Abstract
Papillomavirus DNA replication occurs in the nucleus of infected cells and requires the viral E1 protein, which enters the nuclei of host epithelial cells and carries out enzymatic functions required for the initiation of viral DNA replication. In this study, we investigated the pathway and regulation of the nuclear import of the E1 protein from bovine papillomavirus type 1 (BPV1). Using an in vitro binding assay, we determined that the E1 protein interacted with importins α3, α4, and α5 via its nuclear localization signal (NLS) sequence. In agreement with this result, purified E1 protein was effectively imported into the nucleus of digitonin-permeabilized HeLa cells after incubation with importin α3, α4, or α5 and other necessary import factors. We also observed that in vitro binding of E1 protein to all three α importins was significantly decreased by the introduction of pseudophosphorylation mutations in the NLS region. Consistent with the binding defect, pseudophosphorylated E1 protein failed to enter the nucleus of digitonin-permeabilized HeLa cells in vitro. Likewise, the pseudophosphorylation mutant showed aberrant intracellular localization in vivo and accumulated primarily on the nuclear envelope in transfected HeLa cells, while the corresponding alanine replacement mutant displayed the same cellular location pattern as wild-type E1 protein. Collectively, our data demonstrate that BPV1 E1 protein can be transported into the nucleus by more than one importin α and suggest that E1 phosphorylation by host cell kinases plays a regulatory role in modulating E1 nucleocytoplasmic localization. This phosphoregulation of nuclear E1 protein uptake may contribute to the coordination of viral replication with keratinocyte proliferation and differentiation.
Papillomaviruses are the etiological agents involved in several human cancers such as cervical cancer, anogenital cancer, skin cancer, and cancers of the oral cavity, the larynx, and the esophagus (68). In addition to their importance in clinical disease, papillomaviruses have provided a valuable model system for analyzing the mechanisms regulating eukaryotic DNA replication. The viral E1 protein is the largest open reading frame and is highly conserved among all papillomaviruses, maintaining its size, amino acid composition, and location in the viral genome with respect to other early genes. The E1 protein is expressed during the early stage of virus infection in order to maintain the viral DNA as an episome. The multifunctional E1 protein recognizes and binds to the viral origin of replication in combination with the viral protein E2, recruits host cell replication proteins to the origin, and initiates DNA replication via its ATP-dependent helicase activity (60, 61).
Papillomavirus infection is established in the basal layer of the epithelium, and the complex viral life cycle is coordinated with the differentiation state of the epithelium (13, 14). There are three distinct modes of viral DNA replication: (i) transient amplification, which occurs immediately upon viral infection, (ii) regulated replication for genome maintenance that occurs in cells in the lower levels of the epidermis, and (iii) vegetative replication for genome amplification, which occurs in terminally differentiated cells in the epidermis. In the latter cells, control of copy number appears to be lost, and viral DNA is amplified up to very high copy numbers (13, 15, 50). To meet these replication requirements, the E1 protein is regulated not only at the expression level (44) but also by posttranslational modifications such as sumoylation (48, 49), ubiquitination (34, 38), and phosphorylation. Bovine papillomavirus type 1 (BPV1) E1 protein is phosphorylated at multiple sites and by different kinases, including serines 48 and 584 by CKII (29, 36, 37), threonine 102 by p34cdc2 (9, 30), and serine 109 by protein kinase C (PKC) (64). Mutational analysis indicates that changes in these residues can affect viral replication, but in most cases, mechanistic details are lacking.
One possibility for the control of E1 replicative activity would be to regulate E1 levels in the nucleus. Although the molecular events involved in the nuclear accumulation of BPV1 E1 remain mostly uncharacterized, a nuclear localization signal (NLS) has been mapped to amino acids 84 to 108 (30). There are two clusters of three or four consecutive basic residues within this region, and both clusters contribute to NLS function. This NLS is both necessary and sufficient to mediate nuclear uptake of E1 or reporter proteins, indicating that no other region of E1 is mandatory for nuclear import (28, 30). Phosphorylation is a common mechanism controlling the nuclear transport of many proteins (20), and two of the known E1 phosphorylated residues, threonine 102 and serine 109, are located at the NLS, suggesting the possible regulation of nuclear uptake via E1 phosphorylation. While a mutation of threonine 102 to isoleucine was previously shown to have no observable effect on either the nuclear localization of E1 or viral DNA replication (30), a thorough examination of the contribution of phosphorylation of these residues to nuclear localization has not been performed.
Classic NLS-dependent nuclear import is a two-step process. The first step is the assembly of an importin α/β complex with the NLS of the cargo protein followed by binding to the nuclear pore complex. The second step is translocation across the membrane, where importin β is released from the complex after interaction with Ran-GTP, resulting in the dissociation of the cargo from the transport proteins (16, 33). There are six human importin α proteins (α1, α3, α4, α5, α6, and α7) that fall into three subfamilies, P (α1), Q (α3 and α4), and S (α5, α6, and α7) (19). In this study, we investigated the interaction of BPV1 E1 with the various importin α proteins and identified three importins that bound E1 and supported nuclear import in vitro. Pseudophosphorylation in the NLS region abrogated this in vitro E1-importin α interaction, prevented nuclear uptake in an in vitro import assay, and caused the mislocalization of E1 in vivo. Our results are consistent with the cytoplasmic phosphorylation of E1 being a negative regulator of nuclear entry.
MATERIALS AND METHODS
Plasmids and mutagenesis.
Plasmids pcDNA3.1-HA-E1, pRSET-E1, peGFP-E1, pGEX5X1-E11-311, and pGEX-T-E2 and the pseudophosphorylation mutants (CDK, CKII, ALL, and 1098) were all described in previous studies (48, 49, 51, 62). Targeted mutagenesis to inactivate the E1 NLS or to introduce amino acid replacements (alanines or aspartic acids) at E1 residues 102 and 109 was performed using the QuikChange XL or Multi site-directed mutagenesis kit (Stratagene, La Jolla, CA), and the sequence of each construct was confirmed by sequencing. Importin α clones (pQE70-α1, pQE60-α3, pQE60-α4, pQE60-α5, and pQE70-α7) were provided by Matthias Kohler (26), importin β1 clones (pGEX6p-1/Kapβ1) were provided by Yuh Min Chook (7), pCR3-HPV11-E1 was provided by Jacques Archambault (57), pGEX-2T-Ran was provided by Ian Macara (45), His-Com1 was provided by Guoquan Zhang (66), pGST-NLS was provided by Mark Hannink (52), and pREV1-4(NES3)GFP was provided by Beric Henderson (18).
Expression and purification of protein.
For the His fusion proteins (pQE70-α1, pQE60-α3, pQE60-α4, pQE60-α5, pQE70-α7, and His-Com1), expression was performed in cultures of Escherichia coli M15[pREP4] cells (QIAGEN, Valencia, CA) grown to an optical density at 600 nm of 0.9 and induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 25°C. Phenylmethylsulfonyl fluoride (PMSF) (5 mM) was added immediately before the culture was chilled on ice. After collection by centrifugation, the bacterial pellet was resuspended in cold lysate buffer (500 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, and 5 mM imidazole, pH 7.3) supplemented with a 1/100 volume of protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO) and 1 mg/ml lysozyme, and the suspension was sonicated. After sonication, the lysate was cleared by centrifugation at 15,000 × g for 15 min at 4°C, and the supernatant was loaded onto Ni-nitrilotriacetic acid (NTA) agarose (QIAGEN, Valencia, CA). The beads were washed four times with 10 bead volumes of wash buffer (500 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4) consecutively containing 5 mM, 20 mM, 40 mM, or 50 mM imidazole. Elution from the column was performed four times with 1 bead volume of elution buffer (500 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, and 500 mM imidazole). The eluted fractions were pooled and dialyzed overnight against 1× Tris-buffered saline (TBS) (50 mM Tris-HCl [pH 7.4], 150 mM NaCl) supplemented with 5% glycerol. After sucrose addition to a final concentration of 250 mM, His-tagged proteins were stored at −80°C. The purified the protein concentrations were determined by a Bradford assay (6).
