Abstract
The recent emergence of several new coronaviruses, including the etiological cause of severe acute respiratory syndrome, has significantly increased the importance of understanding virus-host cell interactions of this virus family. We used mouse hepatitis virus (MHV) A59 as a model to gain insight into how coronaviruses affect the type I alpha/beta interferon (IFN) system. We demonstrate that MHV is resistant to type I IFN. Protein kinase R (PKR) and the alpha subunit of eukaryotic translation initiation factor are not phosphorylated in infected cells. The RNase L activity associated with 2′,5′-oligoadenylate synthetase is not activated or is blocked, since cellular RNA is not degraded. These results are consistent with lack of protein translation shutoff early following infection. We used a well-established recombinant vaccinia virus (VV)-based expression system that lacks the viral IFN antagonist E3L to screen viral genes for their ability to rescue the IFN sensitivity of the mutant. The nucleocapsid (N) gene rescued VVΔE3L from IFN sensitivity. N gene expression prevents cellular RNA degradation and partially rescues the dramatic translation shutoff characteristic of the VVΔE3L virus. However, it does not prevent PKR phosphorylation. The results indicate that the MHV N protein is a type I IFN antagonist that likely plays a role in circumventing the innate immune response.
The Coronaviridae family consists of a large number of widespread, medically important viruses that cause primarily respiratory and enteric infections in humans and many animals. Economically important diseases are caused by bovine, porcine, and avian coronaviruses (CoV). Approximately 30% of common colds are caused by human coronaviruses (43). In late 2002 a new coronavirus was identified as the etiological agent that causes severe acute respiratory syndrome (SARS). Almost 9,000 people were infected, with a mortality rate overall of ∼10% and a significantly higher mortality rate of ∼40% in individuals older than 60 years (18). Since SARS-CoV was discovered, at least two new human coronaviruses that are distinct from that virus have been identified from patients with respiratory tract infections. These viruses include HCoV-NL63 and HCoV-HKU1, related to the coronavirus groups I and II, respectively (29). SARS-CoV has not reemerged at this point, but the isolation of related viruses from bats and other animals (26, 35, 37, 48) and the routine circulation of coronaviruses in domesticated animals suggest that animal-to-human transmission of virulent viruses may occur again. Understanding the molecular biology of these viruses and factors that contribute to their pathogenesis is thus important.
Coronaviruses are enveloped and contain single-stranded, positive-sense RNA genomes that range from ∼27 to 31 kb in length. Coronavirus genomes are the largest known among RNA viruses. The RNA genome is capped at the 5′ end and polyadenylated at the 3′ end. Approximately two-thirds of the 5′ end of the genome consists of two overlapping open reading frames (ORF1a and ORF1b) that are translated as two polyproteins that are co- and posttranslationally processed by virus-encoded proteinases into as many as 16 nonstructural proteins (NSPs), including the RNA-dependent RNA polymerase. The genome is encapsidated by the multifunctional phosphorylated N protein. In addition to being the most abundant viral structural protein, N also plays not fully defined roles in viral transcription and/or replication and possibly in translation. At least three proteins are anchored in the envelope, the membrane (M), spike (S), and envelope (E) proteins. The S protein is the viral receptor binding protein, which initiates infection through fusion of the viral and cellular membranes and is the major target of neutralizing antibodies (22). The major envelope component is the M protein, which plays important roles in virus assembly (16). The E protein is a minor component of the viral envelope that also plays an important role in virus assembly (16).
The innate immune response is part of the first line of defense against viruses, which also signals development of the adaptive cellular and humoral immune responses. Type I interferons (IFNs), IFN-α and IFN-β, are key components of the innate immune system that are induced after initial virus-host cell interactions. Type I IFN in turn triggers JAK/STAT-mediated signal transduction pathways that stimulate expression of more than 100 interferon-stimulated gene (ISG) products, which leads to the establishment of an antiviral state (17). A number of the ISGs encode enzymes with antiviral functions, which includes 2′,5′oligoadenylate synthetase (2′-5′ OAS), protein kinase R (PKR), Mx, PML, p56, and many others (58). PKR and 2′-5′ OAS are present in most cells at basal levels even in the absence of IFN (58). PKR synthesis is induced by IFN, but double-stranded RNA (dsRNA) triggers dimerization and activation of PKR, which result in its autophosphorylation. Activated PKR in turn phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (eIF2α). These events can ultimately lead to the inhibition of protein synthesis, thus blocking viral replication and virion progeny production. IFN-induced 2′-5′ OAS is also activated by dsRNA. The enzyme polymerizes ATP into 2′,5′-linked oligoadenylates that, in turn, activate latent RNase L, which results in degradation of mRNA and rRNA. Many, if not all, animal viruses encode gene products that antagonize the antiviral response, thus allowing the viruses to circumvent the early cellular IFN defense (21, 28, 30).
We used the prototypic mouse hepatitis virus A59 (MHV A59), a group 2 coronavirus, as a model system to begin gaining insight into how coronaviruses affect the innate immune system by focusing on two downstream ISG pathways. We have determined that MHV A59 is resistant to type I IFN treatment in tissue culture. Our results show that the PKR and 2′-5′ OAS pathways are not activated in virus-infected cells, resulting in the lack of eIF2α phosphorylation and cellular RNA degradation, consistent with lack of protein translation shutoff during early infection. Most significantly, we have shown that the MHV N protein is an IFN antagonist. Expression of the N gene in a recombinant vaccinia virus in place of the viral antagonist E3L demonstrated that the protein rescues 2′-5′ OAS/RNase L activity, but it does not prevent PKR phosphorylation. Our results indicate that some aspect of the MHV A59 viral life cycle, or possibly multiple viral gene products, are responsible for circumventing the innate immune response through interactions with at least two IFN-stimulated pathways.
MATERIALS AND METHODS
Cells and viruses.
Mouse 17Cl1 (60) and rabbit RK13 (ATCC) cells were grown in Dulbecco's modified Eagle medium (DMEM) containing 5% heat-inactivated fetal calf serum (FCS) and penicillin-streptomycin-glutamine. HeLa MHVR cells expressing the MHV A59 CEACAM (carcinoembryonic antigen cell adhesion molecule isoform 1a) receptor were kindly provided by Tom Gallagher (Loyola University Medical Center, Maywood, IL) and maintained as previously described (50). BHK cells (ATCC) were cultured in Glasgow modified Eagle medium supplemented with 5% heat-inactivated FCS and 10% tryptose phosphate broth. MHV A59 was grown in mouse 17Cl1 cells. Vaccinia virus (VV) Copenhagen strain (63) and VVΔE3L (6) were grown in BHK cells. MHV- and vesicular stomatitis virus (VSV)-infected cells were overlaid with 1.5% low-melting-point agarose (SeaKem) in modified Eagle medium containing 10% FCS for plaque assays. Vaccinia viruses were analyzed for IFN sensitivity by plaque titration in RK-13 cells. Plaques were visualized by crystal violet staining.
