Abstract
H9N2 influenza viruses have become established and maintain long-term endemicity in terrestrial poultry in Asian countries. Occasionally these viruses transmit to other mammals, including humans. Increasing epidemiological and laboratory findings suggest that quail may be an important host, as they are susceptible to different subtypes of influenza viruses. To better understand the role of quail in influenza virus ecology and evolution, H9N2 viruses isolated from quail during 2000 to 2005 were antigenically and genetically characterized. Our results showed that H9N2 viruses are prevalent year-round in southern China and replicate mainly asymptomatically in the respiratory tract of quail. Genetic analysis revealed that both the G1-like and Ck/Bei-like H9N2 lineages were cocirculating in quail since 2000. Phylogenetic analyses demonstrated that most of the isolates tested were double- or multiple-reassortant variants, with four G1-like and 16 Ck/Bei-like genotypes recognized. A novel genotype of G1-like virus became predominant in quail since 2003, while multiple Ck/Bei-like genotypes were introduced into quail, wherein they incorporated G1-like gene segments, but none of them became established in this host. Those Ck/Bei-like reassortants generated in quail have then been introduced into other poultry. These complex interactions form a two-way transmission system between quail and other types of poultry. The present study provides evidence that H9N2 and H5N1 subtype viruses have also exchanged gene segments to generate currently circulating reassortants of both subtypes that have pandemic potential. Continuing influenza virus surveillance in poultry is critical to understanding the genesis and emergence of potentially pandemic strains in this region.
Influenza A H9N2 viruses are present worldwide in poultry populations and derive from two major influenza virus gene pools, the Eurasian and the North American (12, 37). In terrestrial poultry of southern China, two H9N2 virus lineages have become established since the mid-1990s. One virus lineage, represented by Ck/Bei/1/94 or Dk/HK/Y280/97, is prevalent mainly in chicken, while the other one, represented by Qa/HK/G1/97, is predominant in quail (10).
Previous studies have shown that H9N2 viruses were prevalent in different types of poultry in southern China and that some H9N2 viruses from land-based poultry were reverse transmitted back to aquatic poultry, mainly domestic duck, wherein those viruses further reassorted with various viruses resident in duck to generate double and triple reassortants of H9N2 influenza viruses (19). However, the continuing evolutionary pathways of these reassortant H9N2 viruses and whether they could further cross species barriers to other species have not been determined.
The isolation of H9N2 influenza virus in 1999 from two Hong Kong children with mild upper respiratory disease was the first record of human infection with this virus (20, 29), and there have been subsequent cases of H9N2 influenza virus infection reported from mainland China (13). In 2003, a Hong Kong resident was again confirmed to have H9N2 virus infection (2). H9N2 viruses have also been isolated from pigs in Hong Kong in 1998 (28) and in Shandong Province in 2003 (40), raising the possibility of further reassortment with human-like viruses from pigs. Genetic analyses showed that since the late 1990s H9N2 viruses from southern China have profiles that include preferential binding with α-2,6-NeuAcGal, human-like receptors (26). These findings suggested that those H9N2 influenza viruses still had pandemic potential.
The farmed quail population size in southern China has dramatically increased during the last two decades, although it is still considered minor poultry in comparison with chicken, domestic duck, and goose. Quail may have played an important role in facilitating the reassortment events that generated the H5N1 virus (H5N1/97-like virus) responsible for the Hong Kong “bird flu” incident (10, 38). Recent studies have suggested that quail are also susceptible to different subtypes of influenza viruses and that tested viruses replicated mainly in the respiratory tract (24). As H5N1 (4, 18) and H9N2 are both endemic and cocirculate in poultry in this region, it remains to be determined whether H9N2 viruses prevailing in quail have been involved in the generation of recent H5N1 variants currently causing outbreaks in Asia, Europe, and Africa.
Our systematic influenza virus surveillance showed that H9N2 influenza virus continued to be prevalent in quail year-round in southern China from 2000 to 2005. Genetic and antigenic analyses of representative strains revealed that both the G1-like and Ck/Bei-like H9N2 influenza virus lineages cocirculate in quail. Novel Ck/Bei-like genotypes were introduced into quail and further reassorted with G1-like viruses endemic in quail. Those H9N2 reassortants with G1-like gene segments have then transmitted to other poultry, forming a complex system of two-way transmission between quail and other types of poultry. Genetic analysis also provides evidence that H9N2 and H5N1 subtype viruses have a two-way exchange of gene segments to generate current genotypes of both subtypes that have pandemic potential.
MATERIALS AND METHODS
Sampling and virus isolation.
A total of 4,601 quail were sampled at six live-poultry markets from Shantou, Guangdong, between June 2000 and December 2005. Market quail in Shantou were from local farms or imported from adjacent provinces such as Hunan, Jiangxi, and Zhejiang. Of those samples, 3,776 were paired tracheal and cloacal swabs, while for the remaining only a cloacal or tracheal swab was taken. Viruses were isolated in 9- to 11-day-old embryonated chicken eggs as previously described (19).
Antigenic analysis.
Virus isolates were subtyped by standard hemagglutination inhibition (HI) tests using a panel of the World Health Organization reference antisera (http://www.who.int/csr/resources/publications/en/#influenza). Antigenic analysis was performed using three different panels of monoclonal antibodies (MAbs) against Qa/HK/G1/97, Dk/HK/Y280/97, and Ck/HK/G9/97. All MAbs were produced at the Department of Infectious Diseases, St. Jude Children's Research Hospital, TN (7). To visualize similarity between the antigenic reaction patterns of different viruses, numerical analysis of HI titers was conducted using PRIMER version 5.2.9 (PRIMER-E, Plymouth, United Kingdom). The data were standardized and square-root transformed, and the Bray-Curtis coefficient (1) was used to construct a similarity matrix. Hierarchical agglomerative clustering with group-average linking (34) was conducted and a dendrogram produced. Nonmetric multidimensional scaling (16) was also used to produce two- and three-dimensional ordinations over 100 iterations. The two-dimensional configuration with lowest overall stress is presented.
Phylogenetic and molecular analyses.
One or two virus isolates from each positive sampling occasion were selected for sequence analysis. RNA extraction, cDNA synthesis, and PCR were carried out as described previously (19). Sequencing was performed by using a BigDye Terminator v3.1 cycle sequencing kit on an ABI PRISM 3700 DNA analyzer (Applied Biosystems) following the manufacturer's instructions. All eight gene segments of these viruses were characterized and phylogenetically analyzed together with virus sequence data available in GenBank. All sequences were assembled and edited with Lasergene 6.0 (DNASTAR, Madison, WI); BioEdit 7 was used for alignment and residue analysis (14). The program MrModeltest 2.2 (27) was used to determine the appropriate DNA substitution model and γ-rate heterogeneity. The generated model was used in all subsequent analyses. Neighbor-joining and maximum-likelihood trees were constructed by using PAUP* 4.0 (36). Bayesian analysis was conducted with MrBayes 3.1 (15) by using two replicates of 1 million generations with six chains. Estimates of the phylogenies were calculated by performing 1,000 neighbor-joining bootstrap replicates, and Bayesian posterior probabilities were calculated from the consensus of 18,000 trees after excluding the first 2,000 trees as burn-in. All eight genes were sequenced for each virus isolate.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this study are available from GenBank under accession numbers EF154834 to EF155417.