For the glutathione S-transferase (GST) fusion proteins (pGEX5X1-E11-311, pGEX-T-E2, pGST-NLS, pGEX-2T-Ran, pGEX6p-1/β1, and pGEX-mCry2), expression was performed in cultures of E. coli Rossetta(DE3) Single cells (Novagen, San Diego, CA) grown to an optical density at 600 nm of 0.9 and then induced with 0.2 mM IPTG at 25°C for 4 h. After centrifugation, the bacterial pellet was resuspended and sonicated in cold 1× phosphate-buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.3) containing 5 mM dithiothreitol, 1 mg/ml lysozyme, and a 1/100 volume of protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Following sonication, the lysate was cleared by centrifugation at 15,000 × g for 15 min at 4°C, and the supernatant was loaded onto glutathione-Sepharose 4B beads (Amersham, Piscataway, NJ) for 3 h at 4°C. The beads were washed with 10 bead volumes of 1× PBS containing 0.2 mM PMSF, 10 bead volumes of 1× PBS containing 750 mM NaCl and 0.2 mM PMSF, 10 bead volumes of 1× PBS containing 0.2 mM PMSF, and another 10 bead volumes of 1× PBS. Each GST fusion protein was eluted three times with 1 bead volume of 100 mM Tris-HCl (pH 8.0), 120 mM NaCl, and 20 mM reduced glutathione, and the three eluates were pooled. The eluted proteins were dialyzed overnight against 1× TBS supplement with 10% glycerol, the protein concentrations were determined as described above, and the samples were stored at −80°C.
In vitro protein translation and importin binding assay.
For the importin binding assay, 35S-labeled protein was produced in coupled in vitro transcription and translation reactions. Briefly, 2 μg of E1-encoding plasmid DNA was mixed with 32 μCi of Redivue l-[35S]methionine (Amersham, Piscataway, NJ) and 25 μl of the TNT T7 Quick Coupled transcription/translation system (Promega, Madison, WI) in a final volume of 31 μl. After incubation at 30°C for 90 min, the samples were frozen at −80°C or used directly in binding assays.
For the importin α binding assay using affinity-purified His-tagged fusion proteins (His-Com1 or the five α importins), 0.5, 1, or 2 μg of the purified His-tagged proteins was incubated with 40 μl of Ni-NTA agarose (QIAGEN, Valencia, CA) and 3 μl of 35S-labeled in vitro-translated E1 in a total volume of 500 μl containing 1× PBS, 5 mM MgCl2, 0.5% Triton X-100, and 20 mM imidazole. After incubation at 4°C for 3 h, the beads were washed four times with 1× PBS supplemented with 5 mM MgCl2, 0.5% Triton X-100, and 20 mM imidazole. The bound in vitro-translated E1 proteins were eluted with 15 μl of 4× sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer (100 mM Tris-HCl [pH 6.8], 20% glycerol, 8% SDS, 0.02% bromophenol, and 4% 2-mercaptoethanol), resolved on 10% SDS-PAGE gels, and analyzed by phosphordensitometry. As a positive control for importin α7, binding was tested using GST-mCry2 as the substrate.
For importin binding assays supplemented with GST-importin β1, E1/importin α/importin β1 complexes were captured by glutathione affinity chromatography. Each binding reaction mixture contained 2 μg of the appropriate GST fusion protein (GST-importin β1 or GST), one of the His-tagged α importins (0.5, 1, or 2 μg), and 3 μl of 35S-labeled in vitro-translated E1 protein (expressed from pRSET). Reaction mixtures were incubated with 40 μl of glutathione-Sepharose 4B (Amersham, Piscataway, NJ) in a total volume of 500 μl containing 1× TBS supplemented with 0.05% Tween 20, 5 mM MgCl2, and 0.5% bovine serum albumin. After incubation at 4°C for 3 h, the beads were washed three times with 10 bead volumes of loading buffer and then washed one time with 10 bead volumes of the loading buffer without 0.5% bovine serum albumin. The bound in vitro-translated E1 proteins were eluted with 15 μl of 4× SDS-PAGE sample buffer. In vitro binding reactions for BPV1 E1 and E2 were performed as previously described (62).
Cell culture, transfection, and nuclear import assay.
HeLa cells were grown in 10% fetal bovine serum-supplemented Dulbecco's modified eagle's medium at 37°C and 5% CO2 in a humidified incubator. Transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. After transfection, E1 cellular localization was visualized by fluorescence microscopy as previously described (48). Nuclear import assays were performed as reported previously (1, 63). For the nuclear import assay, a 10 μM solution of the E1 protein was incubated with 50 μM of Alexa-555 maleimide (Molecular Probes) for 2 h at room temperature in 1× TBS. Reaction mixtures were quenched by the addition of 2-mercaptoethanol to a final concentration of 18 mM, and the fluorescently labeled E1 proteins were purified with a 30K Nanosep column (Pall, East Hills, NY) to remove free dye. Labeling efficiency for equal microgram amounts of each purified protein sample was assessed spectrophotometrically at an absorbance of 555 nm, and there was less than 10% difference between wild-type (WT) E1 and the various mutant E1 proteins. HeLa cells grown on coverslips in a 12-well plate were permeabilized with 1 ml of 40 μg/ml digitonin on ice for 2 min and incubated at room temperature for 10 min in the dark with 40 μl of an import reaction mixture containing 0.5 μM Alexa-555-labeled BPV1 E1 protein, 0.5 μM importin α, 0.5 μM importin β1, 1 mM GTP, 2 μM Ran, 1 μM NTF2 (kindly provided by Weidong Yang) (63), and 1.5% polyvinylpyrrolidone in 1× transport buffer [20 mM HEPES, 110 mM KAc, 2 mM Mg(Ac)2, 1 mM EGTA, and 5 mM NaAc]. After washing one time with 1 ml of 1× transport buffer supplemented with 1.5% polyvinylpyrrolidone, E1 cellular localization was visualized by fluorescence microscopy.
Coimmunoprecipitation.
HeLa cells (1 × 106 cells) were transfected with 10 μg of peGFP or peGFP-BPV1E1. After 24 h, cells were harvested and lysed with 800 μl of lysis buffer (0.3 M NaCl, 1% Triton X-100, 50 mM HEPES buffer, pH 8.0) containing protease inhibitor cocktail (Pierce, Rockford, IL) and phenylmethylsulfonyl fluoride. For each immunoprecipitation, 1 μg of each anti-importin α (Abcam, Cambridge, MA) and 20 μl of protein G (Santa Cruz, CA) were incubated with the cell lysate for 3 h at 4°C on a circular rotor. The beads were then pelleted, washed with 1× PBS, and eluted with SDS-sample buffer. Eluted polypeptides were resolved on a 10% SDS-polyacrylamide gel and analyzed by immunoblot analysis with anti-green fluorescent protein (GFP) (Santa Cruz, CA) and individual anti-importin antibodies.