Metabolic labeling.
Cells were mock treated or treated with 100 IU/ml of universal type I IFN (recombinant human alpha A/D) (PBL Biomedical Laboratories, Piscataway, NJ) for 24 h before infection with viruses at a multiplicity of infection (MOI) of 5 or 10. Infected cells were further incubated in the presence (100 IU/ml) or absence of IFN at 37°C prior to metabolic labeling. Cells were starved at 6 h postinfection (hpi) in Dulbecco's modified Eagle medium without methionine and cysteine for 30 min prior to being labeled with 50 μCi of Expre35S35S (Perkin-Elmer). Labeled cells were lysed in phosphate-buffered saline containing 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS). Equal amounts of cell lysates were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography. For transient expression of proteins, HeLa MHVR cells were transfected with a pCAGGS-based vector (46) by using Lipofectamine (Invitrogen Life Technologies). Cells were infected 48 h later with VVΔE3L at an MOI of 30. Cells were labeled and analyzed at 6 hpi as described above. Protein products were quantified by densitometric scanning of fluorograms and analyzed using ImageQuant software (Molecular Dynamics).
Western blotting.
Cells were lysed in phosphate-buffered saline containing 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1× Complete Mini EDTA-free Protease Inhibitor Cocktail, and phosphatase inhibitor cocktails 1 and 2 (Sigma). Lysates were analyzed on 10% or 7.5% SDS-PAGE gels for eIF2α and PKR, respectively. Proteins were transferred to nitrocellulose membranes and analyzed with the appropriate antibodies specific for total eIF2α (Santa Cruz Biotechnology), phosphorylated eIF2α (Biosource), and PKR (Santa Cruz Biotechnology) and detected by chemiluminescence (Pierce).
RNA degradation.
Total RNA was isolated from cells with TRIzol reagent (Invitrogen), and 10 μg of RNA was resolved on a 1% formaldehyde gel, followed by staining with ethidium bromide.
Construction of vaccinia virus recombinants.
The coronavirus structural protein genes (N, M, S, and E) were PCR amplified from the MHV G clone (70), a cDNA that encompasses the ∼3′ one-third of the MHV A59 genome that is one of the cassettes used to construct full-length infectious clones of the virus. PCR-amplified products were subcloned into the VV insertion plasmid, pMPΔE3L. Conservative codon changes were introduced to eliminate potential vaccinia virus transcription termination sequences. All sequences were confirmed before recombination into the E3L locus. Recombination and selection of recombinant viruses were carried out as previously described (6). The inserted genes in the recombinant viruses were confirmed by PCR and sequencing following isolation of recombinant viruses. Western blotting was used to confirmed expression of the encoded proteins.
Construction of I ORF deletion mutant.
A mutant virus lacking the I ORF within the N gene was constructed by whole-plasmid PCR using high-fidelity Pfu polymerase (Stratagene). The start codon for the I ORF was eliminated by a T-to-C (underlined) change in the start codon and introduction of a stop codon by a C-to-A (underlined) change in the fourth codon with forward primer 5′-CCTCTGTAAACCGCGCTGGTAACGGAATCCTAAAGAAGACCACTTGGGC-3′ and reverse primer 5′-CGGTTTGGTCAGCCCAAGTGGTCTTCTTTAGGATTCCGTTACCAGCGCGG-3′. Mutations were confirmed by sequencing the entire insert before subcloning into the MHV G clone cassette that is a component of the MHV A59 full-length infectious clone kindly provided by Ralph Baric (University of North Carolina, Chapel Hill) (70). Full-length cDNA clones were assembled as previously described with a few modifications (64). Plasmids containing the cDNA cassettes spanning the MHV genome were purified using QIAfilter Maxi cartridges (QIAGEN) and digested with the appropriate restriction enzymes. The fragments were gel purified and ligated overnight in a reaction volume of 100 to 200 μl. Ligated DNA products were extracted with phenol-chloroform and ethanol precipitated. RNA transcripts were transcribed from the ligated full-length template using the mMessage mMachine T7 transcription reagents supplemented with additional GTP (Ambion). The MHV nucleocapsid gene was transcribed from pMHV-A59 N using T7 RNA polymerase. N gene transcripts were polyadenylated using Ambion's poly(A) tailing system. Full-length genomic transcripts were coelectroporated with the N transcripts into BHK-MHVR cells suspended at a concentration of 107 cells/ml. RNA transcripts were electroporated with three electrical pulses of 850 V at 25 μF using a Bio-Rad Gene Pulser Xcell Electroporator. Cells were seeded and incubated at 37°C. Cells were monitored for appearance of syncytium formation for ∼24 to 48 h postelectroporation.
An aliquot of the medium from electroporated cells was passaged onto L2 cells. The medium was harvested from the infected cells at approximately 24 hpi. Total RNA extracted from cells remaining on the flasks was treated with DNase I prior to being reverse transcribed using oligo(dT) as a primer. PCR was carried out using the forward primer 5′-CCACCTCTACATGCAAGGTGTTAAGC-3′ and reverse primer 5′-GGTCTGCCACAACCTTCTCTATCT-3′ to obtain N subgenomic specific products. PCR products were cleaned up and sequenced directly.
RESULTS
MHV A59 is resistant to type I IFN.
To determine if MHV A59 is sensitive to type I IFN, mouse 17Cl1 cells were pretreated with increasing amounts (0 to 104 IU) of IFN-α/β for 24 h prior to infection. Cells were then infected with MHV or IFN-sensitive VSV and analyzed by plaque assay. IFN treatment with 103 IU had no inhibitory effect on MHV (Fig. 1). Some reduction in plaques was observed at the higher (104-IU) dosage. Under the same conditions a significant reduction in VSV plaques was seen after treatment with as little as 10 IU, and a dramatic inhibitory effect was observed with 100 IU (Fig. 1). The results clearly demonstrated that MHV A59 replication is not sensitive to the action of type I IFN and suggested that the virus might interfere with the innate immune response.
FIG. 1.
Effect of IFN treatment on MHV A59 infection. Monolayers of 17Cl1 cells were treated with increasing amounts (1 to 104 units) of type I IFN 24 h before infection. Cells were infected with MHV A59 or VSV at an MOI of 5.
Protein synthesis is not significantly inhibited in MHV-infected cells.
