Abstract
The human cytomegalovirus (HCMV) IE2 86-kDa protein is an essential transactivator of viral and cellular gene expression. Additional proteins of 60 and 40 kDa are expressed from the IE2 gene at late times postinfection and are identical to the C terminus of IE2 86. We have constructed HCMV recombinants that express wild-type full-length IE2 86 but do not express the IE2 40- and 60-kDa proteins. Each of these recombinants is viable, indicating that neither the 60-kDa nor the 40-kDa protein is required for virus replication, either alone or in combination. Cells infected with the IE2 60 and IE2 40 deletion mutants, however, exhibit decreased expression of selected viral genes at late times. In particular, expression of the viral DNA replication factor UL84 is affected by the deletion of IE2 40, and expression of the tegument protein pp65 (ppUL83) is affected by the deletion of both IE2 40 and IE2 60. IE2 60 and IE2 40 are also required for the production of normal levels of infectious virus. Finally, IE2 40 appears to function as a repressor of major immediate-early transcription in the infected cell. These results begin to define functions for the IE2 60- and IE2 40-kDa proteins and indicate that these products contribute both to the expression of selected viral genes and to the overall progression of the infection.
Human cytomegalovirus (HCMV) gene expression, like that of all herpesviruses, occurs as a tightly controlled series of events beginning with expression of the immediate-early (IE) genes. These go on to activate expression of early viral genes, allowing replication of the viral genome and subsequent transcription of late, primarily structural, genes. Two important IE genes, the UL122 and UL123 genes, comprise the major IE region. This segment of the HCMV genome encodes two predominant products, the IE1 72-kDa (ppUL123) and IE2 86-kDa (ppUL122) proteins. These are expressed from alternatively spliced forms of a five-exon transcript; the IE1 72 mRNA consists of exons 1 to 4, and the IE2 86 mRNA contains exons 1 to 3 and 5. Translation of both proteins begins in exon 2, so that IE1 72 and IE2 86 have identical 85-amino-acid (aa) N termini and unique C-terminal domains. Both proteins have been characterized extensively, most recently in studies that have used recombinant viruses containing deletions in the major IE region to elucidate the functions of IE1 72 and IE2 86 in the HCMV-infected cell. These studies indicate that the IE1 72-kDa protein contributes to virus replication during low-multiplicity, but not high-multiplicity, infections and is therefore nonessential (10, 11, 28). In contrast, IE2 86 is strictly required for HCMV replication, and even small deletions or changes to the sequence of the protein can result in a virus that does not replicate (26, 46).
Both IE1 72 and IE2 86 are transcriptional activators, and their ability to promote viral gene expression has been studied in a number of transient-transfection assays. IE2 86-mediated transactivation of the 1.2- and 2.7-kb RNA and UL112-113 (2.2-kb RNA) early promoters and of promoters driving genes involved in viral DNA replication has been particularly well characterized (5, 18, 36, 37). IE2 86 also functions as a repressor of transcription: it binds to DNA through interactions with the minor groove (20, 44) and downregulates its own expression via site-specific binding to the 14-bp cis repression signal between the TATAA box and transcription start site in the major immediate-early promoter (3, 13, 20, 23, 24, 30). This DNA binding capability allows regulation of early promoters as well as autoregulation, and the DNA binding region comprises aa 290 to 579 of IE2 86 (4, 19, 36). Regions spanning the length of the protein appear to be important for IE2 86 to transactivate heterologous promoters and HCMV early promoters, with the regions between aa 1 to 98 and 170 to 579 required for activation (25, 31, 37, 39, 42, 49). Both proteins have been shown to interact in vitro with multiple viral and cellular factors, although fewer of these interactions have been confirmed in the HCMV-infected cell (for a review, see reference 9). The only viral protein identified to date that interacts with IE2 86 is UL84. The UL84 protein is present in replication centers in the nuclei of infected cells and can promote oriLyt-dependent DNA replication in the presence of core replication proteins from HCMV and Epstein-Barr virus (21, 22, 35, 40, 48). UL84 is required for viral DNA replication and for the production of infectious virus and appears to have UTPase activity (6, 7, 47).
Additional spliced RNAs are expressed from both the IE1 and IE2 genes in infected fibroblasts. Spliced RNAs predicted to encode 19- and 17-kDa forms of IE1 are detectable in infected fibroblasts (1, 38). These contain exons 1, 2, and 3 and portions of exon 4 resulting from splice patterns different from those that generate the IE1 72 RNA. A spliced RNA predicted to encode a 9-kDa form of IE1 has also been detected in infected fibroblasts and contains exons 1 and 2, a 5′ segment of exon 3, and an alternatively spliced portion of exon 4 (1). Reports of protein expression from these transcripts vary. The IE1 17 protein has been detected only following transfection of CV-1 cells, but not in infected fibroblasts. While Shirakata et al. used an antibody directed against the region shared by IE1 and IE2 to detect the IE1 19 protein in infected fibroblasts, studies by Awasthi and colleagues suggest that this protein is in fact a breakdown product of full-length IE1 72 (1, 38). They detected IE1 19 protein only in transfected, not infected, cells and were not able to detect IE1 9 protein in either assay.
Other splicing events in exon 5 generate RNAs from the full-length transcript that could encode 55- and 18-kDa proteins. The IE2 18 RNA is expressed in HCMV-infected monocyte-derived macrophages but not in infected fibroblasts except in the presence of cycloheximide (16). An IE2 55 RNA is expressed only at IE times in HCMV-infected fibroblasts, and the corresponding IE2 55-kDa protein is detected in infected cells only after release from a cycloheximide block (41).
In contrast, other forms of IE2 are translated from unspliced RNAs that are expressed with late kinetics from different, downstream promoters (33). These RNAs encode proteins of 60 and 40 kDa that are detectable at late times in infected fibroblasts. The 40-kDa protein has been more extensively characterized than the 60-kDa form. The IE2 40-kDa protein is colinear with the C terminus of IE2 86 and is predicted to be expressed from a 1.5-kb RNA (32, 33, 41). Other, slightly larger forms of this RNA are present in the cytoplasm of HCMV-infected cells as well (41). The 60-kDa protein can be detected with an antibody that recognizes both IE2 86 and IE2 40, suggesting that IE2 60 is also identical to a C-terminal portion of IE2 86 (32). Translation of IE2 60 is predicted to begin at methionine 170 of IE2 86, and IE2 40 translation initiates at methionine 242 of IE2 86. The numbering used here reflects amino acid numbers in the Towne strain of HCMV. At late times postinfection (p.i.), both IE2 60 and IE2 40 are expressed to higher levels than IE2 86. In transient-transfection assays, IE2 40 represses the major IE promoter in much the same way as IE2 86, and it activates a version of this promoter lacking the cis repression signal when expressed in combination with IE1 72 (15). These smaller forms of IE2 have not yet been characterized in the HCMV-infected cell.
Work from our laboratory previously showed that the IE2 86ΔSX virus, which contains an internal deletion in the IE2 gene spanning codons 136 to 290, is viable. IE2 86ΔSX replicates slowly and to lower titers than wild-type virus but supports early gene expression to approximately wild-type levels. Notable molecular defects in IE2 86ΔSX virus-infected cells include decreased expression of UL83 (pp65) RNA and protein and slightly decreased expression of UL99 (pp28) protein, but not RNA (34). IE2 86 is expressed to lower levels in IE2 86ΔSX virus-infected cells. Also, as a result of the deletion of aa 136 to 290 from IE2 86, the 60-kDa and 40-kDa forms of the protein are not expressed, demonstrating that these forms are not required for the progression of a productive infection. In the present study, we constructed recombinant viruses that do not express the IE2 40 and IE2 60 proteins and made rescued versions of each mutant virus. Using this family of HCMV recombinants, we show that neither the IE2 60 nor the IE2 40 protein is essential for virus replication but that deletion of both forms leads to a 10-fold drop in the production of infectious virus. As predicted by the transient-transfection assays, IE2 60 and IE2 40 contribute to regulation of the major IE promoter. Other functions of these proteins were not predicted by transient-transfection assays. These newly uncovered functions include regulation of pp65 expression via a decrease in transcript levels and control of UL84 protein expression through what appears to be a posttranscriptional mechanism.