RESULTS
Prevalence of H9N2 influenza viruses in quail.
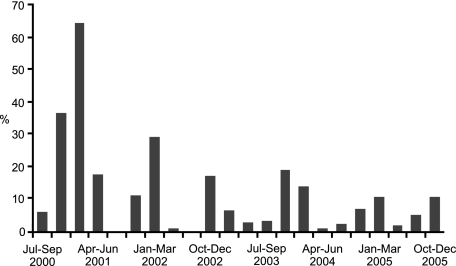
Systematic surveillance of market quail from 2000 to 2005 resulted in 610 influenza isolates from 4,601 samples collected (total isolation rate, 13.3%). Three influenza virus subtypes were identified: H9N2 (n = 414), H6N1 (n = 184), and H5N1 (n = 12). H9N2 influenza virus in quail was prevalent year-round, but an increased isolation rate was usually observed during the winter season (October to March) (Fig. 1 and Table 1). The prevalence of H9N2 viruses in a quail has generally decreased from 2001 (isolation rate, 14.6%) to 2004 (isolation rate, 5.4%) and 2005 (isolation rate, 6.6%). The high isolation rates seen in 2000 may be due to smaller sampling numbers in this year (n = 236) than in subsequent years (Table 1), which may have introduced sampling bias. A total of 396 of 414 (95.7%) of those H9N2 viruses were isolated from tracheal swabs, and only 18 influenza viruses were isolated from the cloacal samples. This information suggests that influenza A virus replicates mainly in the respiratory tract of quail in the field.
FIG. 1.
H9N2 influenza virus isolation rate in quail from southern China, July 2000 to December 2005.
TABLE 1.
Prevalence of H9N2 influenza viruses in quail from southern China from 2000 to 2005
| Month | No. of H9N2 isolates/no. sampled (isolation rate, %) in:
|
|||||
|---|---|---|---|---|---|---|
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | |
| January | 4/10 | 34/95 | 9/92 | 14/60 | 15/80 | |
| February | 11/27 | 0/0 | 0/52 | 3/60 | 0/61 | |
| March | 36/43 | 10/59 | 8/136 | 12/94 | 8/80 | |
| April | 16/69 | 0/65 | 0/134 | 0/66 | 1/80 | |
| May | 12/66 | 1/72 | 4/34 | 1/81 | 2/80 | |
| June | 5/60 | 0/52 | 2/74 | 0/86 | 0/70 | |
| July | 2/61 | 0/31 | 0/107 | 1/97 | 0/72 | 2/90 |
| August | 0/27 | 0/109 | 0/40 | 1/30 | 0/101 | 2/100 |
| September | 6/54 | 0/91 | 0/62 | 4/78 | 5/80 | 9/80 |
| October | 13/27 | 10/74 | 1/36 | 5/108 | 3/80 | 6/80 |
| November | 5/33 | 1/101 | 7/16 | 40/99 | 12/101 | 7/100 |
| December | 16/34 | 19/101 | 6/32 | 9/85 | 1/66 | 13/80 |
| Total | 42/236 (17.8) | 114/782 (14.6) | 59/636 (9.3) | 83/1,019 (8.1) | 51/947 (5.4) | 65/981 (6.6) |
Antigenic analysis.
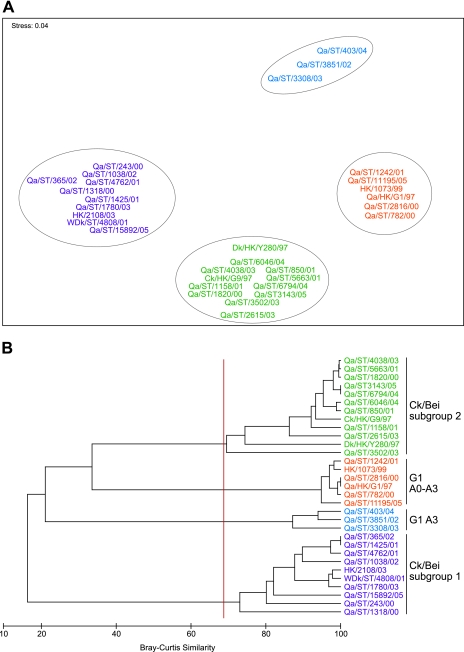
Antigenic analysis using a panel of MAbs raised against Qa/HK/G1/97, Dk/HK/Y280/97, and Ck/HK/G9/97 viruses demonstrated a diversity of reaction patterns that generally corresponded to the phylogenetic relationships of those H9N2 viruses tested (see below). Numerical analysis of HI titers was conducted to visualize similarity between the antigenic reactivities of different viruses and revealed four distinct antigenic subgroups (Fig. 2 and Table 2). Two of them were G1-like viruses and the remaining two were Ck/Bei-like or Y280-like viruses (Fig. 2). One of the G1-like subgroups was comprised of genotype A3 viruses (Qa/ST/3851/02, Qa/ST/3308, and Qa/ST/403/04 [see below]), and the other subgroup contained genotype A0 to A2 viruses plus a genotype A3 virus from 2005 (Qa/ST/11195/05). These G1-like antigenic subgroups were distinguished by their reactivity to MAb 29 against Qa/HK/G1/97 and both MAbs against Ck/HK/G9/97 (Table 2). The two antigenic subgroups of Ck/Bei-like viruses could be distinguished by many MAbs against all three prototype viruses (Table 2).
FIG. 2.
Numerical analysis of HI titers (see Table 2) by using nonmetric multidimensional scaling (A) and hierarchical agglomerative clustering (B).
TABLE 2.