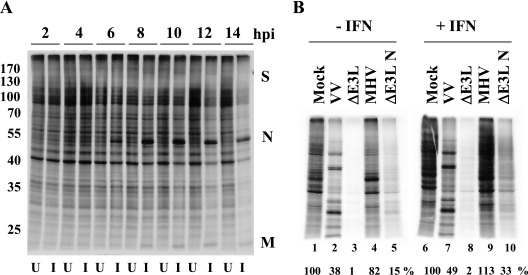
A primary antiviral response of host cells to viral infections is inhibition of translation (53). Since IFN was unable to inhibit MHV replication, this suggested that the virus could be evading antiviral responses that affect translation. We had previously observed that host protein synthesis is not significantly inhibited, at least early, during MHV A59 infection. To confirm this, MHV-infected 17Cl1 cells were labeled with Expre35S35S protein labeling mix for 30 min at different times after infection and cell extracts were analyzed by SDS-PAGE (Fig. 2A). Viral protein synthesis was clearly present by 6 hpi as indicated by the appearance of the N protein. Viral protein synthesis continued through the remainder of the time course without significant effects on global host protein synthesis until ∼12 hpi, the point at which virus replication peaks.
FIG. 2.
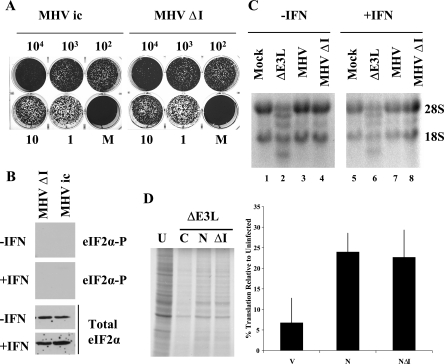
Protein synthesis in MHV A59-infected cells. (A) Time course of uninfected (U) and MHV infected (I) 17Cl1 cells. Cells were infected with WT virus at an MOI of 5. Cells were radiolabeled with [35S]methionine-cysteine for 30 min at the indicated times. Intracellular proteins were analyzed by SDS-PAGE and autoradiography. Positions of molecular weight markers in thousands (left) and of viral proteins (right) are indicated. (B) HeLa MHVR cells were mock pretreated or pretreated with IFN prior to infection with MHV, VV, VVΔE3L, or VVΔE3L N at an MOI of 5. Cells were pulse-labeled at 6 hpi for 30 min. Intracellular proteins were analyzed by SDS-PAGE and autoradiography. Proteins in each lane were quantified by densitometry and analyzed using ImageQuant software. Protein expression levels shown below each lane are expressed as the percentage of that measured for uninfected cells.
The effect of MHV infection on protein synthesis was further examined after IFN treatment. At this point we chose to compare MHV with the well-characterized type I IFN-resistant VV and the IFN-sensitive VV that lacks its viral IFN antagonist gene E3L (VVΔE3L) (7). The effect of vaccinia virus infection on protein synthesis is well characterized in HeLa cells. Thus, HeLa MHVR cells that express the MHV receptor were either mock pretreated or pretreated with IFN before infection with MHV, wild-type (WT) VV, and VVΔE3L. Cells were pulse-labeled at 6 hpi and analyzed by SDS-PAGE and autoradiography (Fig. 2B). In both the absence (lane 4) and presence (lane 9) of IFN, MHV-infected cell protein synthesis exhibited limited inhibition compared with uninfected cells (lanes 1 and 6). IFN-sensitive VVΔE3L exhibited marked inhibition of protein synthesis (lanes 3 and 8), whereas cells infected with WT VV exhibited the normal viral protein synthesis profile (lanes 2 and 7), consistent with the well established phenotypes of these viruses (6). These results confirmed that overall protein synthesis is not shut down during MHV A59 infection, with or without IFN treatment. Additional data for ΔE3L N in Fig. 2 (lanes 5 and 10) and subsequent figures will be discussed below.
PKR and eIF2α are not phosphorylated in MHV A59-infected cells.
The PKR and 2′-5′ OAS pathways are two of the major pathways activated by IFN in response to dsRNA during viral infections (53). Both result in inhibition of protein synthesis that in turn limits virus replication. Activation of PKR results in autophosphorylation and subsequent phosphorylation of eIF2α, which inhibits translation initiation. OAS activates RNase L that degrades rRNA and mRNAs, which results in inhibition of protein synthesis. During CoV replication and transcription, dsRNA intermediates are produced. Therefore, we investigated whether the ability of MHV A59 to maintain protein synthesis is due to its ability to interfere with or block the activation of either pathway. Again, the impact of MHV on the pathway was compared with those of WT VV and the VVΔE3L mutant.
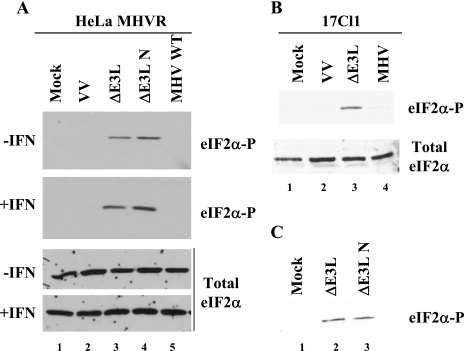
Mock- or IFN-treated HeLa MHVR and mouse 17Cl1 cells were infected with MHV, WT VV, or VVΔE3L. At 6 hpi, cell lysates were analyzed for eIF2α and PKR phosphorylation. No phosphorylated eIF2α was detected in MHV-infected HeLa MHVR (Fig. 3A, lane 5) or mouse 17Cl1 cells (Fig. 3B, lane 4) in the presence or absence of IFN treatment. N protein expression was confirmed in MHV-infected cells (data not shown). Analysis of eIF2α over a time course of up to 8 hpi revealed no change in the phosphorylation block in MHV-infected cells (data not shown). As expected, no eIF2α phosphorylation was detected in mock or WT VV infected cells (Fig. 3A and B, lanes 1 and 2). Consistent with previous studies, eIF2α was phosphorylated in VVΔE3L-infected cells (Fig. 3A and B, lane 3). Blots were stripped and analyzed for total eIF2α to confirm that comparable amounts of protein were loaded and that eIF2α was not degraded.
FIG. 3.
Detection of eIF2α phosphorylation in MHV A59- and VV-infected cells. HeLa MHVR (A) and mouse 17Cl1 (B) cells were infected with viruses as indicated above each lane. Cells were mock or IFN treated prior to infection. At 6 hpi cell lysates were analyzed for eIF2α by Western blotting with antibodies specific for the phosphorylated form of eIF2α and those that recognize both the phosphorylated and unphosphorylated forms (total eIF2α) of the protein. (C) Cell lysates from 17Cl1 cells infected with VVΔE3L or VVΔE3L N viruses were analyzed for eIF2α phosphorylation.