MATERIALS AND METHODS
Cells.
Human foreskin fibroblasts (HFF) were cultured in minimum essential medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and 200 U penicillin, 200 μg streptomycin, 1.5 μg amphotericin B, and 50 μg gentamicin per milliliter and grown as described previously (43).
Bacterial artificial chromosome (BAC) mutagenesis.
Construction of the p40, p60, and p40+60 deletion mutant viruses began with plasmid pSP-J(BglII-StuI), which contains the approximately 2,000-bp fragment generated by digesting the HCMV AD169 genomic EcoRI J region (43) with BglII and StuI. Mutagenic oligonucleotide primers were used in conjunction with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) to create plasmids pSP-J(BglII-StuIΔ40), pSP-J(BglII-StuIΔ60), and pSP-J(BglII-StuIΔ40+60). These are identical to pSP-J(BglII-StuI) but contain changes to the predicted TATAA box for the 1.5-kb RNA (Δ40), an upstream AT-rich region described in Results (Δ60), or both sites (Δ40+60). These constructs were sequenced to verify the presence of the correct mutations. DNA sequencing was performed by Eton Bioscience, San Diego, CA. Sequences of mutagenic primers (Integrated DNA Technologies, Coralville, IA) for the IE2 40 TATAA mutation are as follows: sense, 5′ CCTTTCAAGGTGATCATCAAGCCGCCCGTGCCTCCC 3′; antisense, 5′ GGGAGGCACGGGCGGCTTGATGATCACCTTGAAAGG 3′; those for the upstream AT-rich site mutation are as follows: sense, 5′ GGATCCCACGTCACTATTGCGTACTCATCATCAAGCTCTATGGGACACTCTGTAATCC 3′; antisense, 5′ GGATTACAGAGTGTCCCATAGAGCTTGATGATGAGTACGCAATAGTGACGTGGGATCC 3′.
One or both of the mutations were introduced into the UL122-123 gene coding region using the Counter Selection BAC Modification kit (Gene Bridges, Dresden, Germany). Briefly, oligonucleotide primers were used to amplify a marker cassette containing the neomycin resistance and RpsL genes and to simultaneously introduce 50 nucleotides of homology to the UL122-123 gene region onto either end of the cassette. Sequences of primers (Integrated DNA Technologies, Coralville, IA) are as follows: sense, 5′ GATAGAGGAAGTTGCCCCAGAGGAAGAGGAGGATGGTGCTGAGGAACCCAGGCCTGGTGATGATGGCGGGATCG 3′; antisense, 5′ CCTTCTCGTTGTCCAACTCGGAGATGCGTTTGCTCTTCTTCTTGCGGGGTTCAGAAGAACTCGTCAAGAAGGCG 3′. The linear product was recombined into the UL122-123 gene region contained in the wild-type HCMV strain AD169 BAC pHB5 (gift from M. Messerle) (2), and the resulting intermediate construct, pHB5(IEexon5-RpsLneo), was selected on the basis of resistance to kanamycin. Next, pSP-J(BglII-StuIΔ40), pSP-J(BglII-StuIΔ60), and pSP-J(BglII-StuIΔ40+60) were used as a template in conjunction with sense primer 5′ CAGGAAGAAAGTGAGCAGAGTGATG 3′ and antisense primer 5′ AGCGATTGGTGTTGCGGAAC 3′ (Integrated DNA Technologies, Coralville, IA) to amplify a linear fragment containing the mutated UL122-123 gene region. This fragment was recombined into pHB5(IEexon5-RpsLneo), replacing the RpsLneo cassette, and the resulting IE2 Δ40, IE2 Δ60, and IE2 Δ40+60 BACs were selected on the basis of increased streptomycin resistance. The altered region from each was amplified and sequenced to confirm that the intended deletion had been introduced into the BAC.
A rescued BAC was generated from the each of the mutant BACs by the reverse procedure. The RpsL-neomycin marker cassette was introduced into each of the three mutant BACs using the procedure and primers listed above. pSP-J(BglII-StuI) was then used as a template in conjunction with the primers described above to produce a linear fragment containing the wild-type HCMV sequence. This fragment was inserted into each of the RpsL-neomycin cassette-containing intermediate BACs by homologous recombination. These rescued BACs were designated IE2 40 rescue, IE2 60 rescue, and IE2 40+60 rescue.
Wild-type, mutant, and rescued BAC DNAs were amplified and purified as described previously (34). Each BAC was digested with HindIII and separated by field inversion gel electrophoresis to ensure that no major alterations to the DNA were sustained during the cloning procedure.
Reconstitution of virus, determination of virus titers, and growth curves.
Wild-type, IE2 Δ40, IE2 Δ60, IE2 Δ40+60, IE2 40 rescue, IE2 60 rescue, and IE2 40+60 rescue BACs were transfected into HFF by electroporation as previously described (46) and monitored for plaque development. When all cells in a culture exhibited cytopathic effect, supernatants were harvested and used to infect fresh cells. Stocks of wild-type, mutant, and rescued mutant viruses were harvested and titered by plaque assay. To analyze the kinetics of virus replication, confluence-synchronized HFF were infected at a multiplicity of infection (MOI) of 5 PFU/cell. Supernatant from infected cells was harvested daily, replaced with fresh media, and titered by plaque assay.
Time course of virus infection.
HFF were grown to and maintained at confluence for 3 days prior to infection to allow synchronization in a G0 state. At the time of infection, cells were released from G0 by trypsinization, infected at an MOI of 5 PFU/cell, and replated at a lower density. Cells were refed daily and then harvested by trypsinization at various times p.i.
Quantitative real-time RT-PCR analysis.
Real-time reverse transcription-PCR (RT-PCR) and data analysis were performed essentially as previously described using primers and probes directed against the HCMV IE1 72, IE2 86, UL83, and UL84 genes and the cellular housekeeping glucose-6-phosphate dehydrogenase (G6PD) gene (46). RNA was isolated with a NucleoSpin II kit (Clontech, Mountain View, CA) and subsequently treated with DNase using a Turbo DNA-free kit (Ambion, Austin, TX) to remove any residual DNA contamination and to allow the analysis of unspliced viral transcripts. Sequences of the IE1 72, IE2 86, and G6PD gene primers and TaqMan probes have been previously described (46). The sequences of the primers and probe for the UL83 assay are as follows: sense, 5′ TCTTCCTGGAGGTACAAGCCA 3′; antisense, 5′ CAGCCACGGGATCGTACTG 3′; probe, 5′ [6-carboxyfluorescein (FAM)]-ACGCGAGACCGTGGAACTGCG-[black hole quencher-1] 3′. The sequences of the primers and probe for the UL84 assay are as follows: sense, 5′ AGACATTGGGACCCTCCGTC 3′; antisense, 5′ GCGGTGATTCGTTCGGG 3′; probe, 5′ [6-FAM]-TGGACGATTGGAGCTAG-[black hole quencher-1] 3′.
Western blotting.
