Abstract
To determine the initiation strategy of the hepatitis E virus (HEV) open reading frame 3 (ORF3), we constructed five HEV mutants with desired mutations in the ORF1 and ORF2 junction region and tested their levels of in vivo infectivity in pigs. A mutant with a C-terminally truncated ORF3 is noninfectious in pigs, indicating that an intact ORF3 is required for in vivo infectivity. Mutations with substitutions in the first in-frame AUG in the junction region or with the same T insertion at the corresponding position of HEV genotype 4 did not affect the virus infectivity or rescue, although mutations with combinations of the two affected virus recovery efficiency, and a single mutation at the third in-frame AUG completely abolished virus infectivity in vivo, indicating that the third in-frame AUG in the junction region is required for virus infection and is likely the authentic initiation site for ORF3. A conserved double stem-loop RNA structure, which may be important for HEV replication, was identified in the junction region. This represents the first report of using a unique homologous pig model system to study the molecular mechanism of HEV replication and to systematically and definitively identify the authentic ORF3 initiation site.
Hepatitis E virus (HEV) is an important human pathogen responsible for enterically transmitted acute hepatitis in many regions of the world. A unique feature associated with HEV infection is the relatively high mortality rate in infected pregnant women, which can reach up to 28% (11). In developing countries where sanitation conditions are poor, the disease is transmitted via the fecal-oral route through virus-contaminated water or food (19). In industrialized countries, immunoglobulin G (IgG) anti-HEV antibodies have been detected in a significant proportion of healthy individuals; however, only sporadic cases of acute hepatitis E have been reported. Increasing evidence indicates that hepatitis E is a zoonotic disease, and animal reservoirs for HEV exist (17, 19, 22, 24, 31). We recently discovered and characterized two animal strains of HEV that are genetically and antigenically closely related to human HEV: swine HEV from pigs (15-16) and avian HEV from chickens (1, 7-8). We subsequently demonstrated that swine HEV can cross species barriers and infect nonhuman primates (16) and that pig handlers in the United States and other countries are at increased risk of zoonotic HEV infection (18). All swine HEV isolates thus far identified from pigs belong to either genotype 3 or genotype 4 and in many cases are genetically indistinguishable from human HEV genotypes 3 and 4 (19, 20, 31). Swine HEV infection in specific-pathogen-free pigs provided us a unique animal model to study HEV replication and pathogenesis (6, 10).
HEV is currently classified in the sole genus Hepevirus of the family Hepeviridae (2). The virus is a single-stranded, positive-sense, polyadenylated RNA molecule of approximately 7.2 kb in size with short 5′ and 3′ noncoding regions. The viral genome contains three open reading frames (ORFs). ORF1 at the 5′ end encodes nonstructural proteins that are involved in virus replication and viral protein processing. ORF2 at the 3′ end encodes a 660-amino-acid capsid protein. The AUG start codon of ORF3 was predicted to overlap with the UGA stop codon of ORF1, thus encoding a small protein of 123 amino acid residues. ORF3 also overlaps with ORF2 at its 3′ end since ORF2 begins 38 nucleotides (nt) downstream of ORF1 (19, 23).
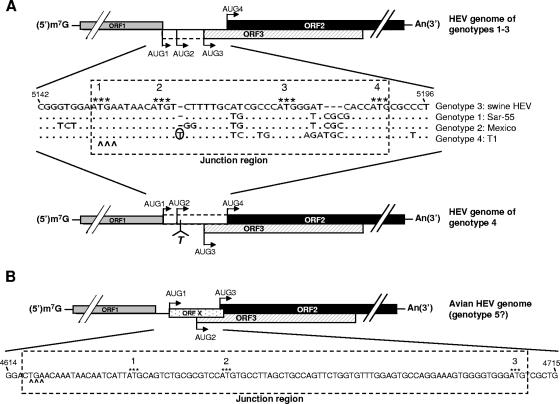
At least four major genotypes of HEV have been identified in mammalian species (8-9, 21). The genomic organization among HEV genotypes 1, 2, and 3 is conserved (Fig. 1A). However, the T1 strain of the prototype HEV genotype 4 has a unique genomic organization due to a frameshift mutation caused by a single nucleotide insertion (Fig. 1A) (30): ORF3 does not overlap with ORF1 and starts 28 bases downstream of ORF1, whereas ORF2 utilizes the AUG start codon, overlapping ORF1 as its putative initiation site, and thus extends 14 amino acids at the 5′ end. Sequence analyses of additional HEV genotype 4 isolates showed that they all have similar types of genomic organization, with only 114 amino acid residues in the predicted ORF3 protein (19, 21). The genomic organization of the recently identified avian HEV (putative genotype 5) is quite different from those of the four mammalian HEV genotypes: ORF3 also overlaps ORF2, but neither ORF3 nor ORF2 overlaps ORF1 (8-9) (Fig. 1B).
FIG. 1.