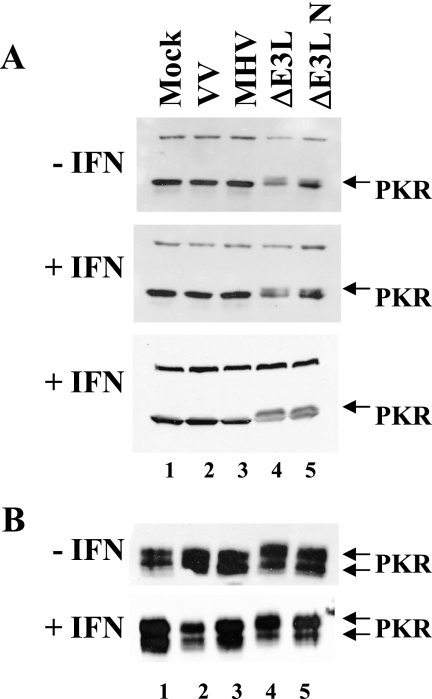
To determine whether the lack of eIF2α phosphorylation in MHV-infected cells results from lack of PKR activation, we assayed for PKR phosphorylation in both HeLa MHVR and mouse 17Cl1 cells in the presence or absence of IFN (Fig. 4). Cell lysates from mock-infected and infected cells were analyzed by SDS-PAGE, followed by Western blotting with a PKR-specific antibody. Phosphorylated PKR exhibits a slightly slower electrophoretic mobility compared with the nonphosphorylated protein when analyzed by SDS-PAGE (25, 27). No differential mobility was seen with PKR from MHV-infected HeLa MHVR cell lysates, since the protein comigrated with that from mock-infected and WT VV-infected cells (Fig. 4A, lanes 1 to 3). WT VV is known to inhibit PKR phosphorylation (6). In contrast, a slower-migrating PKR band, indicative of phosphorylation, was present in the HeLa cells infected with VVΔE3L (Fig. 4A, lane 4). The same results were seen in mouse 17Cl1 cells (Fig. 4B). However, mouse PKR consistently migrated as two bands. No decrease in gel mobility was seen with either WT VV or MHV (lanes 1 to 3), but both forms of mouse PKR exhibited slower mobility in the VVΔE3L-infected cells (lane 4). Taken together, these results demonstrate that MHV A59 fails to activate or inhibits the IFN-induced dsRNA-activated PKR pathway. The results are consistent with the lack of significant protein synthesis inhibition in the virus-infected cells. This provides further support for a mechanism by which the virus counteracts the antiviral effect of IFN.
FIG. 4.
PKR phosphorylation in MHV A59- and VV-infected cells. Immunoblot analysis of PKR phosphorylation in HeLa MHVR (A) and mouse 17Cl1 (B) cells that were pretreated or not with IFN prior to infection with the viruses indicated above each lane is shown. PKR was detected by Western blotting. The slower-migrating phosphorylated PKR is indicated by arrows. Results from two independent experiments are shown for HeLa MHVR-infected cells with IFN treatment in panel A.
MHV A59 prevents RNase L activity.
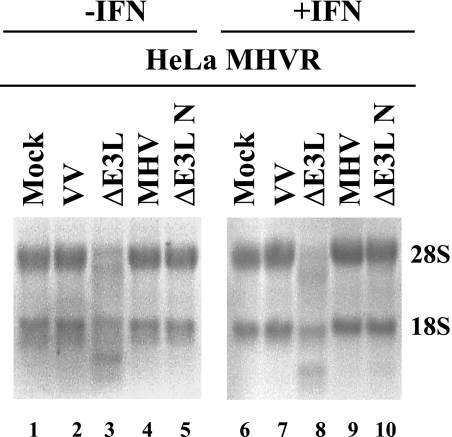
To determine if MHV interferes with the 2′-5′ OAS pathway, we monitored rRNA degradation in virus-infected cells. RNase L activation results in rRNA cleavage (67). RNA extracts were prepared from HeLa MHVR cells that were infected in both the presence and absence of IFN. Uninfected cells and cells infected with either WT VV or MHV had intact 28S and 18S rRNAs (Fig. 5, lanes 1, 2, 4, 6, 7, and 9). In contrast, RNase L-induced RNA degradation was clearly evident in VVΔE3L-infected cells in both the absence and presence of IFN, consistent with earlier observations (lanes 3 and 8) (51). These results suggest that MHV somehow blocks the IFN-induced dsRNA-dependent 2′-5′ OAS pathway. rRNA integrity was monitored over a time course of up to 24 hpi, and no significant degradation was observed at 6 and 12 hpi. Some degradation of 28S RNA was observed at 24 hpi (data not shown). The latter is consistent with the RNase L-independent cleavage of 28S rRNA observed in MHV-infected DBT cells (2), which is distinct from the RNase L activity we describe here.
FIG. 5.
Regulation of 2′-5′ OAS activity in MHV A59- and VV-infected cells. HeLa MHVR cells were infected with the viruses as indicated above each lane. Equal amounts of total RNA from cells at 6 hpi were analyzed by formaldehyde agarose gel electrophoresis and visualized by ethidium bromide staining. The positions of 18S and 28S rRNAs are indicated.
IFN antagonist activity is associated with the MHV A59 N gene.
Having determined that MHV fails to activate or interferes with both the PKR and 2′-5′ OAS pathways, it was reasonable to hypothesize that MHV might express an innate immune response antagonist. To test this idea, we made use of an approach that was previously developed and used to identify genes from other viruses that counteract the antiviral effects of IFN (6, 27, 34). VVΔE3L recombinant viruses with genes of interest recombined into the E3L locus in place of the IFN antagonist are generated. The recombinant viruses are then used to identify genes that rescue or complement the VVΔE3L IFN sensitivity phenotype.
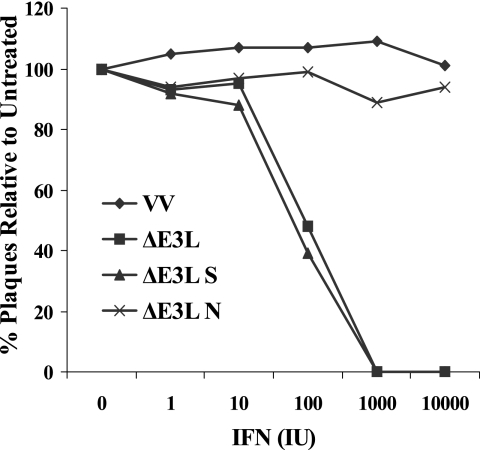
We inserted the N, M, S, and E structural genes from MHV A59 into the shuttle vector and selected for recombinant viruses. Only N and S recombinants, designated VVΔE3L-N and VVΔE3L-S, were ultimately isolated. To screen the recombinant viruses for their ability to rescue the IFN-sensitive ΔE3L phenotype, rabbit kidney RK-13 cells were pretreated with 0 to 104 IU IFN-α/β prior to being infected with VVΔE3L-N, VVΔE3L-S, parental VVΔE3L, or WT VV. VVΔE3L replicates in RK-13 cells, but the virus is sensitive to the effects of IFN in these cells. The cells do exhibit PKR activity after treatment with IFN (31). During single-step growth experiments VVΔE3L-N exhibited IFN resistance similar to that of WT VV, whereas VVΔE3L-S exhibited sensitivity to IFN like VVΔE3L (Fig. 6). A dramatic decrease in VVΔE3L and VVΔE3L-S growth was observed in the presence of only 100 IU of IFN. These results suggested that the MHV A59 N gene encodes an IFN antagonist.