Cells were lysed in reducing sample buffer (50 mM Tris [pH 6.8], 0.2% sodium dodecyl sulfate, 10% glycerol, 5% 2-mercaptoethanol, 25 mM sodium fluoride, 1 mM sodium orthovanadate, 5 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 50 μM leupeptin, and 100 μM pepstatin A), and protein content was determined by a Bradford assay. Equal amounts of protein were separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose. Membranes were stained with amido black to ensure equal protein loading. After being blocked in 5% nonfat dried milk in TBS-T (Tris-buffered saline [pH 7.4] with 0.05% Tween 20), blots were incubated with primary antibodies in 5% nonfat dried milk in TBS-T, diluted as follows: CH16.0 monoclonal antibody (MAb), 1:10,000; IE2 MAb 8140, 1:1,000; UL44 MAb, 1:5,000; UL57 MAb, 1:10,000; pp65 MAb, 1:10,000; UL84 MAb, 1:2,000 to 1:10,000; pp28 MAb, 1:10,000; β-actin MAb AC-15, 1:10,000. CH16.0, anti-UL44, anti-UL57, anti-pp65, and anti-pp28 were purchased from the Goodwin Institute (Plantation, FL) and from Virusys (Sykesville, MD). Anti-IE2 was purchased from Clontech (Mountain View, CA). Anti-β-actin was purchased from Sigma-Aldrich (St. Louis, MO). Anti-UL84 was a gift from Greg Pari (University of Nevada, Reno). Membranes were washed in TBS-T and incubated in horseradish peroxidase-coupled anti-mouse antibody (Calbiochem, San Diego, CA), diluted 1:2,000. After being washed in TBS-T, proteins were detected using SuperSignal chemiluminescent substrate (Pierce, Rockford, IL) according to the manufacturer's instructions.
RESULTS
Construction of IE2 60, IE2 40, and IE2 60+40 deletion mutant viruses.
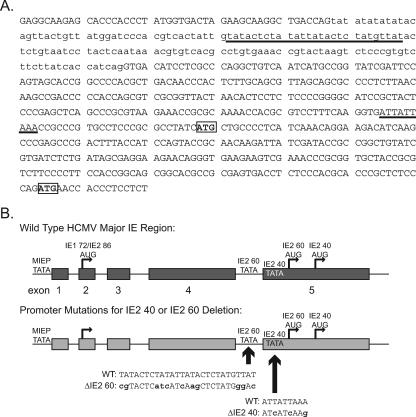
In addition to full-length IE2 86, the UL122 gene encodes additional, smaller proteins that are identical to the C terminus of IE2 86. Two of these proteins have molecular masses of 40 kDa and 60 kDa and are expressed at late times p.i. from one or more transcripts distinct from the full-length IE2 86 mRNA (32, 33, 41). The 1.5-kb transcript expressed from this region is the predominant IE2 RNA present in the cytoplasm of infected cells at late times p.i. (41). The experiment that mapped the 1.5-kb species identified several other IE2 transcripts also present in the cytoplasm at late times, and these were approximately 130 to 280 bases longer than the 1.5-kb RNA. Translation of the 60-kDa protein initiates at methionine 170 of IE2 86, while translation of the 40-kDa protein begins at methionine 242, and codons for both of these residues are present in the 1.5-kb RNA as well as in the longer species. The sequence of the promoter region and the sites that were changed to eliminate the expression of these proteins are shown in Fig. 1.
FIG. 1.
Construction of the IE2 40 and IE2 60 deletion mutant viruses. (A) Partial sequence of the wild-type HCMV major IE region from the 3′ end of exon 4 through the 5′ portion of exon 5. Exon sequences are capitalized, and intron sequences are in lowercase. The regions that were changed to eliminate expression of the IE2 40 and IE2 60 proteins are underlined. The codons for the initiating methionines of each protein are boxed. (B) Schematic of the major IE region. The relative locations of the exons and the TATAA boxes and initiating methionines for IE1 72, IE2 86, IE2 60, and IE2 40 are indicated. The sequences that were altered to remove the predicted TATAA boxes for the late RNAs are indicated. For the mutations, nucleotides that are identical to the wild-type sequence are capitalized and altered nucleotides are in lowercase. The diagram is not to scale. MIEP, major immediate-early promoter.
Previous work from our laboratory described the construction and characterization of IE2 86ΔSX-EGFP, a viable HCMV recombinant with a deletion spanning aa 136 to 290 of IE2 86 (34). The initiating methionines for both the IE2 40- and 60-kDa proteins are not encoded by this virus, and the mutant virus replicates slowly, grows to lower titers than wild-type virus, and exhibits distinct defects in viral gene expression. At the outset, the present study had two goals. First, we aimed to delineate the functions of the smaller, C-terminal 40-kDa and 60-kDa forms of IE2 and their roles in HCMV-infected cells at late times p.i. Second, we wanted to determine which of the effects observed in the IE2 86ΔSX virus-infected cell were due to the deletion of aa 136 to 290 in full-length IE2 86 versus the elimination of IE2 40 and IE2 60 expression.
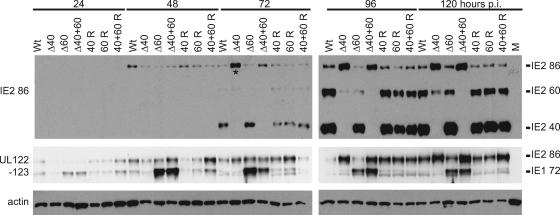
To answer these questions, we constructed recombinant HCMV BACs encoding viruses that do not express one or both of the IE2 40 and IE2 60 proteins. These BACs were generated as described in Materials and Methods using the AD169 BAC pHB5 (gift from Martin Messerle) and homologous recombination in Escherichia coli. Initially, we constructed a recombinant BAC, designated IE2 Δ40, in which the TATAA box driving expression of the predominant 1.5-kb RNA is rendered nonfunctional using silent mutations at the wobble bases. The amino acid sequence of the full-length IE2 86 protein expressed by this recombinant is identical to the wild type. We noted that, although this mutation eliminates all IE2 40 protein expression in the IE2 Δ40 virus-infected cell, some residual IE2 60 protein is still expressed (Fig. 2, top panel). In order to characterize HCMV replication in the absence of all IE2 40 and IE2 60 protein, we introduced a second mutation into the BAC. This change is in an AT-rich region that could serve as a TATAA box to drive the expression of the slightly longer transcripts that were also detected in the study that mapped the start site of the 1.5-kb RNA (Fig. 1A) (41). We introduced this change into the HCMV BAC either alone (IE2 Δ60) or in combination with the initial mutation (IE2 Δ40+60). The altered sequence is in the intron between exons 4 and 5, and while several residues have been altered, neither the size of the intron nor the coding sequence of any form of IE1 or IE2 protein has been changed. Introducing both TATAA mutations into the HCMV BAC (IE2 Δ40+60) eliminated all detectable expression of IE2 40 and IE2 60 proteins (Fig. 2, top panel). When only the change to the 5′ TATAA site was included (IE2 Δ60), we still detected some expression of IE2 60 protein. This is consistent with the behavior of the IE2 Δ40 recombinant, which suggested that some IE2 60 protein was translated from the predominant 1.5-kb RNA.
FIG. 2.
Major immediate-early protein expression is altered following infection with IE2 Δ40 and IE2 Δ60 viruses. G0-synchronized HFF cells were infected with 5 PFU/cell of wild-type (Wt), mutant (Δ40, Δ60, or Δ40+60), and rescued mutant (40 R, 60 R, or 40+60 R) virus or mock infected (M) and harvested at the indicated times p.i. Equal amounts of cell lysates, in micrograms, were separated by SDS-PAGE and transferred to nitrocellulose. IE1 72, IE2 86, IE2 60, and IE2 40 protein levels were analyzed by Western blotting as described in Materials and Methods. Cellular actin levels were analyzed as a control for protein loading. The asterisk indicates that the Δ40 72-h lane is slightly overloaded in the IE2 Western blot (top panel) relative to the IE2 86 and IE1 72 Western blot (middle panel).
To further ensure the integrity of the three recombinant BACs, we constructed a rescued version of each BAC in which the altered sequences were returned to wild type. The resulting six recombinant BACs and the wild-type pHB5 parent were digested with HindIII, and fragments were separated by field inversion gel electrophoresis to confirm that no large-scale deletions or rearrangements had occurred during the cloning procedure. The restriction digest pattern of each of the recombinant BACs was the same as the restriction digest pattern of the wild-type BAC (data not shown). The wild-type, mutant, and rescued mutant BACs were transfected into HFF, and virus stocks were reconstituted as described in Materials and Methods.