Schematic diagram of the predicted genomic organization and nucleotide sequence alignment of the noncoding junction region between ORF1 and ORF2 of HEV. (A) Swine HEV (genotype 3), Sar-55 strain (genotype 1), Mexico strain (genotype 2), and T1 strain (genotype 4). Nucleotide position numbers correspond to the sequence of clone pSHEV-3 of genotype 3 swine HEV. Dots denote sequence identity, and dashes denote deletions. The stop codon of ORF1 is indicated by carets, and the four AUGs are indicated by asterisks. AUG1, -2, and -3 are in-frame and serve as the potential ORF3 start codons, and AUG4 serves as the ORF2 start codon among HEV genotypes 1 to 3. In genotype 4, a single-nucleotide T insertion 2 nt downstream of AUG2 (marked with circle) results in the predicted ORF3 initiation at AUG3 and the predicted ORF2 initiation at AUG1, -2, and/or -4. The presumed coding regions between AUG1 and AUG3 for ORF3 in HEV genotypes 1 to 3 and between AUG1 and AUG4 for ORF2 in genotype 4 HEV are denoted by the dashed-line box based on our hypothesis that AUG3 is the authentic ORF3 initiation site and that AUG4 is the authentic ORF2 initiation site for all HEV genotypes. (B) Avian HEV (putative genotype 5). There exist three AUGs that could potentially serve as the initiation sites for different ORFs. In addition to AUG2 and AUG3 as the potential ORF3 and ORF2 start codons, respectively, a novel small ORF X is predicted to start at AUG1.
The function of ORF3 protein is unknown, but recent studies based upon genotype 1 HEV suggested that it may be involved in HEV virion morphogenesis and viral pathogenesis (25-26). It has been reported that recombinant ORF3 protein forms dimers through amino acid residues 81 to 123 (28); interacts with ORF2 protein mediated by amino acid residues 57 to 81 (23); binds various SH3 domain-containing proteins via amino acid residues 75 to 113 and activates mitogen-activated protein kinase (12); and interacts with a liver-specific protein, α1-microglobulin/bikunin precursor (AMBP), and its two processed proteins, α1m (26) and bikunin (27), through the C-terminal region (amino acid residues 81 to 123). These data indicated that the C-terminal region of the HEV ORF3 protein is a multifunctional domain.
We recently developed a reverse genetics system for genotype 3 HEV using pigs as a model (10). To demonstrate the utility of this unique system, which bypasses the traditional cell culture system that is inefficient for HEV (3), for studying the molecular mechanism of HEV replication and to determine the function of the ORF3 protein in viral replication and pathogenesis in vivo, in this study we utilized the newly developed genotype 3 HEV reverse genetics system and the unique pig model to identify the authentic initiation site of the HEV ORF3 protein.
Predicted initiation site of HEV ORF3 protein based upon sequence analyses.
It has been a common assumption in the HEV scientific community that the ORF3 start codon overlaps the ORF1 stop codon in HEV genotypes 1 through 3 and that the ORF2 start codon begins 35 (genotype 3) or 38 bases (genotypes 1 and 2) downstream from ORF1. The recent discoveries of HEV genotype 4 and the avian HEV (putative genotype 5) with different predicted ORF2 and ORF3 initiation codons as well as the recent finding that the functional domains of the ORF3 protein are all located in the C-terminal region led us to question whether the common assumptions are correct.
For convenience, in this study, the region between the ORF1 stop codon and the putative ORF2 initiation codon that contains the presumed N terminus of ORF3 was designated the “junction region.” Sequence analyses of the junction regions from the four recognized HEV genotypes revealed the existence of three in-frame AUGs in all genotype 1 through 3 HEVs, designated AUG1, -2, and -3, from 5′ to 3′ ends, which could each potentially serve as the ORF3 start codon (Fig. 1A). The first AUG (AUG1) overlapping the ORF1 stop codon is the current presumptive ORF3 initiation site, while AUG4 is the start codon for ORF2. Since there is a single-nucleotide T insertion between AUG2 and AUG3 in HEV genotype 4, AUG1 and AUG2 in HEV genotypes 1 through 3 could not be used as alternative ORF3 initiation sites. Consequently, the ORF3 initiation site for HEV genotype 4 is predicted to start at only AUG3 but the ORF2 could potentially start at AUG1, -2, or -4 (Fig. 1A).
One of the critical factors affecting translation initiation and efficiency is the context of the AUG start codon sequence (13). The Kozak consensus sequence, GCCGCC(A/G)CCAUGG (underlining indicates the start codon), is the optimal sequence, where the −3 and +4 nucleotides are important for controlling initiation efficiency. Sequence analyses of the junction region revealed that the context of AUG3 in HEV genotypes 1 through 4 is more consistent with the Kozak sequence (Fig. 1A). In addition, AUG3 exists at the same position in all four genotypes of mammalian HEV; it exists as well in avian HEV but at a different position. We thus hypothesize that all genotypes of HEV utilize AUG3 instead of the current commonly presumed AUG1 for the ORF3 initiation and that HEV genotype 4 utilizes the same ORF2 start codon as do HEV genotypes 1 through 3. Therefore, the AUG1 overlapping the ORF1 stop codon is neither the presumed ORF3 initiation site for HEV genotypes 1 through 3 nor the predicted ORF2 initiation site for genotype 4 HEV. We believe that all HEVs, including avian HEV, have the same genomic organization in that ORF3 overlaps with ORF2, but neither ORF2 nor ORF3 overlaps with ORF1.
