itself did not have the ability to synthesize ToMV-related RNAs, it formed an RNA replication complex, which synthesized negative- and positive-strand ToMV RNAs, when mixed with P30BYL membranes (Fig. 1 to 4). These results suggest that PMTC is an intermediate of ToMV RNA replication complex formation.
The 130K and 180K replication proteins synthesized by translation of ToMV RNA in mdBYL were fractionated not only into the P100 fraction but also into the S100 (mdS100[ToMV]) fraction. The replication proteins in the mdS100[ToMV] fraction were not able to replicate exogenously added ToMV RNA (Fig. 2) or D1 RNA (data not shown) even in the presence of membranes and rNTPs, which suggests that PMTC formation occurs only when coupled with the translation of ToMV RNA. It is possible that ToMV replication proteins recognize ToMV RNA either during or immediately after translation or that ToMV replication proteins recognize a specific RNA structure that is transiently formed on ToMV RNA after the passage of ribosomes.
When the mdP100[TL130] and mdS100[TL180] fractions were mixed and incubated with P30BYL membranes and rNTPs, the replication of TL130 RNA occurred (Fig. 5, lane 7). This result suggests that one or more 130K protein molecules bind cotranslationally to a TL130 RNA molecule and that the 180K protein (and perhaps an additional 130K protein) binds posttranslationally to the 130K protein-TL130 RNA complex to form PMTC. We propose to call this cotranslationally formed, 130K protein-viral RNA complex the “core PMTC.” On the other hand, when the mdP100[TL180] fraction alone was incubated with P30BYL membranes and rNTPs, the replication of TL180 RNA was observed (Fig. 5, lane 4). When the mdP100[TL180] fraction was mixed with the mdP100[TL130] fraction, followed by incubation with P30BYL membranes and rNTPs, not only the replication of TL180 RNA but also more active replication of TL130 RNA was observed (Fig. 5, lane 8). These results suggest that the 180K protein alone can form core PMTC at lower efficiency than the 130K protein. Thus, in the wild-type context, in which both the 130K and 180K proteins are expressed from the same translation template, PMTC appears to be established mainly through the cotranslational formation of the 130K protein-ToMV RNA complex (core PMTC), followed by posttranslational binding of the 180K protein (and perhaps an additional 130K protein) to the core PMTC. The 180K protein may acquire the ability to synthesize (−)RNA through binding to core PMTC and the subsequent interactions with membranes and host proteins.
In keeping with these possibilities, Lewandowski and colleagues found that some deletion derivatives of TMV RNAs (defective RNAs) that encode the 130K protein alone can be replicated in plant cells when helper TMV RNA derivatives that express the 180K protein are coinoculated; however, frame-shift and other deleterious mutations in the 130K protein ORF in the defective RNA greatly reduce the efficiency of defective RNA replication. From these observations, these authors proposed a model in which the 130K protein first binds to its translation template (i.e., defective RNA) in cis, the 180K protein is then incorporated into this complex, and finally, the synthesis of (−)RNA occurs (19, 20). It is reasonable to assume that the replication proteins preferentially select their translation template as the replication template (i.e., in cis), since the closest replication template for nascent replication protein is its own translation template for positive-strand RNA viruses with nonsegmented genomes. Taking into consideration the fact that core PMTC is cotranslationally formed, it is assumed that the cis preference is much stronger.
We have also demonstrated that a deletion derivative of ToMV RNA, D1, is replicated when D1 RNA is mixed with PMTC and then with P30BYL membranes and rNTPs. In the in vitro system, translation of D1 RNA was not required for D1 RNA to be replicated (Fig. 6B, lane 4). In contrast, translation of at least a certain part of the 130K protein ORF is necessary for defective RNAs to be replicated efficiently by helper viruses in vivo (20). To explain this contradiction, we propose two alternative pathways of defective RNA replication: (i) through posttranslational, transrecognition of defective RNA by PMTC supplied by helper RNA and (ii) through cotranslational formation of core PMTC (or core PMTC-like complex) on the defective RNA molecule, followed by recruitment of the 180K protein (and perhaps the 130K protein) supplied by helper RNA. In vivo, pathway ii would be more active than pathway i. The presence of membranes during translation may contribute to this effect. In vitro, where D1 RNA was incubated with helper RNA-derived PMTC in the presence of puromycin and in the absence of membranes, only pathway i would occur. Considering that core PMTC is formed cotranslationally, it seems unlikely that the replication protein in the core PMTC detaches from the translation template and then binds again to D1 RNA. Rather, it is plausible to postulate that independently of the translation template RNA-binding site of the replication protein in core PMTC, the PMTC has one or more additional template RNA-binding sites that are open for exogenous templates.