Hemagglutination inhibition titers from antigenic analysis of influenza A H9N2 viruses
| Virus | Genotype | Titer with the indicated MAb raised against:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Qa/HK/G1/97
|
Dk/HK/Y280/97
|
Ck/HK/G9/97
|
||||||||||||||
| 1073-9 | 26 | 29 | 7B10 | 8C4 | 15F1 | 18G4 | 3D11 | 4G3 | 19A10 | 18B10 | 2F4 | 18B1 | G9-6 | G9-25 | ||
| Qa/HK/G1/97 | A0 | <a | >12,800 | >12,800 | < | < | < | < | < | < | < | < | < | < | >12,800 | >12,800 |
| Dk/HK/Y280/97 | B0 | < | 400 | >12,800 | >12,800 | >12,800 | >12,800 | 100 | 100 | 100 | 100 | 6,400 | >12,800 | >12,800 | >12,800 | >12,800 |
| Ck/HK/G9/97 | Bn | < | 200 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | 1,600 | >12,800 | 6,400 | >12,800 | >12,800 | >12,800 | >12,800 |
| WDk/ST/4808/01 | B7 | < | < | < | 200 | >12,800 | < | < | 100 | < | < | 3,200 | < | < | < | < |
| HK/1073/99 | A0 | 100 | >12,800 | >12,800 | < | < | < | 200 | < | < | < | 100 | < | < | >12,800 | >12,800 |
| HK/2108/03 | B7 | < | < | < | 200 | >12,800 | < | < | < | < | < | 3,200 | < | < | < | < |
| Qa/ST/782/00 | A0 | 100 | >12,800 | >12,800 | < | < | < | < | < | < | < | < | < | < | >12,800 | >12,800 |
| Qa/ST/2816/00 | A1 | < | >12,800 | >12800 | < | < | < | < | < | < | < | < | < | < | >12800 | >12800 |
| Qa/ST/1242/01 | A2 | 800 | >12,800 | >12,800 | < | < | < | 100 | < | < | < | 100 | < | < | >12,800 | >12,800 |
| Qa/ST/11195/05 | A3 | 3200 | >12,800 | >12,800 | < | < | < | < | < | < | < | 200 | < | < | >12,800 | >12,800 |
| Qa/ST/3851/02 | A3 | 100 | < | >12,800 | < | < | < | < | < | < | < | 100 | < | < | < | < |
| Qa/ST/3308/03 | A3 | 800 | < | >12,800 | 200 | < | < | < | < | < | < | 200 | < | < | < | < |
| Qa/ST/403/04 | A3 | < | < | 12,800 | < | < | < | < | < | < | < | < | < | < | < | < |
| Qa/ST/243/00 | B1 | 200 | < | < | 200 | >12,800 | < | < | < | < | < | 400 | < | < | 100 | < |
| Qa/ST/1318/00 | B2 | < | < | < | < | 1,600 | < | < | < | < | < | 1,600 | < | < | < | < |
| Qa/ST/1425/01 | B5 | 200 | < | < | 400 | >12,800 | < | < | < | < | < | 6,400 | < | 200 | < | < |
| Qa/ST/4762/01 | B6 | 200 | < | < | 400 | >12,800 | < | < | < | < | < | 6,400 | < | < | < | < |
| Qa/ST/365/02 | B8 | 200 | < | < | 400 | >12,800 | < | < | < | < | < | 6,400 | < | 200 | < | < |
| Qa/ST/1038/02 | B9 | 3200 | < | < | 400 | >12,800 | < | < | < | < | < | 6,400 | < | < | < | < |
| Qa/ST/1780/03 | B10 | 100 | < | < | 200 | >12,800 | < | < | < | < | < | 3,200 | < | < | < | < |
| Qa/ST/15892/05 | B16 | < | < | 400 | 200 | >12,800 | < | 100 | 100 | < | 100 | 6,400 | 100 | 400 | < | < |
| Qa/ST/1820/00 | B3 | 400 | 1,600 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | 6,400 | >12,800 | >12,800 | >12,800 | >12,800 |
| Qa/ST/850/01 | B3 | 1600 | 3,200 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 |
| Qa/ST/1158/01 | B4 | 100 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 |
| Qa/ST/5663/01 | B7 | 800 | 800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | 6,400 | >12,800 | >12,800 | >12,800 | >12,800 |
| Qa/ST/2615/03 | B11 | 400 | < | 800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | 3,200 | >12,800 | >12,800 | 3,200 | 12,800 |
| Qa/ST/3502/03 | B12 | < | 400 | 3,200 | 800 | >12,800 | >12,800 | >12,800 | 100 | 100 | >12,800 | 1,600 | 3,200 | 1,600 | >12,800 | >12,800 |
| Qa/ST/4038/03 | B13 | 200 | 800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | 6,400 | >12,800 | >12,800 | >12,800 | >12,800 |
| Qa/ST/6046/04 | B14 | 800 | 800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 |
| Qa/ST/6794/04 | B15 | < | 100 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | 3,200 | >12,800 | >12,800 | >12,800 | >12,800 |
| Qa/ST/3143/05 | B8 | < | 100 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | >12,800 | 3,200 | >12,800 | >12,800 | >12,800 | >12,800 |
<, HI titer lower than 100.
Phylogenetic analysis of the surface genes.
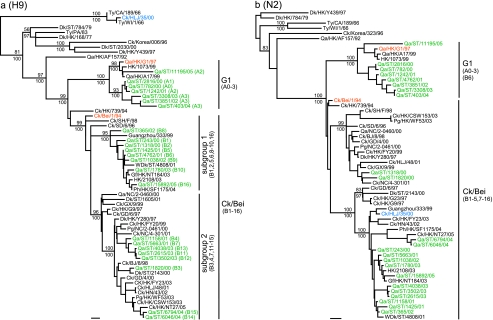
To understand the evolution and ecology of H9N2 viruses in quail, 73 representative viruses isolated during 2000 to 2005 were genetically characterized. Phylogenetic analysis of the H9 hemagglutinin (HA) gene revealed that 33 of those viruses belonged to the G1-like lineage, while the remaining 40 isolates were closely related to Ck/Bei-like viruses (Fig. 3a). The G1-like viruses gave rise to a stable lineage reflecting their long-term endemicity and evolution in this host. Among those Ck/Bei-like viruses two subgroups were recognized in quail since 2000 (Fig. 3a). Subgroup 1 consisted of 19 viruses, represented by Qa/ST/243/00, and subgroup 2 consisted of 21 viruses, represented by Dk/HK/Y280/97 (Fig. 3a). It is noteworthy that a single virus, Ck/Heilongjiang/35/2000 (Ck/HLJ/35/00), is almost identical (99.6% homology) to Ty/Wisconsin/1/1966 (Ty/WI/1/66) (17), an early H9N2 subtype reference strain from the North American lineage.
FIG. 3.
Phylogenetic relationships of the HA (a) and NA (b) genes of representative influenza A viruses isolated in Asia. Trees were generated by the neighbor-joining method with the PAUP* program (Bayesian analysis revealed similar relationships.) Numbers above and below branches indicate neighbor-joining bootstrap values and Bayesian posterior probabilities, respectively. Not all supports are shown because of space constraints. Analysis was based on nucleotides 129 to 1042 of the HA gene and 231 to 1297 of the NA gene. The HA and NA trees were rooted to Qa/Arkansas/29209-1/93 (H9N2) and Ck/Pennsylvania/8125/83 (H5N2), respectively. Viruses characterized in this study are highlighted in green. Genotypes characterized in this study was shown in brackets. Bars, 0.01 substitution per site. BJ and Bei, Beijing; Ck, chicken; Dk, duck; GD, Guangdong; Gf, guinea fowl; GX, Guangxi; HLJ, Heilongjiang; HN, Henan; HK, Hong Kong; NC, Nanchang; Pg, pigeon; Ph, pheasant; Qa, quail; SCk, silky chicken; SD, Shandong; SH, Shanghai; ST, Shantou; Ty, turkey; WDk, wild duck.
