Abstract
Chronic hepatitis C virus (HCV) infection is a significant worldwide health problem with limited therapeutic options. A number of novel, small molecular inhibitors of HCV replication are now entering early clinical trials in humans. Resistance to small molecular inhibitors is likely to be a significant hurdle to their use in patients. A systematic assessment of combinations of interferon and/or novel anti-hepatitis C virus agents from several different mechanistic classes was performed in vitro. Combinations of inhibitors with different mechanisms of action consistently demonstrated more synergy than did compounds with similar mechanisms of action. These results suggest that combinations of inhibitors with different mechanisms of action should be prioritized for assessment in clinical trials for chronic hepatitis C virus infection.
Chronic hepatitis C virus (HCV) infection is a major worldwide health problem; in the United States, an estimated 3 million persons are chronically infected (4). Estimates of the health care burden of chronic HCV infection predict a drastic increase in hospitalizations and medical costs related to complications such as cirrhosis and hepatocellular carcinoma over the next 1 to 2 decades (3). Effective and better-tolerated therapy for HCV could effectively stem this tide (7).
Current interferon-based therapy for chronic HCV infection results in sustained responses in roughly 55% of patients and is accompanied by significant toxicity. Genotype 1 HCV, the most prevalent genotype in the United States, responds less well to therapy with pegylated interferon plus ribavirin, with response rates of 42 to 46% (11, 23). These limitations have spurred an intense drug discovery effort, resulting in a number of promising compounds (8).
Hepatitis C virus replication takes place in the cytoplasm, with the replication complex being tightly associated with lipid membranes (1). Key components of the replication complex include several promising antiviral targets, including the NS3/4A protease and the NS5B RNA-dependent RNA polymerase. A number of candidate protease inhibitors (PIs) which have excellent potency in vitro have been developed (2, 17, 20); several of these compounds have also been evaluated in phase I/II trials, with encouraging results (15, 16, 29, 36). Resistance to this class of inhibitors has been described, with some mutations conferring cross-resistance to several compounds (17, 18, 21, 34, 35).
The NS5B RNA polymerase is also essential for viral replication, and a number of nucleoside inhibitors and nonnucleoside inhibitors (NNIs) of the HCV polymerase with potent activity in vitro and in early clinical trials have been described (5, 12, 13, 27, 30). Resistance to both nucleoside and nonnucleoside inhibitors in vitro has been described (22, 24, 26). We have assessed a number of combinations of HCV inhibitors with several molecular targets currently in development, using an HCV genotype 1 replicon-based luciferase reporter system.
Replicon constructs.
The BM4-5 replicon is a subgenomic HCV genotype 1b replicon which contains a deletion of a serine in NS5A and has been previously described (14). The firefly luciferase gene was inserted in the BM4-5 replicon, in a manner previously described (33), to create a luciferase/neomycin phosphotransferase fusion protein (FEO) and the replicon (BM4-5 FEO). Briefly, the Photinus pyralis luciferase gene was amplified using primers coding for the AscI restriction site. Following amplification, both the BM4-5 plasmid and luciferase PCR product were restriction digested with AscI. Ligation was then carried out to insert the luciferase gene in phase with the neomycin phosphotransferase gene, creating the desired BM4-5 FEO replicon. The sequence of the replicon was verified by DNA sequencing.
Cell culture.
Human hepatoma Huh-7.5.1 cells (a kind gift from Francis Chisari, Scripps Research Institute, La Jolla, CA) and BM4-5 FEO cells stably expressing the BM4-5 FEO replicon were grown at 37°C and 5% CO2 in Dulbecco's modification of Eagle's medium supplemented with 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal bovine serum. BM4-5 FEO cells were additionally grown in the presence of 500 μg/ml of G-418.
Transfection and clone selection.
