Abstract
Rhesus rhadinovirus (RRV) is closely related to Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus 8 (HHV-8) and causes KSHV-like diseases in immunocompromised rhesus macaques (RM) that resemble KSHV-associated diseases including multicentric Castleman's disease and non-Hodgkin's lymphoma. RRV retains a majority of open reading frames (ORFs) postulated to be involved in the pathogenesis of KSHV and is the closest available animal model to KSHV infection in humans. Here we describe the generation of a recombinant clone of RRV strain 17577 (RRV17577) utilizing bacterial artificial chromosome (BAC) technology. Characterization of the RRV BAC demonstrated that it is a pathogenic molecular clone of RRV17577, producing virus that behaves like wild-type RRV both in vitro and in vivo. Specifically, BAC-derived RRV displays wild-type growth properties in vitro and readily infects simian immunodeficiency virus-infected RM, inducing B cell hyperplasia, persistent lymphadenopathy, and persistent infection in these animals. This RRV BAC will allow for rapid genetic manipulation of the RRV genome, facilitating the creation of recombinant versions of RRV that harbor specific alterations and/or deletions of viral ORFs. This system will provide insights into the roles of specific RRV genes in various aspects of the viral life cycle and the RRV-associated pathogenesis in vivo in an RM model of infection. Furthermore, the generation of chimeric versions of RRV containing KSHV genes will allow analysis of the function and contributions of KSHV genes to viral pathogenesis by using a relevant primate model system.
Kaposi's sarcoma-associated herpesvirus (KSHV)/human herpesvirus-8 (HHV-8) is the causative agent of Kaposi's sarcoma (KS), as well as B cell lymphoproliferative disorders, primary effusion lymphoma, and multicentric Castleman's disease (7, 9, 29). These diseases frequently occur in human immunodeficiency virus (HIV)/KSHV-coinfected individuals, with KS being the most common AIDS-associated neoplasm. Unfortunately, understanding how KSHV is involved in these diseases is complicated by the absence of a relevant animal model that is susceptible to KSHV infection (14, 27) and by the fact that lytic growth of KSHV in vitro is poor. An alternative is to utilize an animal model that is naturally infected with a herpesvirus closely related to KSHV and which is capable of growth to high titers in vitro. Rhesus rhadinovirus (RRV) is a herpesvirus that naturally infects rhesus macaques (RM), and based on genomic sequence and structural analyses (4, 10, 11, 26, 28, 39), RRV has been shown to be the most closely related herpesvirus to KSHV identified to date. More importantly, coinfection of simian immunodeficiency virus (SIV)-infected RM with RRV strain 17577 (RRV17577) results in the development of lymphoproliferative disorders, in which the primary display of disease is B cell hyperplasia and a multicentric Castleman's disease-like disease similar to that seen in HIV/KSHV-coinfected humans. In addition, RRV has also been found to establish a persistent infection in B cells in vivo (5).
Identification of viral determinants involved in RRV-associated disease is critical to understanding the contributions of individual viral open reading frames (ORFs) to KSHV pathogenesis. The majority of RRV ORFs are shared with ORFs in KSHV, including many thought to be involved in viral pathogenesis. These include, but are not limited to, homologues of viral G protein-coupled receptor (vGPCR), viral CD200 (vCD200), viral interferon regulatory factors (vIRFs), viral macrophage inflammatory protein (vMIP), viral interleukin 6 (vIL-6), and viral complement control protein (vCCP). Studies in our laboratory have shown that vIL-6, vGPCR, and vCD200 of RRV behave similarly to their counterparts in KSHV (15, 17, 20), supporting the use of RRV as a model system to study the role and function of these and other ORFs in virus-mediated pathogenesis in vivo. Thus, given the high similarity between these viruses and their associated diseases, RRV possesses great potential for the analysis of the contribution of specific viral ORFs to pathogenesis of KS and B cell malignancies associated with KSHV infection in humans.
In order to determine the function of viral ORFs in the context of a viral infection, and to assess their potential involvement in disease development, modification of viral genomes to create altered forms of a virus is commonly performed. Traditionally, this has been achieved by utilizing homologous recombination in eukaryotic cells to introduce specific changes into the viral genome, a technique that can be both time consuming and labor intensive. An alternative approach that has recently become the preferred method for genetic manipulation of viruses involves the cloning of an entire viral genome as an infectious bacterial artificial chromosomes (BAC). This approach allows for rapid and efficient alteration of the viral genome in Escherichia coli, followed by the subsequent isolation of recombinant virus in cell culture. Numerous herpesvirus genomes, including human cytomegalovirus, herpes simplex virus type 1, varicella-zoster virus, Epstein-Barr virus, and KSHV, as well as several animal herpesviruses, including mouse cytomegalovirus, rhesus cytomegalovirus, murine gammaherpesvirus 68, and herpesvirus saimiri, have been cloned as BACs (1, 2, 8, 18, 24, 25, 30, 33, 37, 38, 40). BACs provide an invaluable tool for the genetic manipulation of herpesvirus genomes and for the production of altered forms of viruses important to deciphering the roles of individual ORFs in the virus life cycle and pathogenesis. In this report, we describe the generation of an infectious BAC of RRV17577, and the experimental infection of RM with virus derived from this molecular clone. The construction of the RRV BAC will facilitate the examination of the role of individual viral ORFs in the development of RRV-associated disease and also provide important insight into the function of homologous ORFs in KSHV and their potential roles in disease associated with KSHV infection in humans.
MATERIALS AND METHODS
Cells and virus.
Primary rhesus fibroblasts were grown in Dulbecco's modification of Eagle's medium (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT). All viral stocks utilized were grown in primary rhesus fibroblasts and were purified and concentrated by centrifugation through a 30% sorbitol cushion. Titers of viral stocks were assessed by standard plaque assay on primary rhesus fibroblasts.
Cloning of RRV17577 as a BAC.
The 7.2-kb BAC vector pKSO-gpt (24) was utilized to generate a recombination plasmid with flanking RRV sequence and was derived from the parental plasmid pHA1 (a generous gift from Martin Messerle, Institute for Virology, Hannover, Germany) by digestion with PacI. pKSO-gpt contains a bacterial origin of replication, a chloramphenicol resistance gene, and the E. coli guanine phosphoribosyltransferase (gpt) selection marker and is flanked on both ends by 34-bp loxP recombination sites. A fragment of the RRV17577 genome from nucleotide (nt) 78376 to 80000 was isolated from a cosmid containing RRV17577 genomic DNA by digestion with XhoI and KpnI and inserted into plasmid pSP73 (Promega, Madison, WI), digested with the same enzymes to create pSP73-RRV (nt 78376 to nt 80000). This XhoI/KpnI RRV genomic fragment contains a single HindIII site at nt 79010, which is located within the intergenic region between ORF57 and R6. The HindIII site at nt 79010 was converted to a PacI site by digestion with HindIII, followed by treatment with Klenow polymerase to fill in digested ends and ligation of PacI linkers. pKSO-gpt isolated from pHA1 by digestion with PacI was then ligated into pSP73-RRV (nt 78376 to nt 80000) using the newly generated PacI site. This plasmid therefore contains pKSO-gpt flanked by 638 bp of RRV genomic sequence on the 5′ end and 989 bp of RRV genomic sequence on the 3′ end.
The pSP73-RRV (nt 78376 to nt 80000)/pKSO-gpt plasmid was linearized by digestion with AatII, and 5 μg was used to transfect primary rhesus fibroblasts in a 60-mm dish using TransitLT1 transfection reagent (Mirus, Madison, WI). The next day, cells were infected with RRV17577 at a multiplicity of infection (MOI) of 0.1 and incubated until the development of cytopathic effect (CPE). Supernatant from these cells was then collected and used to infect primary rhesus fibroblasts seeded in a 6-well dish, and medium containing 100 μM mycophenolic acid and 25 μM xanthine was added to the cells to select for recombinant RRV containing pKSO-gpt. After the development of CPE, supernatants were collected and passed to new cells. This selection process was performed six times, after which a pool of potential recombinant BAC virus was identified in infected cell supernatants by PCR analysis using primers for a chloramphenicol sequence found in pKSO-gpt (primer 1, 5′ ATGGAGAAAAAAATCACTGGATA 3′; and primer 2, 5′ TTACGCCCCGCCCTGCCACTCAT 3′). The selected virus was then used to infect primary rhesus fibroblasts, and circular DNA was isolated from infected cells 18 h postinfection, using the method described by Hirt (16). Circular DNA was electroporated into a GeneHogs strain of E. coli (Invitrogen, Carlsbad, CA), using a Bio-Rad GenePulser (Bio-Rad, Hercules, CA) with 0.1-cm cuvettes, and recombinant RRV BAC-containing clones were selected by growth on LB plates containing chloramphenicol at 30 μg/ml. Resistant bacterial clones were isolated and grown overnight in LB medium containing chloramphenicol, and DNA was extracted by alkaline lysis.