Phylogenetic analysis of the neuraminidase (NA) gene revealed an evolutionary patterns similar to that for the HA gene tree, with the NA genes of all but one virus corresponding with the lineage of the HA gene (Fig. 3b). The exception was that Qa/ST/4762/01, with a Ck/Bei-like HA gene, had a G1-like NA gene, revealing reassortment between the two virus sublineages. Taken together, these findings show that G1-like viruses have remained endemic in quail and that Ck/Bei-like viruses were introduced into this species in 2000, where they have cocirculated with G1-like viruses.
Phylogenetic analysis of the internal genes.
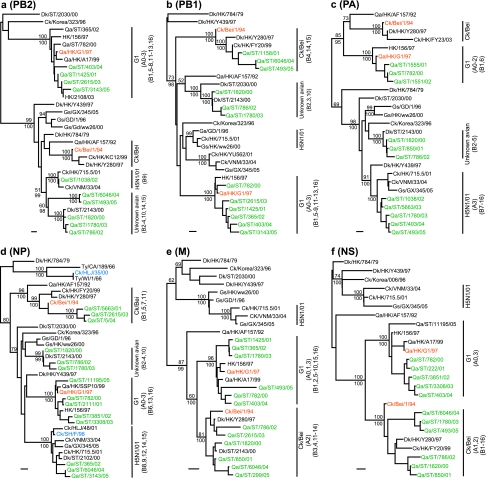
Phylogenetic analysis of the six internal genes revealed that H9N2 viruses from quail in southern China have undergone extensive reassortment to generate multiple novel genotypes. In the PB2 gene tree, representative H9N2 viruses clustered into three different lineages; 58 were G1-like, while 14 formed a group from an unknown avian source, likely derived from aquatic birds in the region (Fig. 4a). One virus, Qa/ST/1038/02, clustered with the H5N1/01-like viruses.
FIG. 4.
Phylogenetic relationships of the PB2 (a), PB1 (b), PA (c), NP (d), M (e), and NS (f) genes of representative influenza A viruses isolated in Asia. Trees were generated by the neighbor-joining method with the PAUP* program (Bayesian analysis revealed similar relationships.) Numbers above and below branches indicate neighbor-joining bootstrap values and Bayesian posterior probabilities, respectively. Not all supports are shown because of space constraints. Analysis was based on the following nucleotides: PB2, 1079 to 2138; PB1, 42 to 1217; PA, 1429 to 2127; NP, 31 to 917; M, 49 to 864; and NS, 88 to 815. The PB2, PA, NP, and M trees were rooted to A/equine/Prague/1/56 (H7N7), the PB1 tree to Qa/Arkansas/29209-1/93 (H9N2), and the NS tree to A/swine/Hong Kong/168/93 (H1N1). Viruses characterized in this study are highlighted in green. Bars, 0.01 substitution per site. Virus names and abbreviations are in the legend to Fig. 3.
Analysis of the PB1 gene showed that those H9N2 quail isolates formed three distinct lineages, including G1-like (n = 59), Ck/Bei-like (n = 9), and unknown avian (n = 5) (Fig. 4b). Their PA genes also fall into three different groups. Most of the PA genes of those quail viruses isolated since 2002 were closely related to H5N1/01-like viruses. The PA genes from the remaining viruses were either G1-like or unknown avian (Fig. 4c).
The NP genes of the H9N2 viruses were separated into four groups: G1-like (n = 37), Ck/Bei-like (n = 14), H5N1/01-like (n = 16), and unknown avian (n = 6) (Fig. 4d). It is interesting to note that the virus Ck/Shanghai/F/1998 (Ck/SH/F/98) contains an NP gene segment that was first detected in H5N1 virus in 2001 (23). Furthermore, the NP gene of Ck/HLJ/35/06 is almost identical (99.9% homology) to that of Ty/WI/1/66. Those H5N1/01-like PB2, PA, and NP genes are closely related to those of reassortant H5N1 variants isolated since 2001, including the dominant H5N1 genotype Z (9, 18).
The M and NS genes showed less diversity than the other internal genes, belonging to either the G1-like or Ck/Bei-like lineage (Fig. 4e and f). The M genes of 11 viruses grouped with Ck/Bei-like viruses, and the remaining viruses were joined to the G1-like lineage. The NS genes of 45 viruses were Ck/Bei-like, while the other 28 viruses were closely related to G1-like H9N2 viruses, which generally corresponded to the lineages of their HA genes.
Genotyping.
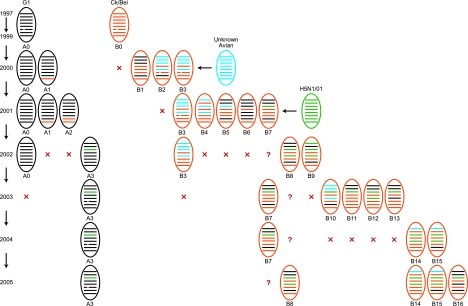
With the accumulated genetic information from H9N2 influenza viruses, it is necessary to provide a systematic nomenclature for identification of viruses with different sources and gene constellations. Here we propose to assign H9N2 virus genotypes as follows: viruses with a G1-like HA will be designated the genotype A series, and those with a Ck/Bei-like HA will be designated the genotype B series. Therefore, nonreassortant G1-like viruses are designated A0, while reassortant G1-like viruses are designated sequentially A1, A2, and so on, according to when the novel genotype was first identified. In the same manner, nonreassortant Ck/Bei-like viruses are designated B0, and novel reassortants then numbered sequentially B1, B2, and so on.
Phylogenetic analysis revealed 20 different reassortant H9N2 genotypes isolated from 2000 to 2005 in quail. Four genotypes were G1-like (genotypes A0 to A3), while 16 genotypes were Ck/Bei-like (genotypes B1 to B16) (Fig. 5 and Table 3). In the G1-like lineage, nonreassortant G1-like virus (genotype A0) was detected from 2000 to 2002. A G1-like reassortant incorporating a Ck/Bei-like NS gene (genotype A1) was first detected in late 2000, and during 2001 a double reassortant between G1-like and Ck/Bei-like viruses (genotype A2) was also identified (Fig. 5).
FIG. 5.
Genotypes of H9N2 influenza viruses from quail in southern China. The eight gene segments (horizontal bars, starting at the top) are PB2, PB1, PA, HA, NP, NA, M, and NS. Each color represents a virus lineage. Genotypes were defined by gene phylogeny (Fig. 3 and 4); a distinct phylogenetic lineage with bootstrap support of ≥80% (≥60% for PB2 genes) indicated a common origin. Question marks indicate that the corresponding genotype was not detected in that year, and red crosses indicate the genotype was not detected in two consecutive years.