The hypothesized ORF2 and ORF3 initiation codons for HEV are based upon sequence analyses and computer predictions. Recently, Graff et al. (5) identified a bicistronic, subgenomic mRNA of human HEV genotype 1 from which both ORF2 and ORF3 were translated using an in vitro HEV replicon system. The start site of this subgenomic RNA lies between AUG2 and AUG3, and thus, it is deduced that AUG3 is the site for ORF3 translation (5). However, with the lack of an efficient cell culture system for HEV (3), the results obtained from an artificial in vitro HEV replicon system (in which viral replication is not in the context of complete viral genome) may not reflect what really happens in vivo. Importantly, major differences in HEV replication and pathogenesis have been reported among different HEV genotypes: levels of replication of genotypes 1 and 2 are restricted in humans, whereas HEV genotypes 3 and 4 infect both humans and pigs (and possibly other animal species). Therefore, it is important to define the role of ORF3 protein in HEV replication in a homologous animal model system using a different HEV genotype with a broader host tropism. In this study, we take advantage of the HEV genotype 3 reverse genetics system and the unique homologous swine model system (pigs are the natural host of genotype 3 swine HEV) we recently developed to systematically delineate the roles of the three in-frame AUGs of the junction region in ORF3 initiation in vivo and to evaluate the utility of this unique homologous pig model system for studying the molecular mechanism of HEV replication.
Construction of HEV mutants and determination of their levels of in vivo infectivity in pigs.
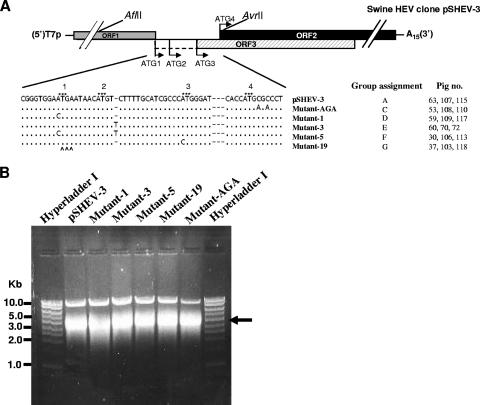
The construction and characterization of an infectious cDNA clone of genotype 3 swine HEV, pSHEV-3, has been previously described (10). In this study, pSHEV-3 was used as the parental genomic template for the construction of HEV mutants. The pSHEV-3 infectious cDNA clone contains two unique AflII and AvrII restriction enzyme sites, located outside the junction region, that were used to facilitate the cloning steps in this study. By using standard recombinant DNA techniques, we introduced mutations into various mutants by PCR-directed mutagenesis with a sense primer (5′-GTCTCTTAAGGGTTTCTGGAAGAAGC-3′) containing an AflII site (underlined) and reverse primers containing the respective desired mutation (Fig. 2A) as well as an AvrII site. Pfu Ultra high-fidelity DNA polymerase (Stratagene) was used in PCR amplification to minimize unwanted mutations. Each amplified PCR fragment was digested with the two restriction enzymes and subsequently replaced the corresponding region in the pSHEV-3 clone to produce respective mutants (Fig. 2A). Five HEV mutants, designated mutant-AGA, mutant-1, mutant-3, mutant-5, and mutant-19, were constructed. The full-length genome of each mutant was sequenced to verify the introduced mutations.
FIG. 2.
Construction of HEV mutants derived from the wild-type genotype 3 swine HEV consensus cDNA clone pSHEV-3. (A) Alignment of nucleotide sequences in the junction regions among the mutants and their group assignments for the in vivo infectivity study in pigs. T7p represents a T7 RNA polymerase core promoter, and A15 represents a 15-nt poly(A) tail. The unique AflII and AvrII restriction sites on pSHEV-3 used for the cloning purpose are also shown. For an explanation of the symbols, see the legend for Fig. 1, panel A. (B) In vitro transcription of full-length capped RNAs from the wild-type infectious cDNA clone pSHEV-3 and five HEV mutant clones. Five microliters of the transcription mixture was separated in a 1% agarose gel to check the quality of the transcription before being intrahepatically inoculated into the liver of pigs. An arrow indicates the in vitro RNA transcripts of expected size.
To generate RNA transcripts for the in vivo infectivity study of the mutants, the pSHEV-3 wild-type infectious cDNA clone and the five mutant clones were each linearized with XbaI. In vitro transcriptions of capped RNA were performed for each construct at 37°C in a 750-μl reaction mixture containing approximately 43 μg of each linearized plasmid DNA, 75 μl of 10× reaction buffer, 375 μl of 2× nucleoside triphosphate-CAP analog, 75 μl of enzyme mix, and an additional 37.5 μl of a 30 mM GTP stock for capping (mMessage mMachine T7 transcription kit; Ambion). The RNA transcripts were analyzed by electrophoresis of 5 μl of the reaction mixture in a 1% agarose gel. A strong RNA band of expected size was observed for each construct, and the yields from each construct were similar (Fig. 2B). The remaining RNA reaction mixture from each construct was diluted with 2.25 ml of cold RNase-, DNase-, and proteinase-free phosphate-buffered saline (PBS) buffer, aliquoted in three vials of 1 ml each, and immediately frozen on dry ice until used for the intrahepatic inoculation of pigs.