DNA isolated from bacterial clones was digested with HindIII and BamHI and resolved on a 1% agarose gel and stained with ethidium bromide for visualization, and the observed digestion pattern was compared to that of wild-type RRV17577 DNA digested with the same enzymes. BAC clones with the desired digestion patterns were also confirmed by Southern blotting analysis of digested DNA using a BAC vector-specific DNA probe (chloramphenicol resistance gene) and an RRV-specific DNA probe (ORFR9). For Southern blotting analysis, digested DNA was transferred to Duralon-UV membranes (Stratagene, La Jolla, CA) and DNA probes were labeled using a digoxigenin-High Prime kit (Roche, Indianapolis, IN), with all hybridization, washing, and stripping steps carried out per the kit protocol. BAC DNA of individual clones was also subjected to direct sequence analysis to further identify the insertion of the BAC cassette at the correct location in the RRV genome, utilizing primers that bind outside of the region of recombination used for BAC insertion (forward primer, 5′ ACGCGCAAAAACACGCTAGCGTCT 3′; and reverse primer, 5′ TCTGAATACACGGTAAAGCAGCTG 3′).
Generation of infectious virus from RRV BAC DNA.
BAC-derived virus was generated by transfecting BAC DNA isolated from a single bacterial clone into primary rhesus fibroblasts using TransitLT1 reagent. Briefly, cells were transfected in a 35-mm dish with 1 μg of BAC DNA and incubated until the development of CPE. Then, cells and supernatants were collected, freeze-thawed once, and sonicated to release cell-associated virus. This virus, which still contained the pKSO-gpt BAC vector, was then used to infect primary rhesus fibroblasts that had been previously transfected with pCDNA3.1(−) expressing cyclization recombination (CRE) recombinase, allowing for the removal of the BAC plasmid pKSO-gpt by recombination of the loxP sites located in the terminal regions of the vector. Cells were transfected with 3 μg of pCDNA3.1(−) CRE and infected 2 days later with virus. After the development of CPE, supernatants and cells were collected, freeze-thawed, and sonicated to release virus, and the resulting virus was passaged once more through CRE-expressing cells before being used in a plaque assay with primary rhesus fibroblasts. Next, individual isolates of BAC-derived RRV lacking the BAC plasmid pKSO-gpt were identified by picking single plaques and passing virus to individual wells of a 24-well dish containing primary rhesus fibroblasts. Clonal isolates lacking pKSO-gpt were screened by performing PCR on supernatants from wells of infected cells displaying CPE, using primers specific for the chloramphenicol resistance gene in the pKSO-gpt plasmid. An isolate identified as lacking the BAC vector by PCR analysis was harvested and plaque purified once more, and viral stocks were then generated from individual plaque isolates.
3′ RACE and Northern blotting analysis.
Primary rhesus fibroblasts were infected at an MOI of 5, and RNA was collected 72 h postinfection. 3′ rapid amplification of cDNA ends (RACE) analysis was performed with a GeneRacer RACE kit (Invitrogen). Gene-specific primers used in each reaction bind the sequence upstream of the predicted stop codon for ORF57 or R6 (ORF57 RACE primer, 5′ ACGCGCAAAAACACGCTAGCGTCT 3′; and R6 RACE primer, 5′ TCTGAATACACGGTAAAGCAGCTG 3′). RACE reactions were analyzed on a 1% agarose gel, and the identified products were purified and cloned into vector pCR4-TOPO (Invitrogen) for direct sequence analysis. At least three individual clones of each RACE product were sequenced. Band quantification was performed using an Image Station 440 CF and Molecular Imaging Software (Eastman Kodak Co., Rochester, NY). For Northern blotting analysis, 10 μg of total RNA per lane was loaded on 1.0% agarose gels containing formaldehyde and then run and transferred to nitrocellulose membranes. Next, the membranes were probed with [γ-32P]dCTP-labeled double-stranded DNA (dsDNA) probes specific for RRV ORF57 or -R6, using standard protocols. As with loading a control, membranes were stripped using 0.1% boiling sodium dodecyl sulfate and reprobed with a [γ-32P]dCTP-labeled dsDNA probe specific for GAPDH (glyceraldehyde-3-phosphate dehydrogenase).
Viral DNA isolation and restriction digest analysis.
Primary rhesus fibroblasts were seeded in 850-cm2 roller bottles, infected at an MOI of 0.01, and incubated until the appearance of full CPE. Supernatants and cells were collected and pooled and treated with proteinase K (0.4 mg/ml) overnight at 37°C; DNA was centrifuged through a cesium chloride gradient at 7,7400 × g for 72 h in a Beckman 75 Ti rotor; and fractions containing viral DNA were collected, pooled, and dialyzed against Tris-EDTA. For restriction digest analysis, 0.5 μg to 1 μg of viral DNA was digested overnight at 37°C and analyzed on a 0.7% agarose gel containing ethidium bromide.
Comparative genomic hybridization of BAC and viral DNA.
Comparative genomic hybridization (CGH) of viral DNA or BAC DNA was performed using a microarray hybridization-based method with services provided by NimbleGen Systems, Inc. (Madison, WI). Oligonucleotides for use in the generation of the RRV CGH array were generated using published sequence data for RRV17577 (28) and were designed to be 29 bp in length with overlapping sequence of 8 bases, with coverage spanning both strands of the entire RRV genome. Viral DNA for hybridization was isolated as already described. Bacterium-derived BAC DNA for CGH was isolated from recombinogenic E. coli strain EL250 (21) that had been transformed with the RRV BAC, using a BAC MAX DNA isolation kit (Epicenter, Madison, WI). RRV17577 viral DNA was utilized as the reference genome for all comparative analyses. Hybridization data were analyzed using SignalMap software (NimbleGen Systems, Inc.) to identify regions containing mismatches. The exact changes associated with mismatches were analyzed by direct sequencing of PCR products obtained from amplification of the region encompassing the identified mismatch. The primers utilized for PCR included those designed to amplify the ORF4 sequence (primer 1, 5′ CGTTTTTTACGTAAAACATAT 3′; and primer 2, 5′ TTACAGTAATTGATTGTCCAT 3′) and those used to amplify the sequence encompassing both the R6 sequence and the intergenic region between ORF57 and R6 containing the HindIII site used for BAC vector insertion (primer 1, 5′ GGACTTTTTTTCTTTCCAGTGTAA 3′; and primer 2, 5′ TCTGAATACACGGTAAAGCAGC TG 3′).
In vitro viral growth curves.
Growth curves were obtained with primary rhesus fibroblasts seeded in culture tubes (Corning, Acton, MA). Cells were seeded the day before infection at 2 × 105 cells per tube and infected the next day at an MOI of 2.5. After 2 h, cells were washed twice with phosphate-buffered saline to remove the original inoculum, and fresh medium was added to each tube. Cells were then collected at 2 h, 12 h, 24 h, 48 h, 72 h, and 96 h postinfection, freeze-thawed once, and sonicated to release cell-associated virus. Each time point was performed in duplicate, and the titers of each sample were assessed by plaque assay using primary rhesus fibroblasts.
Experimental inoculation of RM with BAC-derived RRV.