Loading of Ran with GTP.
Purified Ran (33 μg; cleaved from GST-Ran) was incubated with guanine nucleotide exchange buffer (100 mM Tris-HCl [pH 7.5], 250 mM NaCl, 2 mM dithiothreitol, 10 mM EDTA, and 5 mM GTP) in a total volume of 66 μl on ice overnight. After loading, the reaction was stopped by adding MgCl2 to a final concentration of 20 mM. Ran-GTP was purified with a BioGel P-6 Micro Biospin chromatography column (Bio-Rad, Hercules, CA) according to the user's manual.
RESULTS
BPV1 E1 protein binds to importins α3, α4, and α5 in vitro via its nuclear localization signal but not to importins α1 and α7.
BPV1 E1 protein contains a well-defined NLS (28, 30), and such elements typically interact with importin α. Six importin α proteins have been described in humans, but no information about the relationship between different importin α proteins and the nuclear import of BPV1 E1 has been reported. To identify relevant importin α proteins, we tested complex formation between in vitro-translated E1 (pcDNA3.1-HA-E1) and His-tagged importin proteins using a pull-down assay (Fig. 1). The in vitro transcription/translation reactions produced full-length E1 and a variable amount of a 55-kDa form. As the precise sequences present in the shorter form are unknown, our conclusions are based entirely on results with full-length E1. His-tagged Com1, a bacterial protein (66), was used as the negative control for nonspecific binding. Full-length wild-type E1 bound importins α3, α4, and α5 in a dose-dependent fashion but had minimal association with importin α1 or α7 (Fig. 1B to D). Importin α6 was not examined in our studies, as its expression is restricted to the testis (24). The percentage of input E1 bound at the highest quantity of importin tested was approximately 15 to 20% for α3 to α5 and was less than 1% for α1 and α7 (Fig. 1D). The NLS dependence of these interactions was verified using an NLS-minus mutant (Fig. 1A) constructed based on previous data showing that a mutation of both clusters of basic amino acids was necessary and sufficient to eliminate E1 nuclear import (28, 30). A third cluster of basic residues (amino acids 112 to 114) adjacent to the NLS was also mutated to ensure that it would have no possible contribution to the E1-importin interaction. The NLS mutant had significantly reduced binding to importins α3, α4, and α5 (Fig. 1B), confirming that the observed E1-importin interactions were mediated via this NLS motif. NLS-dependent E1 binding to these same importins was also observed using a yeast two-hybrid system (data not shown). The low-level binding of E1 to importins α1 and α7 was not affected by the NLS mutations and likely reflects nonspecific interactions under these in vitro conditions.
FIG. 1.
Papillomavirus E1 proteins bind α importins in vitro. (A) The sequence of WT BPV1 E1 protein in the region of the NLS is shown, with the clusters of basic residues underlined. The amino acid changes present in three mutant E1 proteins (NLS−; a double pseudophosphorylation mutant, MD; and the uncharged double-alanine mutant MA) constructed for this study are indicated below the WT E1 sequence. (B) Autoradiograph of the in vitro-translated HA-E1-WT and HA-E1-NLS− proteins bound to His-Com1 (2.0 μg) or increasing amounts (0.5, 1.0, and 2.0 μg) of different importin α proteins (indicated by the hollow triangles). The input panel shows a sample of the in vitro-translated E1 proteins used for the binding assay. (C) Immunoblot of the His-importin α fusion proteins used in the pull-down assay shown in B. His-importins were detected with an anti-His antibody. (D) Quantification of the full-length WT E1 product (75 kDa) bound to His-Com1 and the different importin α proteins by phosphordensitometry. The amount of bound NLS− E1 was subtracted from the amount of bound WT E1 for Com1 and each of the five α importins tested, and the difference is presented as the percentage of the input WT E1 protein. Error bars represent standard errors obtained in two or three experiments. (E) Autoradiograph of the in vitro-translated HPV11 E1 bound to His-Com1 (2.0 μg) or increasing amounts (0.5, 1.0, and 2.0 μg) of different importin α proteins (upper panel). The assay was performed as described above for BPV1 E1. The lower panel shows the immunoblot of the importins as in C. Numbers to the left of the blots and autoradiographs in B, C, and E show the positions of molecular mass markers (in kilodaltons). (F) Immunoblot of GST-mCry2 bound to 1.0 μg of importin α7. Equal amounts of GST or GST-mCry2 were incubated with α7, the complexes were collected on Ni-NTA agarose, and the bound material was detected with anti-GST. (G) Coimmunoprecipitation of E1 with α importin. Extracts from cells expressing either eGFP or eGFP-E1 were immunoprecipitated with anti-importin α4 and then immunoblotted with anti-GFP or anti-importin α4 as indicated.
In addition to BPV1 E1, we also tested importin α binding by human papillomavirus type 11 (HPV11) E1 (Fig. 1E), which has a bipartite cluster of basic amino acids comparable to the NLS sequence of BPV1 E1. As for BPV1 E1, HPV11 E1 interacted effectively with importins α3 to α5 but not α7. Surprisingly, HPV11 E1 also bound importin α1 but to a slightly lower extent than the other importins. The significance of the importin α1 binding by HPV11 E1 is unknown and has not been investigated further. Since importin α7 bound neither BPV nor HPV E1, its functionality was assessed by testing its interaction with a known substrate, mCry2 (Fig. 1F). Importin α7 bound GST-mCry2, but not GST, indicating that the purified protein was functional yet unable to bind E1 proteins. Lastly, the ability of BPV E1 to bind importins in vivo was tested in HeLa cells. When expressed via transfection, E1 was coimmunoprecipitated with importin α4 (Fig. 1G) and importin α3 (data not shown), confirming the intracellular interaction between E1 and these importins.
BPV1 E1 is transported into the nucleus by importins α3, α4, and α5 but not by importins α1 and α7 in an in vitro nuclear import assay.
Having established that BPV1 E1 protein binds to importins α3, α4, and α5 in vitro, we examined the functionality of this interaction using an in vitro digitonin-permeabilized cell nuclear import assay. This system requires the reconstitution of the permeabilized cells with exogenous transport factors, including α importins, in order to restore nuclear uptake. The transport substrates GST-E11-311-WT, GST-E11-311-NLS−, and GST-NLS (a positive control with the simian virus 40 T-antigen NLS) were expressed in E. coli cells, and the affinity-purified fusion proteins are shown in Fig. 2A. The truncated GST-E11-311 form of E1 was used in this assay, since this fragment is more easily expressed and purified than full-length E1, it contains the NLS sequence for nuclear transport, it is conformationally intact, as indicated by its functionality for binding origin DNA and the E2 protein (27), and the presence of the GST moiety results in a fusion protein similar in size to that of full-length E1. All the purified fusion proteins were labeled with Alexa-555 for monitoring by fluorescence microscopy.
FIG. 2.