We previously developed an in vivo transfection system to bypass the cell culture system, which is inefficient for HEV (3), for testing the infectivity of the swine HEV infectious clone and its derivatives in pigs (10). The procedure we used to determine the infectivity of the five HEV mutants in this study is essentially the same as that we previously reported (10). Briefly, 3-week-old, specific-pathogen-free piglets (n = 21) that were seronegative for HEV were randomly assigned into seven groups of three pigs each (groups A to G). Each group of pigs was housed separately and maintained under conditions meeting ICAUC requirements. The capped RNA transcript inocula from each mutant as well as the wild-type swine HEV infectious cDNA clone were intrahepatically injected into the liver of each piglet via the ultrasound-guided intrahepatic inoculation technique (10). The three group A pigs (identification [ID] numbers 63, 107, and 115) were each injected with 1 ml of the RNA transcripts from the wild-type pSHEV-3 clone as positive controls. Group B pigs (ID numbers 19, 67, and 120) were each injected with 1 ml of PBS buffer as negative controls. Pigs in groups C (ID numbers 53, 108, and 110), D (ID numbers 59, 109, and 117), E (ID numbers 60, 70, and 72), F (ID numbers 30, 106, and 113), and G (ID numbers 37, 103, and 118) were each similarly injected with capped RNA transcripts from mutant-AGA, mutant-1, mutant-3, mutant-5, and mutant-19, respectively (Fig. 2A). Pigs in all groups were monitored daily for clinical signs of disease for a total of 10 weeks. Fecal and serum samples from each pig were collected prior to inoculation. Fecal samples were collected every 3 days postinoculation, and serum samples were collected every 6 days postinoculation. None of the pigs had clinical signs of hepatitis during the course of the study, which is consistent with the results of our previous study (6, 10). Fecal and serum samples were tested for the presence of swine HEV RNA by a nested reverse transcription-PCR (RT-PCR) as described previously (10), and serum samples were also tested by an enzyme-linked immunosorbent assay (ELISA) for IgG anti-HEV antibodies (6, 15-16).
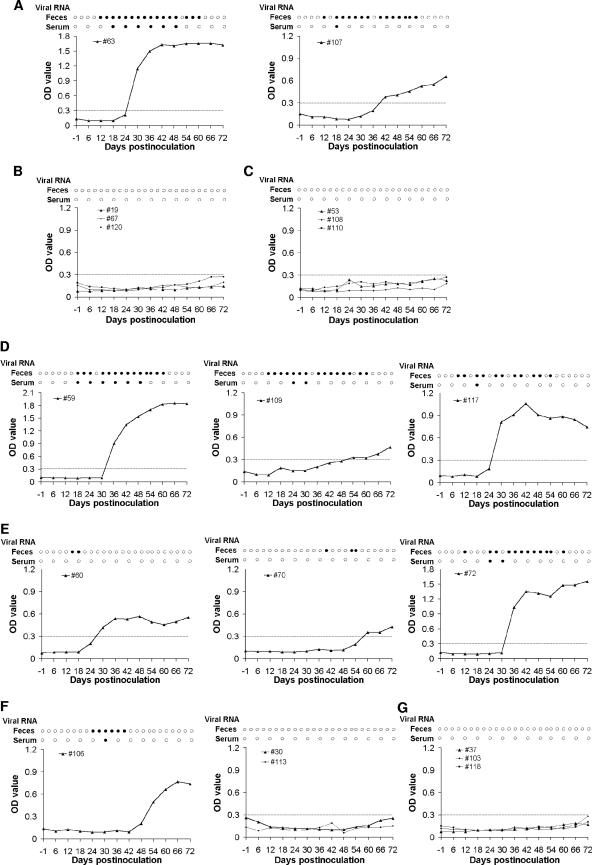
Pig 115 in group A died 1 day after inoculation, likely from the inoculation procedure. The remaining two pigs in group A seroconverted to IgG anti-HEV antibodies at 30 (pig 63) and 42 (pig 107) days postinfection (dpi) (Fig. 3A), whereas the three pigs in group B remained seronegative throughout the study (Fig. 3B). Since the ELISA to detect HEV antibodies was based on a recombinant HEV capsid ORF2 protein, seroconversion to anti-ORF2 antibody implies that the ORF2 protein was produced in vivo in group A pigs. Both of the remaining pigs in group A shed virus in feces in a pattern consistent with that associated with the wild-type swine HEV infectious cDNA clone (10) (Fig. 3A). Viremia was also detected variably in the two group A positive control pigs (Fig. 3A). As expected, viremia and fecal virus shedding were not detected in the three group B negative control pigs.
FIG. 3.
Determination of the in vivo infectivity of wild-type and mutant cDNA clones of genotype 3 HEV in pigs. Fecal viral shedding, viremia, and seroconversion to IgG anti-HEV antibodies in pigs intrahepatically inoculated with capped RNA transcripts from wild-type swine HEV clone pSHEV-3 (A), PBS buffer (B), mutant-AGA (C), mutant-1 (D), mutant-3 (E), mutant-5 (F), and mutant-19 (G). Results of RT-PCR for the presence (closed circles) or absence (open circles) of HEV RNA in fecal and serum samples are indicated. IgG anti-HEV antibody is plotted as the ELISA optical density (A405). The ELISA cutoff value, indicated by a dashed line, is 0.30.
Expression of an intact ORF3 is required for in vivo HEV infectivity.