All aspects of the experimental animal studies were performed according to institutional guidelines for animal care and use at the Oregon National Primate Research Center, Beaverton, OR. Four specific-pathogen-free RM seronegative for RRV were inoculated intravenously with 5 ng of SIV strain mac239 (SIVmac239) p27 antigen containing cell-free supernatants, and 8 weeks later these animals were inoculated intravenously with 5 × 106 PFU of BAC-derived RRV isolate number 13. Blood samples were collected prior to RRV infection (day 0) and every 7 days postinfection for the first 11 weeks and later at the indicated times for analysis. Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood using Histopaque (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's specifications. To measure viremia, 2 × 105 PBMCs were serially diluted 1:3, and dilutions were cocultured on monolayers of primary rhesus fibroblasts in 24-well dishes in duplicate and observed for the development of an RRV-specific CPE. Viremia was defined as the limiting dilution factor where virus was detected, with a viremia scale of 1 representing the starting concentration of PBMCs (2 × 105 PBMCs) utilized for coculture. A viremia scale of 2 represents the first dilution where virus was detected in duplicate, and a maximum viremia scale of 5 indicates that all duplicate dilutions were positive for RRV by coculture analysis. A half scale indicates that virus was detected in one of the duplicates of the higher dilution. To identify BAC-derived virus in coculture samples, PCR was performed using primers flanking the intergenic region between ORF57 and R6, which includes the site located at nt 79010 used for BAC insertion (forward primer, 5′ ACGCGCAAAAACACGCTAGCGTCT 3′; reverse primer, 5′ TCTGAATACACGGTAAAGCAGCTG 3′), and resulting products were directly sequenced. For the determination of total B cell numbers, 1 × 106 PBMCs were stained with 20 μl of anti-CD20 PE-Cy7 (E-Bioscience, San Diego, CA) according to the manufacturer's recommendations and analyzed by fluorescence-activated cell sorter (FACS) on a BD LSRII (Becton Dickinson, Franklin Lakes, NJ). Data analysis was performed using FlowJo software (version 8.0.1; Tree Star Inc., Ashland, OR).
RRV load determination by real-time PCR.
For viral load determination, PBMCs were isolated at scheduled blood draws and DNA purified as described previously (35). Approximately 100 ng of purified total cellular DNA was subsequently analyzed by real-time PCR on an ABI Prism 7700 DNA sequence detector (Applied Biosystems, Foster City, CA). DNA primers utilized for the analysis were designed to amplify a segment of RRV ORF3, which encodes the vMIP. The sequence of the TaqMan primers are as follows: vMIP-1, 5′ CCTATGGGCTCCATGAGC 3′; and vMIP-2, 5′ ATCGTCAATCAGGCTGCG 3′. The probe sequence is 5′ TCATCTGCCGCCACCCGGTTTA 3′.
Antibody response to BAC-derived RRV infection.
Serological responses to BAC-derived RRV infection were determined by enzyme-linked immunosorbent assay (ELISA) using animal plasma obtained from defined blood draws essentially as previously described (35). Plasma from immunocompetent animal 305-19092, which was experimentally infected only with the wild-type RRV strain 17577, was utilized as a positive control.
RESULTS
Cloning of RRV17577 as a bacterial artificial chromosome.
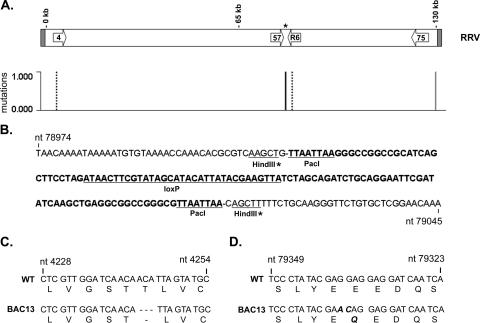
To construct a bacterial artificial chromosome of RRV, we utilized the BAC plasmid pKSO-gpt, isolated from the parental plasmid pHA1 by digestion with PacI. The site of insertion in the RRV genome chosen was a unique HindIII site located at nt 79010, which is located in the intergenic region between ORF57 and R6 (Fig. 1A). ORF57 encodes an immediate-early protein, while R6 encodes one of 8 vIRFs found in RRV. This site was chosen for the BAC vector insertion due to the fact that these ORFs have opposing directions of transcription and since the predicted poly(A) signals for both ORFs are located prior to this restriction site. To confirm that the predicted poly(A) signals for both ORFs were in fact those utilized during transcription of both ORFs during infection, we performed 3′ RACE analysis with gene-specific primers for ORF57 or R6 to assess the exact location of transcript termination for each ORF. In both cases, single discrete products were obtained from 3′ RACE reactions, and the products were of the correct size to be associated with the use of predicted poly(A) signals for each ORF (Fig. 1B). Furthermore, sequence analysis of multiple clones of purified RACE products confirmed that the transcripts for both ORFs do indeed utilize the predicted poly(A) signals located after each ORF and that transcripts terminate prior to the HindIII site used for the BAC vector insertion (Fig. 1C). Therefore, insertion of the BAC vector into the HindIII site between ORF57 and R6 should not significantly alter transcription of either gene.
FIG. 1.
Insertion of BAC vector pKSO-gpt into the RRV genome. (A) The RRV genome is shown with the region of BAC vector insertion enlarged to show the relative location and orientation of ORFs. The genome of RRV17577 consists of a long unique region of 131,364 bp with flanking terminal repeat sequences. The BamHI and HindIII digestion patterns for the enlarged region are noted, with the predicted size (in kb) of restriction fragments listed above each fragment. The BAC vector pKSO-gpt was inserted into the HindIII site at nt 79010 (*), which is located in the intergenic region between ORF57 and R6, by performing homologous recombination with a 1,600-bp RRV genomic fragment (nt 78376 to 80000) engineered to contain pKSO-gpt. pKSO-gpt contains a chloramphenicol resistance gene (CAMr) and the selection marker guanosine phosphoribosyl transferase (gpt) and is flanked by loxP sites. (B) 3′ RACE analysis of RNA from wild-type RRV-infected primary rhesus fibroblasts results in single discrete products for R6 and ORF57. (C) Sequencing of 3′ RACE products identified the poly(A) signal utilized by ORF57 and R6 and the location of the poly(A) tail addition for ORF57 at nt 79001 (>) and for R6 at nt 79194 (<). ORFs are denoted by boldface type, with arrows indicating the direction of transcription. The HindIII site utilized for pKSO-gpt insertion (*) is located after the transcript termination site for both ORFs. Three clones of each RACE product were sequenced.
The first step in the construction of a recombinant RRV BAC was the generation of a plasmid containing the 7.2-kb BAC vector pKSO-gpt inserted into an RRV genomic fragment between ORF57 and R6, such that the BAC vector is flanked on either side by the RRV genomic sequence. Thus, we generated a plasmid containing a XhoI-KpnI fragment of the RRV genome (from nt 78376 to 79980), converted the HindIII site located at nt 79010 in the intergenic region between ORF57 and R6 to a PacI site, and inserted the BAC vector pKSO-gpt into this location (Fig. 1A). This plasmid therefore contains pKSO-gpt flanked on the 5′ end by 638 bp of the RRV sequence and on the 3′ end by 989 bp of the RRV sequence, with no deletions to any genomic sequence surrounding the site of insertion. For recombination with the RRV genome, this plasmid was linearized by restriction digestion and transfected into primary rhesus fibroblasts. The following day, cells were infected with RRV17577, allowing for recombination to occur between the BAC vector flanked by viral sequence and the RRV genome. To select for recombinant RRV containing the BAC vector, cells were incubated in medium containing xanthine and mycophenolic acid, allowing only for the propagation of viruses expressing the E. coli gpt gene carried by pKSO-gpt. Several rounds of selection were performed to enrich for recombinant viruses, and supernatants were tested periodically by PCR for the presence of pKSO-gpt. After six rounds of drug selection, supernatants tested positive for pKSO-gpt (data not shown), indicating the presence of recombinant BAC virus in the culture.