The BPV1 E1 protein is imported into the nuclei of permeabilized cells by importins α3, α4, and α5 but not by importins α1 and α7. (A) Coomassie blue staining of 0.5 μg of the purified GST-NLS, GST-E11-311-WT, and GST-E11-311-NLS− proteins used for the in vitro uptake assay. The positions of molecular mass markers (in kilodaltons) are indicated on the left side of each panel. (B) Nuclear import assay of the fluorescence-labeled GST-NLS fusion protein (positive control) tested in the absence or presence of different importin α proteins as indicated. The assay was performed as described in Materials and Methods, and the labeled GST-NLS protein was detected by fluorescence microscopy. (C) Nuclear import assay as in B with fluorescence-labeled GST-E11-311-WT and GST-E11-311-NLS−.
In the presence of importin α, the control GST-NLS protein was efficiently imported into the nucleus (Fig. 2B). In contrast, nuclear fluorescence was not observed when importin α was omitted from the reconstitution mixture, confirming the specificity of this system. Similarly, GST-E11-311-WT protein was not localized to the nucleus in the absence of an importin α, but a nuclear fluorescence signal was readily detected in the presence of importin α3, α4, or α5 (Fig. 2C). The GST-E11-311-NLS− protein, which was unable to bind any of the importins, was not imported. Furthermore, the nuclear signal was greatly reduced for GST-E11-311-WT when assayed with either importin α1 or α7, both of which exhibited only low levels of nonspecific binding to E1. Thus, the binding and nuclear uptake results are in complete concordance and indicate that the complexes formed between E1 and importins α3, α4, and α5 are fully functional for NLS-dependent nuclear import.
Pseudophosphorylation inhibits the interaction between BPV1 E1 and importin α.
The presence of two known phosphorylated residues, threonine 102 and serine 109, in the vicinity of the BPV1 E1 NLS raised the possibility that phosphorylation might regulate the interaction between E1 and importin α. To address this possibility, a double pseudophosphorylation mutant (E1-MD) was constructed by converting both threonine 102 and serine 109 to aspartic acid. A second double-replacement mutant containing an uncharged alanine at positions 102 and 109 (E1-MA) was also constructed (Fig. 1A). In vitro-translated E1-WT, E1-MA, and E1-MD proteins were assessed for importin α interactions with a variation of the pull-down binding assay described in the legend of Fig. 1. It is known that α importins are partially autoinhibited for substrate binding and that this autoinhibition is relieved by conformational changes that occur upon importin β1 binding (23). Endogenous importin β1 present in the reticulocyte lysate used for in vitro translation of the E1 proteins was presumably sufficient to allow the effective binding seen in Fig. 1, but to ensure a completely adequate supply of β1 in this experiment, we supplemented the E1 and importin α reaction mixtures with purified GST-importin β1 and captured the complexes on glutathione beads. No significant differences were observed in binding assays performed in the absence or presence of exogenous importin β1.
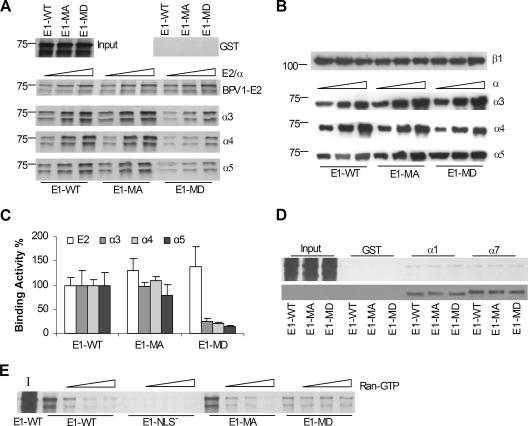
Under the above-described conditions, E1-WT exhibited dose-dependent binding to importins α3, α4, and α5, similar to what was observed in the absence of exogenous importin β (Fig. 3A). The binding of the E1-MA mutant protein to the importins was comparable to that of E1-WT (Fig. 3A and C), indicating that neither threonine 102 nor serine 109 was critical for E1-importin interactions. In contrast, the pseudophosphorylation mutant E1-MD showed greatly reduced interactions with all three α importins. Quantitation by phosphordensitometry indicated that the E1-MD mutant had only 20 to 25% of wild-type binding activity. It is unlikely that the reduced interaction of the E1-MD protein with the α importins was due to a global conformational defect, since E1-MD bound E2 protein just as effectively as E1-WT and E1-MA (Fig. 3A and C). None of the three E1 proteins showed any significant binding to control GST protein (Fig. 3A). Overall, these results implicate phosphorylation as a negative regulator of E1-importin α interactions.
FIG. 3.
Pseudophosphorylation of the BPV1 E1 protein at T102 and S109 reduces its binding activity to importin α proteins in vitro. (A) Autoradiograph of the in vitro-translated His-E1-WT, His-E1-MA, and His-E1-MD proteins (expressed from pRSET) bound to α importins in the presence of GST (upper right panel) or GST-importin β1 (α3, α4, and α5 panels) or to GST-E2 alone (BPV1-E2 panel) as described in Materials and Methods. Hollow triangles indicate increasing amounts of α importins or BPV1 E2 used in the binding reaction mixtures (0.5, 1.0, and 2.0 μg). The input panel (upper left) shows a sample of the in vitro-translated WT and mutant E1 proteins. The position of a 75-kDa molecular mass marker is indicated to the left. (B) Immunoblots of importins α and β1 used in pull-down assay shown in A. Proteins were detected with anti-His (α importins) or anti-GST (β importin) antibody. (C) Phosphordensitometric quantification of the full-length E1 product (75 kDa) bound to α importins in the presence of importin β1 or bound to BPV1 E2 protein. Data shown in the graph are derived from binding reactions shown in A using 2.0 μg of wild-type or mutant E1 proteins. The binding activities of the E1-MA and E1-MD mutants are relative to that of the WT BPV1 E1 protein, which was assigned as 100%. Error bars represent standard errors obtained in two or three experiments. (D) The upper panel is an autoradiograph of in vitro-translated WT or mutant E1 protein bound to complexes of α1/GST-β1, α7/GST-β1, or GST alone. The input lanes show portions of the original in vitro translation reaction for the WT, MA, and MD E1 proteins as indicated. The lower panel is an immunoblot of α importins used in the pull-down assay shown in the upper panel. The importins were detected with anti-His antibody. (E) The E1 binding assay was performed with WT and mutant E1 proteins as in A except with the inclusion of 0.5, 1.0, or 2.0 μg of Ran-GTP in the reaction mixture, as indicated by the hollow triangles. The lane marked I shows the input WT E1 protein.
Unlike importins α3, α4, and α5, importins α1 and α7 did not interact effectively with unmodified BPV1 E1 protein. Consequently, it is possible that importins α1 and α7 might require an NLS modification, such as phosphorylation, to facilitate their interaction with E1. However, the E1-MD pseudophosphorylation mutant showed no increased binding to either of these importins compared to E1-WT or E1-MA (Fig. 3D). While a positive regulatory role for posttranslational modifications at other amino acids cannot be ruled out from this experiment, phosphorylation at residues 102 and 109 is unlikely to be involved in targeting importin α1 or α7 for the E1 interaction.
In the nucleus, importin β1 is released from the cargo/importin complex after an interaction with Ran-GTP (16, 33). As a further measure of the authenticity of our E1/α importin/β importin complexes, we sought to test if Ran-GTP had the same releasing activity in the in vitro binding assay. As shown for importin α5 in Fig. 3E, the interaction between importin β1 and the importin α5/E1-WT or E1-MA complexes was reduced in the presence of increasing amounts of Ran-GTP, as expected. However, Ran-GTP did not affect the low level of binding seen with the E1-MD complex, suggesting that this binding does not reflect an authentically assembled transport complex. Similar results were obtained with α3 and α4 (data not shown).