The BM4-5 FEO plasmid was linearized with ScaI. In vitro transcription (Megascript; Ambion) was carried out according to the manufacturer's instruction to yield BM4-5 FEO RNA. Transfection was performed as previously described (32). Four hundred microliters of a Huh-7.5.1 cell suspension (107 cells/ml) was placed in a 0.4-cm cuvette with 10 μg of BM4-5 FEO RNA. The mixture was electroporated (Bio-Rad Gene Pulser) at 270 V and 975 μF and transferred to a 10-cm tissue culture dish. G-418 at 500 μg/ml was added at 24 h, and the medium was changed every 3 to 4 days. Individual G-418-resistant colonies were visible within 2 to 3 weeks. Individual colonies were harvested and expanded for characterization of luciferase expression.
Luciferase compound assay.
BM4-5 FEO cells were seeded into 96-well plates at a density of 10,000 cells per well in 100 μl medium. After allowing 4 h for attachment, compounds were added to wells at the specified concentrations. All conditions were run in triplicate. Cells and compounds were incubated for 48 h. The luciferase assay (Bright-Glo; Promega) was carried out according to the manufacturer's instructions. Luciferase activity was determined using a microplate luminometer (Veritas microplate luminometer; Turner Biosystems). The relative light units (RLU) for each condition were reported as the mean ± the standard error of the mean for the three wells.
Compounds tested.
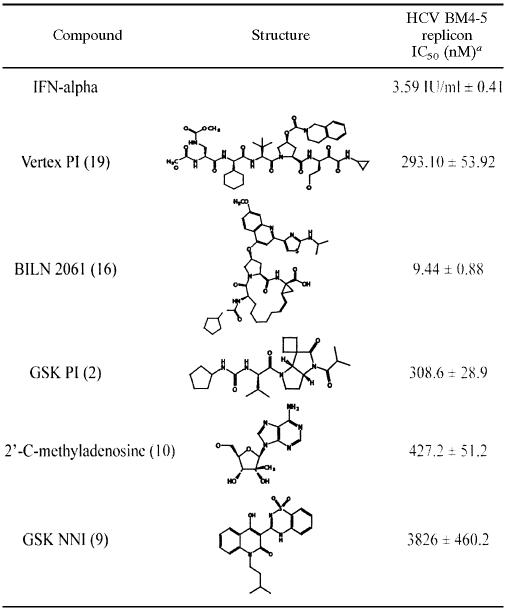
Compounds tested included two peptidomimetic HCV PIs, BILN 2061 (16) and a Vertex PI (19) (Vicki Sato, Vertex Pharmaceuticals, Cambridge, MA); a GlaxoSmithKline trans-lactam PI active-site mimic (2) (Karen Romines, GlaxoSmithKline, Research Triangle Park, NC); one nucleoside analog HCV RNA-dependent RNA polymerase inhibitor (RdRpI), 2′-C-methyladenosine (10) (William Lee, Gilead Sciences, Foster City, CA); one nonnucleoside GSK benzo-thiadiazine RNA polymerase inhibitor directed at the “thumb” region of the polymerase (Karen Romines, GlaxoSmithKline) (9); and alpha interferon (Interferon-αA; Sigma-Aldrich).
The 50% inhibitory concentration (IC50) of each compound was determined independently and used to set the range of concentrations used for the synergy experiments. Each compound was tested singly and in combination at two twofold serial dilutions above and below the IC50. The ratio of the two compounds tested remained fixed across the dosing range. Potential cytotoxicity of individual compounds and all combinations was assessed using a luminescent ATP-based cell viability assay (Cell Titer-Glo; Promega).
Data analysis.
Determinations of compound interactions were based on the median-effect principle and the multiple drug effect equation as described by Chou and Talalay (6). Combination indices (CIs) were determined using Calcusyn (Biosoft) for each experiment at the IC50, IC70, and IC90 levels. In total, 15 combinations were evaluated with from three to five replicates per condition; this yielded a total of 61 data points per CI level analyzed. A CI of <0.9 was considered synergistic, a CI of ≥0.9 or ≤1.1 was considered additive, and a CI of >1.1 was deemed antagonistic.