The pool of recombinant virus obtained after selection was then used to infect rhesus fibroblasts, and circular recombinant RRV BAC DNA was isolated from these cells. Next, E. coli was transformed with the isolated DNA and grown on selective chloramphenicol LB-agar plates overnight. Several colonies were obtained from this procedure, which were then isolated and grown in liquid LB cultures. BAC DNA was isolated from liquid cultures, and this DNA was analyzed by restriction digest analysis (Fig. 2A). Based on the known restriction digest patterns for RRV17577 (28), the proper insertion of pKSO-gpt into the RRV genome was examined. The insertion of pKSO-gpt into the modified HindIII site between ORF57 and R6 results in the loss of the 17.2-kb BamHI fragment B1 and the generation of two new BamHI fragments of approximately 7.1 kb and 17.3 kb in size, consisting of a ∼6.3-kb fragment (containing the ORF57 sequence) fused to ∼0.8 kb of pKSO-gpt and a ∼10.9-kb fragment (containing the R6 sequence) fused to 6.4 kb of pKSO-gpt, respectively. HindIII fragments H1 (8.5 kb) and H2 (3.4 kb) were also joined due to the insertion of the BAC vector, resulting in the loss of HindIII fragments H1 and H2 and the creation of a larger approximately 19.7-kb HindIII fragment consisting of H1, H2, and pKSO-gpt (8.5 kb + 3.3 kb + 7.9 kb, respectively, see Fig. 1A). Based on restriction digest analysis of three isolated RRV BAC clones, two were found to have the desired pattern for proper insertion of the BAC vector into the RRV genome (clones B1 and B6). Southern blotting analysis of BamHI-digested DNA using probes specific for either pKSO-gpt or the RRV genome further confirmed the presence of the correctly sized DNA fragments in these two clones (Fig. 2B). A single clone (clone B1) was then chosen for further analysis and for the production of recombinant virus. The insertion of pKSO-gpt into the RRV genome was also confirmed by direct sequence analysis of clone B1 BAC DNA, which demonstrated the proper insertion of the BAC cassette with no observed sequence alterations in the region of the RRV genome surrounding the insertion site (data not shown).
FIG. 2.
Identification of bacterial clones containing an RRV BAC. Primary rhesus fibroblasts were infected with a pool of recombinant RRV BAC virus, and circular DNA was isolated from infected cells by Hirt extraction and then used to transform E. coli. DNA isolated from three CAMr bacterial clones was purified and subjected to restriction digest and Southern blotting analysis. (A) BamHI and HindIII digests of RRV BAC clones. *, (in lanes B1 and B6) 17.3-kb fragment which consists of a 10.9-kb fragment of BamHI restriction fragment B1 (containing R6 sequence) joined to 6.4 kbp of pKSO-gpt (containing the CAMr sequence); Δ, 6.3-kb fragment of BamHI restriction fragment B1 (containing ORF57 sequence) joined to 0.8 kb of pKSO-gpt; >, BamHI fragment in RRV that is known to possess size variability due to the presence of tandem repeat sequence; #, in the wild-type (WT) HindIII digest, indicate fragments H1 and H2, which are absent in BAC clones due to their fusion with pKSO-gpt (creating an ∼19.7-kb fragment). The size variability of the largest band seen in each digest is due to the presence of terminal repeat sequences in these fragments. (B) Southern blotting of BamHI-digested BAC DNA. A chloramphenicol-specific probe detects the 17.3-kb fragment (*) present only in BAC clones, while an ORF57-specific probe detects the intact 17.2-kb BamHI restriction fragment B1 (+) in wild-type RRV17577 DNA and the 7.1-kb fragment (Δ) present only in BAC clones with the correct insertion of pKSO-gpt. BAC clones B1 and B6 possess the correct overall digestion pattern, while BAC clone B5 has unknown alterations in genomic sequence.
Generation of BAC-derived RRV.
To generate virus from the isolated recombinant RRV BAC DNA, rhesus fibroblasts were transfected with RRV BAC DNA and cultured until the development of CPE. Supernatants and cells were then collected and pooled, and virus was harvested from each sample. This pool of BAC-derived virus was then used to infect rhesus fibroblasts transfected with a plasmid encoding CRE recombinase, such that the BAC vector in the virus could be removed by recombination of the loxP sites flanking each side of the vector. After two passages of the virus through CRE-expressing cells, the resulting virus was used to infect rhesus fibroblasts for plaque purification of individual viral isolates. Numerous plaques were obtained from the initial pool of virus, and individual plaques were isolated and passed to single wells of a 24-well plate. After the development of CPE in individual wells, the supernatants were tested for the absence of pKSO-gpt by performing PCR assays for the chloramphenicol resistance gene present in the vector. A plaque isolate found to lack the BAC vector was identified by this approach and was then subjected to a second round of plaque purification, ultimately resulting in four individual isolates (isolates 1, 6, 13, and 15) of BAC-derived RRV. All four isolates were confirmed to no longer contain the BAC vector pKSO-gpt by PCR analysis (Fig. 3A).
FIG. 3.
Isolation and analysis of BAC-derived RRV isolates. After passage of a pool of BAC-derived RRV through primary rhesus fibroblasts expressing CRE recombinase, four individual plaque isolates (1, 6, 13, and 15) of BAC-derived RRV lacking pKSO-gpt were identified by PCR analysis of supernatants from infected primary rhesus fibroblasts (A) using primers specific for pKSO-gpt chloramphenicol sequence (Chlor). As a control for chloramphenicol reactions (lane, Ctrl), a sample of infected cell supernatants was spiked with pKSO-gpt plasmid. PCR of the same supernatants using primers for a virus- specific gene (R9) demonstrates the presence of virus in all samples. After identification, viral DNA was purified from each BAC-derived isolate and was analyzed by restriction digestion with BamHI (B). Comparison of the restriction digest pattern of BAC-derived viral isolate DNA to RRV BAC DNA and wild-type RRV DNA demonstrates that the genomic DNA of BAC-derived viruses is similar to that of wild-type RRV 17577. Southern blotting analysis of the same digests (C), with a probe specific for the chloramphenicol sequence in pKSO-gpt (Chlor) or a viral gene specific probe (R9), demonstrates the removal of pKSO-gpt vector from BAC-derived isolates and the associated restoration of BamHI restriction fragment B1 to nearly its original size of ∼17.2 kb.
To further analyze the DNA of BAC-derived RRV isolates, viral DNA of each isolate was purified from infected rhesus fibroblasts and analyzed by restriction digestion and Southern blotting analysis. Comparison of digested BAC-derived viral DNA to wild-type RRV17577 DNA indicates that the BAC-derived viral isolates maintain the correct restriction digestion pattern overall, with no unexpected changes to the viral genome (Fig. 3B), while Southern analysis confirms the loss of the chloramphenicol sequence after removal of the BAC vector by CRE recombinase (Fig. 3C). Removal of the BAC vector leaves only 120 bp of pKSO-gpt sequence remaining at the site of insertion and essentially restores BamHI fragment B1 nearly to its original size. Although BamHI fragment B1 is ∼17.2 kb and therefore not readily distinguishable from the ∼17.3-kb BAC-containing BamHI fragment by restriction digestion alone, Southern analysis of the digested DNA for R9 (located to the right of the BAC insertion site) demonstrates the presence of the restored ∼17.2-kb BamHI B1 restriction fragment in all BAC-derived isolates. The highest molecular weight band seen in BamHI-digested RRV BAC DNA is absent in digests of wild-type and all BAC-derived viral DNA and is the result of the circularization of the BAC within E. coli and the potential variability in the size of the terminal repeat regions of the RRV genome (28).
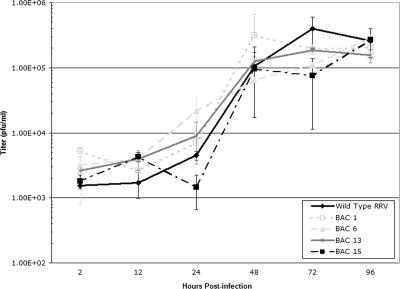
To test the growth properties of the BAC-derived RRV isolates in vitro, a single-step growth curve was performed with rhesus fibroblasts (Fig. 4). The titers for all four BAC-derived RRV isolates were similar to that of wild-type RRV17577 at all time points postinfection, with only slight and likely insignificant variations from wild-type levels seen at some time points. Overall, however, the BAC-derived viral isolates all appear to behave like wild-type RRV17577 in terms of in vitro growth, suggesting that there are no major genomic changes that alter viral growth properties.
FIG. 4.