Pseudophosphorylation regulates E1 nuclear import both in vitro and in vivo.
Based on our results showing that pseudophosphorylation inhibited the interaction between BPV1 E1 and importin α, we attempted to determine if this inhibition regulates E1 nuclear localization. For in vitro nuclear import studies, the MA and MD mutations were constructed into the pGEX5X1-E11-311 backbone for expression in E. coli cells and subsequent purification. Purified recombinant proteins GST-E11-311-WT, GST-E11-311-MA, and GST-E11-311-MD are shown in Fig. 4B. When added to the permeabilized cells, the GST-E11-311-MA protein accumulated in the nucleus as effectively as wild-type GST-E11-311 protein when incubated with importin α3, α4, or α5 (Fig. 4A). These results confirm that neither threonine 102 nor serine 109 is intrinsically critical for nuclear transport of the E1 protein. In contrast to the GST-E11-311-MA results, none of these three importin α proteins could transport GST-E11-311-MD into the nucleus, consistent with the conclusion that the reduced importin binding due to the pseudophosphorylation of E1 was sufficient to prevent the nuclear localization of E1.
FIG. 4.
Pseudophosphorylation of BPV1 E1 at T102 and S109 affects BPV1 E1 protein nuclear import. (A) Nuclear uptake of GST-E11-311-WT, GST-E11-311-MA, and GST-E11-311-MD in permeabilized HeLa cells supplemented with importins α3, α4, and α5 as indicated. The assay was performed as described in the legend of Fig. 2, and the localization of the E1 protein was detected by fluorescence microscopy. (B) Coomassie blue staining of 0.5 μg of GST-E11-311-WT, GST-E11-311-MA, and GST-E11-311-MD proteins used in the in vitro import assay, with the molecular mass markers (in kilodaltons) indicated on the left. (C) HeLa cells were transfected with peGFP-E1-WT, peGFP-E1-MA, peGFP-E1-MD, and pREV1-4(NES3)GFP (positive control). The cellular localization of the E1 protein was detected 24 h after transfection by fluorescence microscopy for the eGFP signal.
To examine the pseudophosphorylation phenotype in vivo, we developed enhanced GFP (eGFP)-E1 fusions for the overexpression of wild-type, MA, and MD E1 proteins in HeLa cells. The pREV1-4(NES3)GFP vector served as a positive control expressing a predominantly nuclear protein. After transfection, the eGFP-E1-WT protein showed a cellular localization pattern consisting of a diffuse nuclear signal and considerable cytoplasmic fluorescence (Fig. 4C). The degree of cytoplasmic signal was somewhat unexpected but appears to be due to a high level of expression from this vector and nuclear export signal-dependent active nuclear export of BPV1 E1 protein (our unpublished data). Nonetheless, the MA mutant was indistinguishable from wild-type E1 in its localization, again confirming that there is no intrinsic requirement for threonine and serine at positions 102 and 109, respectively, in order for E1 to be transported into the nucleus. However, the MD mutant gave a completely different pattern, with accumulation on the nuclear envelope of the transfected cells (Fig. 4C), indicating that the pseudophosphorylation of these two residues disrupted the normal nuclear localization of E1. The mechanistic basis for this aberrant localization pattern is unclear, but perhaps the Ran-GTP-unresponsive importin β1/importin α/E1-MD complex seen in Fig. 3E also forms in vivo and becomes dsyfunctionally associated with the nuclear pores. Alternatively, the failure to observe nuclear envelope accumulation of E1 in vitro may simply reflect a limitation in the in vitro assay system. The in vitro system was reconstituted with the minimal components required for import and likely lacks additional factors that contribute to overall transport regulation in vivo. For example, our reconstituted import system does not support nuclear export, and we have recently determined that BPV1 E1 contains a Crm1-dependent nuclear export signal sequence (G. Rosas-Acosta et al., unpublished data). Regardless of the mechanism involved, these results are consistent with authentic phosphorylation being a negative regulator of E1 nuclear import. Whether or not the aberrant distribution of the MD mutant E1 protein would be ameliorated in the presence of other viral proteins, such as E2, has not been tested.
Effect of other possible modifications on BPV1 E1 nuclear import.
Besides threonine 102 and serine 109 located at the NLS sequence, the BPV1 E1 protein is phosphorylated or predicted to be phosphorylated at several additional amino acid residues by the action of different host kinases (61). There are two reported CKII sites, serines 48 and 584 (29, 36, 37), and two putative CDK sites, threonine 126 and serine 283, in addition to the known CDK site at threonine 102. To test if phosphorylation at these other four sites might regulate E1 nuclear import, we examined the importin α binding capacities of three additional sets of pseudophosphorylation mutants: CKII (S48D/S584D), CDK (T102D/T126D/S283D), and ALL (S48D/T102D/T126D/S283D/S584D) (51). As shown in Fig. 5, the importin α5 binding activities of the pseudophosphorylation mutants CKII, CDK, and ALL were 107%, 76%, and 16% of those of wild-type E1, respectively. From these results, we conclude that phosphorylation at the CKII sites is unlikely to contribute to the regulation of E1 nuclear uptake. The modest decrease in binding activity seen with the CDK mutant, which includes threonine 102, suggested that phosphorylation at the CDK sites is relatively unimportant for importin binding and that serine 109 phosphorylation might be the major contributor to E1-importin α interactions. To assess this possibility, a single pseudophosphorylation mutant at serine 109 (designated 1098) was tested. The 1098 mutation reduced importin α binding to 36% of wild-type levels (Fig. 5C) compared to the double pseudophosphorylation mutant E1-MD, which reduced the binding activity to 25% for importin α5 (Fig. 3C). We conclude from these combined results that phosphorylation at threonine 102 and other CDK sites would have only a minor effect on importin binding, whereas serine 109 phosphorylation would significantly reduce importin binding and the subsequent transport of E1 into the nucleus. The somewhat more severe binding defect observed for the ALL mutant (16% of wild-type binding versus 36% for 1098) may indicate modest cumulative contributions of the other pseudophosphorylations combined with the S109D effect. Alternatively, the extensive mutational burden in the ALL mutant protein may simply result in structural perturbations that further reduce the ability of E1 to bind importins rather than a specific contribution through the introduced charged residues.
FIG. 5.
Examination of other possible posttranslational modification sites of BPV1 E1 for involvement in E1 binding to importin α. Wild-type E1 and the four pseudophosphorylation mutants were all expressed in vitro from pRSET. (A) Autoradiograph of the in vitro-translated WT and mutant E1 proteins bound to importin β1/α5 complexes or to GST alone as described in the legend of Fig. 3. Hollow triangles indicate increasing amounts of importin α5 (0.5, 1.0, and 2.0 μg) used in the binding reaction mixtures (for GST, only the results with 2 μg are shown). The input lanes show samples of the original in vitro-translated WT and mutant E1 proteins used for the binding reactions. Two independent experiments (upper and lower panels) are shown, with the position of the 75-kDa molecular mass marker indicated on the left. (B) Phosphordensitometry quantification of the bound WT and mutant E1 proteins calculated as described in the legend of Fig. 3C. (C) Tabular presentation of the quantitative data shown graphically in B.