IE2 60- and 40-kDa proteins contribute to virus replication.
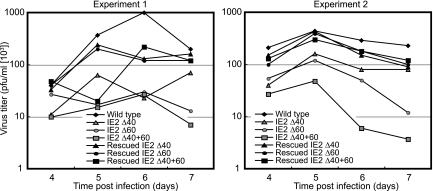
The ability to reconstitute virus from each of these mutant BACs indicated that neither the 60- nor 40-kDa form of IE2 86 was strictly required for HCMV replication in cultured cells. To understand how these proteins contribute to virus replication, we first constructed single-step growth curves for the recombinant viruses. Following high-multiplicity infection (MOI = 5 PFU/cell) of HFF, the amount of virus produced reached its peak 5 to 6 days p.i. for wild-type virus and for each of the three deletion mutant viruses (Fig. 3). The peak titers for the mutant viruses were lower than for the wild-type and rescued viruses. This indicated that, during high-MOI growth, the 60- and 40-kDa proteins did not contribute to the timing of virus production but were required for the virus to replicate to maximal titers. In particular, cells infected with IE2 Δ40+60 virus produced an average of 12 times less virus than wild-type at 5 days p.i. and up to 40 times less virus at 6 days p.i.
FIG. 3.
Deletion of IE2 40 and IE2 60 reduces virus production following high-multiplicity infection. HFF were infected with 5 PFU/cell of wild-type, mutant, or rescued mutant virus. Infected-cell supernatants were collected at the indicated times, and titers were determined as described in Materials and Methods. The two graphs indicate the titers determined in two independent experiments. All plaque assays were performed in duplicate.
IE1 72 and IE2 86 protein levels are altered by deletion of IE2 60- and 40-kDa proteins.
Since the replication of the IE2 60 and IE2 40 deletion mutant viruses is impaired, we began to look for defects in viral gene expression that might contribute to reduced production of infectious virus when IE2 60- or 40-kDa proteins are not expressed. We infected HFF cells at an MOI of 5 PFU/cell with wild-type virus, mutant viruses, and rescued mutant viruses and harvested infected cells at the indicated times (Fig. 2) p.i. Cells were processed for Western blots, and blots were probed with antibodies that recognize the HCMV major IE proteins. Having already characterized the expression of IE2 40 and IE2 60 proteins in the recombinant virus-infected cells (Fig. 2, top panel), we next assayed the expression of both IE1 72 and IE2 86 proteins in the mutant-virus-infected cells using an antibody directed against the shared region of these proteins encoded by exon 2 (Fig. 2, middle panel). We detected expression of both IE1 72 and IE2 86 in cells infected with wild-type virus, the three deletion mutant viruses, and the three rescued mutant viruses. Using either antibody, we detected increased expression of full-length IE2 86 relative to wild-type when IE2 40 was deleted, either alone or in combination with IE2 60. Furthermore, the expression of IE1 72 increased relative to wild type when IE2 60 was deleted, either alone or in combination with IE2 40.
UL122 and UL123 RNAs increase upon deletion of IE2 40.
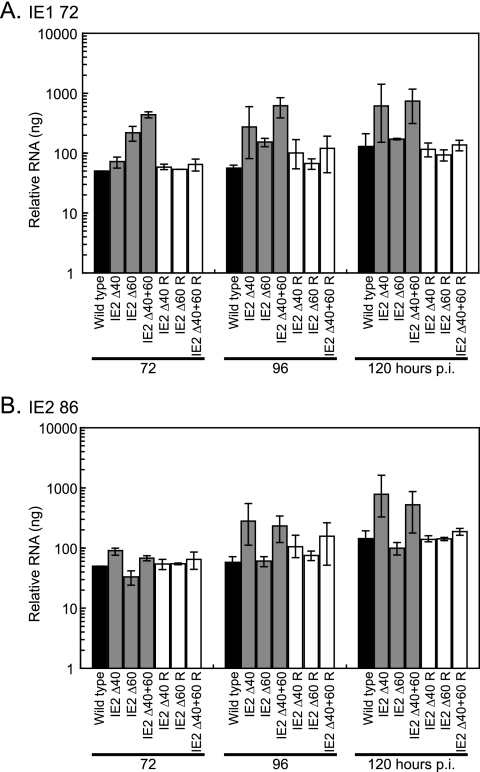
To understand which of these changes in protein expression resulted from differences at the RNA level, we next examined the levels of UL123 (IE1 72) and UL122 (IE2 86) transcripts following a high-multiplicity (MOI = 5 PFU/cell) infection. Since the differences in the amount of protein were most apparent at late times p.i., we examined RNA levels at 72, 96, and 120 h p.i. Total RNA was isolated from cells infected with wild-type, mutant, and rescued mutant viruses and assayed by real-time RT-PCR using primers and TaqMan probes that detect IE1 72 and IE2 86 transcripts (46). The TaqMan probe used in the IE1 assay was complementary to the splice junction between exons 3 and 4, and the TaqMan probe used in the IE2 assay was complementary to the splice junction between exons 3 and 5. Neither probe, therefore, detected the unspliced late transcripts.
The differences in IE1 72 and IE2 86 RNA levels in the wild-type and deletion mutant virus-infected cells were consistent with the hypothesis that IE2 40 can function to repress transcription from the major IE promoter. Although there was variability between experiments, at 96 and 120 h p.i., cells infected with IE2 Δ40 and IE2 Δ40+60 mutant viruses expressed more IE1 72 RNA and IE2 86 RNA than did wild-type-virus-infected cells (Fig. 4). There was up to 11 times more IE1 72 transcript and 4 times more IE2 86 transcript present in IE2 Δ40+60 virus-infected cells than in wild-type-virus-infected cells. We believe that in infected cells, as in transient-transfection assays, the IE2 40 protein functions at late times to downregulate expression of IE1 72 and IE2 86 transcripts. When IE2 40 is not expressed, we therefore observe an increase in the levels of these major IE transcripts.
FIG. 4.
IE1 72 and IE2 86 RNA levels are altered in deletion mutant virus-infected cells. G0-synchronized HFF cells were infected with 5 PFU/cell of wild-type virus, mutant viruses (IE2 Δ40, IE2 Δ60, or IE2 Δ40+60), and rescued mutant viruses (IE2 Δ40 R, IE2 Δ60 R, or IE2 Δ40+60 R) or mock infected and harvested at the indicated times p.i. Total RNA was analyzed by quantitative real-time RT-PCR as described in Materials and Methods to measure the relative levels of (A) IE1 72 or (B) IE2 86 transcripts. The values plotted on the graphs are the averages of two or three independent experiments, and range bars indicating the highest and lowest values obtained in the independent experiments are shown. To ensure that an equal amount of RNA was included in each reaction, samples were analyzed with G6PD-specific primers and probe. Values shown in the graphs have been standardized to G6PD levels. When mock-infected cell RNA was analyzed with either one of the TaqMan probes, amplification was near or below the limit of detection. In addition, there was no amplification when the samples were treated with RNase prior to PCR analysis.
These results are consistent with the increase in IE2 86 protein observed when IE2 40 is not expressed but differ from the observation that cells infected with IE2 Δ60 (and IE2 Δ40+60) mutant virus overexpress IE1 72 protein compared to wild type. Cells that are infected with the IE2 Δ60 virus express slightly more IE1 72 RNA than do wild-type-virus-infected cells, particularly at 72 h p.i., but the change in IE1 72 transcript levels does not appear sufficient to explain the difference in protein expression. Possible reasons for this discrepancy are discussed below.
Early viral protein expression is not altered by the deletion of IE2 60 or IE2 40 proteins.