Statistical analysis.
At each of the three inhibitory concentrations evaluated (IC50, IC70, and IC90) the CIs in the three synergy groups were compared using a linear mixed-effects model allowing for different means in the three synergy groups and random effects for the individual drug combinations. The random effects were not significant (likelihood ratio test), indicating no statistical difference in CI values between the antiviral compound combinations in the same synergy group. The CI replicates were further compared between synergy groups by using the Wilcoxon rank test.
Synergy of small molecular inhibitors.
Transfection of Huh-7.5.1 cells with BM4-5 FEO RNA yielded numerous (>50) G-418-resistant clones. Individual clones were expanded and assessed using the luciferase assay to determine the individual clones with highest RLU per cell. Four clones yielded from 30,000 to 50,000 RLU per 10,000 cells at 48 h (data not shown); these clones were expanded and used for all subsequent studies.
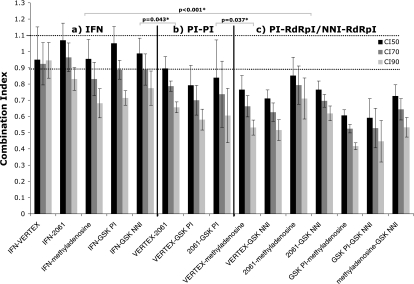
The IC50 for each of the individual compounds is listed in Table 1. CI50, CI70, and CI90 refer to the combination index at the IC50, IC70, and IC90, respectively, of each drug. All compounds tested were additive (CI50 and CI70) or mildly synergistic (CI90) with alpha interferon. Antagonism was not demonstrated for any combination of small molecular inhibitors, including compounds targeting the same viral protein. Significantly more synergy was demonstrated between compounds in the group combining two small molecular inhibitors targeting the same viral enzyme (in this case NS3 protease) than between the group of compounds combined with alpha interferon at both the CI70 (P = 0.043) and CI90 (P = 0.017) levels. There was no significant difference between the groups at the CI50 level (P = 0.108) (Fig. 1). Similarly, the group consisting of two inhibitors with different viral targets showed significantly lower combination indices than either of the other two groups, i.e., compounds with interferon (P < 0.001 at all levels) or compounds with same mechanism of action (P = 0.038 and 0.037 at the CI50 and CI70 levels, respectively). The comparison of CI90s between small molecular inhibitors with the same and different viral targets showed a trend toward a lower combination index in the group with two compounds with different viral targets (P = 0.056 at the CI90 level) (Fig. 1). None of the compounds or combinations showed cytotoxicity at the concentrations tested in the activity and synergy studies (data not shown).
TABLE 1.
Activities of different small molecular inhibitors in the BM4-5 replicon
a The IC50 is the average ± standard error of mean of the results from at least three independent experiments.
FIG. 1.
CI50s, CI70s, and CI90s for the compound combinations evaluated. Dotted lines at combination index values of 0.9 and 1.1 indicate the boundaries of an additive interaction. The P values displayed (*) are for analyses at the CI70 level.
Small molecular inhibitors of the HCV protease and polymerase show antiviral activity in our genotype 1 replicon system. Most importantly, no combination of small molecular inhibitors of HCV replication demonstrated antagonism in our system, including those with the same mechanism of action or viral target. Combinations of inhibitors targeting different viral proteins (PI-RdRpI or PI-NNI) or with different mechanisms of inhibiting the same viral protein (RdRpI-NNI) were strongly synergistic and had significantly lower combination indices than the other two groups. Combinations targeting the same site within a viral protein showed lesser degrees of synergy or were additive, but they still possessed significantly lower combination indices than the group composed of the same compounds with alpha interferon. It is important to remember that the definition of synergy as a CI of less than 0.9 is an arbitrary distinction (along a continuum) and thus does not preclude two inhibitors which occupy the same site from being “synergistic” according to a CI of <0.9. Additionally, metabolic interactions between compounds or the impact of divergent resistance pathways on different compounds may also affect the appearance of drug-drug interactions as assessed by the combination index.