FIG. 6.
IFN sensitivity of VV recombinant viruses in RK-13 cells. Monolayers of RK-13 cells were treated with increasing amounts (0 to 104 units) of type I IFN 24 h before infection. Plaque counts are expressed as a percentage of the untreated parallel control value for each virus. The data represent the averages of duplicate infections.
The MHV N gene partially rescues protein synthesis and inhibits 2′-5′ OAS/RNase L activity but does not inhibit PKR pathway activation.
To gain insight into how the N gene antagonizes IFN activity, we used the VVΔE3L-N recombinant virus to examine the effects on global protein synthesis, as well as PKR and eIF2α phosphorylation and 2′-5′ OAS/RNase L activity. All analyses were carried out in IFN-pretreated and parallel untreated cells infected with MHV A59, WT VV, VVΔE3L, and VVΔE3L-N viruses. Expression of the N protein was confirmed by Western blotting in all experiments (data not shown).
VVΔE3L-N partially rescued viral and cellular protein translation (Fig. 2B, lanes 5 and 10). This indicated that expression of the N gene likely interferes with activation of PKR and/or the 2′-5′ OAS pathways, since protein synthesis shutoff in VVΔE3L-infected cells is known to be due to activation of these pathways (6, 7, 12, 51). However, in both the absence and presence of IFN treatment, PKR phosphorylation was not inhibited during VVΔE3L-N infection (Fig. 4A and B, lane 5). This suggests that the N gene does not prevent activation of PKR. Consistent with the PKR phosphorylation results, eIF2α was phosphorylated in the VVΔE3L-N infected cells (Fig. 3C).
We then examined the status of rRNA in VVΔE3L-N infected cells. Interestingly, the RNA degradation seen in the parental VVΔE3L virus-infected cells was clearly not present in cells infected with the N recombinant virus (Fig. 5, lanes 5 and 10). Altogether, the results strongly indicate that a product(s) of the MHV N gene interferes with the 2′-5′ OAS RNase L-induced activity but that it is not able to prevent PKR phosphorylation.
The I ORF within the N gene is not responsible for the antagonist activity.
In addition to the N ORF, the N gene carries a second, internal (I) ORF. The encoded ∼23-kDa I protein is expressed from the ORF in MHV 59- as well as bovine CoV-infected cells (20, 54). To determine if the I gene product is required for the antagonist activity observed with the N gene, we used reverse genetics to construct an MHV I ORF knockout virus. The I ORF start codon was eliminated, and the fourth codon was changed to a stop codon by introduction of conservative changes that do not change the amino acids encoded in the N ORF. Recovered plaque-purified viruses were analyzed for their ability to resist type I IFN treatment and their impact on the downstream PKR and 2′-5′ OAS pathways as described above. The MHV ΔI virus exhibited resistance to IFN treatment comparable to that of the parental WT infectious clone virus (Fig. 7A). Translation factor eIF2α was not phosphorylated in cells infected with the ΔI virus (Fig. 7B). RNA degradation was not observed in cells infected with the virus, consistent with what was observed for the WT MHV and ΔE3L N recombinant viruses (Fig. 7C). This clearly demonstrated that the I ORF is not required for the antagonist activity that prevents PKR and RNase L activation during MHV infection. However, since there are possibly other viral genes or redundant processes used by the virus to antagonize these pathways, the contribution of the I ORF may be masked in the knockout virus background. To further address this, we transiently expressed the WT N and N ΔI proteins in VVΔE3L virus-infected cells. The N protein expressed from the gene lacking the I ORF rescued translation comparably to the WT N gene (Fig. 7C). This indicates that the N protein is the antagonist.
FIG. 7.
N protein is a type I IFN antagonist. (A) Monolayers of 17Cl1 cells were treated with increasing amounts (1 to 104 units) of type I IFN 24 h before infection. Cells were infected with the parental infectious clone-derived MHV A59 or ΔI virus at an MOI of 5. Plaques were stained with crystal violet at 48 hpi. (B) HeLa MHVR cells were mock or IFN treated prior to infection with MHV ΔI and WT MHV. At 6 hpi cell lysates were analyzed for eIF2α by Western blotting with antibodies specific for the phosphorylated form of eIF2α (two upper panels) and those that recognize both the phosphorylated and unphosphorylated forms of the protein for total eIF2α (lower two panels). The lanes shown were analyzed in parallel with results shown in Fig. 3A. (C) HeLa MHVR cells were infected with the viruses as indicated above each lane. Equal amounts of total RNA from cells at 6 hpi were analyzed by formaldehyde agarose gel electrophoresis and visualized by ethidium bromide staining. The positions of 18S and 28S rRNAs are indicated. (D) HeLa MHVR cells were transfected with either empty pCAGGS vector (C) or vector containing the WT N and ΔI N genes. Transfected cells were infected with VVΔE3L at 48 hpi and labeled 6 h later. Intracellular proteins were analyzed by SDS-PAGE and autoradiography. Proteins in each lane were quantified by densitometry. Measurements from three independent experiments were averaged. Error bars represent the standard deviations of the individual measurements.
DISCUSSION
The present study clearly demonstrates that MHV A59 circumvents the effects of type I IFN. Our results indicate that the virus interferes with both the IFN-induced PKR and 2′-5′ OAS pathways (Fig. 8). PKR is not phosphorylated, and the RNase L activity associated with induction of 2′-5′ OAS is not observed in MHV-infected cells. We have identified the N protein as a contributor to the resistance of MHV to type I IFN treatment. The results are significant since this is one of the first coronavirus genes to be identified that antagonizes the antiviral effects of type I IFN. While this paper was in revision, three SARS-CoV protein antagonists (ORF 3b, ORF 6, and N) that inhibit IFN-β production were reported (32). The recent report demonstrates that SARS-CoV N inhibits IFN production, while the ORF 3b and ORF 6 proteins are able to inhibit both production and interferon signaling, but the mechanisms are not known. Our results clearly demonstrate that MHV A59 N protein mediates its function by interfering with the RNase L activity associated with the induction of 2′-5′ OAS.
FIG. 8.