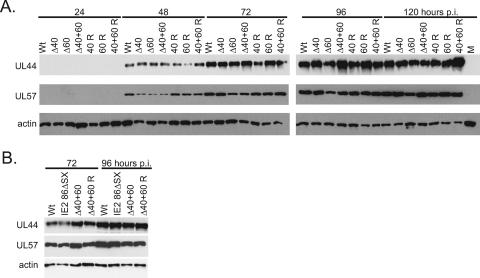
IE2 40 and IE2 60 proteins are expressed with late kinetics, and the low titers observed for the deletion mutants are therefore unlikely to be due to defects in early gene expression. To confirm that defective early gene expression was not the source of the lower titers produced by the mutant viruses, we examined the levels of two early proteins by Western blotting (Fig. 5A). The expression of UL44 and UL57 proteins did not appear to be altered significantly by deletion of IE2 40 or IE2 60, particularly at the later time points examined. UL44 is the HCMV polymerase processivity factor and is expressed with early kinetics (8, 14, 45). UL57 is a protein that binds single-stranded DNA and is also expressed with early kinetics (17). This result suggests that even late in the infection, when the effects of the lack of IE2 40 and IE2 60 might be most apparent, the expression of viral early proteins proceeds normally.
FIG. 5.
Early viral protein expression is not altered following infection with IE2 Δ40 and IE2 Δ60 viruses. (A) G0-synchronized HFF cells were infected with 5 PFU/cell of wild-type (Wt), mutant (Δ40, Δ60, or Δ40+60), and rescued mutant (40 R, 60 R, or 40+60 R) virus or mock infected (M) and harvested at the indicated times p.i. Equal amounts of cell lysates, in micrograms, were separated by SDS-PAGE and transferred to nitrocellulose. UL44 and UL57 protein levels were analyzed by Western blotting as described in Materials and Methods. Cellular actin levels were analyzed as a control for protein loading. (B) G0-synchronized HFF cells were infected with 5 PFU/cell of wild-type (Wt), IE2 86ΔSX-EGFP, IE2 Δ40+60 (Δ40+60), or rescued IE2 Δ40+60 (40+60 R) virus and harvested at the indicated times p.i. Equal amounts of cell lysates, in micrograms, were separated by SDS-PAGE and transferred to nitrocellulose. UL44 and UL57 protein levels were analyzed by Western blotting as described in Materials and Methods. Cellular actin levels were analyzed as a control for protein loading.
The IE2 86ΔSX-EGFP recombinant virus does not express IE2 40 or IE2 60 and also lacks the segment of the open reading frame that codes for aa 136 to 290 of IE2 86. IE2 86ΔSX-EGFP also expresses enhanced green fluorescent protein (EGFP) fused to the C terminus of the mutated IE2 86 protein. IE2 86ΔSX-EGFP and its wild-type parent virus, WT IE2 86-EGFP, have been previously characterized (34). IE2 86ΔSX-EGFP grows indistinguishably from IE2 86ΔSX, a recombinant virus in which the mutated IE2 86 protein has not been fused to EGFP, and WT IE2 86-EGFP grows indistinguishably from the wild-type pHB5-BAC-derived virus used in this study (E. A. White and D. H. Spector, unpublished results). We wanted to distinguish between the effects observed in IE2 86ΔSX-EGFP-infected cells that are due to the lack of aa 136 to 290 versus those that result from the absence of IE2 60 and IE2 40. In a separate experiment, we compared early gene expression in cells infected with IE2 86ΔSX-EGFP, IE2 Δ40+60 virus, and the rescued IE2 Δ40+60 virus. As previously reported, the deletion of aa 136 to 290 in the IE2 86ΔSX-EGFP virus did not affect early gene expression. At the 72 h and 96 h p.i. time points, where the effect of deleting the late forms of IE2 might be most apparent, UL44 protein levels were comparable in IE2 Δ40+60 virus-infected cells and IE2 86ΔSX-EGFP-infected cells. This suggested that neither IE2 40 and IE2 60 nor aa 136 to 290 of IE2 86 contribute directly to UL44 expression (Fig. 5B).
To further verify that viral early gene expression proceeds normally in IE2 40 and IE2 60 deletion mutant virus-infected cells, we examined the expression of representative early proteins in cells infected with wild-type, IE2 Δ40, IE2 Δ60, and IE2 Δ40+60 viruses by immunofluorescence analysis. Gene expression levels in cells infected with any one of the four viruses appeared comparable; in each case, all the cells that expressed IE1 72 protein also expressed UL44 protein by 48 h p.i. (data not shown). Consistent with the Western blot results, this observation suggested that early gene expression, as judged by UL44 positivity in individual infected cells, was not affected by the deletion of IE2 40 or IE2 60.
IE2 60- and 40-kDa proteins contribute to the expression of some late viral genes.
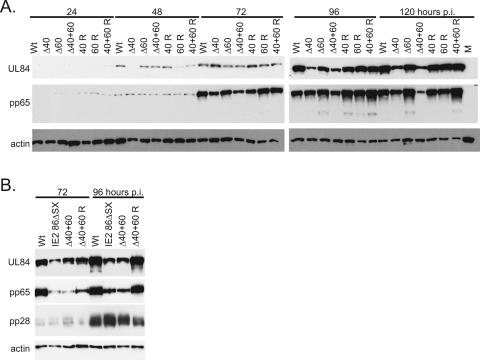
Since IE2 40 and IE2 60 proteins are expressed with late kinetics, we predicted that the deletion mutants would be most likely to show defects in the expression of late viral genes. We therefore examined the expression of several viral genes that are expressed with delayed early or late viral kinetics. First, we measured the levels of UL84 and pp65 proteins by Western blotting following a time course of infection with wild-type virus, deletion mutant viruses, and rescued mutant viruses, in HFF (Fig. 6A). UL84 protein binds to IE2 86 and participates in replication of the viral DNA (6, 29, 40). Although UL84 is initially expressed at early times in the virus-infected cell, the early rise in UL84 RNA is not sustained and UL84 transcript levels are low between 6 to 24 h p.i. Maximum UL84 gene transcription is observed after 72 h p.i., and the study that initially characterized this expression suggested that the peak production of UL84 might depend on DNA replication (12). The UL83 gene encodes the pp65 protein, which is a component of the HCMV tegument (27).
FIG. 6.
UL84 and pp65 protein expression is reduced following infection with IE2 Δ40 and IE2 Δ60 viruses. (A) G0-synchronized HFF cells were infected with 5 PFU/cell of wild-type (Wt), mutant (Δ40, Δ60, or Δ40+60), and rescued mutant (40 R, 60 R, or 40+60 R) virus or mock infected (M) and harvested at the indicated times p.i. Equal amounts of cell lysates, in micrograms, were separated by SDS-PAGE and transferred to nitrocellulose. UL84 and pp65 protein levels were analyzed by Western blotting as described in Materials and Methods. Cellular actin levels were analyzed as a control for protein loading. (B) G0-synchronized HFF cells were infected with 5 PFU/cell of wild-type (Wt), IE2 86ΔSX-EGFP, IE2 Δ40+60 (Δ40+60), or rescued IE2 Δ40+60 (40+60 R) virus and harvested at the indicated times p.i. Equal amounts of cell lysates, in micrograms, were separated by SDS-PAGE and transferred to nitrocellulose. UL84, pp65, and pp28 protein levels were analyzed by Western blotting as described in Materials and Methods. Cellular actin levels were analyzed as a control for protein loading.
Of the three mutant viruses, the IE2 Δ40+60 mutant virus was the most impaired in the expression of UL84 and pp65. Cells infected with the IE2 Δ40+60 mutant virus began to exhibit decreased expression of UL84 and pp65 proteins as early as 72 h p.i. compared to wild-type-virus-infected cells. This lower level of expression was also apparent at 96 and 120 h p.i. In the experiment shown in Fig. 6A, there is approximately twofold more pp65 in wild-type-virus-infected cells than in IE2 Δ40+60 mutant virus-infected cells at 72 h and 96 h p.i. and approximately four times more at 120 h p.i. The deletion of IE2 40 alone also resulted in a small decrease in pp65 protein, but this effect was more modest.