TABLE 3.
Emergence and prevalence of different genotypes of H9N2 influenza viruses in quails from southern China
| Yr | G1-like viruses (n)
|
Ck/Bei-like viruses (n)
|
||||
|---|---|---|---|---|---|---|
| Prevailing | Emerging | Representative | Prevailing | Emerging | Representative | |
| 1997 | A0 | Qa/HK/G1/97 | ||||
| 1999 | A0 | |||||
| 2000 | A0 (3) | Qa/ST/782/00 | B1 (1) | B1 | Qa/ST/243/00 | |
| A1 (1) | A1 | Qa/ST/2816/00 | B2 (1) | B2 | Qa/ST/1318/00 | |
| B3 (1) | B3 | Qa/ST/1820/00 | ||||
| 2001 | A0 (4) | B3 (1) | ||||
| A1 (3) | B4 (1) | B4 | Qa/ST/1158/01 | |||
| A2 (1) | A2 | Qa/ST/1242/01 | B5 (3) | B5 | Qa/ST/1425/01 | |
| B6 (1) | B6 | Qa/ST/4762/01 | ||||
| B7 (2) | B7 | Qa/ST/5663/01 | ||||
| 2002 | A0 (2) | B3 (1) | ||||
| A3 (3) | A3 | Qa/ST/3851/02 | B8 (3) | B8 | Qa/ST365/02 | |
| B9 (1) | B9 | Qa/ST/1038/02 | ||||
| 2003 | A3 (7) | B7 (6) | ||||
| B10 (1) | B10 | Qa/ST/1780/03 | ||||
| B11 (1) | B11 | Qa/ST/2615/03 | ||||
| B12 (1) | B12 | Qa/ST/3502/03 | ||||
| B13 (1) | B13 | Qa/ST/4038/03 | ||||
| 2004 | A3 (5) | B7 (1) | ||||
| B14 (2) | B14 | Qa/ST/6046/04 | ||||
| B15 (3) | B15 | Qa/ST/6794/04 | ||||
| 2005 | A3 (4) | B8 (3) | ||||
| B14 (1) | ||||||
| B15 (2) | ||||||
| B16 (2) | B16 | Qa/ST/15892/05 | ||||
Since 2002, H9N2 genotype A3 virus emerged and became predominant in quail, and it is the only G1-like virus detected in this host since 2003. This genotype has seven G1-like gene segments and an H5N1/01-like PA gene, which was originally detected in H5N1 reassortant viruses isolated at live-poultry markets in Hong Kong in 2001 and is still present in current H5N1 genotype Z viruses (Fig. 4c and 5 and Table 2).
For Ck/Bei-like viruses, a different complement of reassortant H9N2 genotypes was detected in quail each year, but none of them became established in this host. These genotypes were all double or triple reassortants of Ck/Bei-like, G1-like, H5N1/01-like, and unknown avian viruses, with the exception of genotypes B7, B10, and B15, which are four-way reassortants (Fig. 5 and Table 2). Nonreassortant Ck/Bei-like virus (genotype B0) was not detected in this study. Interestingly, since 2003 all Ck/Bei-like genotype viruses detected in quail contained the H5N1/01-like PA gene also present in genotype A3 (Fig. 4c and 5 and Table 2).
Given the diversity of H9N2 viruses, to understand the interrelationships between the viruses characterized in present study and previously published H9N2 viruses, we analyzed all H9N2 viruses with eight gene sequence available and genotyped them using the system outlined above (Table 4). The data showed that all H9N2 viruses published since 2003 were Ck/Bei-like, which belong to the genotype B series. Of the genotypes that we isolated from quail, genotypes B1 to B5 and B7 have also been isolated from chicken, duck, pigeon, and Guinea fowl, while genotype B4 has previously been isolated from quail (Table 4). An additional 11 genotypes, designated B-n1 to B-n11, from other types of poultry were not detected in this study.
TABLE 4.
Genotype comparison of H9N2 viruses in China reported in different publications
| Isolation yr | Reference | Previous genotype designation | Representative | Genotype | No. of viruses characterized |
|---|---|---|---|---|---|
| 1995-1999 | 21 | Ck/BJ/1/95 | B0 | 10 | |
| 1996-2002 | 17 | A and F | Ck/SD/6/96 | B0 | 18 |
| B | Ck/GD/6/97 | B-n1 | 2 | ||
| H and I | Ck/SH/10/01 | B-n2 | 3 | ||
| C and D | Ck/GX/9/99 | B-n3 | 2 | ||
| E | Ck/HLJ/35/00 | 1 | |||
| G | Ck/HN/26/00 | B-n4 | 1 | ||
| 1997-2002 | 25 | I and VI | Ck/Os/aq48/97 | B0 | 4 |
| II | Ck/Os/aq19/01 | B-n5 | 3 | ||
| III | Ck/Ko/aq26/01 | B-n6 | 2 | ||
| IV | Ck/Os/aq58/01 | B3 | 1 | ||
| V | Ck/Os/aq69/01 | B-n2 | 1 | ||
| 1998 | 23 | Ck/SH/F/98 | B-n2 | 1 | |
| 2000-2001 | 22 | Group 3 | Pg/NC/2-0461/00 | B0 | 7 |
| Group 1 | Qa/NC/2-0460/00 | B4 | 5 | ||
| Group 4 | Pg/NC/11-145/00 | B-n7 | 2 | ||
| Group 2 | Ck/NC/4-301/01 | B3 | 2 | ||
| 2000-2001 | 19 | Dk/ST1042/00 | B1 | 2 | |
| Dk/ST1796/00 | B3 | 5 | |||
| Dk/ST/830/00 | B4 | 1 | |||
| Dk/ST/2134/00 | B5 | 1 | |||
| Dk/ST/2102/00 | B-n8 | 1 | |||
| Dk/ST/4808/01 | B7 | 1 | |||
| Dk/ST/1605/01 | B-n6 | 1 | |||
| 2003 | 7 | C | Ck/HK/FY23/03 | B0 | 7 |
| A | Gf/HK/NT184/03 | B7 | 1 | ||
| B | Ck/HK/NT142/03 | B-n9 | 1 | ||
| D and F | Ck/CSW/153/03 | B-n10 | 8 | ||
| E | Pg/HK/WF53/03 | B-n11 | 1 | ||
| 2003 | 2 | Y280 | Ck/HK/WF126/03 | B0 | 2 |
| B | Ck/HK/NT142/03 | B-n9 | 1 | ||
| A | Gf/HK/NT184/03 | B7 | 4 | ||
| D | Ck/HK/YU463/03 | B-n10 | 1 | ||
| D+ | Ck/HK/YU577/03 | B-n10 | 4 |
As none of those genotype B series viruses have become established in quail and they have been isolated from other types of poultry, it is suggested that Ck/Bei-like viruses isolated in quail resulted from repeated introduction from other type of poultry. Also, 13 of 16 B genotypes viruses had some gene segment with a G1-like origin. These findings suggested that two-way transmission between quail and other type of poultry occurred frequently in southern China.