HCV, like human immunodeficiency virus type 1, possesses an error-prone RNA polymerase, and it replicates to levels 10- to 100-fold higher than those of human immunodeficiency virus type 1 in chronically infected individuals (25, 28). These characteristics suggest that selection of drug-resistant viral variants will be a challenge to the use of small molecular inhibitors. In fact, resistance to these compounds both in vitro and in vivo has already been described (17, 24, 26, 31, 34).
Synergistic combinations of HCV inhibitors may produce greater viral load decreases in vivo and could potentially delay the appearance of multiply drug-resistant virus. This system provides a useful approach for the in vitro testing of antiviral combinations in anticipation of rationally designed clinical studies of combination chemotherapy directed at HCV. Our results support the evaluation of combinations of small molecular inhibitors in human clinical trials and further suggest that combinations with different mechanisms of action may be particularly attractive.
Acknowledgments
This work was funded in part by a 2005 developmental grant from the UC San Diego Center for AIDS Research, an NIH-funded program (no. 5P30 AI-36214).
Footnotes
Published ahead of print on 20 December 2006.
REFERENCES
- 1.Aizaki, H., K. J. Lee, V. M. Sung, H. Ishiko, and M. M. Lai. 2004. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology 324:450-461. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, D. M., M. C. Barnes, M. D. Dowle, S. L. Hind, M. R. Johnson, P. S. Jones, G. Mills, A. Patikis, T. J. Pateman, T. J. Redfern, J. E. Robinson, M. J. Slater, and N. Trivedi. 2003. Pyrrolidine-5,5-trans-lactams. 5. Pharmacokinetic optimization of inhibitors of hepatitis C virus NS3/4A protease. Org. Lett. 5:4631-4634. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong, G. L., M. J. Alter, G. M. McQuillan, and H. S. Margolis. 2000. The past incidence of hepatitis C virus infection: implications for the future burden of chronic liver disease in the United States. Hepatology 31:777-782. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong, G. L., A. Wasley, E. P. Simard, G. M. McQuillan, W. L. Kuhnert, and M. J. Alter. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705-714. [DOI] [PubMed] [Google Scholar]
- 5.Chan, L., T. J. Reddy, M. Proulx, S. K. Das, O. Pereira, W. Wang, A. Siddiqui, C. G. Yannopoulos, C. Poisson, N. Turcotte, A. Drouin, M. H. aoui-Ismaili, R. Bethell, M. Hamel, L. L'Heureux, D. Bilimoria, and N. Nguyen-Ba. 2003. Identification of N,N-disubstituted phenylalanines as a novel class of inhibitors of hepatitis C NS5B polymerase. J. Med. Chem. 46:1283-1285. [DOI] [PubMed] [Google Scholar]
- 6.Chou, T. C., and P. Talalay. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22:27-55. [DOI] [PubMed] [Google Scholar]
- 7.Davis, G. L., J. E. Albright, S. F. Cook, and D. M. Rosenberg. 2003. Projecting future complications of chronic hepatitis C in the United States. Liver Transpl. 9:331-338. [DOI] [PubMed] [Google Scholar]
- 8.De Francesco. R., L. Tomei, S. Altamura, V. Summa, and G. Migliaccio. 2003. Approaching a new era for hepatitis C virus therapy: inhibitors of the NS3-4A serine protease and the NS5B RNA-dependent RNA polymerase. Antiviral Res. 58:1-16. [DOI] [PubMed] [Google Scholar]