The deletion of IE2 40 also affected the expression of UL84. Quantification of the UL84 Western blots shown indicates that, in the absence of IE2 40 or both IE2 40 and IE2 60, there is a two- to fourfold drop in UL84 protein expression at 96 to 120 h p.i. These results indicated that the absence of IE2 40 protein was sufficient to reduce the expression of UL84 protein compared to wild type. In contrast, decreased levels of pp65 were observed primarily when both IE2 60 and IE2 40 were not expressed. It is possible, then, that pp65 expression does not depend on IE2 40 and that the small amount of IE2 60 protein expressed in the IE2 Δ40 virus-infected cell is sufficient to mediate wild-type levels of pp65 expression.
Decreased expression of pp65 protein had been previously identified as a hallmark of the IE2 86ΔSX infection, but in the previous study the expression of UL84 had not been examined (34). We therefore wanted to compare levels of expression of these delayed early and late factors in cells infected with the IE2 86ΔSX-EGFP and IE2 Δ40+60 viruses. Again, we compared these two mutants at the 72-h- and 96-h-p.i. time points, which had shown some of the most significant changes in the experiment described above. We observed that UL84 and pp65 expression was equally affected by the IE2 86ΔSX mutation and the IE2 Δ40+60 deletions (Fig. 6B). This result suggested that the defect in late protein expression observed for the IE2 86ΔSX-EGFP virus was due to the lack of expression of IE2 40 and IE2 60 proteins, not due to the additional deletion of aa 136 to 290 of IE2 86. IE2 86ΔSX was previously shown to express levels of pp28 protein slightly lower than the wild-type level in a low-multiplicity infection (34). To see whether pp28 protein levels differed in IE2 86ΔSX versus IE2 Δ40+60 virus-infected cells, we examined the expression of pp28 protein in this high-multiplicity experiment. At the higher multiplicity, we did not detect significant differences in pp28 protein expression in wild-type-, IE2 86ΔSX, IE2 Δ40+60, or rescued IE2 Δ40+60 virus-infected cells.
UL83 expression, but not UL84 expression, is affected at the transcriptional level.
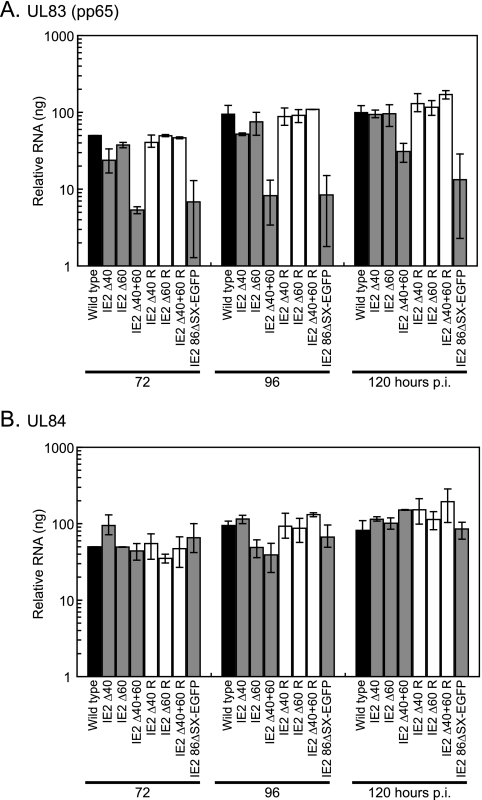
Again, to determine which of the observed changes in protein expression at late times p.i. were due to changes in RNA levels, we measured the levels of UL83 (pp65) and UL84 transcripts by real-time RT-PCR following high-multiplicity infection (Fig. 7A). As a control for RNA contamination, samples were also treated with RNase prior to analysis by real-time RT-PCR. No amplification was observed in the RNase-treated samples. It had previously been shown that, following low-multiplicity infection, decreased pp65 protein levels in IE2 86ΔSX-EGFP virus-infected cells corresponded to a decrease in the UL83 transcript, which encodes pp65 (34). Here, UL83 levels following a high-multiplicity infection behaved in the same way and were between 7- and 12-fold lower in IE2 86ΔSX-EGFP virus-infected cells than in wild-type-virus-infected cells. Using the IE2 Δ40+60 mutant virus, we saw that this effect could be entirely due to the lack of IE2 40 and IE2 60 expression in IE2 86ΔSX-EGFP-infected cells. IE2 Δ40+60 virus-infected cells expressed up to 12-fold less UL83 RNA than did wild-type-virus-infected cells, and this result is consistent with our observation that pp65 protein levels were lower in IE2 Δ40+60 virus-infected cells than in wild-type-virus-infected cells.
FIG. 7.
UL83, but not UL84, RNA levels are altered in deletion mutant virus-infected cells. G0-synchronized HFF cells were infected with 5 PFU/cell of wild-type virus, IE2 86ΔSX-EGFP virus, mutant viruses (IE2 Δ40, IE2 Δ60, or IE2 Δ40+60), and rescued mutant viruses (IE2 Δ40 R, IE2 Δ60 R, or IE2 Δ40+60 R) or mock infected and harvested at the indicated times p.i. Total RNA was analyzed by quantitative real-time RT-PCR as described in Materials and Methods to measure the relative levels of (A) UL83 or (B) UL84 transcripts. The values plotted on the graphs are the averages of two or three independent experiments, and range bars indicating the highest and lowest values obtained in the independent experiments are shown. To ensure that an equal amount of RNA was included in each reaction, samples were analyzed with G6PD-specific primers and probe. Values shown in the graphs have been standardized to G6PD levels. When mock-infected cell RNA was analyzed with either one of the TaqMan probes, amplification was near or below the limit of detection. In addition, there was no amplification when the samples were treated with RNase prior to PCR analysis.
Finally, we wanted to determine whether changes in UL84 transcript levels might similarly cause the differences we observed in UL84 protein abundance. In contrast to the UL83 result, UL84 RNA levels were not significantly affected by infection with any of the deletion mutant viruses or with IE2 86ΔSX-EGFP (Fig. 7B). In the experiment shown, there were small fluctuations in transcript levels, but the mutant-virus-infected cells expressed an amount of RNA that was at most 2.4-fold different than the level in the wild-type-virus-infected cells, and this difference was not observed at every time point. This result suggested that the lower levels of UL84 protein observed in IE2 Δ40 and IE2 Δ40+60 virus-infected cells did not result from decreased expression of the UL84 transcript.
DISCUSSION
The IE2 60- and 40-kDa proteins are abundantly expressed in the HCMV-infected cell and are colinear with the C terminus of the essential viral regulatory protein IE2 86, but their contributions to the HCMV infection have not been well understood. An earlier study showed that the IE2 40 protein could function both as an activator and a repressor of the major IE promoter in a transient-transfection assay (15). Due to its methodology, this work could not identify functions of IE2 40 that are separate from the functions provided by full-length IE2 86 or confirm the role of IE2 40 in the HCMV-infected cell, but it did raise interesting questions about how IE2 40 and IE2 60 might contribute to virus replication. A study from our laboratory suggested that IE2 40 and IE2 60 were not essential for HCMV growth in cultured cells, but the recombinant virus used in this study carried an additional deletion of aa 136 to 290 of IE2 86 that complicated attempts to assign functions to the smaller proteins (34). The slow growth of this recombinant also suggested that IE2 40 and IE2 60 might, individually or in combination, contribute to the virus's ability to replicate to wild-type levels and with wild-type kinetics. Finally, the IE2 40 and IE2 60 proteins are unique among the factors expressed from the major IE region. Unlike the proteins that arise from splice variant transcripts under unique conditions (such as release from a cycloheximide block) and are expressed in cell types other than fibroblasts, the expression of IE2 40 and IE2 60 is robust and easily detectable in infected fibroblasts and occurs with late kinetics. These observations suggested to us that these proteins might provide functions at late times during HCMV infection that had not been previously identified. This idea was supported by our previous observation that mutations in IE2 86 led to altered regulation of late viral genes (46), suggesting that IE2 86 or related proteins could directly regulate late viral gene expression in HCMV-infected cells.