DISCUSSION
The location and sequence features of the BPV1 E1 NLS are well defined (28, 30, 56); however, the molecular events involved in the nuclear transport of E1 remain mostly undefined. For the first time, we demonstrate that BPV1 E1 interacts with multiple α importins and that this interaction is functional for in vitro nuclear import, suggesting that the classical importin α/β pathway serves to transport E1 in vivo as well (Fig. 1 and 2). Utilization of the α/β transport pathway by E1 is consistent with previous observations that several other papillomavirus proteins, including L1 (HPV11, HPV16, and HPV45), L2 (HPV16), and E6 (HPV16), also enter the nucleus via the importin α1/β1 pathway (11, 32, 39, 41-43). In addition, we found that HPV11 and HPV16 E2 proteins, which both contain a well-characterized NLS (4, 55, 67), also displayed in vitro binding to importins α3 and α5 (unpublished data). Thus, it appears likely that papillomaviruses have evolved to utilize the classical NLS-dependent α/β transport pathway as a general mechanism for nuclear entry of the larger viral proteins. However, it is also clear that some papillomavirus proteins can enter the nucleus via importin α-independent pathways, such as direct binding to β importins for HPV16 L2 (11) or E6 (32), or a nonclassical Ran-dependent pathway for HPV16 E7 (3). BPV1 E1 was unable to be imported into the nuclei of permeabilized cells by β importins alone (unpublished data), supporting an obligate role for α importins in E1 nuclear transport, but further in vivo study will be required to fully eliminate other possible entry mechanisms.
The importin α family is encoded by six genes in humans, namely, importin α1 (Rch1 or KPNA2) (10, 59), importin α3 (Qip1 or KPNA4) (24, 53), importin α4 (hSRP1γ or KPNA3) (24, 40), importin α5 (hDRP1 or KPNA5) (8), importin α6 (24), and importin α7 (26). These six importin α proteins differ in their primary structures, substrate-specific import efficiencies, and tissue/cell-specific expression patterns (24, 26, 40) yet can be grouped into three subfamilies based on sequence similarities: α-P (importin α1), α-Q (importins α3 and α4), and α-S (importins α5 to α7) (19). Expression of importin α6 is restricted to the testis (24), while the other five α importins are widely expressed in most tissues and cell types, including keratinocytes (21, 25, 58; our unpublished observations). Our results indicate that BPV1 E1 bound to members of both the α-Q (α3 and α4) and α-S (α5) families but not to members of the α-P family. The significance of this binding pattern is not yet known but may reflect the biology of importin α expression in the epithelium. Several recent studies have begun to examine the relationship between importin expression and cell proliferation or differentiation. Levels of importin α proteins appear to be coordinated with cellular proliferation, and the down-regulation of importin α3, α5, or α7 inhibits HeLa cell proliferation, while the down-regulation of importins α1 and α4 had only a minor or no effect (47). In two different cell culture models of differentiation, levels of importins α1, α4, and α7 were strongly decreased during differentiation, while α3 and α5 levels were stable or upregulated (25). These observations have not yet been extended to stratified keratinocytes but suggest that intracellular levels of different α importins could vary widely across the epithelial layers. The effective binding and transport of E1 protein by multiple α importins would provide efficient nuclear entry irrespective of the differentiation stage of the cell. Additionally, there could be regulatory implications for viral genome replication if differentiation-dependent changes in importin α levels influence the overall abundance of nuclear E1 protein.
Many proteins contain a phosphorylation site within or adjacent to their NLS sequence that contributes to the regulation of nuclear localization. For example, the phosphorylation of serine 385 near the EBNA-1 NLS up-regulates nuclear transport efficiency by increasing the binding affinity of EBNA-1 for NPI-1 (importin α5), while the phosphorylation of serine 386 and serine 383 down-regulates uptake (22). Numerous other studies have also shown that phosphorylation near or within NLS elements can regulate nuclear transport in either a positive (STAT1 [35] or cytomegalovirus ppUL44 [2]) or negative (p27Kip1 [54], Swi6p [17], or APC [65]) fashion. Our results demonstrate that the binding of BPV1 E1 protein to α importins was significantly decreased when a pseudophosphorylation mutation was introduced at serine 109. Furthermore, the pseudophosphorylation mutant E1 protein failed to enter the nucleus of digitonin-permeabilized HeLa cells in vitro and showed aberrant nuclear envelope accumulation in transfected HeLa cells. In contrast, an alanine replacement at serine 109 had no effect on importin binding or E1 nuclear uptake. Why the pseudophosphorylation mutant exhibits this pattern of nuclear envelope association as opposed to simply cytoplasmic distribution is unclear but likely reflects a more complex interplay between E1 and nucleocytoplasmic transport systems than is present in our reconstituted in vitro system. Nonetheless, these results imply that E1 phosphorylation at serine 109 in vivo would negatively regulate E1 nuclear uptake and are consistent with a previous study that examined the replication function of serine 109 mutants (64). That study found that a BPV1 viral genome carrying the E1 S109A mutation replicated to slightly higher levels than the WT genome in an in vivo transient assay, while an E1 S109E mutant genome replicated less effectively than the WT. At least a portion of the replicative defect in the S109E mutant might have resulted from the pseudophosphorylation mutation interfering with importin α-dependent E1 nuclear uptake, resulting in lower intranuclear E1 levels than those with WT E1. Conversely, the S109A mutant that would be constitutively unphosphorylated at this residue may accumulate to higher intranuclear concentrations, leading to increased genome replication.
The BPV1 E1 serine 109 is located within a consensus sequence for PKC (31) and can be phosphorylated by this enzyme in vitro (64). PKC is known to comprise a large family of at least 12 isozymes that differ in structures, cofactor requirements, and functions. Multiple PKC isoforms are expressed in the epidermis and make different contributions to keratinocyte proliferation, differentiation, and apoptotic death (12). In the proliferative basal layer, highly levels of PKCɛ are detected, suggesting that PKCɛ may help drive keratinocyte proliferation in the basal layer, while PKCα, δ, and η are associated with differentiation (12). The PKC pathway is also an important regulator of differentiation-dependent HPV31 replication, and viral genome amplification depends on both PKCα and PKCδ activity as well as tyrosine kinases (5). Based on these observations, it is interesting to speculate that diverse PKC isoforms present in the epithelial layers could have a differential capacity for phosphorylating E1 residue 109 and that this could provide control of E1 nuclear levels by modulating E1-importin α interactions and the subsequent E1 uptake, thus coordinating viral genome replication with a keratinocyte differentiation stage.
After the completion of most of our study, a new phosphorylation map of the BPV1 E1 protein showing four additional phosphorylation sites adjacent to or within NLS sequence was reported (32). Therefore, there are now seven total known phosphorylation sites in the immediate vicinity of the NLS sequence (S90, S94, S95, S100, T102, S109, and T126), suggesting that the phosphoregulation of BPV1 E1 nuclear import may be exceedingly complex. While we have shown that the pseudophosphorylation of residues T102 and T126 makes at best a small contribution to E1-importin interactions in the context of S109D, all three of these residues must now be reevaluated in conjunction with S90, S94, S95, and S100. Residues S94, S95, and S100 are all predicted to be phosphorylated by CKII, which is also known to be involved in the regulation of keratinocyte proliferation (46).
Acknowledgments
We thank Weidong Yang and Siegfried M. Musser for help with the nuclear import assay.