Acknowledgments
We thank Tetsuo Meshi for critical reading of the manuscript. We also thank the members of our laboratory for helpful discussions and Kumi Fujiwara and Akemi Kikuchi for general assistance.
This work was supported by the Core Research for Evolutional Science and Technology of the Japan Science and Technology Agency. K.K. is supported by the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 15 November 2006.
REFERENCES
- 1.Benicourt, C., and A. L. Haenni. 1974. Recognition of TMV RNA by the tRNA nucleotidyltransferase. FEBS Lett. 45:228-231. [DOI] [PubMed] [Google Scholar]
- 2.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, K. W. 1999. Replication of tobacco mosaic virus RNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 354:613-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiba, Y., R. Sakurai, M. Yoshino, K. Ominato, M. Ishikawa, H. Onouchi, and S. Naito. 2003. S-adenosyl-L-methionine is an effector in the posttranscriptional autoregulation of the cystathionine γ-synthase gene in Arabidopsis. Proc. Natl. Acad. Sci. USA 100:10225-10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Díez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goregaoker, S. P., and J. N. Culver. 2003. Oligomerization and activity of the helicase domain of the tobacco mosaic virus 126- and 183-kilodalton replicase proteins. J. Virol. 77:3549-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goregaoker, S. P., D. J. Lewandowski, and J. N. Culver. 2001. Identification and functional analysis of an interaction between domains of the 126/183-kDa replicase-associated proteins of tobacco mosaic virus. Virology 282:320-328. [DOI] [PubMed] [Google Scholar]
- 9.Hagiwara, Y., K. Komoda, T. Yamanaka, A. Tamai, T. Meshi, R. Funada, T. Tsuchiya, S. Naito, and M. Ishikawa. 2003. Subcellular localization of host and viral proteins associated with tobamovirus RNA replication. EMBO J. 22:344-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishibashi, K., K. Komoda, and M. Ishikawa. 2006. In vitro translation and replication of tobamovirus RNA in a cell-free extract of evacuolated tobacco BY-2 protoplasts, p. 183-194. In T. Nagata, K. Matsuoka, and D. Inzé (ed.), Biotechnology in agriculture and forestry, vol. 58. Tobacco BY-2 cells: from cellular dynamics to omics. Springer, Berlin, Germany. [Google Scholar]
- 11.Ishikawa, M., T. Meshi, F. Motoyoshi, N. Takamatsu, and Y. Okada. 1986. In vitro mutagenesis of the putative replicase genes of tobacco mosaic virus. Nucleic Acids Res. 14:8291-8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa, M., T. Meshi, T. Ohno, and Y. Okada. 1991. Specific cessation of minus-strand RNA accumulation at an early stage of tobacco mosaic virus infection. J. Virol. 65:861-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa, M., S. Naito, and T. Ohno. 1993. Effects of the tom1 mutation of Arabidopsis thaliana on the multiplication of tobacco mosaic virus RNA in protoplasts. J. Virol. 67:5328-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishikawa, M., and Y. Okada. 2004. Replication of tobamovirus RNA. Proc. Jpn. Acad. Ser. B 80:215-224. [Google Scholar]
- 15.Komoda, K., S. Naito, and M. Ishikawa. 2004. Replication of plant RNA virus genomes in a cell-free extract of evacuolated plant protoplasts. Proc. Natl. Acad. Sci. USA 101:1863-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 17.Kubota, K., S. Tsuda, A. Tamai, and T. Meshi. 2003. Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 77:11016-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, W.-M., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewandowski, D. J., and W. O. Dawson. 1998. Deletion of internal sequences results in tobacco mosaic virus defective RNAs that accumulate to high levels without interfering with replication of the helper virus. Virology 251:427-437. [DOI] [PubMed] [Google Scholar]