In vitro growth analysis of BAC-derived RRV isolates. A single-step growth curve of four BAC-derived viral isolates (BAC isolates 1, 6, 13, and 15) was performed with primary rhesus fibroblasts infected at an MOI of 2.5, using wild-type RRV17577 as a control (Wild Type RRV). Supernatants and cells were collected at the indicated time points postinfection, and the amount of virus in each sample was assessed by plaque assay. Each time point assessment was performed in duplicate, and the average value is indicated. Error bars represent standard deviations.
Whole-genome analysis of RRV BAC and BAC-derived RRV isolates.
The cloning of a viral genome such as a BAC, as with any genetic manipulation of a viral genome, raises the possibility of the introduction of random mutations and genomic reorganization that may not be easily discernible by restriction digestion and Southern blotting analysis of viral DNA alone. Sequencing an entire BAC clone or a BAC-derived virus would be both costly and labor intensive, especially if multiple viruses are to be made simultaneously. To circumvent the issue of overlooking potentially important sequence variations in the RRV BAC itself, or any viruses derived from it, we employed the array-based method of CGH to analyze the entire genome of the parental RRV BAC, as well as viral isolates obtained from the RRV BAC. Utilizing CGH analysis, any change that alters the genomic sequence of RRV will result in incomplete hybridization with oligonucleotides that overlap that region, resulting in the detection of a mismatch by using the array. This technique is sensitive enough to detect a single base change, as well as more major changes to the genome such as insertions, deletions, and rearrangements. CGH has been used successfully for the analysis of entire bacterial genomes (3), as well as viral genomes (34), and provides a rapid and accurate method to examine the complete sequence of DNA from BAC clones or viral DNA from any BAC-derived viruses.
An array was designed containing oligonucleotides spanning the entire RRV17577 genome (both forward and reverse strands), and BAC DNA or viral DNA from BAC-derived RRV isolates was utilized for hybridization to the array. In all hybridizations, wild-type RRV17577 DNA was used as a reference sample, allowing for the direct comparison between each sample and the parental virus utilized to generate the RRV BAC. Employing this technique, we were able to analyze the sequence of the entire RRV BAC and all four BAC-derived viral isolates (data not shown). Upon examination of the results, we found that the RRV BAC and all four BAC-derived isolates contained a single common mutation, which was located in the intergenic region between ORF57 and R6, near nt 79010 (Fig. 5A). This was an expected result, which correlates exactly with the location of the pKSO-gpt vector in the RRV BAC and the presence of loxP-containing sequence in BAC-derived RRV isolates that remains after BAC vector removal by CRE recombinase. Essentially, this common mutation represents an insertion of exogenous bacterial sequence (pKSO-gpt vector) into the RRV genome. Direct sequencing of RRV BAC DNA confirmed the presence of the entire pKSO-gpt vector in this location, while sequencing of PCR products from BAC-derived viral DNA identified the 120 bp of the pKSO-gpt sequence (including a single loxP site) which remains after BAC vector removal (Fig. 5B).
FIG. 5.
CGH analysis and identification of mutations in the RRV BAC and BAC-derived viral isolate DNA. RRV BAC DNA and BAC-derived RRV isolate 13 viral DNA were used in CGH analysis with RRV17577 as the reference genome. Differences in genomic sequence were identified based on variations in hybridization intensities, indicating locations of potential mutations in the RRV sequence (A). The mutation found only in the RRV BAC is indicated by a solid gray line, mutations found only in BAC-derived isolate 13 are indicated by a dashed black line, and the common mutation found in both samples is indicated by a solid black line. PCR of the region encompassing the BAC vector insertion was performed using BAC-derived RRV isolate 13, and the resulting product was directly sequenced. (B) The sequence indicates that the mutation identified near nt 79010 is due to the presence of 120 bp of pKSO-gpt sequence at the site of the BAC vector insertion that remains after BAC vector removal by CRE recombinase. The pKSO-gpt sequence is indicated in bold type, with the PacI sites used for cloning and the loxP site underlined. The partial HindIII sites remaining after the insertion of pKSO-gpt into RRV are also noted, with an asterisk. PCR was also performed using BAC-derived RRV isolate 13 as a template to amplify regions encoding ORF4 and R6, and the resulting products were directly sequenced. The sequence alterations detected include a 3-bp deletion in the ORF4 coding sequence (nt 4243 to 4245), which results in the deletion of Thr190 from the vCCP encoded by ORF4 (C) and the conversion of nt 79338 and 79337 in the R6 coding sequence from GG to AC, ultimately causing the conversion of Glu393 to Gln in the encoded vIRF (D).
Besides the BAC vector, the RRV BAC contained only one other apparent mutation, which was located in a noncoding sequence near the terminal repeat region at the right end of the genome (Fig. 5A). This difference is likely due to the variation in terminal repeat sequences or the circularization of the BAC, since the mutation was not present in the genomes of any of the BAC-derived viruses we analyzed. Surprisingly, besides the mismatch associated with BAC vector insertion, all of the BAC-derived viral isolates contained few other mismatches compared to wild-type RRV17577. Isolate 13 contains only two other mutations (Fig. 5A), which were found to be common to all four BAC-derived isolates, while the remaining isolates each have one additional mutation at different locations in the genome. The two common mutations in these viruses arose after transfecting purified RRV BAC DNA into rhesus fibroblasts, and their presence in all four BAC-derived viruses is likely due to their common origin from a single plaque isolate, with the development of other mutations occurring during further purification of these viruses in vitro.
The mutations common to all four BAC-derived RRV isolates are located within sequences for ORF4 and R6, which encode the 646-amino-acid (aa) vCCP and a 416-aa vIRF, respectively. To determine the exact sequence changes associated with the observed mismatches, the regions encompassing ORF4 and R6 were amplified by PCR and directly sequenced. The sequence alteration detected in the ORF4 sequence is a 3-bp deletion of nt 4243 to 4245, which results in the deletion of Thr190 from vCCP (Fig. 5C). The alteration detected in the ORFR6 coding sequence consists of the conversion of nt 79338 and nt 79337 from GG to AC, ultimately resulting in the conversion of Glu393 to Gln. Although the genomic mutations detected in the BAC-derived viral isolates do alter the proteins encoded by ORF4 and vIRF, the changes appear to be minor and are presumed to have no significant effects on protein function. The potential effects these mutations might have on protein function are being investigated.
In order to confirm the accuracy of CGH assays at detecting differences in the RRV genome, we performed CGH using RRV17577 as the reference DNA sample and viral DNA isolated from RRV strain 26-95 as the test sample. RRV strain 26-95 is an independent isolate of RRV (11), which shares significant genomic sequence homology with RRV17577 (4) but differs in pathogenic potential (23). CGH analysis with these two RRV strains detected ∼175 mismatches between the viral genomes (data not shown), demonstrating that this technique is sensitive enough to detect the numerous genetic variations between two very closely related strains of RRV and further strengthening the fact that the RRV BAC and BAC-derived viral isolates are essentially genetically identical to the parental RRV17577. Thus, no major changes to viral genomic sequence developed during the construction of the RRV BAC, and very few mutations appeared to spontaneously arise during the production of BAC-derived RRV isolates. Based on the presence of the least number of mutations in BAC-derived RRV isolate 13 and on the similar growth properties of all isolates in vitro, we chose to utilize this virus as a representative of wild-type BAC-derived RRV for further studies.