This work was supported by a grant from the National Cancer Institute (RO1 CA089298) to V.G.W.
Footnotes
Published ahead of print on 27 December 2006.
REFERENCES
- 1.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvisi, G., D. A. Jans, J. Guo, L. A. Pinna, and A. Ripalti. 2005. A protein kinase CK2 site flanking the nuclear targeting signal enhances nuclear transport of human cytomegalovirus ppUL44. Traffic 6:1002-1013. [DOI] [PubMed] [Google Scholar]
- 3.Angeline, M., E. Merle, and J. Moroianu. 2003. The E7 oncoprotein of high-risk human papillomavirus type 16 enters the nucleus via a nonclassical Ran-dependent pathway. Virology 317:13-23. [DOI] [PubMed] [Google Scholar]
- 4.Blachon, S., S. Bellanger, C. Demeret, and F. Thierry. 2005. Nucleo-cytoplasmic shuttling of high risk human papillomavirus E2 proteins induces apoptosis. J. Biol. Chem. 280:36088-36098. [DOI] [PubMed] [Google Scholar]
- 5.Bodily, J. M., S. Alam, and C. Meyers. 2006. Regulation of human papillomavirus type 31 late promoter activation and genome amplification by protein kinase C. Virology 348:328-340. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Chook, Y. M., A. Jung, M. K. Rosen, and G. Blobel. 2002. Uncoupling Kapbeta2 substrate dissociation and ran binding. Biochemistry 41:6955-6966. [DOI] [PubMed] [Google Scholar]
- 8.Cortes, P., Z. S. Ye, and D. Baltimore. 1994. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl. Acad. Sci. USA 91:7633-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cueille, N., R. Nougarede, F. Mechali, M. Philippe, and C. Bonne-Andrea. 1998. Functional interaction between the bovine papillomavirus virus type 1 replicative helicase E1 and cyclin E-Cdk2. J. Virol. 72:7255-7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuomo, C. A., S. A. Kirch, J. Gyuris, R. Brent, and M. A. Oettinger. 1994. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl. Acad. Sci. USA 91:6156-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darshan, M. S., J. Lucchi, E. Harding, and J. Moroianu. 2004. The L2 minor capsid protein of human papillomavirus type 16 interacts with a network of nuclear import receptors. J. Virol. 78:12179-12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning, M. F. 2004. Epidermal keratinocytes: regulation of multiple cell phenotypes by multiple protein kinase C isoforms. Int. J. Biochem. Cell Biol. 36:1141-1146. [DOI] [PubMed] [Google Scholar]
- 13.Doorbar, J. 2005. The papillomavirus life cycle. J. Clin. Virol. 32(Suppl. 1):S7-S15. [DOI] [PubMed] [Google Scholar]
- 14.Flores, E. R., and P. F. Lambert. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 71:7167-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, D. M., and S. N. Cohen. 1987. Bovine papilloma virus plasmids replicate randomly in mouse fibroblasts throughout S phase of the cell cycle. Cell 50:59-68. [DOI] [PubMed] [Google Scholar]
- 16.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 17.Harreman, M. T., T. M. Kline, H. G. Milford, M. B. Harben, A. E. Hodel, and A. H. Corbett. 2004. Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J. Biol. Chem. 279:20613-20621. [DOI] [PubMed] [Google Scholar]
- 18.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 19.Hogarth, C. A., S. Calanni, D. A. Jans, and K. L. Loveland. 2006. Importin alpha mRNAs have distinct expression profiles during spermatogenesis. Dev. Dyn. 235:253-262. [DOI] [PubMed] [Google Scholar]
- 20.Jans, D. A., and S. Hubner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651-685. [DOI] [PubMed] [Google Scholar]
- 21.Kamei, Y., S. Yuba, T. Nakayama, and Y. Yoneda. 1999. Three distinct classes of the alpha-subunit of the nuclear pore-targeting complex (importin-alpha) are differentially expressed in adult mouse tissues. J. Histochem. Cytochem. 47:363-372. [DOI] [PubMed] [Google Scholar]
- 22.Kitamura, R., T. Sekimoto, S. Ito, S. Harada, H. Yamagata, H. Masai, Y. Yoneda, and K. Yanagi. 2006. Nuclear import of Epstein-Barr virus nuclear antigen 1 mediated by NPI-1 (importin α5) is up- and down-regulated by phosphorylation of the nuclear localization signal for which Lys379 and Arg380 are essential. J. Virol. 80:1979-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobe, B. 1999. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 6:388-397. [DOI] [PubMed] [Google Scholar]
- 24.Kohler, M., S. Ansieau, S. Prehn, A. Leutz, H. Haller, and E. Hartmann. 1997. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417:104-108. [DOI] [PubMed] [Google Scholar]
- 25.Kohler, M., A. Fiebeler, M. Hartwig, S. Thiel, S. Prehn, R. Kettritz, F. C. Luft, and E. Hartmann. 2002. Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cell. Physiol. Biochem. 12:335-344. [DOI] [PubMed] [Google Scholar]
- 26.Kohler, M., C. Speck, M. Christiansen, F. R. Bischoff, S. Prehn, H. Haller, D. Gorlich, and E. Hartmann. 1999. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 19:7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leng, X., J. H. Ludes-Meyers, and V. G. Wilson. 1997. Isolation of an amino-terminal region of bovine papillomavirus type 1 E1 protein that retains origin binding and E2 interaction capacity. J. Virol. 71:848-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leng, X., and V. G. Wilson. 1994. Genetically defined nuclear localization signal sequence of bovine papillomavirus E1 protein is necessary and sufficient for the nuclear localization of E1-beta-galactosidase fusion proteins. J. Gen. Virol. 75:2463-2467. [DOI] [PubMed] [Google Scholar]
- 29.Lentz, M., T. Zanardi, R. Filzen, J. Carter, and M. Hella. 2002. Functional analysis of a carboxyl-terminal phosphorylation mutant of the bovine papillomavirus E1 protein. J. Mol. Biol. 316:599-609. [DOI] [PubMed] [Google Scholar]
- 30.Lentz, M. R., D. Pak, I. Mohr, and M. R. Botchan. 1993. The E1 replication protein of bovine papillomavirus type 1 contains an extended nuclear localization signal that includes a p34cdc2 phosphorylation site. J. Virol. 67:1414-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lentz, M. R., S. M. Stevens, Jr., J. Raynes, and N. Elkhoury. 2006. A phosphorylation map of the bovine papillomavirus E1 helicase. Virol. J. 3:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Roux, L. G., and J. Moroianu. 2003. Nuclear entry of high-risk human papillomavirus type 16 E6 oncoprotein occurs via several pathways. J. Virol. 77:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malcles, M. H., N. Cueille, F. Mechali, O. Coux, and C. Bonne-Andrea. 2002. Regulation of bovine papillomavirus replicative helicase E1 by the ubiquitin-proteasome pathway. J. Virol. 76:11350-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride, K. M., G. Banninger, C. McDonald, and N. C. Reich. 2002. Regulated nuclear import of the STAT1 transcription factor by direct binding of importin-alpha. EMBO. J. 21:1754-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McShan, G. D., and V. G. Wilson. 1997. Casein kinase II phosphorylates bovine papillomavirus type 1 E1 in vitro at a conserved motif. J. Gen. Virol. 78:171-177. [DOI] [PubMed] [Google Scholar]