Molecular characterization.
The deduced amino acid sequences of the HA and NA proteins were aligned and compared with those of other H9N2 reference viruses. All of the H9N2 viruses characterized, with the exception of Qa/ST/2061/00, had the same R-S-S-R amino acid motif at the connecting peptide, representing low pathogenicity in chicken (Table 5). It is noteworthy that the virus Qa/ST/2061/00 had an S→R substitution at position −2 of the HA1 protein, giving an R-S-R-R motif at the connecting peptide, which satisfies the basic requirement for high pathogenicity in chicken (35); however, pathogenicity tests in chicken revealed that it is still a low-pathogenicity virus (data not shown). The HAs of G1-like viruses isolated since 2004 maintained the avian-like motif 226-Gln (H3 numbering) at the receptor binding site (Table 5) (26). All Ck/Bei-like viruses had human-like motif 226-Leu at the receptor binding site, except for Qa/ST/1318/00, which maintained 226-Gln. However, both G1-like and Ck/Bei-like viruses had 228-Gly (Table 5).
TABLE 5.
Comparison of amino acid sequences of HAs and NAs of representative viruses from southern China
| Virus | Genotype | RBSa
|
NA deletion (aa) | Connecting peptide | |
|---|---|---|---|---|---|
| 226 | 228 | ||||
| Qa/HK/G1/97 | A0 | L | G | 45-46 | R-S-S-R |
| Dk/HK/Y280/97 | B0 | L | G | 62-64 | R-S-S-R |
| Dk/HK/Y439/97 | Q | G | A-S-N-R | ||
| Dk/HK/289/78 | Q | G | A-S-N-R | ||
| Qa/ST/243/00 | B1 | L | G | R-S-S-R | |
| Qa/ST/1318/00 | B2 | Q | G | 62-64 | R-S-S-R |
| Qa/ST/782/00 | A0 | L | G | R-S-S-R | |
| Qa/ST/2061/00 | A0 | L | G | R-S-R-R | |
| Qa/ST/1551/02 | A0 | M | G | R-S-S-R | |
| Qa/ST/3851/02 | A3 | Q | G | R-S-S-R | |
| Qa/ST/3008/03 | A3 | Q | G | 50-55 | R-S-S-R |
| Qa/ST/11195/05 | A3 | Q | G | 38-39 | R-S-S-R |
Amino acid at the receptor binding site (RBS).
The original 2-amino-acid (aa) deletion in the NA stalk of G1-like viruses (12), at positions 45 to 46, was not detected in any viruses that were characterized. However, novel 6-aa (positions 50 to 55) and 2-aa (positions 38 and 39) NA deletions, were recognized in G1-like genotype A3 viruses isolated from 2003 to 2004 and in 2005, respectively. These deletions do not affect any potential NA glycosylation sites. For Ck/Bei-like viruses, only genotypes B2 and B3 had the 3-aa NA deletion (positions 62 to 64) that was originally observed in nonreassortant Dk/HK/Y280/97 (12). All viruses with a G1-like NS gene had 92-Glu, which is reportedly associated with increased virulence in pigs (30).
DISCUSSION
Characterization of H9N2 influenza viruses isolated from quail from 6 years of influenza virus surveillance revealed that both G1-like and Ck/Bei-like viruses were cocirculating in this host in southern China since 2000. These viruses were prevalent year-round and replicate mainly asymptomatically in the respiratory tract of the quail. Genetic and antigenic studies demonstrated that a single H9N2 G1-like reassortant (genotype A3) had become established and predominant in this host since 2003, while previous “pure” G1-like influenza viruses (genotype A0) that had caused previous human infections (20, 29) were replaced by this novel virus.
In the present study, the evolutionary behaviors of the two H9N2 virus lineages from quail were markedly different. The G1-like virus lineage (genotype A series) was stable in quail, with limited reassortment events and a single dominant genotype at one time, indicating that these viruses were genetically stable and well adapted to quail. However, the Ck/Bei-like virus lineage (genotype B series) appeared to be very unstable, with new, short-lived reassortants emerging each year, none of which had become established, indicating those viruses were not well adapted to this host.
Phylogenetic analyses demonstrated that H9N2 genotype B viruses were regularly introduced into quail from other poultry between 2000 and 2005, as most of those viruses were detected in other types of poultry. Thirteen of 16 H9N2 genotype B viruses detected in quail from the present study were reassortants that contained G1-like gene segments, while a further 5 of 11 previously reported Ck/Bei-like viruses (the genotype B-n series) from other types of poultry also incorporated G1-like genes (2, 7, 17, 18) (Table 4). As H9N2 G1-like viruses have so far been recognized only in quail (11), these findings suggest that after being introduced into quail, Ck/Bei-like viruses had further reassorted with G1-like viruses endemic in quail and subsequently transmitted to other poultry. These complex interactions formed a two-way transmission system between quail and other types of poultry. Through this system, quail severed as a “mixing vessel” to facilitate many reassortment events in the current influenza virus ecosystem.
It was noted that three genotypes of Ck/Bei-like viruses, which do not include G1-like gene segments, are also double and triple reassortants of Ck/Bei-like, H5N1/01-like, and other unknown avian viruses. As the Ck/Bei-like viruses are predominantly found in chicken (11), those reassortants probably were originally generated in chicken and then transmitted to quail. Therefore, it is apparent that an ecosystem has evolved in which reassortment in different hosts occurs, followed by frequent interspecies transmission of Ck/Bei-like but not G1-like viruses.
Despite experimental evidence suggesting the susceptibility of quail to multiple subtypes of influenza virus (24), only three influenza virus subtypes (H9N2, H5N1, and H6N1) have been recognized in quail in the past 6 years, which have also been prevalent in other poultry in this region over the same period, and Ck/Bei-like H9N2 viruses could not become established in this host. Therefore, the present findings suggest that there is still an interspecies transmission barrier between quail and other types of birds and that quail may not be susceptible to all influenza virus subtypes under field conditions. Another possibility for why quail host only limited subtypes of influenza viruses in this region is that quail are not raised as “backyard” poultry but are kept caged and have limited contact with other poultry.