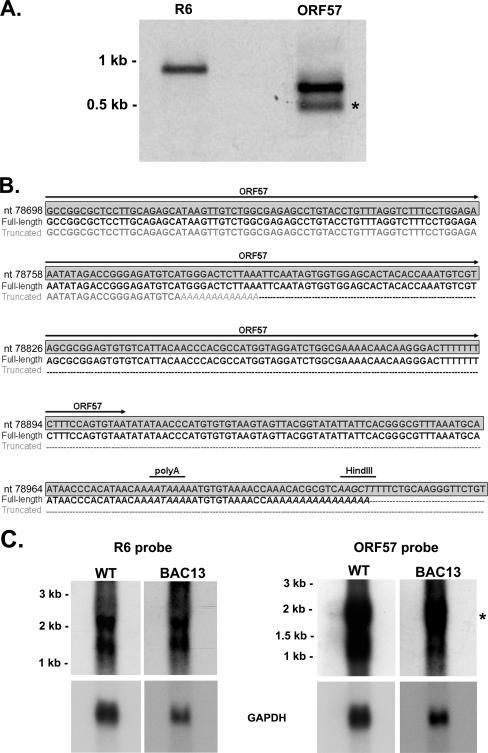
To determine whether the loxP-containing sequence remaining in the BAC-derived virus after BAC vector removal has any effects on transcription of the neighboring ORF57 or R6 genes, RNA isolated from primary rhesus fibroblasts infected with the BAC-derived viral isolate 13 was used in 3′ RACE with primers targeting ORF57 and R6. As was seen with wild-type RRV17577, a single product of the expected size was obtained for R6 (Fig. 6A). However, ORF57 3′ RACE analysis resulted in two products, one of the predicted size for transcript termination after ORF57 and the other a less abundant truncated product, indicative of possible premature transcript termination. Sequence analysis of these products indicated that, as with wild-type RRV, all BAC-derived virus R6 transcripts terminate at nt 79194 and that the majority of ORF57 transcripts terminate at nt 79001, both prior to the site of BAC insertion. However, a small fraction of BAC-derived virus ORF57 transcripts appear to terminate prematurely at nt 78778, 128 bp before the predicted stop codon for ORF57 (Fig. 6B). This alteration in ORF57 transcription could be due to the presence of the 120 bp of loxP containing sequence in BAC-derived viruses that remains after pKSO-gpt removal by CRE recombinase, although given the lower overall abundance of the truncated RACE product, it is likely that most transcripts from ORF57 do not terminate prematurely.
FIG. 6.
3′ RACE and Northern blotting analysis of BAC-derived RRV. (A) 3′ RACE analysis of RNA from BAC-derived RRV isolate 13-infected primary rhesus fibroblasts. A single full-length product is obtained for R6, while full-length and truncated products (*) are detected for ORF57. Quantification of relative band intensities indicates that the full-length and truncated products represent 61.4% and 38.6% of the total ORF57 transcripts, respectively. Cloning and sequencing of the R6 3′ RACE product identified the poly(A) signal utilized by R6 as that identified for wild-type RRV. (B) The consensus sequence of both the full-length and the truncated ORF57 3′ RACE products was determined by cloning each product and sequencing multiple clones. The genomic sequence for RRV is highlighted by a gray box, the full-length product consensus sequence is in black, and the truncated product consensus sequence is in gray. The ORF57 coding sequence is denoted by arrows above the sequence. The poly(A) sequence addition for each transcript is noted by italics, and dashes represent sequences not in the truncated transcript. The poly(A) signal utilized by the full-length transcripts corresponds to that identified in wild-type RRV and is noted by a line above the italicized sequence. Truncated ORF57 transcripts terminate 128 bp prior to the ORF57 stop codon. The HindIII site utilized for pKSO-gpt insertion is also noted. (C) Northern blotting analysis of ORF57 and R6 transcription during infection with wild-type or BAC-derived RRV. Rhesus fibroblasts were infected with either wild-type RRV17577 or BAC-derived RRV isolate 13 (MOI, 5), and RNA was isolated 72 h postinfection for use in Northern blotting analysis with dsDNA probes specific for the R6 or ORF57 sequence. The size of the predicted coding sequence for R6 is 1,247 bp and 1,328 bp for ORF57. Using both wild-type and BAC-derived RRV, two R6-specific transcripts were detected in similar amounts (∼1.5 kb and ∼2.0 kb, respectively). A major ∼1.8-kb ORF57 transcript (denoted by an asterisk) was detected during infection with both wild-type and BAC-derived viruses, to comparable levels. A smaller transcript of ∼1 kb was also detected during infection, with both viruses using an ORF57 probe, although this transcript is too small to encode full-length ORF57.
To further analyze the effects of BAC vector insertion on the transcription of ORF57 and R6 during infection with BAC-derived virus, Northern blotting analysis was performed on RNA isolated from rhesus fibroblasts infected with either wild-type RRV17577 or BAC-derived viral isolate 13. Although a slight reduction in levels of ORF57 transcription may occur in the BAC-derived virus (Fig. 6C), the overall pattern of ORF57 transcription observed with both wild-type and BAC-derived viruses is similar to what has been observed in previous studies with RRV (13), with the detection of a major ∼1.8-kb transcript that is thought to encode full-length ORF57. A smaller ∼1-kb transcript is also detected with the ORF57 probe and appears to display slightly reduced expression during infection with BAC-derived RRV; however, this transcript is smaller than the predicted coding sequence for ORF57 (1,328 bp) and, thus, is likely not important for ORF57 protein expression. The transcription profiles for R6 are also similar for both the wild-type and the BAC-derived RRV, with the detection of comparable amounts of two transcripts of ∼2 kb and ∼1.5 kb in both viruses, either of which possesses the potential to encode R6, whose predicted coding sequence is 1,247 bp. Therefore, the presence of the loxP-containing sequence in the genome of BAC-derived RRV does not appear to significantly alter the transcription of either ORF57 or R6.
BAC-derived RRV is pathogenic in vivo.
To determine the growth kinetics and pathogenic properties of BAC-derived RRV in vivo, we utilized our established SIV/RM model of infection (35) to assess the ability of a BAC-derived RRV isolate to replicate and cause disease in SIV-infected RM. For these studies, four juvenile specific-pathogen-free RM (n, 4) were inoculated with SIVmac239 to provide a background of immunosuppression before experimental inoculation with BAC-derived RRV isolate 13 approximately 8 weeks later. Sequential blood samples were collected from animals directly prior to RRV inoculation (day 0) and every 7 days postinfection thereafter or as indicated. PBMCs were isolated from whole blood and cocultured with primary rhesus fibroblasts to assess levels of viremia utilized for DNA isolation to define viral load by real-time PCR, and total B cell numbers were determined by staining PBMCs for CD20. Plasma was subsequently utilized to determine the presence of antibodies against whole RRV by ELISA.
Viremia was first observed in coculture samples 2 weeks post-RRV infection, with peak viremia occurring between weeks 3 and 6 for all four animals (Fig. 7A), which is consistent with previous studies of RRV infection (35). Viral load as assessed by real-time PCR was shown to coincide with viremia (Fig. 7B), with peak loads detected at week 3. However, the amounts of virus detected by this method were slightly different among the four animals, with animal 23430 having the greatest load.
FIG. 7.
BAC-derived RRV is infectious and induces disease in vivo. Four RM (animal numbers 23413, 23430, 23545, 23639) were inoculated with SIVmac239 and 8 weeks later were inoculated with 5 × 106 PFU of BAC-derived RRV isolate 13. Blood was collected from RM prior to RRV infection (day 0) and every 7 days postinfection and analyzed at indicated time points. Sample time points vary as sample volume may not have been sufficient for all analyses. (A) Viremia was assessed by coculture of PBMCs with primary rhesus fibroblasts, with five dilutions of PBMCs each tested in duplicate. Viremia is thus defined as the limiting dilution factor where virus isolation was detected, with a viremia scale of 1 representing the starting concentration of PBMCs (2 × 105 PBMCs) utilized for coculture. A viremia scale of 2 represents the first dilution where virus is detected in duplicate, while a viremia scale of 5 represents that the level of virus is detected in duplicate in all five dilutions. A half scale indicates that virus was detected in one of the duplicates of the higher dilution. Peak viremia was observed in all animals between weeks 3 and 6. (B) Real-time PCR analysis of viral DNA loads using DNA isolated from PBMCs and RRV-specific primers. (C) Plasma from infected RM was analyzed for the presence of RRV-specific antibodies using an ELISA specific for whole RRV. (D) Total B cell numbers (CD20+ cells) were obtained by performing FACS analysis of PBMCs stained with anti-CD20 PE-Cy7. The observed increase in CD20+ cells in all four animals demonstrates that BAC-derived RRV induces B cell hyperplasia in SIV-infected macaques, which is similar to what is seen in SIV/RRV17577 coinfected macaques (35).