- 37.McShan, G. D., and V. G. Wilson. 2000. Contribution of bovine papillomavirus type 1 E1 protein residue 48 to replication function. J. Gen. Virol. 81:1995-2004. [DOI] [PubMed] [Google Scholar]
- 38.Mechali, F., C. Y. Hsu, A. Castro, T. Lorca, and C. Bonne-Andrea. 2004. Bovine papillomavirus replicative helicase E1 is a target of the ubiquitin ligase APC. J. Virol. 78:2615-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merle, E., R. C. Rose, L. LeRoux, and J. Moroianu. 1999. Nuclear import of HPV11 L1 capsid protein is mediated by karyopherin alpha2beta1 heterodimers. J. Cell. Biochem. 74:628-637. [PubMed] [Google Scholar]
- 40.Nachury, M. V., U. W. Ryder, A. I. Lamond, and K. Weis. 1998. Cloning and characterization of hSRP1 gamma, a tissue-specific nuclear transport factor. Proc. Natl. Acad. Sci. USA 95:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson, L. M., R. C. Rose, L. LeRoux, C. Lane, K. Bruya, and J. Moroianu. 2000. Nuclear import and DNA binding of human papillomavirus type 45 L1 capsid protein. J. Cell. Biochem. 79:225-238. [PubMed] [Google Scholar]
- 42.Nelson, L. M., R. C. Rose, and J. Moroianu. 2003. The L1 major capsid protein of human papillomavirus type 11 interacts with Kap beta2 and Kap beta3 nuclear import receptors. Virology 306:162-169. [DOI] [PubMed] [Google Scholar]
- 43.Nelson, L. M., R. C. Rose, and J. Moroianu. 2002. Nuclear import strategies of high risk HPV16 L1 major capsid protein. J. Biol. Chem. 277:23958-23964. [DOI] [PubMed] [Google Scholar]
- 44.Ozbun, M. A., and C. Meyers. 1998. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology 248:218-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plafker, K., and I. G. Macara. 2002. Fluorescence resonance energy transfer biosensors that detect Ran conformational changes and a Ran · GDP-importin-β-RanBP1 complex in vitro and in intact cells. J. Biol. Chem. 277:30121-30127. [DOI] [PubMed] [Google Scholar]
- 46.Praskova, M., S. Kalenderova, L. Miteva, Y. Poumay, and V. Mitev. 2002. The ornithine decarboxylase inhibitor, difluoromethylornithine, inhibits casein kinase II activity, c-Myc expression and normal human keratinocyte proliferation. Arch. Dermatol. Res. 293:590-593. [DOI] [PubMed] [Google Scholar]
- 47.Quensel, C., B. Friedrich, T. Sommer, E. Hartmann, and M. Kohler. 2004. In vivo analysis of importin alpha proteins reveals cellular proliferation inhibition and substrate specificity. Mol. Cell. Biol. 24:10246-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rangasamy, D., and V. G. Wilson. 2000. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J. Biol. Chem. 275:30487-30495. [DOI] [PubMed] [Google Scholar]
- 49.Rangasamy, D., K. Woytek, S. A. Khan, and V. G. Wilson. 2000. SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275:37999-38004. [DOI] [PubMed] [Google Scholar]
- 50.Ravnan, J. B., D. M. Gilbert, K. G. Ten Hagen, and S. N. Cohen. 1992. Random-choice replication of extrachromosomal bovine papillomavirus (BPV) molecules in heterogeneous, clonally derived BPV-infected cell lines. J. Virol. 66:6946-6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosas-Acosta, G., M. A. Langereis, A. Deyrieux, and V. G. Wilson. 2005. Proteins of the PIAS family enhance the sumoylation of the papillomavirus E1 protein. Virology 331:190-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sachdev, S., S. Bagchi, D. D. Zhang, A. C. Mings, and M. Hannink. 2000. Nuclear import of IκBα is accomplished by a Ran-independent transport pathway. Mol. Cell. Biol. 20:1571-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seki, T., S. Tada, T. Katada, and T. Enomoto. 1997. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem. Biophys. Res. Commun. 234:48-53. [DOI] [PubMed] [Google Scholar]
- 54.Shin, I., J. Rotty, F. Y. Wu, and C. L. Arteaga. 2005. Phosphorylation of p27Kip1 at Thr-157 interferes with its association with importin alpha during G1 and prevents nuclear re-entry. J. Biol. Chem. 280:6055-6063. [DOI] [PubMed] [Google Scholar]
- 55.Skiadopoulos, M. H., and A. A. McBride. 1996. The bovine papillomavirus type 1 E2 transactivator and repressor proteins use different nuclear localization signals. J. Virol. 70:1117-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun, S., L. Thorner, M. Lentz, P. MacPherson, and M. Botchan. 1990. Identification of a 68-kilodalton nuclear ATP-binding phosphoprotein encoded by bovine papillomavirus type 1. J. Virol. 64:5093-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Titolo, S., A. Pelletier, F. Sauve, K. Brault, E. Wardrop, P. W. White, A. Amin, M. G. Cordingley, and J. Archambault. 1999. Role of the ATP-binding domain of the human papillomavirus type 11 E1 helicase in E2-dependent binding to the origin. J. Virol. 73:5282-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsuji, L., T. Takumi, N. Imamoto, and Y. Yoneda. 1997. Identification of novel homologues of mouse importin alpha, the alpha subunit of the nuclear pore-targeting complex, and their tissue-specific expression. FEBS Lett. 416:30-34. [DOI] [PubMed] [Google Scholar]
- 59.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 60.Wilson, V. G., and G. Rosas-Acosta. 2003. Molecular targets for papillomavirus therapy. Curr. Drug Targets Infect. Disord. 3:221-239. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, V. G., M. West, K. Woytek, and D. Rangasamy. 2002. Papillomavirus E1 proteins: form, function, and features. Virus Genes 24:275-290. [DOI] [PubMed] [Google Scholar]
- 62.Woytek, K. J., D. Rangasamy, C. Bazaldua-Hernandez, M. West, and V. G. Wilson. 2001. Effects of mutations within two hydrophilic regions of the bovine papillomavirus type 1 E1 DNA-binding domain on E1-E2 interaction. J. Gen. Virol. 82:2341-2351. [DOI] [PubMed] [Google Scholar]
- 63.Yang, W., J. Gelles, and S. M. Musser. 2004. Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. USA 101:12887-12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zanardi, T. A., C. M. Stanley, B. M. Saville, S. M. Spacek, and M. R. Lentz. 1997. Modulation of bovine papillomavirus DNA replication by phosphorylation of the viral E1 protein. Virology 228:1-10. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, F., R. L. White, and K. L. Neufeld. 2000. Phosphorylation near nuclear localization signal regulates nuclear import of adenomatous polyposis coli protein. Proc. Natl. Acad. Sci. USA 97:12577-12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, G. Q., and J. E. Samuel. 2003. Identification and cloning potentially protective antigens of Coxiella burnetii using sera from mice experimentally infected with Nine Mile phase I. Ann. N. Y. Acad. Sci. 990:510-520. [DOI] [PubMed] [Google Scholar]
- 67.Zou, N., B. Y. Lin, F. Duan, K. Y. Lee, G. Jin, R. Guan, G. Yao, E. J. Lefkowitz, T. R. Broker, and L. T. Chow. 2000. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J. Virol. 74:3761-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]