The HEV genotype 3 mutant-AGA was designed to test whether the expression of an intact ORF3 is required for viral infectivity in vivo. Since both triplets CGC and AGA encode arginine, the second codon of ORF2 was silently mutated from CGC to AGA to introduce a TGA stop codon in the ORF3 leading to the C-terminal truncation of ORF3 (Fig. 2A). In this mutant-AGA, the authentic capsid protein encoded by the ORF2 gene remains unchanged. The three pigs in group C that were intrahepatically inoculated with capped RNA transcripts from the mutant-AGA had no evidence of seroconversion, fecal virus shedding, or viremia throughout the experiment (Fig. 3C), indicating the lack of infectivity for the mutant-AGA. Under the in vivo experimental conditions, it is difficult to directly detect whether ORF2 capsid protein was synthesized in infected porcine cells. However, since the recombinant ORF2-derived polypeptide was used as the antigen in the ELISA to detect the production of anti-ORF2 antibody from the sera of inoculated pigs, it is reasonable to assume that the seronegative results imply the lack of ORF2 protein production due to lack of infection. A genotype 1 HEV mutant with an introduced stop codon (ORF3-null mutant) derived from the Sar-55 strain of human HEV was noninfectious in rhesus macaques (4); however, the ORF3-null mutant was able to synthesize ORF2 protein in the absence of ORF3 in cultured Huh-7 cells, suggesting that the deficiency of ORF3 expression resulted in the loss of infectivity for the mutant (4). Therefore, it is reasonable to believe that the failure to recover the HEV genotype 3 mutant-AGA in inoculated pigs was due to the deficiency of ORF3 protein expression. Since the intact ORF3 expression is required for HEV in vivo infectivity, one of the three in-frame AUGs of ORF3 in the junction region must be utilized for translation initiation.
Initiation at AUG1 of ORF3 is not required for viral infectivity.
HEV mutant-1 was constructed with a single point mutation at nt 5150, from A to C, which knocked out AUG1 of ORF3 (Fig. 2A). The introduced mutation is a structurally conserved substitution for ORF1, from glutamic acid (GAA) to aspartic acid (GAC). Mutant-1 was designed to test whether AUG1 serves as the initiation codon of ORF3. All three group D pigs inoculated with the capped RNA transcripts from mutant-1 had fecal virus shedding and viremia (Fig. 3D). Pigs 59, 109, and 117 all seroconverted to IgG anti-HEV antibodies at 36, 54, and 30 dpi, respectively. To determine whether the introduced mutation was retained in the virus recovered from pigs, we amplified and sequenced a genomic fragment containing the junction region by using a sense primer (5′-CAGCGAAGGGGTTGGTTGGATG-3′) and a reverse primer (5′-CAGCGAAGGGGTTGGTTGGATG-3′) from selected fecal samples of the three group D pigs. Sequence analyses showed that the sequences recovered from the three pigs all retained the introduced mutation C at nt 5150, indicating that the infectious virus originated from mutant-1. The results indicated that AUG1 of ORF3 is not required for viral infectivity in vivo and is likely not utilized as the ORF3 initiation codon.
Initiation at AUG2 of ORF3 is not required for viral infectivity in vivo.
To test whether the AUG2 is required for viral infectivity, a single-nucleotide T insertion downstream from AUG2 was introduced into the pSHEV-3 clone of HEV genotype 3 (at the same position as that of HEV genotype 4) to produce mutant-3 (Fig. 2A). Mutant-3 would abolish the potential function of AUG2 and mimic the AUG1 and AUG2 frameshift mutants in the junction region of HEV genotype 4. All three group E pigs inoculated with capped RNA transcripts from mutant-3 seroconverted to IgG anti-HEV antibodies (Fig. 3E). Fecal virus shedding was detected in the samples from pig 60 at 18 and 21 dpi as well as in samples from pig 70 at 39, 51, and 54 dpi. Pig 72 also had detectable viremia at 24 and 30 dpi. A similar construct with a T insertion in the backbone of a genotype 1 human HEV strain was shown to be infectious in vitro and produced both ORF2 and ORF3 proteins (5), which is consistent with our finding in this study with the T insertion mutant of the genotype 3 swine HEV.
HEV mutant-5 was constructed to further verify the results obtained from mutant-1 and mutant-3, in which the single point mutation at AUG1 and the single-nucleotide T insertion downstream from AUG2 were introduced simultaneously, leading to a double mutant (Fig. 2A). Mutant-5 was intended not only to verify the dispensable nature of AUG1 and AUG2 as the initiation sites of ORF3 for viral infectivity for HEV genotypes 1 to 3 but also to determine whether AUG1 is used as the ORF2 initiation codon for HEV genotype 4. Of the three group F pigs inoculated with the RNA transcripts from mutant-5, only pig 106 had fecal virus shedding, viremia, and seroconversion; pigs 30 and 113 did not show any of the three markers (Fig. 3F), suggesting that the recovery of mutant-5 was less efficient than those of mutant-1 and mutant-3.
The junction region was amplified by RT-PCR from selected fecal samples of pig 72 in group E and pig 106 in group F and subsequently sequenced. The nucleotide T insertion was retained in the recovered viruses from pig 72, and the double mutations were also retained in the recovered viruses from pig 106. Additional mutations were not identified in this region.