Quail was the hypothetical host for generating the H5N1/97-like virus, as seven of eight gene segments were identified in this host (6, 10). However, the donors of internal genes present in H5N1 variants isolated since 2001 have still not been identified. Based on the sequence of reassortment events, the present study suggests that H5N1/01-like internal genes were first incorporated into Ck/Bei-like viruses in 2001 and then into G1-like viruses in 2002 (Fig. 5). However, the phylogenetic relationships of the NP genes suggest that a H9N2 virus, Ck/SH/F/98-like, may be a possible donor of H5N1/01-like internal genes. If this is the case, then the gene flow between those subtypes may be in the reverse direction. Furthermore, the dominant G1-like virus found in quail (genotype A3) along with all Ck/Bei-like viruses isolated in this study since 2003 all incorporate an H5N1/01-like PA gene that is also found in current H5N1 genotype Z viruses. In this regard, quail may serve as a “mixing vessel” to facilitate reassortment events for H9N2, H5N1, and H6N1 viruses and the emergence of pandemic potential virus in this region (6, 18, 19).
Interestingly, H9N2 influenza viruses introduced into terrestrial poultry in different regions, such as Korea, Pakistan, and Iran, have undergone only limited further reassortment (3, 8). The same situation was observed with H5N1 virus in Southeast Asia, where multiple reassortants have emerged in poultry in southern China with transmission to other countries. Those H5N1 viruses have continually circulated in poultry since at least 2003 and are now endemic in these countries, but there is still no evidence that those viruses have undergone further reassortment with “local” viruses (4, 33, 39). This raises the question as to why so many reassortant viruses were detected in China and how they were generated. The usual explanation has been the unique ecology of southern China (31, 32); however, a similar interaction between different types of poultry and humans also exists in H5N1-affected areas such as Indonesia and Vietnam. Therefore, as part of pandemic preparedness, this question needs to be further explored.
The HA and NP genes of another virus, Ck/HLJ/35/00 (17), have ≥99.6% homology to those of Ty/WI/1/66, while the remaining genes are either Ck/Bei- or G1-like, suggesting that this virus may be evolving independently in China. However, such high homology of the HA and NP genes (just one and six nucleotide differences, respectively) from viruses isolated 34 years apart is exceptional and suggests that this virus has not been evolving under natural conditions. There are a number of possibilities that may explain these results. One of these is that the virus Ck/HLJ/35/00 is derived from the use of Ty/WI/1/66 as an H9N2 poultry vaccine, thereby introducing this North American virus into the Eurasian gene pool. Since 1998 a “pure” Ck/Bei-like virus, Ck/SD/6/96, has been used as a vaccine for the control of H9N2 virus infection in poultry in China (17). However, there is little information available regarding other vaccines used before this time, as H9N2 virus outbreaks were first recorded in China in 1994 (12). It is of note that an incompletely inactivated vaccine was probably responsible for the low-pathogenicity H5N2 outbreak in Taiwan in 2004 (5). Thus, it is not impossible that similar incidents may occur in other regions, especially areas where vaccination is the main influenza control measure.
This study demonstrates the continued circulation of H9N2 influenza viruses from quail in southern China and shows that both the G1-like and Ck/Bei-like lineages are now present in this host. Furthermore, reassortment between H9N2 and H5N1 subtype viruses has contributed to the generation of those currently circulating reassortants of both subtypes, including H5N1 genotype Z. Therefore, continuing influenza virus surveillance in poultry is critical to understanding the genesis and emergence of coming pandemic strains in this region.
Acknowledgments
This work was supported by the Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau of the Hong Kong SAR Government (project number 06060722), the Li Ka Shing Foundation, and the Ellison Foundation.
Footnotes
Published ahead of print on 27 December 2006.
REFERENCES
- 1.Bray, R. J., and J. T. Curtis. 1957. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 27:325-349. [Google Scholar]
- 2.Butt, K. M., G. J. D. Smith, H. Chen, L. J. Zhang, Y. H. Leung, K. M. Xu, W. Lim, R. G. Webster, K. Y. Yuen, J. S. M. Peiris, and Y. Guan. 2005. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 43:5760-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameron, K. R., V. Gregory, J. Banks, I. H. Brown, D. J. Alexander, A. J. Hay, and Y. P. Lin. 2000. H9N2 subtype influenza A viruses in poultry in Pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology 278:36-41. [DOI] [PubMed] [Google Scholar]
- 4.Chen, H., G. J. D. Smith, K. S. Li, J. Wang, X. H. Fan, J. M. Rayner, D. Vijaykrishna, J. X. Zhang, L. J. Zhang, C. T. Guo, C. L. Cheung, K. M. Xu, L. Duan, K. Huang, K. Qin, Y. H. Leung, W. L. Wu, H. R. Lu, Y. Chen, N. S. Xia, T. S. Naipospos, K. Y. Yuen, S. S. Hassan, S. Bahri, T. D. Nguyen, R. G. Webster, J. S. Peiris, and Y. Guan. 2006. Establishment of multiple sublineages of H5N1 influenza virus in Asia: implications for pandemic control. Proc. Natl. Acad. Sci. USA 103:2845-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, M. C., M. S. Lee, and C. H. Wang. 2005. Molecular characterization of H5N2 low pathogenic avian influenza viruses from Taiwan poultry in 2004, p. 30. In Proceedings of the APEC Conference on Avian Influenza. National Chung Hsing University, Taichung, Taiwan.