To further investigate these questions, we constructed and characterized recombinant viruses that lack the ability to express one or both of the IE2 40 and IE2 60 proteins. Neither small form of IE2 is required for HCMV replication to proceed, but both contribute to efficient virus growth. We began by measuring replication of the mutant viruses following high-multiplicity infections and demonstrated that their growth is impaired. The IE2 Δ40+60 mutant virus, which does not express either small protein, shows the largest defect in replication. IE2 Δ40+60 replicates to titers that are about 10-fold lower than the titers of wild-type or rescued mutant virus in a matched infection. However, the kinetics of virus replication are the same for the mutant and wild-type viruses under these conditions.
Further analysis of the events in the viral life cycle that might be impaired and therefore contribute to this decrease in replication revealed two main effects: altered expression of full-length IE2 86 and IE1 72 proteins in deletion mutant virus-infected cells and reduced levels of two viral proteins that are produced late in the infection. These two effects and their proposed causes are discussed more fully below.
First, we observed that, in mutant-virus-infected cells, IE1 72 and IE2 86 expression was altered at both the RNA and protein levels. The most striking difference is that, when changes were made to the 1.5-kb RNA promoter and IE2 40 protein is not expressed, we observed an increase in IE2 86 and IE1 72 RNA levels. This behavior is consistent with the prediction that IE2 40 functions as a repressor of the major IE promoter (15). When IE2 40 is not expressed, we expect that overall levels of the major IE transcript will increase and that there will be a corresponding increase in IE1 72 and IE2 86 RNAs. Indeed, we observed these predicted changes in RNA levels.
Unexpectedly, the increase in the major IE RNA is reflected at the protein level for IE2 86, but not for IE1 72. IE1 72 protein levels are more affected by changes to the upstream AT-rich site, when the change is part of either the IE2 Δ60 virus or the IE2 Δ40+60 virus. Although it is not yet clear why the change to the upstream AT-rich site more directly impacts IE1 72, we note that the mutation that has altered that site has changed not only a possible promoter for the ∼1.7-kb late IE2 RNAs but also the sequence of the 3′ untranslated region for the IE1 72 message. It is possible that introducing this change into the IE1 72 RNA has increased its translation relative to wild type but that its transcription has not been significantly altered. Experiments to address these questions are in progress.
Two viral genes that are expressed with delayed early kinetics behaved differently in cells infected with the mutant viruses. These effects were most apparent in cells infected with the IE2 Δ40+60 mutant virus, which does not express any detectable IE2 40 or IE2 60 protein. The expression of the UL83 gene, which encodes pp65, was decreased in IE2 Δ40+60 virus-infected cells at the level of both RNA and protein synthesis. This is the same result that had previously been reported for the IE2 86ΔSX-EGFP virus and therefore demonstrates that full expression of pp65 depends on the IE2 40 and IE2 60 proteins and not on aa 136 to 290 of IE2 86. There are two ways in which these proteins could be responsible for pp65 expression. It is possible that some expression of either IE2 40 or IE2 60 protein is required for full expression of pp65, and so we detect the drop in pp65 RNA and protein levels only when neither late form of IE2 is expressed. However, we cannot rule out the alternative possibility that a small amount of IE2 60 protein alone is sufficient to support the full expression of pp65 and that we therefore detect wild-type levels of pp65 protein and RNA in both IE2 Δ40 and IE2 Δ60 virus-infected cells.
In contrast, IE2 40 and IE2 60 must contribute differently to the regulation of UL84 expression. While UL84 protein is less abundant in cells infected with IE2 Δ40+60 mutant virus or with IE2 86ΔSX-EGFP virus relative to wild-type-virus-infected cells, we do not see a significant drop in UL84 RNA in mutant-virus-infected cells. A similar drop in UL84 protein, but not RNA, is also observed for the IE2 Δ40 virus, indicating that deletion of the 40-kDa protein is sufficient to mediate this effect. Reducing levels of the IE2 60 protein alone does not alter UL84 expression, although whether this is due to the higher abundance of IE2 40 than IE2 60 protein or due to the difference in their sequences is not clear. It appears that, while UL83 expression is regulated by IE2 40 or IE2 60 at the level of transcription (or RNA stability), UL84 is regulated by IE2 40 at the level of translation or protein stability. Experiments to distinguish between these alternatives are in progress.
It is intriguing that two HCMV-encoded proteins that are identical to 50% or more of the essential viral protein IE2 86 and are more abundant than IE2 86 are not themselves required for productive HCMV infection of cultured fibroblasts. These factors clearly contribute to specific events in the HCMV replicative cycle, and no doubt there are additional events dependent on IE2 40 and IE2 60 that have not yet been identified. Further studies will aim to demonstrate how these proteins contribute to the progression of the HCMV infection by specific activation and repression of viral and cellular gene expression at late times in the virus-infected cell.
Acknowledgments
We are grateful to Edward Mocarski for the initial suggestion of sequence changes for the IE2 40 TATAA mutation. We thank Martin Messerle for providing the pHB5 BAC.
This work was supported by NIH grants CA73490 and CA34729. E.A.W. was supported by NIH training grant GM07240.
Footnotes
Published ahead of print on 3 January 2007.