Taken together from the results with mutant-3 and -5, we showed that AUG1 and AUG2 are not necessary for viral infectivity in vivo and thus provide indirect evidence that the third in-frame AUG (AUG3) is the authentic initiation site for all four genotypes of HEV. The results with mutant-5 also indicated that the common presumption of AUG1 as the initiation codon of ORF2 for genotype 4 HEV is likely incorrect.
Initiation at AUG3 of the ORF3 is required for viral infectivity.
The AUG3 of the ORF3 was changed to a leucine (CUG) by a point mutation from A to C to produce the HEV genotype 3 mutant-19 (Fig. 2A). The three group G pigs inoculated with the capped RNA transcripts from mutant-19 had no viremia, fecal virus shedding, or seroconversion throughout the experiment (Fig. 3G), indicating a lack of viral infectivity as we expected. Based upon the results from mutant-AGA and mutant-19, it is likely that the failure to recover mutant-19 was due to the ORF3 expression deficiency. Since AUG1 and AUG2 remain unchanged in mutant-19, we thus conclude that the AUG3 plays a key role in the ORF3 translation initiation which is essential for viral infectivity in vivo and that AUG3 is the authentic initiation site of ORF3 not only in HEV genotype 4, but also in HEV genotypes 1 to 3. Most recently, Graff et al. (5) reported that human HEV genotype 1 mutant pHEV5131Ala, in which AUG3 was mutated to GCA, did not produce ORF3 protein in the presence of ORF2 synthesis in transfected Huh-7 cells in vitro (5), although the infectivity of the genotype 1 pHEV5131Ala was not tested in primates. The results of an in vitro study of a genotype 1 HEV from Graff et al. (5) and the in vivo results of a genotype 3 HEV presented in this paper provided convincing evidence that AUG3 is the authentic ORF3 initiation site and that ORF3 expression is required for in vivo infectivity.
By analogy with alpha superfamily viruses, the noncoding junction region should comprise cis-acting elements that are critical for viral subgenomic RNA synthesis. Tzeng et al. (29) predicted the subgenomic start site of HEV at nt 5 or 6 upstream from AUG3 based upon results with the rubella virus subgenomic promoter and the conservative nature between the two viruses. Recently, a continuous 12 nucleotides between AUG1 and AUG2 were recognized as the cis-acting element in HEV genotype 1 since mutations in this region eliminated the production of both ORF2 and ORF3 proteins (4). In an early study, Tam et al. (23) reported the detection of two subgenomic RNA transcripts of approximately 3.7 and 2.0 kb in HEV-infected liver. Since the N termini of HEV ORF2 and -3 are in close proximity, they could be translated from a single subgenomic RNA instead of two separate mRNAs, and indeed, this was confirmed recently by Graff et al. (5). By using an artificial in vitro HEV replicon system containing the neomycin resistance gene that was able to synthesize viral RNA, Graff et al. (5) identified a bicistronic subgenomic mRNA that encodes both ORF3 and ORF2 (5). The 5′ terminus of the subgenomic mRNA starts at nt 5122 between AUG 2 and AUG3, suggesting that the first two in-frame AUGs are not included in the subgenomic RNA, and therefore, they are not utilized as the initiation sites for ORF3. This in vitro observation with a genotype 1 HEV is consistent with our in vivo results with the genotype 3 HEV reported in this study. The systematic mutagenesis data from this in vivo study also imply that it is likely all four genotypes of mammalian HEV share the same characteristics of ORF3 initiation.
Identification of a conserved double stem-loop structure in the junction regions of all five genotypes of HEV that may be important for virus replication.
RNA secondary structures often play important roles in viral replication, viral subgenomic RNA synthesis, and translation efficiency (14, 20). Therefore, to further understand how mutations in the junction region may affect virus infectivity, we performed RNA secondary structure prediction by using the program RNA FOLD (http://www.bioinfo.rpi.edu/∼zukerm/rna/) for the junction regions of the wild-type swine HEV, mutant-1, mutant-3, and mutant-5, all of which were successfully recovered in pigs but with different efficiencies.
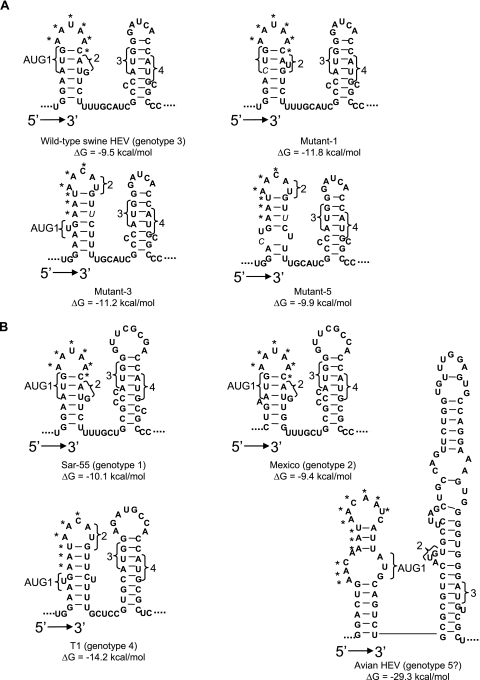
In wild-type genotype 3 swine HEV, a double stem-loop structure of approximately 50 nt was identified (Fig. 4A). Despite the introduced mutations in the junction region, mutant-1, -3, and -5 still maintain the double stem-loop structure with subtle differences in the first stem-loop (Fig. 4A). The cis-acting element between AUG1 and AUG2 in HEV genotype 1 comprised 12 continuous nucleotides (4). Aside from the two AUGs, a conserved 6-nt sequence, AAUAAC, was found in the loop part of the first stem-loop. The single substitution in mutant-1 did not alter the nucleotide sequence on the loop. Accordingly, the infection profile of pigs inoculated with mutant-1 (Fig. 3D) was indistinguishable from that of the wild-type virus (Fig. 3A). Compared to that in mutant-1, however, mutations in mutant-3 and mutant-5 altered the sequence on the loop from AAUAAC to AACAUG, thus resulting in less-efficient mutant virus recovery (Fig. 3E and F). Therefore, it appears that the presence of the exact nucleotides of the cis-acting element on the loop part of the first stem-loop is critical for viral infectivity in vivo.