MHV A59 interference with type I IFN pathways. MHV A59 interferes with the downstream 2′-5′ OAS/RNase L and PKR pathways following treatment of cells with type I IFN. The N protein blocks the RNase L activity associated with 2′-5′ OAS. Recent results indicate that MHV A59 does not induce IFN-β expression (71).
Viral antagonists that modulate the innate immune response have been identified for other RNA viruses, including influenza viruses, paramyxoviruses, rhabdoviruses, filoviruses, bornaviruses, flaviviruses, and picornaviruses. Many of the viral antagonists are nonstructural proteins. Influenza virus NS1 (23), respiratory syncytial viruses NS1 and NS2 (38, 56), rotavirus NSP1 (4), and hepatitis C NSP5 (27) are examples of nonstructural proteins that modulate the innate immune response. Among the structural proteins of RNA viruses that have been implicated as IFN antagonists, only one nucleocapsid protein, the core protein of hepatitis C virus, is known at this time to play a role in immune evasion. Expression of hepatitis C virus core protein is associated with increase in suppressor of cytokine signaling 3 (SOCS-3) in cultured cells (9). Recently, induction of SOCS-1 and SOCS-3 was shown to be involved in hepatitis C virus core-gc1qR complex induction of T-cell inhibition (68).
Our results demonstrate that the N protein, not the internal ORF protein, is responsible for interference with the RNase L activity associated with the 2′-5′ OAS pathway. Essentially all of the group II coronaviruses, including a large number of MHV strains, bovine coronavirus, and human coronavirus OC43, have an internal open reading frame in the N gene. In the case of MHV A59, the I ORF is in the +1 reading frame relative to the N ORF. The ∼23-kDa I protein is expressed in virus-infected cells and is present in purified virions (20, 55), but the function of the protein is not understood. It was previously shown that expression of the I ORF is not essential for replication of MHV in tissue culture or in mice, and the mutant virus was reported to be equivalent to the parental MHV A59 virus in sensitivity to the effects of low doses of IFN, even though data were not shown for the latter (20). This suggested to us that the I ORF gene product was probably not the antagonist. Nonetheless, it was important to show that this is the case, since there is precedent for internally encoded proteins that function as IFN antagonists. The phosphoprotein (P) genes of a number of viruses in the Paramyxovirinae subfamily encode type I IFN antagonists (reviewed in reference [28]). The P protein is a cofactor of the viral polymerase and, like the coronavirus N protein, is involved in RNA replication and/or transcription. The V and C proteins are encoded within the P gene and are expressed by mRNA editing. Examination of the MHV ΔI virus lacking the ORF suggested that the I ORF product is not responsible for the interferon-resistant phenotype of MHV A59 and that the N protein is responsible for interference with the RNase L activity. The virus may have redundant processes by which it antagonizes the 2′-5′ OAS pathway which would mask any contribution of the I ORF in the MHV background. However, our results demonstrating that the N gene without the I ORF rescues translation comparably to the WT N gene indicates that the N protein is responsible for the antagonist activity.
The coronavirus N proteins are multifunctional viral gene products that encapsidate the viral genome. The protein is a major structural component of virions that plays a role in virus assembly through interactions with the viral RNA and the M protein and through N-N interactions (19, 33, 44). The protein also plays a not yet fully understood role in viral RNA synthesis. Recovery of infectious cloned coronaviruses is increased when N protein transcripts are included during RNA transfection (1, 10, 69). Recent data strongly support earlier studies by providing direct evidence that N is involved in viral RNA replication and/or transcription (52). N may also be involved in translation of viral mRNAs (62).
Coronavirus N proteins share functional and structural features, and some motifs within the proteins are conserved. All N proteins are phosphorylated and highly basic, with isoelectric points of 10.3 to 10.7 (36). However, the proteins are not highly conserved at the amino acid level. MHV A59 N protein shares 93% identity with the proteins from the closely related JHM and MHV 1 strains. The homologies between MHV A59 N and the proteins from two other well-characterized group II viruses, bovine CoV and human CoV OC43, are 68% and 69%, respectively. The homology with SARS-CoV N is 34%, similar to the 31 to 32% homology to group I transmissible gastroenteritis virus, human coronavirus 229E, and group III infectious bronchitis virus. It remains to be determined whether the N proteins from other coronaviruses function as IFN antagonists, but the expectation is that some will, especially in light of the recent identification of the SARS-CoV N antagonist.
Coronavirus N proteins bind RNA (3, 13, 42, 59). Aside from encapsidating the genome that is packaged into virions (14, 39), MHV N interacts with both genomic RNA and subgenomic mRNAs, as well as RNA intermediates containing both positive- and negative-stranded RNAs in virus-infected cells, since all of these can be coimmunoprecipitated with N-specific antibodies (3). RNA binding is not absolutely restricted to viral RNA, since N also binds non-MHV RNA transcripts when expressed in virus-infected cells and in in vitro binding assays (13, 42, 45). However, the protein binds coronavirus RNAs more efficiently than nonviral RNAs (13). Because of its RNA binding properties, it is attractive to speculate that N might exert its interference with the PKR and 2′-5′ OAS pathways through its binding to dsRNA. Both the PKR and 2′-5′ OAS pathways are activated by dsRNA.
The argument that the N protein may exert its antagonist activity through binding to dsRNA is suggested by the fact that the N gene rescues the IFN-sensitive ΔE3L vaccinia virus mutant, which lacks the E3L gene, that exerts its antagonist activity in part by binding to dsRNA (12). The MHV N gene clearly blocks the RNase L activity both in MHV-infected cells and in the VV background when replacing the E3L gene. However, expression of the N gene alone only partially restores protein synthesis and does not appear to prevent phosphorylation of PKR in the VV background. This suggests either that N does not antagonize the PKR pathway or that it may not be a sufficiently strong enough dsRNA binding protein to strongly inhibit PKR, at least in the VVΔE3L background.
Many viruses have evolved mechanisms to interfere with the function of PKR. Viral antagonists have been identified that target PKR function by interfering with dsRNA activation, preventing dimerization, blocking kinase activity or downstream substrate interactions, promoting degradation or protein expression, or interfering with downstream substrates or processes (21). PKR does not appear to be degraded in MHV-infected cells, since the amount of total PKR is not diminished. We think that it is unlikely that PKR dimerization is disrupted. PKR in MHV-infected cell lysates was heavily phosphorylated in an in vitro kinase assay, though we cannot exclude the possibility that an inhibitor(s) might have been lost during immunoprecipitation (data not shown). It is possible that PKR is phosphorylated during MHV infection but that it is dephosphorylated by a phosphatase. A phosphatase might not be coimmunoprecipitated or nonfunctional in the in vitro kinase reaction. We have coimmunoprecipitated N with PKR from virus-infected cells, but the significance of this remains to be determined.