- 9.Dhanak, D., K. J. Duffy, V. K. Johnston, J. Lin-Goerke, M. Darcy, A. N. Shaw, B. Gu, C. Silverman, A. T. Gates, M. R. Nonnemacher, D. L. Earnshaw, D. J. Casper, A. Kaura, A. Baker, C. Greenwood, L. L. Gutshall, D. Maley, A. DelVecchio, R. Macarron, G. A. Hofmann, Z. Alnoah, H. Y. Cheng, G. Chan, S. Khandekar, R. M. Keenan, and R. T. Sarisky. 2002. Identification and biological characterization of heterocyclic inhibitors of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 277:38322-38327. [DOI] [PubMed] [Google Scholar]
- 10.Eldrup, A. B., M. Prhavc, J. Brooks, B. Bhat, T. P. Prakash, Q. Song, S. Bera, N. Bhat, P. Dande, P. D. Cook, C. F. Bennett, S. S. Carroll, R. G. Ball, M. Bosserman, C. Burlein, L. F. Colwell, J. F. Fay, O. A. Flores, K. Getty, R. L. Lafemina, J. Leone, M. MacCoss, D. R. McMasters, J. E. Tomassini, L. D. Von, B. Wolanski, and D. B. Olsen. 2004. Structure-activity relationship of heterobase-modified 2′-C-methyl ribonucleosides as inhibitors of hepatitis C virus RNA replication. J. Med. Chem. 47:5284-5297. [DOI] [PubMed] [Google Scholar]
- 11.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 12.Gopalsamy, A., A. Aplasca, G. Ciszewski, K. Park, J. W. Ellingboe, M. Orlowski, B. Feld, and A. Y. Howe. 2006. Design and synthesis of 3,4-dihydro-1H-[1]-benzothieno[2,3-c]pyran and 3,4-dihydro-1H-pyrano[3,4-b]benzofuran derivatives as non-nucleoside inhibitors of HCV NS5B RNA dependent RNA polymerase. Bioorg. Med. Chem. Lett. 16:457-460. [DOI] [PubMed] [Google Scholar]
- 13.Gopalsamy, A., K. Lim, G. Ciszewski, K. Park, J. W. Ellingboe, J. Bloom, S. Insaf, J. Upeslacis, T. S. Mansour, G. Krishnamurthy, M. Damarla, Y. Pyatski, D. Ho, A. Y. Howe, M. Orlowski, B. Feld, and J. O'Connell. 2004. Discovery of pyrano[3,4-b]indoles as potent and selective HCV NS5B polymerase inhibitors. J. Med. Chem. 47:6603-6608. [DOI] [PubMed] [Google Scholar]
- 14.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinrichsen, H., Y. Benhamou, H. Wedemeyer, M. Reiser, R. E. Sentjens, J. L. Calleja, X. Forns, A. Erhardt, J. Cronlein, R. L. Chaves, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2004. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127:1347-1355. [DOI] [PubMed] [Google Scholar]
- 16.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, G. R. St, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 17.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y. P. Luong, J. D. Frantz, K. Lin, S. Ma, Y. Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 18.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 19.Lin, K., A. D. Kwong, and C. Lin. 2004. Combination of a hepatitis C virus NS3-NS4A protease inhibitor and alpha interferon synergistically inhibits viral RNA replication and facilitates viral RNA clearance in replicon cells. Antimicrob. Agents Chemother. 48:4784-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, K., R. B. Perni, A. D. Kwong, and C. Lin. 2006. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCv replicon cells. Antimicrob. Agents Chemother. 50:1813-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludmerer, S. W., D. J. Graham, E. Boots, E. M. Murray, A. Simcoe, E. J. Markel, J. A. Grobler, O. A. Flores, D. B. Olsen, D. J. Hazuda, and R. L. LaFemina. 2005. Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay. Antimicrob. Agents Chemother. 49:2059-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 24.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, F. R. De, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 25.