FIG. 4.
A conserved double stem-loop RNA secondary structure is predicted by the mfold program in the junction region between ORF1 and ORF2 of HEV. (A) Genotype 3 HEV and its derived mutants. (B) The other four genotypes of HEV. The four AUGs (AUG1, -2, -3, and 4) in genotypes 1 through 4 HEV strains and the three AUGs in avian HEV putative genotype 5 are indicated. The contiguous AAC/U triplets are marked with asterisks. The mutations introduced in mutant-1, -3, and -5 are indicated by italic letters.
We further compared the predicted secondary structures of the corresponding junction regions of HEV strains Sar-55 (genotype 1), Mexico (genotype 2), and T1 (genotype 4). All were shown to fold into a conserved double stem-loop structure (Fig. 4B). The newly identified start site for the single subgenomic RNA of genotype 1 HEV was located in the linking region between the two stem-loops (5). The structure conservation among four genotypes of HEV supported that the subgenomic RNA of HEV genotype 3 starts at the guanine following the four uracils in the linking region (Fig. 4A), which is the same start site for the human HEV genotype 1 (5).
Interestingly, the junction region of avian HEV putative genotype 5 (8), which is very divergent in sequence from HEV genotypes 1 to 4 and consists of more nucleotides in the junction region, also forms a double stem-loop structure without the linking nucleotides between the two stem-loops (Fig. 4B). The contiguous AAC/U triplets also reside on the loop of the first stem-loop following the UGA stop codon of ORF1, suggesting that this motif may play an important role in viral replication similar to that in mammalian HEVs. It will now be interesting to determine whether the first guanine in the second stem-loop is the start site for the subgenomic RNA of avian HEV based upon structure and sequence analogy with the mammalian HEVs. Future in-depth mutagenesis experiments in this junction region, which are beyond the scope of this study, are warranted to definitively understand the role of the stem-loop structures in viral replication.
Acknowledgments
We thank Denis Guenette for his technical assistance.
This study is supported by a grant from the National Institutes of Health (AI065546) and in part by another grant (AI50611) from NIH.
Footnotes
Published ahead of print on 3 January 2007.
REFERENCES
- 1.Billam, P., F. F. Huang, Z. F. Sun, F. W. Pierson, R. B. Duncan, F. Elvinger, D. K. Guenette, T. E. Toth, and X. J. Meng. 2005. Systematic pathogenesis and replication of avian hepatitis E virus in specific-pathogen-free adult chickens. J. Virol. 79:3429-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerson, S. U., D. Anderson, A. Arankalle, X. J. Meng, M. Purdy, G. G. Schlauder, and S. A. Tsarev. 2004. Hepevirus, p. 851-855. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. VIIIth report of the ICTV. Elsevier/Academic Press, London, United Kingdom.
- 3.Emerson, S. U., H. Nguyen, U. Torian, and R. H. Purcell. 2006. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J. Virol. 80:10457-10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Graff, J., H. Nguyen, C. Yu, W. R. Elkins, M. St. Claire, R. H. Purcell, and S. U. Emerson. 2005. The open reading frame 3 gene of hepatitis E virus contains a cis-reactive element and encodes a protein required for infection of macaques. J. Virol. 79:6680-6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graff, J., U. Torian, H. Nguyen, and S. U. Emerson. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J. Virol. 80:5919-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halbur, P. G., C. Kasorndorkbua, C. Gilbert, D. Guenette, M. B. Potters, R. H. Purcell, S. U. Emerson, T. E. Toth, and X. J. Meng. 2001. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. J. Clin. Microbiol. 39:918-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haqshenas, G., H. L. Shivaprasad, P. R. Woolcock, D. H. Read, and X. J. Meng. 2001. Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis-splenomegaly syndrome in the United States. J. Gen. Virol. 82:2449-2462. [DOI] [PubMed] [Google Scholar]
- 8.Huang, F. F., Z. F. Sun, S. U. Emerson, R. H. Purcell, H. L. Shivaprasad, F. W. Pierson, T. E. Toth, and X. J. Meng. 2004. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J. Gen. Virol. 85:1609-1618. [DOI] [PubMed] [Google Scholar]
- 9.Huang, F. F., F. W. Pierson, T. E. Toth, and X. J. Meng. 2005. Construction and characterization of infectious cDNA clones of a chicken strain of hepatitis E virus (HEV), avian HEV. J. Gen. Virol. 86:2585-2593. [DOI] [PubMed] [Google Scholar]