In addition to VV, a number of other viruses have also evolved strategies to counteract the 2′-5′ OAS pathway. An RNase L inhibitor protein which down regulates the 2′-5′ OAS/RNase L pathway is induced during human immunodeficiency virus type 1 infection of T cells (40). Encephalomyocarditis virus also down regulates RNase L activity through increased expression of the RNase L inhibitor (41). Herpes simplex virus type 1 infection induces synthesis of 2′-5′ OAS derivatives that antagonize RNase L activation (11). Studies to determine the mechanism by which the N protein interferes with the pathway are under way.
One possible mechanism by which MHV may circumvents the effects of IFN is through the inaccessibility of dsRNA intermediates to activate the PKR and 2′-5′ OAS pathways during MHV infection. CoV RNA replication takes place in the cytoplasm in double-membrane vesicles that contain the viral replicase proteins and N (49). These complexes could sequester dsRNA and prevent PKR and 2′-5′ OAS activation. Since expression of the N protein alone does not inhibit phosphorylation of PKR, it is possible that another viral protein(s) is involved. One or more of the 15 or 16 replicase proteins that result from processing of the gene 1 polyprotein early in infection are potentially good candidates that may provide a second viral antagonist function that also plays a role in helping subvert the antiviral response.
Little is known about how coronaviruses affect the innate immune system. A large number of murine coronavirus isolates, variants, and subtypes exist, which exhibit different tropism and pathogenesis (for recent reviews, see references 8, 47, and 66). Some coronaviruses are known to induce IFN expression, and the type of glycosylation linkage on the M protein affects induction (5, 15). Early studies indicated that sensitivity to type I IFN is strain specific (24, 61). A recent report indicates that MHV JHM and MHV A59 do not induce expression of IFN in infected cultured cells and that IFN-β transcription factors NF-κB and IRF-3 are not activated (71). Microarray analysis suggest that NF-κB mRNA levels decreased during MHV infection (65). Minimal transcriptional activation of the PKR and 2′-5′ OAS genes was recently reported for MHV 1 (72).
SARS-CoV inhibits IFN-β expression through interference with IRF-3 activation (32, 57). The three SARS virus antagonists that were recently identified inhibit the IFN response at different levels (32). NF-κB is inhibited by the SARS N protein. The ORF 3b and ORF 6 gene products interfere with expression from an IFN-stimulated response element. ORF 6 protein inhibits STAT1 nuclear translocation.
Understanding the molecular mechanism by which coronaviruses interfere with type I IFN activity and identification of other viral parameters and/or genes that are involved are obviously important. Our results provide insight into one part of the picture, the response of MHV A59 to IFN treatment and the impact of the virus on two downstream pathways (Fig. 8). The innate immune response is complex, involving induction of IFN expression and many potential downstream signaling pathways. It is likely that MHV, as well as other coronaviruses, use a combination of mechanisms that target multiple steps to avoid the antiviral effects of the IFN system. Ultimate understanding of the overall interactions between coronaviruses and the host innate response will provide insight into the pathogenesis of the viruses. Additionally, the information may aid in the design of antiviral reagents and attenuated virus vaccines.
Acknowledgments
We thank members of the Hogue and Jacobs labs for helpful discussions and Connie Chamberlain for technical assistance.
The work was supported by Public Health Service grants AI53704 to B.G.H. and AI52347 to B.L.J.
Footnotes
Published ahead of print on 20 December 2006.
REFERENCES
- 1.Almazan, F., C. Galan, and L. Enjuanes. 2004. The nucleoprotein is required for efficient coronavirus genome replication. J. Virol. 78:12683-12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, S., S. An, A. Zhou, R. H. Silverman, and S. Makino. 2000. RNase L-independent specific 28S rRNA cleavage in murine coronavirus-infected cells. J. Virol. 74:8793-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric, R. S., G. W. Nelson, J. O. Fleming, R. J. Deans, J. G. Keck, N. Casteel, and S. A. Stohlman. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 62:4280-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. USA 102:4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beattie, E., K. L. Denzler, J. Tartaglia, M. E. Perkus, E. Paoletti, and B. L. Jacobs. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 69:499-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beattie, E., E. Paoletti, and J. Tartaglia. 1995. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L− and E3L− mutant viruses. Virology 210:254-263. [DOI] [PubMed] [Google Scholar]
- 8.Bergmann, C. C., T. E. Lane, and S. A. Stohlman. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4:121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bode, J. G., S. Ludwig, C. Ehrhardt, U. Albrecht, A. Erhardt, F. Schaper, P. C. Heinrich, and D. Haussinger. 2003. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 17:488-490. [DOI] [PubMed] [Google Scholar]
- 10.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cayley, P. J., J. A. Davies, K. G. McCullagh, and I. M. Kerr. 1984. Activation of the ppp(A2′p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2′p)nA-dependent RNase. Eur. J. Biochem. 143:165-174. [DOI] [PubMed] [Google Scholar]
- 12.Chang, H. W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cologna, R., J. F. Spagnolo, and B. G. Hogue. 2000. Identification of nucleocapsid binding sites within coronavirus-defective genomes. Virology 277:235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies, H. A., R. R. Dourmashkin, and M. R. Macnaughton. 1981. Ribonucleoprotein of avian infectious bronchitis virus. J Gen. Virol. 53:67-74. [DOI] [PubMed] [Google Scholar]
- 15.de Haan, C. A., M. de Wit, L. Kuo, C. Montalto-Morrison, B. L. Haagmans, S. R. Weiss, P. S. Masters, and P. J. Rottier. 2003. The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. Virology 312:395-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Haan, C. A., and P. J. Rottier. 2005. Molecular interactions in the assembly of coronaviruses. Adv. Virus Res. 64:165-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donnelly, C. A., A. C. Ghani, G. M. Leung, A. J. Hedley, C. Fraser, S. Riley, L. J. Abu-Raddad, L. M. Ho, T. Q. Thach, P. Chau, K. P. Chan, T. H. Lam, L. Y. Tse, T. Tsang, S. H. Liu, J. H. Kong, E. M. Lau, N. M. Ferguson, and R. M. Anderson. 2003. Epidemiological determinants of spread of causal agent of severe acute respiratory syndrome in Hong Kong. Lancet 361:1761-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer, F., D. Peng, S. T. Hingley, S. R. Weiss, and P. S. Masters. 1997. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 71:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gale, M., Jr., and M. G. Katze. 1998. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol. Ther. 78:29-46. [DOI] [PubMed] [Google Scholar]
- 22.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 24.Garlinghouse, L. E., Jr., A. L. Smith, and T. Holford. 1984. The biological relationship of mouse hepatitis virus (MHV) strains and interferon: in vitro induction and sensitivities. Arch. Virol. 82:19-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorchakov, R., E. Frolova, B. R. Williams, C. M. Rice, and I. Frolov. 2004. PKR-dependent and -independent mechanisms are involved in translational shutoff during Sindbis virus infection. J. Virol. 78:8455-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. Peiris, and L. L. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. [DOI] [PubMed] [Google Scholar]
- 27.He, Y., S. L. Tan, S. U. Tareen, S. Vijaysri, J. O. Langland, B. L. Jacobs, and M. G. Katze. 2001. Regulation of mRNA translation and cellular signaling by hepatitis C virus nonstructural protein NS5A. J. Virol. 75:5090-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengel, H., U. H. Koszinowski, and K. K. Conzelmann. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol. 26:396-401. [DOI] [PubMed] [Google Scholar]
- 29.Kahn, J. S. 2006. The widening scope of coronaviruses. Curr. Opin. Pediatr. 18:42-47. [DOI] [PubMed] [Google Scholar]
- 30.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 31.Kibler, K. V., T. Shors, K. B. Perkins, C. C. Zeman, M. P. Banaszak, J. Biesterfeldt, J. O. Langland, and B. L. Jacobs. 1997. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J. Virol. 71:1992-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. SARS coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langland, J. O., S. Pettiford, B. Jiang, and B. L. Jacobs. 1994. Products of the porcine group C rotavirus NSP3 gene bind specifically to double-stranded RNA and inhibit activation of the interferon-induced protein kinase PKR. J. Virol. 68:3821-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laude, H., and P. S. Masters. 1995. The coronavirus nucleocapsid protein, p. 141-163. In S. G. Siddell (ed.), The Coronaviridae. Plenum, New York, NY.