REFERENCES
- 1.Awasthi, S., J. A. Isler, and J. C. Alwine. 2004. Analysis of splice variants of the immediate-early 1 region of human cytomegalovirus. J. Virol. 78:8191-8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherrington, J. M., E. L. Khoury, and E. S. Mocarski. 1991. Human cytomegalovirus IE2 negatively regulates α gene expression via a short target sequence near the transcription start site. J. Virol. 65:887-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiou, C.-J., J. Zong, I. Waheed, and G. S. Hayward. 1993. Identification and mapping of dimerization and DNA-binding domains in the C terminus of the IE2 regulatory protein of human cytomegalovirus. J. Virol. 67:6201-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colberg-Poley, A. M., L. D. Santomenna, P. P. Harlow, P. A. Benfield, and D. J. Tenney. 1992. Human cytomegalovirus US3 and UL36-38 immediate-early proteins regulate gene expression. J. Virol. 66:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colletti, K. S., Y. Xu, S. A. Cei, M. Tarrant, and G. S. Pari. 2004. Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication. J. Virol. 78:9203-9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colletti, K. S., Y. Xu, I. Yamboliev, and G. S. Pari. 2005. Human cytomegalovirus UL84 is a phosphoprotein that exhibits UTPase activity and is a putative member of the DExD/H box family of proteins. J. Biol. Chem. 280:11955-11960. [DOI] [PubMed] [Google Scholar]
- 8.Ertl, P. F., and K. L. Powell. 1992. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J. Virol. 66:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortunato, E. A., and D. H. Spector. 1999. Regulation of human cytomegalovirus gene expression. Adv. Virus Res. 54:61-128. [DOI] [PubMed] [Google Scholar]
- 10.Gawn, J. M., and R. F. Greaves. 2002. Absence of IE1 p72 protein function during low-multiplicity infection by human cytomegalovirus results in a broad block to viral delayed-early gene expression. J. Virol. 76:4441-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greaves, R. F., and E. S. Mocarski. 1998. Defective growth correlates with reduced accumulation of a viral DNA replication protein after low-multiplicity infection by a human cytomegalovirus ie1 mutant. J. Virol. 72:366-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He, Y. S., L. Xu, and E. S. Huang. 1992. Characterization of human cytomegalovirus UL84 early gene and identification of its putative protein product. J. Virol. 66:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang, L., and M. F. Stinski. 1995. Binding of cellular repressor protein or the IE2 protein to a cis-acting negative regulatory element upstream of a human cytomegalovirus early promoter. J. Virol. 69:7612-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hwang, E. S., J. Kim, H. S. Jong, J. W. Park, C. G. Park, and C. Y. Cha. 2000. Characteristics of DNA-binding activity of human cytomegalovirus ppUL44. Microbiol. Immunol. 44:827-832. [DOI] [PubMed] [Google Scholar]
- 15.Jenkins, D. E., C. L. Martens, and E. S. Mocarski. 1994. Human cytomegalovirus late protein encoded by ie2: a transactivator as well as a repressor of gene expression. J. Gen. Virol. 75:2337-2348. [DOI] [PubMed] [Google Scholar]
- 16.Kerry, J. A., A. Sehgal, S. W. Barlow, V. J. Cavanaugh, K. Fish, J. A. Nelson, and R. M. Stenberg. 1995. Isolation and characterization of a low-abundance splice variant from the human cytomegalovirus major immediate-early gene region. J. Virol. 69:3868-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiehl, A., L. Huang, D. Franchi, and D. G. Anders. 2003. Multiple 5′ ends of human cytomegalovirus UL57 transcripts identify a complex, cycloheximide-resistant promoter region that activates oriLyt. Virology 314:410-422. [DOI] [PubMed] [Google Scholar]
- 18.Klucher, K. M., M. Sommer, J. T. Kadonaga, and D. H. Spector. 1993. In vivo and in vitro analysis of transcriptional activation mediated by the human cytomegalovirus major immediate-early proteins. Mol. Cell. Biol. 13:1238-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang, D., and T. Stamminger. 1993. The 86-kilodalton IE-2 protein of human cytomegalovirus is a sequence-specific DNA-binding protein that interacts directly with the negative autoregulatory response element located near the cap site of the IE-1/2 enhancer-promoter. J. Virol. 67:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang, D., and T. Stamminger. 1994. Minor groove contacts are essential for an interaction of the human cytomegalovirus IE2 protein with its DNA target. Nucleic Acids Res. 22:3331-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lischka, P., C. Rauh, R. Mueller, and T. Stamminger. 2006. Human cytomegalovirus UL84 protein contains two nuclear export signals and shuttles between the nucleus and the cytoplasm. J. Virol. 80:10274-10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lischka, P., G. Sorg, M. Kann, M. Winkler, and T. Stamminger. 2003. A nonconventional nuclear localization signal within the UL84 protein of human cytomegalovirus mediates nuclear import via the importin alpha/beta pathway. J. Virol. 77:3734-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, B., T. W. Hermiston, and M. F. Stinski. 1991. A cis-acting element in the major immediate-early (IE) promoter of human cytomegalovirus is required for negative regulation by IE2. J. Virol. 65:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macias, M. P., and M. F. Stinski. 1993. An in vitro system for human cytomegalovirus immediate early 2 protein (IE-2)-mediated site-dependent repression of transcription and direct binding of IE2 to the major immediate-early promoter. Proc. Natl. Acad. Sci. USA 90:707-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malone, C. L., D. H. Vesole, and M. F. Stinski. 1990. Transactivation of a human cytomegalovirus early promoter by gene products from the immediate-early gene IE2 and augmentation by IE1: mutational analysis of the viral proteins. J. Virol. 64:1498-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mocarski, E. S., and C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 28.Mocarski, E. S., G. W. Kemble, J. M. Lyle, and R. F. Greaves. 1996. A deletion mutant in the human cytomegalovirus gene encoding ie1(491aa) is replication defective due to a failure in autoregulation. Proc. Natl. Acad. Sci. USA 93:11321-11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pizzorno, M. C., and G. S. Hayward. 1990. The IE2 gene products of human cytomegalovirus specifically down-regulate expression from the major immediate-early promoter through a target sequence located near the cap site. J. Virol. 64:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pizzorno, M. C., M.-A. Mullen, Y.-N. Chang, and G. S. Hayward. 1991. The functionally active IE2 immediate-early regulatory protein of human cytomegalovirus is an 80-kilodalton polypeptide that contains two distinct activator domains and a duplicated nuclear localization signal. J. Virol. 65:3839-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plachter, B., W. Britt, R. Vornhagen, T. Stamminger, and G. Jahn. 1993. Analysis of proteins encoded by IE regions 1 and 2 of human cytomegalovirus using monoclonal antibodies generated against recombinant antigens. Virology 193:642-652. [DOI] [PubMed] [Google Scholar]
- 33.Puchtler, E., and T. Stamminger. 1991. An inducible promoter mediates abundant expression from the immediate-early 2 gene region of human cytomegalovirus at late times after infection. J. Virol. 65:6301-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 76:2973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz, R., M. H. Sommer, A. Scully, and D. H. Spector. 1994. Site-specific binding of the human cytomegalovirus IE2 86-kilodalton protein to an early gene promoter. J. Virol. 68:5613-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scully, A. L., M. H. Sommer, R. Schwartz, and D. H. Spector. 1995. The human cytomegalovirus IE2 86-kilodalton protein interacts with an early gene promoter via site-specific DNA binding and protein-protein associations. J. Virol. 69:6533-6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirakata, M., M. Terauchi, M. Ablikim, K. Imadome, K. Hirai, T. Aso, and Y. Yamanashi. 2002. Novel immediate-early protein IE19 of human cytomegalovirus activates the origin recognition complex I promoter in a cooperative manner with IE72. J. Virol. 76:3158-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sommer, M. H., A. L. Scully, and D. H. Spector. 1994. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J. Virol. 68:6223-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spector, D. J., and M. J. Tevethia. 1994. Protein-protein interactions between human cytomegalovirus IE2-580aa and pUL84 in lytically infected cells. J. Virol. 68:7549-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenberg, R. M., A. S. Depto, J. Fortney, and J. A. Nelson. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J. Virol. 63:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stenberg, R. M., J. Fortney, S. W. Barlow, B. P. Magrane, J. A. Nelson, and P. Ghazal. 1990. Promoter-specific trans activation and repression by human cytomegalovirus immediate-early proteins involves common and unique protein domains. J. Virol. 64:1556-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamashiro, J. C., L. J. Hock, and D. H. Spector. 1982. Construction of a cloned library of the EcoRI fragments from the human cytomegalovirus genome (strain AD169). J. Virol. 42:547-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waheed, I., C. Chiou, J. Ahn, and G. Hayward. 1998. Binding of the human cytomegalovirus 80-kDa immediate-early protein (IE2) to minor groove A/T-rich sequences bounded by CG dinucleotides is regulated by protein oligomerization and phosphorylation. Virology 252:235-257. [DOI] [PubMed] [Google Scholar]
- 45.Weiland, K. L., N. L. Oien, F. Homa, and M. W. Wathen. 1994. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 34:191-206. [DOI] [PubMed] [Google Scholar]
- 46.White, E. A., C. L. Clark, V. Sanchez, and D. H. Spector. 2004. Small internal deletions in the human cytomegalovirus IE2 gene result in nonviable recombinant viruses with differential defects in viral gene expression. J. Virol. 78:1817-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu, Y., S. A. Cei, A. R. Huete, and G. S. Pari. 2004. Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth. J. Virol. 78:10360-10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu, Y., K. S. Colletti, and G. S. Pari. 2002. Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments. J. Virol. 76:8931-8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeung, K. C., C. M. Stoltzfus, and M. F. Stinski. 1993. Mutations of the human cytomegalovirus immediate-early 2 protein defines regions and amino acid motifs important in transactivation of transcription from the HIV-1 LTR promoter. Virology 195:786-792. [DOI] [PubMed] [Google Scholar]