- 10.Huang, Y. W., G. Haqshenas, C. Kasorndorkbua, P. G. Halbur, S. U. Emerson, and X. J. Meng. 2005. Capped RNA transcripts of full-length cDNA clones of swine hepatitis E virus are replication competent when transfected into Huh7 cells and infectious when intrahepatically inoculated into pigs. J. Virol. 79:1552-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussaini, S. H., S. J. Skidmore, P. Richardson, L. M. Sherratt, B. T. Cooper, and J. G. O'Grady. 1997. Severe hepatitis E infection during pregnancy. J. Viral Hepat. 4:51-54. [DOI] [PubMed] [Google Scholar]
- 12.Korkaya, H., S. Jameel, D. Gupta, S. Tyagi, R. Kumar, M. Zafrullah, M. Mazumdar, S. K. Lal, L. Xiaofang, D. Sehgal, S. R. Das, and D. Sahal. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 276:42389-42400. [DOI] [PubMed] [Google Scholar]
- 13.Kozak, M. 1992. Regulation of translation in eukaryotic systems. Annu. Rev. Cell Biol. 8:197-225. [DOI] [PubMed] [Google Scholar]
- 14.Maia, I. G., K. Seron, A. L. Haenni, and F. Bernardi. 1996. Gene expression from viral RNA genomes. Plant Mol. Biol. 32:367-391. [DOI] [PubMed] [Google Scholar]
- 15.Meng, X. J., R. H. Purcell, P. G. Halbur, J. R. Lehman, D. M. Webb, T. S. Tsareva, J. S. Haynes, B. J. Thacker, and S. U. Emerson. 1997. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 94:9860-9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng, X. J., P. G. Halbur, M. S. Shapiro, S. Govindarajan, J. D. Bruna, I. K. Mushahwar, R. H. Purcell, and S. U. Emerson. 1998. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J. Virol. 72:9714-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng, X. J. 2000. Novel strains of hepatitis E virus identified from humans and other animal species: is hepatitis E a zoonosis? J. Hepatol. 33:842-845. [DOI] [PubMed] [Google Scholar]
- 18.Meng, X. J., B. Wiseman, F. Elvinger, D. K. Guenette, T. E. Toth, R. E. Engle, S. U. Emerson, and R. H. Purcell. 2002. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. J. Clin. Microbiol. 40:117-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng, X. J. 2003. Swine hepatitis E virus: cross-species infection and risk in xenotransplantation. Curr. Top. Microbiol. Immunol. 278:185-216. [DOI] [PubMed] [Google Scholar]
- 20.Pappas, C. L., W. P. Tzeng, and T. K. Frey. 2006. Evaluation of cis-acting elements in the rubella virus subgenomic RNA that play a role in its translation. Arch. Virol. 151:327-346. [DOI] [PubMed] [Google Scholar]
- 21.Schlauder, G. G., and I. K. Mushahwar. 2001. Genetic heterogeneity of hepatitis E virus. J. Med. Virol. 65:282-292. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi, M., T. Nishizawa, H. Miyajima, Y. Gotanda, T. Iita, F. Tsuda, and H. Okamoto. 2003. Swine hepatitis E virus strains in Japan form four phylogenetic clusters comparable with those of Japanese isolates of human hepatitis E virus. J. Gen. Virol. 84:851-862. [DOI] [PubMed] [Google Scholar]
- 23.Tam, A. W., M. M. Smith, M. E. Guerra, C. C. Huang, D. W. Bradley, K. E. Fry, and G. R. Reyes. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tei, S., N. Kitajima, K. Takahashi, and S. Mishiro. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362:371-373. [DOI] [PubMed] [Google Scholar]
- 25.Tyagi, S., H. Korkaya, M. Zafrullah, S. Jameel, and S. K. Lal. 2002. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 277:22759-22767. [DOI] [PubMed] [Google Scholar]
- 26.Tyagi, S., M. Surjit, A. K. Roy, S. Jameel, and S. K. Lal. 2004. The ORF3 protein of hepatitis E virus interacts with liver-specific α1-microglobulin and its precursor α1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 279:29308-29319. [DOI] [PubMed] [Google Scholar]
- 27.Tyagi, S., M. Surjit, and S. K. Lal. 2005. The 41-amino-acid C-terminal region of the hepatitis E virus ORF3 protein interacts with bikunin, a Kunitz-type serine protease inhibitor. J. Virol. 79:12081-12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyagi, S., S. Jameel, and S. K. Lal. 2001. Self-association and mapping of the interaction domain of hepatitis E virus ORF3 protein. J. Virol. 75:2493-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzeng, W. P., and T. K. Frey. 2002. Mapping the rubella virus subgenomic promoter. J. Virol. 76:3189-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Y., H. Zhang, R. Ling, H. Li, and T. J. Harrison. 2000. The complete sequence of hepatitis E virus genotype 4 reveals an alternative strategy for translation of open reading frames 2 and 3. J. Gen. Virol. 81:1675-1686. [DOI] [PubMed] [Google Scholar]
- 31.Yazaki, Y., H. Mizuo, M. Takahashi, T. Nishizawa, N. Sasaki, Y. Gotanda, and H. Okamoto. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J. Gen. Virol. 84:2351-2357. [DOI] [PubMed] [Google Scholar]