- 20.Lewandowski, D. J., and W. O. Dawson. 2000. Functions of the 126- and 183-kDa proteins of tobacco mosaic virus. Virology 271:90-98. [DOI] [PubMed] [Google Scholar]
- 21.Mas, A., I. Alves-Rodrigues, A. Noueiry, P. Ahlquist, and J. Díez. 2006. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J. Virol. 80:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Más, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikiori, M., K. Dohi, M. Mori, T. Meshi, S. Naito, and M. Ishikawa. 2006. Membrane-bound tomato mosaic virus replication proteins participate in RNA synthesis and are associated with host proteins in a pattern distinct from those that are not membrane bound. J. Virol. 80:8459-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno, T., M. Aoyagi, Y. Yamanashi, H. Saito, S. Ikawa, T. Meshi, and Y. Okada. 1984. Nucleotide sequence of the tobacco mosaic virus (tomato strain) genome and comparison with the common strain genome. J. Biochem. 96:1915-1923. [DOI] [PubMed] [Google Scholar]
- 25.Osman, T. A. M., and K. W. Buck. 1997. The tobacco mosaic virus RNA polymerase complex contains a plant protein related to the RNA-binding subunit of yeast eIF-3. J. Virol. 71:6075-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osman, T. A. M., and K. W. Buck. 2003. Identification of a region of the tobacco mosaic virus 126- and 183-kilodalton replication proteins which binds specifically to the viral 3′-terminal tRNA-like structure. J. Virol. 77:8669-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panavas, T., C. M. Hawkins, Z. Panaviene, and P. D. Nagy. 2005. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of cucumber necrosis tombusvirus. Virology 338:81-95. [DOI] [PubMed] [Google Scholar]
- 28.Pogany, J., K. A. White, and P. D. Nagy. 2005. Specific binding of tombusvirus replication protein p33 to an internal replication element in the viral RNA is essential for replication. J. Virol. 79:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajendran, K. S., and P. D. Nagy. 2006. Kinetics and functional studies on interaction between the replicase proteins of Tomato Bushy Stunt Virus: requirement of p33:p92 interaction for replicase assembly. Virology 345:270-279. [DOI] [PubMed] [Google Scholar]
- 30.Schwartz, M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 31.Serva, S., and P. D. Nagy. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 80:2162-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomita, Y., T. Mizuno, J. Díez, S. Naito, P. Ahlquist, and M. Ishikawa. 2003. Mutation of host dnaJ homolog inhibits brome mosaic virus negative-strand RNA synthesis. J. Virol. 77:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsujimoto, Y., T. Numaga, K. Ohshima, M. Yano, R. Ohsawa, D. B. Goto, S. Naito, and M. Ishikawa. 2003. Arabidopsis TOBAMOVIRUS MULTIPLICATION (TOM) 2 locus encodes a transmembrane protein that interacts with TOM1. EMBO J. 22:335-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe, T., A. Honda, A. Iwata, S. Ueda, T. Hibi, and A. Ishihama. 1999. Isolation from tobacco mosaic virus-infected tobacco of a solubilized template-specific RNA-dependent RNA polymerase containing a 126K/183K protein heterodimer. J. Virol. 73:2633-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamanaka, T., T. Ohta, M. Takahashi, T. Meshi, R. Schmidt, C. Dean, S. Naito, and M. Ishikawa. 2000. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 97:10107-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeenko, V. V., L. A. Ryabova, A. S. Spirin, H. M. Rothnie, D. Hess, K. S. Browning, and T. Hohn. 2002. Eukaryotic elongation factor 1A interacts with the upstream pseudoknot domain in the 3′ untranslated region of tobacco mosaic virus RNA. J. Virol. 76:5678-5691. [DOI] [PMC free article] [PubMed] [Google Scholar]