- 6.Chin, P. S., E. Hoffmann, R. Webby, R. G. Webster, Y. Guan, M. Peiris, and K. F. Shortridge. 2002. Molecular evolution of H6 influenza viruses from poultry in southeastern China: prevalence of H6N1 influenza viruses possessing seven A/Hong Kong/156/97 (H5N1)-like genes in poultry. J. Virol. 76:507-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, Y. K., H. Ozaki, R. J. Webby, R. G. Webster, J. S. Peiris, L. Poon, C. Butt, Y. H. Leung, and Y. Guan. 2004. Continuing evolution of H9N2 influenza viruses in southeastern China. J. Virol. 78:8609-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, Y. K., S. H. Seo, J. A. Kim, R. J. Webby, and R. G. Webster. 2005. Avian influenza viruses in Korean live poultry markets and their pathogenic potential. Virology 332:529-537. [DOI] [PubMed] [Google Scholar]
- 9.Guan, Y., J. S. Peiris, A. S. Lipatov, T. M. Ellis, K. C. Dyrting, S. Krauss, L. J. Zhang, R. G. Webster, and K. F. Shortridge. 2002. Emergence of multiple genotypes of H5N1 avian influenza viruses in Hong Kong SAR. Proc. Natl. Acad. Sci. USA 99:8950-8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan, Y., K. F. Shortridge, S. Krauss, and R. G. Webster. 1999. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong. Proc. Natl. Acad. Sci. USA 16:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan, Y., K. F. Shortridge, S. Krauss, P. S. Chin, K. C. Dyrting, T. M. Ellis, R. G. Webster, and M. Peiris. 2000. H9N2 influenza viruses possessing H5N1-like internal genomes continue to circulate in poultry in southeastern China. J. Virol. 74:9372-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo, Y. J., S. Krauss, D. A. Senne, I. P. Mo, K. S. Lo, X. P. Xiong, M. Norwood, K. F. Shortridge, R. G. Webster, and Y. Guan. 2000. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267:279-288. [DOI] [PubMed] [Google Scholar]
- 13.Guo, Y., J. Dong, M. Wang, Y. Zhang, J. Guo, and K. Wu. 2001. Characterization of hemagglutinin gene of influenza A virus subtype H9N2. Chin. Med. J. 114:76-79. [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 15.Huelsenbeck, J. P., and F. R. Ronquist. 2001. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 16.Kruskal, J. B. 1964. Nonmetric multidimensional scaling: a numerical method. Psychometrika 29:115-129. [Google Scholar]
- 17.Li, C., K. Yu, G. Tian, D. Yu, L. Liu, B. Jing, J. Ping, and H. Chen. 2005. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology 340:70-83. [DOI] [PubMed] [Google Scholar]
- 18.Li, K. S., Y. Guan, J. Wang, G. J. D. Smith, K. M. Xu, L. Duan, A. P. Rahardjo, P. Puthavathana, C. Buranathai, T. D. Nguyen, A. T. Estoepangestie, A. Chaisingh, P. Auewarakul, H. T. Long, N. T. Hanh, R. J. Webby, L. L. Poon, H. Chen, K. F. Shortridge, K. Y. Yuen, R. G. Webster, and J. S. M. Peiris. 2004. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature 430:209-213. [DOI] [PubMed] [Google Scholar]
- 19.Li, K. S., K. M. Xu, J. S. Peiris, L. L. Poon, K. Z. Yu, K. Y. Yuen, K. F. Shortridge, R. G. Webster, and Y. Guan. 2003. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans. J. Virol. 77:6988-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, Y. P., M. Shaw, V. Gregory, K. Cameron, W. Lim, A. Klimov, K. Subbarao, Y. Guan, S. Krauss, K. Shortridge, R. Webster, N. Cox, and A. Hay. 2000. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc. Natl. Acad. Sci. USA 97:9654-9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, J., K. Okazaki, H. Ozaki, Y. Sakoda, Q. Wu, F. Chen, and H. Kida. 2003. H9N2 influenza viruses prevalent in poultry in China are phylogenetically distinct from A/quail/Hong Kong/G1/97 presumed to be the donor of the internal protein genes of the H5N1 Hong Kong/97 virus. Avian Pathol. 32:551-560. [DOI] [PubMed] [Google Scholar]
- 22.Liu, M., S. He, D. Walker, N. Zhou, D. R. Perez, B. Mo, F. Li, X. Huang, R. G. Webster, and R. J. Webby. 2003. The influenza virus gene pool in a poultry market in South central china. Virology 305:267-275. [DOI] [PubMed] [Google Scholar]
- 23.Lu, J. H., X. F. Liu, W. X. Shao, Y. L. Liu, D. P. Wei, and H. Q. Liu. 2005. Phylogenetic analysis of eight genes of H9N2 subtype influenza virus: a mainland China strain possessing early isolates' genes that have been circulating. Virus Genes 31:163-169. [DOI] [PubMed] [Google Scholar]
- 24.Makarova, N. V., H. Ozaki, H. Kida, R. G. Webster, and D. R. Perez. 2003. Replication and transmission of influenza viruses in Japanese quail. Virology 310:8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mase, M., M. Eto, K. Imai, K. Tsukamoto, and S. Yamaguchi. 7 July 2006, posting date. Characterization of H9N2 influenza A viruses isolated from chicken products imported into Japan from China. Epidemiol. Infect. doi: 10.1017/S0950268806006728. [DOI] [PMC free article] [PubMed]
- 26.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156-162. [DOI] [PubMed] [Google Scholar]
- 27.Nylander, J. A. A. 2004. MRMODELTEST 2. Evolutionary Biology Centre, Uppsala University, Uppsala, Sweden.
- 28.Peiris, J. S., Y. Guan, D. Markwell, P. Ghose, R. G. Webster, and K. F. Shortridge. 2001. Cocirculation of avian H9N2 and contemporary “human” H3N2 influenza A viruses in pigs in southeastern China: potential for genetic reassortment? J. Virol. 75:9679-9686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peiris, M., K. Y. Yuen, C. W. Leung, K. H. Chan, P. L. Ip, R. W. Lai, W. K. Orr, and K. F. Shortridge. 1999. Human infection with influenza H9N2. Lancet 354:916-917. [DOI] [PubMed] [Google Scholar]
- 30.Seo, S. H., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed] [Google Scholar]
- 31.Shortridge, K. F. 1992. Pandemic influenza: a zoonosis? Semin. Respir. Infect. 7:11-25. [PubMed] [Google Scholar]
- 32.Shortridge, K. F., and C. H. Stuart-Harris. 1982. An influenza epicentre? Lancet ii:812-813. [DOI] [PubMed] [Google Scholar]
- 33.Smith, G. J. D., T. S. Naipospos, T. D. Nguyen, M. D. de Jong, D. Vijaykrishna, T. B. Usman, S. S. Hassan, T. V. Nguyen, T. V. Dao, N. A. Bui, Y. H. Leung, C. L Cheung, J. M. Rayner, J. X. Zhang, L. J. Zhang, L. L. Poon, K. S. Li, V. C. Nguyen, T. T. Hien, J. Farrar, R. G. Webster, H. Chen, J. S. Peiris, and Y. Guan. 2006. Evolution and adaptation of H5N1 influenza virus in avian and human hosts in Indonesia and Vietnam. Virology 350:258-268. [DOI] [PubMed] [Google Scholar]
- 34.Sokal, R. R., and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. Univ. Kansas Sci. Bull. 38:1409-1438. [Google Scholar]
- 35.Steinhauer, D. A.. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1-20. [DOI] [PubMed] [Google Scholar]
- 36.Swofford, D. L. 2001. PAUP*: phylogenetic analysis using parsimony (and other methods) 4.0 beta. Sinauer Associates, Sunderland, MA.
- 37.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webster, R. G., Y. Guan, M. Peiris, D. Walker, S. Krauss, N. N. Zhou, E. A. Govorkova, T. M. Ellis, K. C. Dyrting, T. Sit, D. R. Perez, and K. F. Shortridge. 2002. Characterization of H5N1 influenza viruses that continue to circulate in geese in southeastern China. J. Virol. 76:118-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.World Health Organization Global Influenza Program Surveillance Network. 2005. Evolution of H5N1 avian influenza viruses in Asia. Emerg. Infect. Dis. 11:1515-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, C., W. Fan, R. Wei, and H. Zhao. 2004. Isolation and identification of swine influenza recombinant A/Swine/Shandong/1/2003 (H9N2) virus. Microbes Infect. 10:919-925. [DOI] [PubMed] [Google Scholar]