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen, T. T., A. T. Gates, L. L. Gutshall, V. K. Johnston, B. Gu, K. J. Duffy, and R. T. Sarisky. 2003. Resistance profile of a hepatitis C virus RNA-dependent RNA polymerase benzothiadiazine inhibitor. Antimicrob. Agents Chemother. 47:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olsen, D. B., A. B. Eldrup, L. Bartholomew, B. Bhat, M. R. Bosserman, A. Ceccacci, L. F. Colwell, J. F. Fay, O. A. Flores, K. L. Getty, J. A. Grobler, R. L. LaFemina, E. J. Markel, G. Migliaccio, M. Prhavc, M. W. Stahlhut, J. E. Tomassini, M. MacCoss, D. J. Hazuda, and S. S. Carroll. 2004. A 7-deaza-adenosine analog is a potent and selective inhibitor of hepatitis C virus replication with excellent pharmacokinetic properties. Antimicrob. Agents Chemother. 48:3944-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 29.Reesink, H., S. Zeuzem, C. Weegink, N. Forestier, A. van Vilet, J. van de Wetering de Rooij, L. McNair, S. Purdy, J. Chu, and P. Jansen. 2005. Final results of a phase 1B, multiple-dose study of VX-950, a hepatitis C virus protease inhibitor. Hepatology 42:234A-235A. [Google Scholar]
- 30.Roberts, S., G. Cooksley, G. Dore, R. Robson, D. Shaw, H. Berns, M. Brandl, S. Fettner, G. Hill, E. Ipe, K. Klumpp, M. Mannino, E. O'Mara, I. Najera, Y. Tu, and C. Washingtion. 2006. Results of a phase 1B, multiple dose study of R1626, a novel nucleoside analog targeting HCV polymerase in chronic HCV genotype 1 patients. Hepatology 44:692A. [Google Scholar]
- 31.Sarrazin, C., T. Kieffer, D. Bartels, B. Hanzelka, U. Muh, M. Welker, D. Wincheringer, C. Lin, T. Grossman, S. Purdy, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2005. Characterization of viral variants in the HCV NS3 protease domain of genotype 1 patients that are selected during 14 days of dosing with VX-950. Hepatology 42:751A. [Google Scholar]
- 32.Shimakami, T., M. Hijikata, H. Luo, Y. Y. Ma, S. Kaneko, K. Shimotohno, and S. Murakami. 2004. Effect of interaction between hepatitis C virus NS5A and NS5B on hepatitis C virus RNA replication with the hepatitis C virus replicon. J. Virol. 78:2738-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanabe, Y., N. Sakamoto, N. Enomoto, M. Kurosaki, E. Ueda, S. Maekawa, T. Yamashiro, M. Nakagawa, C. H. Chen, N. Kanazawa, S. Kakinuma, and M. Watanabe. 2004. Synergistic inhibition of intracellular hepatitis C virus replication by combination of ribavirin and interferon-alpha. J. Infect. Dis. 189:1129-1139. [DOI] [PubMed] [Google Scholar]
- 34.Trozzi, C., L. Bartholomew, A. Ceccacci, G. Biasiol, L. Pacini, S. Altamura, F. Narjes, E. Muraglia, G. Paonessa, U. Koch, F. R. De, C. Steinkuhler, and G. Migliaccio. 2003. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77:3669-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yi, M., X. Tong, A. Skelton, R. Chase, T. Chen, A. Prongay, S. L. Bogen, A. K. Saksena, F. G. Njoroge, R. L. Veselenak, R. B. Pyles, N. Bourne, B. A. Malcolm, and S. M. Lemon. 2006. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J. Biol. Chem. 281:8205-8215. [DOI] [PubMed] [Google Scholar]
- 36.Zeuzem, S., C. Sarrazin, R. Rouzier, A. Tarral, N. Brion, N. Forestier, S. Gupta, D. Deckman, K. Fellows, M. Hussain, D. Cutler, and J. Zhang. 2005. Anti-viral activity of SCH 503034, a HCV protease inhibitor, administered as monotherapy in hepatitis C genotype-1 patients refractory to pegylated interferon. Hepatology 42:233A-234A. [Google Scholar]