To positively identify the virus obtained from coculture samples as the input BAC-derived virus, virus isolated from week-3 coculture samples from all four animals was plaque purified and then used to infect primary rhesus fibroblasts. After development of CPE, PCR for BAC-derived RRV was performed with supernatants of infected cells using primers spanning the region of BAC vector insertion between ORF57 and R6, and individual PCR products were then purified and directly sequenced. Virus isolated from each animal was positively identified as BAC-derived RRV isolate 13 based on the presence of the remaining 120 bp of vector sequence at the site of the BAC vector insertion (data not shown).
The humoral response to BAC-derived RRV infection was assessed sequentially using a whole-virus ELISA. All four animals developed a measurable antibody response to BAC-derived RRV (Fig. 7C), which was comparable to the response we previously reported for one RRV17577/SIV-infected animal but still lower than the response seen with immunocompetent animals infected with the wild-type RRV17577 (35). This demonstrates that BAC-derived RRV possesses similar, if not identical, immunogenic properties as the parental RRV17577 isolate in SIV-infected RM.
Analysis of total CD20+ cells in blood from infected animals demonstrates that inoculation of SIV-infected RM with BAC-derived RRV induces B cell hyperplasia in these animals (Fig. 7D). A relatively small increase in B cell numbers is seen in the first 2 weeks after BAC-derived RRV inoculation, followed by a more robust increase in total B cell numbers by week 3. Peak B cell numbers were observed at weeks 4 and 5 in all animals, with a significant increase of B cell numbers in the peripheral blood ranging from 2.5-fold to 7.0-fold increases over preinoculation levels. B cell numbers exhibited a relatively rapid decline in animals 23413 and 23430 from weeks 6 to 8, whereas animals 23545 and 23639 demonstrated a more gradual decline in B cell numbers and maintained a relatively steady level of elevated B cell numbers. Overall, this indicates that BAC-derived RRV possesses wild-type growth properties in vivo and is capable of promoting B cell hyperplasia development, as previously reported for infection of the wild-type RRV17577 (35).
DISCUSSION
Recently, cloning of herpesvirus genomes as BACs has become the established method for rapidly generating mutant viruses (2). Previously, the manipulation of herpesvirus genomes was accomplished in eukaryotic cells, relying on homologous recombination to generate altered viruses, a process which can be both lengthy and inefficient. By propagation of a viral BAC in E. coli, many of these difficulties are overcome, since the use of prokaryotic recombination machinery provides a much more reliable and rapid method for manipulating viral genomes. In addition, this approach provides the added benefit of being able to confirm and analyze the presence of any engineered changes in the viral genome before virus is even produced, further decreasing the time required to isolate a mutant virus.
The work presented here describes the construction of a pathogenic BAC of RRV17577, a gamma-2 herpesvirus that is closely related to KSHV (28). A fragment of RRV17577 containing the BAC vector pKSO-gpt was used to perform homologous recombination with RRV17577, and a stable BAC clone containing the entire viral genome was isolated and verified. The BAC vector was inserted between two opposing ORFs in the RRV genome (ORF57 and R6), a site that was found suitable for BAC insertion based on 3′ RACE analysis that demonstrated that transcripts for both ORFs terminate prior to this location. In addition, CGH analysis allowed us to determine the integrity of the sequence of the entire RRV BAC, confirming that no significant changes in viral genomic sequence exist, other than BAC vector insertion, and indicating that the RRV BAC represents a near identical bacterial clone of RRV17577. We have also found that the RRV BAC is stable in E. coli, since it was subjected to numerous passages without any observed changes in genomic organization (data not shown).
Virus was readily produced from the RRV BAC, and analysis of viral DNA indicates that the genomes of BAC-derived virus are essentially identical to wild-type RRV17577 and that these viruses grow with properties similar to wild-type virus in vitro and in vivo. CGH analysis of DNA from BAC-derived viral isolates was also performed and allowed us to rapidly assess the entire sequence of each virus for the presence of any alterations in genomic sequence. Although some undesired mutations were identified, these mutations do not appear to have any effect on the growth properties of BAC-derived RRV in vitro or in vivo and likely have no significant effects on the activity of any gene products. Indeed, the presence of mutations in BAC-derived viruses is not desired, yet it is inevitable that some random changes in viral genomic sequence may arise during either the production of a BAC or during production and propagation of viruses derived from a BAC. In previous studies describing the construction of BACs for various herpesviruses (1, 2, 8, 18, 24, 25, 30, 33, 37, 38, 40), we know of no instance where the entire BAC or any BAC-derived viruses have been subjected to complete sequence analysis, and it is extremely likely that some or all of these BAC clones or viruses may contain unknown mutations. Thus, our approach to the analysis of BAC and BAC-derived viral DNA is advantageous, given that we will have prior knowledge of any mutations and can thus examine the effects mutations may have on the resulting virus, if found necessary.
Removal of the BAC vector from any BAC-derived virus is ideal, since it has been suggested that the presence of the entire BAC vector sequence could potentially have some effect on the growth properties of BAC-derived viruses, especially in vivo (32). Thus, the removal of the BAC vector from BAC-derived RRV was accomplished by CRE-mediated recombination between the terminal loxP sites located within the vector. This recombination event results in the deletion of the majority of the BAC vector, leaving only 120 bp of vector sequence in these viruses. Also, 3′ RACE analysis of transcripts from BAC-derived RRV infected cells was performed to confirm that BAC vector insertion does not affect the transcription of either neighboring ORF. Though we did observe that ORF57 produces a truncated transcript in addition to the normal full-length message, this message is much less abundant. Therefore, the transcription of ORF57 is thought not to be significantly affected due to the presence of the remaining BAC vector sequence.
RRV is a well-characterized primate homologue of KSHV, and infection of RM with both SIV and RRV results in disease development that resembles disease seen in humans infected with HIV and KSHV (35). Based on this knowledge, we sought to determine whether BAC-derived RRV maintains wild-type growth properties in vivo and whether this virus was capable of inducing pathogenic sequelae similar to those seen with RRV17577 infection. Therefore, RM were coinfected with SIV and a BAC-derived RRV isolate and observed for the presence of virus and the development of B cell proliferation. Analysis of PBMCs indicates that, similar to RRV17577, BAC-derived RRV induces B cell hyperplasia, as indicated by a rapid increase in total CD20+ cell numbers following infection with this virus. Importantly, the observed B cell hyperplasia also parallels closely the level of viremia and viral DNA load detected in each animal. In addition, two animals developed significant lymphadenopathy after infection with BAC-derived RRV, which histological examination revealed is similar to that originally reported for SIV/RRV-infected animals (data not shown). Furthermore, the humoral response to BAC-derived RRV in infected RM is similar to that observed with infection by parental RRV17577. Therefore, BAC-derived RRV closely mimics wild-type RRV17577 in vivo in terms of growth and pathogenesis, indicating that overall, the RRV BAC will provide a valuable tool to assess the effects of various viral genes on the development of KSHV-like diseases.
Although a KSHV BAC has been developed (40) and used in several studies (19, 22, 36, 37), the major drawback of this system is the lack of an available animal model to study the infection and pathogenesis of BAC-derived viruses. This is due to the fact that KSHV does not appear to infect and produce disease in any animal system tested thus far, including primates (14, 27). Thus, without any method to test the effects of BAC-derived viral mutants on disease development, the assessment of pathogenic determinants of KSHV is severely hampered.
BACs of gammaherpesviruses other than RRV that are also related to KSHV have been created as well, including murine gammaherpesvirus 68 (2) and herpesvirus saimiri (33). However, it is important to note that these viruses, although extremely valuable for investigating aspects of gammaherpesvirus pathogenesis and immunity, are not as closely related to KSHV as is RRV (28) and lack some genes found in both RRV and KSHV that are thought to be important in KSHV-associated disease development. Therefore, RRV stands as an ideal model for studying KSHV, and the development of an RRV BAC provides a valuable tool for dissecting the determinants of pathogenesis for both viruses.
Recently, an approach to making recombinant RRV strain 26-95 using cosmids was described (6). This approach is also useful for making recombinant RRV but is significantly different from the BAC system, which allows for the utilization of E. coli to rapidly and efficiently perform recombination (21). Additionally, a BAC clone of a virus will essentially always produce the same virus, while utilizing a cosmid approach, it would be necessary to analyze the genomic structure and sequence of each viral isolate in complete detail to assure that no major structural changes or gross mutations have been introduced into the genome due to the number of recombination events required to produce virus.