- 37.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 38.Lo, M. S., R. M. Brazas, and M. J. Holtzman. 2005. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 79:9315-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macneughton, M. R., and H. A. Davies. 1978. Ribonucleoprotein-like structures from coronavirus particles. J. Gen. Virol. 39:545-549. [DOI] [PubMed] [Google Scholar]
- 40.Martinand, C., C. Montavon, T. Salehzada, M. Silhol, B. Lebleu, and C. Bisbal. 1999. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J. Virol. 73:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinand, C., T. Salehzada, M. Silhol, B. Lebleu, and C. Bisbal. 1998. RNase L inhibitor (RLI) antisense constructions block partially the down regulation of the 2-5A/RNase L pathway in encephalomyocarditis-virus-(EMCV)-infected cells. Eur. J. Biochem. 254:248-255. [DOI] [PubMed] [Google Scholar]
- 42.Masters, P. S. 1992. Localization of an RNA-binding domain in the nucleocapsid protein of the coronavirus mouse hepatitis virus. Arch. Virol. 125:141-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McIntosh, K., R. K. Chao, H. E. Krause, R. Wasil, H. E. Mocega, and M. A. Mufson. 1974. Coronavirus infection in acute lower respiratory tract disease of infants. J. Infect. Dis. 130:502-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narayanan, K., K. H. Kim, and S. Makino. 2003. Characterization of N protein self-association in coronavirus ribonucleoprotein complexes. Virus Res. 98:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narayanan, K., and S. Makino. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 47.Perlman, S., and A. A. Dandekar. 2005. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5:917-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poon, L. L., D. K. Chu, K. H. Chan, O. K. Wong, T. M. Ellis, Y. H. Leung, S. K. Lau, P. C. Woo, K. Y. Suen, K. Y. Yuen, Y. Guan, and J. S. Peiris. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao, P. V., and T. M. Gallagher. 1998. Intracellular complexes of viral spike and cellular receptor accumulate during cytopathic murine coronavirus infections. J. Virol. 72:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivas, C., J. Gil, Z. Melkova, M. Esteban, and M. Diaz-Guerra. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 243:406-414. [DOI] [PubMed] [Google Scholar]
- 52.Schelle, B., N. Karl, B. Ludewig, S. G. Siddell, and V. Thiel. 2005. Selective replication of coronavirus genomes that express nucleocapsid protein. J. Virol. 79:6620-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 54.Senanayake, S. D., and D. A. Brian. 1997. Bovine coronavirus I protein synthesis follows ribosomal scanning on the bicistronic N mRNA. Virus Res. 48:101-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senanayake, S. D., M. A. Hofmann, J. L. Maki, and D. A. Brian. 1992. The nucleocapsid protein gene of bovine coronavirus is bicistronic. J. Virol. 66:5277-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78:4363-4369. (Erratum, 78:6705.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 59.Stohlman, S. A., R. S. Baric, G. N. Nelson, L. H. Soe, L. M. Welter, and R. J. Deans. 1988. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J. Virol. 62:4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sturman, L. S., and K. K. Takemoto. 1972. Enhanced growth of a murine coronavirus in transformed mouse cells. Infect. Immun. 6:501-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taguchi, F., and S. G. Siddell. 1985. Difference in sensitivity to interferon among mouse hepatitis viruses with high and low virulence for mice. Virology 147:41-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tahara, S. M., T. A. Dietlin, C. C. Bergmann, G. W. Nelson, S. Kyuwa, R. P. Anthony, and S. A. Stohlman. 1994. Coronavirus translational regulation: leader affects mRNA efficiency. Virology 202:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tartaglia, J., M. E. Perkus, J. Taylor, E. K. Norton, J. C. Audonnet, W. I. Cox, S. W. Davis, J. van der Hoeven, B. Meignier, M. Riviere, and. 1992. NYVAC: a highly attenuated strain of vaccinia virus. Virology 188:217-232. [DOI] [PubMed] [Google Scholar]
- 64.Verma, S., V. Bednar, A. Blount, and B. G. Hogue. 2006. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 80:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Versteeg, G. A., O. Slobodskaya, and W. J. Spaan. 2006. Transcriptional profiling of acute cytopathic murine hepatitis virus infection in fibroblast-like cells. J. Gen. Virol. 87:1961-1975. [DOI] [PubMed] [Google Scholar]
- 66.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69:635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wreschner, D. H., T. C. James, R. H. Silverman, and I. M. Kerr. 1981. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 9:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yao, Z. Q., S. N. Waggoner, M. W. Cruise, C. Hall, X. Xie, D. W. Oldach, and Y. S. Hahn. 2005. SOCS1 and SOCS3 are targeted by hepatitis C virus core/gC1qR ligation to inhibit T-cell function. J. Virol. 79:15417-15429. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou, H., and S. Perlman. 2007. Mouse hepatitis virus does not induce beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 81:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zorzitto, J., C. L. Galligan, J. J. Ueng, and E. N. Fish. 2006. Characterization of the antiviral effects of interferon-alpha against a SARS-like coronoavirus infection in vitro. Cell Res. 16:220-229. [DOI] [PMC free article] [PubMed] [Google Scholar]