We have transferred the RRV BAC into the recombinogenic E. coli strain EL250 (21) and are currently in the process of generating viruses with deletions of selected ORFs that are believed to be involved in the pathogenesis of both RRV and KSHV, including vGPCR (ORF74), vCD200(ORFR15), vIRFs (ORFR6-R13), vIL-6 (ORFR2), and vCCP (ORF4). The initial characterization of these viruses is ongoing and should give insight into the contributions of these genes to virus-associated disease development. Another long-term goal is to utilize the RRV BAC to generate chimeric viruses containing KSHV genes to determine what affects specific KSHV ORFs may have both in vitro and in vivo. One example is the construction of an RRV BAC-derived virus in which the ORF encoding the latency-associated nuclear antigen (LANA) is replaced with that from KSHV. LANA is encoded by ORF73 in both viruses, and in KSHV, this gene product is known to be involved in the establishment and maintenance of latency in infected cells as well as transformation (31). Less is known about RRV LANA, although it is presumed to play a similar role during RRV infection (12). Interestingly, the RRV ORF73 is severely truncated compared to the KSHV ORF73 (28), encoding a 342-aa protein versus a 1,162-aa protein, respectively, an observation that could possibly help explain the differences in the propensity of these viruses for lytic versus latent infections. Numerous other chimeric viruses could also be generated and could provide invaluable information regarding the roles of specific HHV-8 genes or genomic regions in disease development.
Acknowledgments
This work was supported by NIH grants CA75922 and RR00163.
We thank Michael Jarvis, Beata Orzechowska, and Bridget Robinson for helpful discussions and Colin Powers for reagents and technical advice.
Footnotes
Published ahead of print on 10 January 2007.
REFERENCES
- 1.Adler, H., M. Messerle, and U. H. Koszinowski. 2003. Cloning of herpesviral genomes as bacterial artificial chromosomes. Rev. Med. Virol. 13:111-121. [DOI] [PubMed] [Google Scholar]
- 2.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951-953. [DOI] [PubMed] [Google Scholar]
- 4.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergquam, E. P., N. Avery, S. M. Shiigi, M. K. Axthelm, and S. W. Wong. 1999. Rhesus rhadinovirus establishes a latent infection in B lymphocytes in vivo. J. Virol. 73:7874-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilello, J. P., J. S. Morgan, B. Damania, S. M. Lang, and R. C. Desrosiers. 2006. A genetic system for rhesus monkey rhadinovirus: use of recombinant virus to quantitate antibody-mediated neutralization. J. Virol. 80:1549-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 8.Chang, W. L. W., and P. A. Barry. 2003. Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. J. Virol. 77:5073-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Damania, B., and R. C. Desrosiers. 2001. Simian homologues of human herpesvirus 8. Philos. Trans. R. Soc. Lond. B 356:535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeWire, S. M., and B. Damania. 2005. The latency-associated nuclear antigen of rhesus monkey rhadinovirus inhibits viral replication through repression of Orf50/Rta transcriptional activation. J. Virol. 79:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeWire, S. M., M. A. McVoy, and B. Damania. 2002. Kinetics of expression of rhesus monkey rhadinovirus (RRV) and identification and characterization of a polycistronic transcript encoding the RRV Orf50/Rta, RRV R8, and R8.1 genes. J. Virol. 76:9819-9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmer, D., C. Stoddart, R. Renne, V. Linquist-Stepps, M. E. Moreno, C. Bare, J. M. McCune, and D. Ganem. 1999. Experimental transmission of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) to SCID-hu Thy/Liv mice. J. Exp. Med. 190:1857-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estep, R. D., M. K. Axthelm, and S. W. Wong. 2003. A G protein-coupled receptor encoded by rhesus rhadinovirus is similar to ORF74 of Kaposi's sarcoma-associated herpesvirus. J. Virol. 77:1738-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 17.Kaleeba, J. A. R., E. P. Bergquam, and S. W. Wong. 1999. A rhesus macaque rhadinovirus related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 encodes a functional homologue of interleukin-6. J. Virol. 73:6177-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanda, T., M. Yajima, N. Ahsan, M. Tanaka, and K. Takada. 2004. Production of high-titer Epstein-Barr virus recombinants derived from Akata cells by using a bacterial artificial chromosome system. J. Virol. 78:7004-7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnan, H. H., N. Sharma-Walia, L. Zeng, S.-J. Gao, and B. Chandran. 2005. Envelope glycoprotein gB of Kaposi's sarcoma-associated herpesvirus is essential for egress from infected cells. J. Virol. 79:10952-10967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langlais, C. L., J. M. Jones, R. D. Estep, and S. W. Wong. 2006. Rhesus rhadinovirus R15 encodes a functional homologue of human CD200. J. Virol. 80:3098-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 22.Luna, R. E., F. Zhou, A. Baghian, V. Chouljenko, B. Forghani, S.-J. Gao, and K. G. Kousoulas. 2004. Kaposi's sarcoma-associated herpesvirus glycoprotein K8.1 is dispensable for virus entry. J. Virol. 78:6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansfield, K. G., S. V. Westmoreland, C. D. DeBakker, S. Czajak, A. A. Lackner, and R. C. Desrosiers. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J. Virol. 73:10320-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagaike, K., Y. Mori, Y. Gomi, H. Yoshii, M. Takahashi, M. Wagner, U. Koszinowski, and K. Yamanishi. 2004. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine 22:4069-4074. [DOI] [PubMed] [Google Scholar]
- 26.O'Connor, C. M., B. Damania, and D. H. Kedes. 2003. De novo infection with rhesus monkey rhadinovirus leads to the accumulation of multiple intranuclear capsid species during lytic replication but favors the release of genome-containing virions. J. Virol. 77:13439-13447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renne, R., D. Dittmer, D. Kedes, K. Schmidt, R. C. Desrosiers, P. A. Luciw, and D. Ganem. 2004. Experimental transmission of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) to SIV-positive and SIV-negative rhesus macaques. J. Med. Primatol. 33:1-9. [DOI] [PubMed] [Google Scholar]
- 28.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 30.Tanaka, M., H. Kagawa, Y. Yamanashi, T. Sata, and Y. Kawaguchi. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verma, S. C., and E. S. Robertson. 2003. Molecular biology and pathogenesis of Kaposi sarcoma-associated herpesvirus. FEMS Microbiol. Lett. 222:155-163. [DOI] [PubMed] [Google Scholar]
- 32.Wagner, M., S. Jonjíc, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White, R. E., M. A. Calderwood, and A. Whitehouse. 2003. Generation and precise modification of a herpesvirus saimiri bacterial artificial chromosome demonstrates that the terminal repeats are required for both virus production and episomal persistence. J. Gen. Virol. 84:3393-3403. [DOI] [PubMed] [Google Scholar]
- 34.Wong, C. W., T. J. Albert, V. B. Vega, J. E. Norton, D. J. Cutler, T. A. Richmond, L. W. Stanton, E. T. Liu, and L. D. Miller. 2004. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 14:398-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong, S. W., E. P. Bergquam, R. M. Swanson, F. W. Lee, S. M. Shiigi, N. A. Avery, J. W. Fanton, and M. K. Axthelm. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J. Exp. Med. 190:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, Y., D. P. AuCoin, A. R. Huete, S. A. Cei, L. J. Hanson, and G. S. Pari. 2005. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 79:3479-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye, F.-C., F.-C. Zhou, S. M. Yoo, J.-P. Xie, P. J. Browning, and S.-J. Gao. 2004. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 78:11121-11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, X.-K., C. M. O'Connor, I. Atanasov, B. Damania, D. H. Kedes, and Z. H. Zhou. 2003. Three-dimensional structures of the A, B, and C capsids of rhesus monkey rhadinovirus: insights into gammaherpesvirus capsid assembly, maturation, and DNA packaging. J. Virol. 77:13182-13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, F.-C., Y.-J. Zhang, J.-H. Deng, X.-P. Wang, H.-Y. Pan, E. Hettler, and S.-J. Gao. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]