Abstract
Viruses have evolved a multitude of strategies to subvert the innate immune system by interfering with components of the alpha/beta interferon (IFN-α/β) induction and signaling pathway. It is well established that the pestiviruses prevent IFN-α/β induction in their primary target cells, such as epitheloidal and endothelial cells, macrophages, and conventional dendritic cells, a phenotype mediated by the viral protein Npro. Central players in the IFN-α/β induction cascade are interferon regulatory factor 3 (IRF3) and IRF7. Recently, it was proposed that classical swine fever virus (CSFV), the porcine pestivirus, induced the loss of IRF3 by inhibiting the transcription of IRF3 mRNA. In the present study, we show that endogenous IRF3 and IRF3 expressed from a cytomegalovirus (CMV) promoter are depleted in the presence of CSFV by means of Npro, while CSFV does not inhibit CMV promoter-driven protein expression. We also demonstrate that CSFV does not reduce the transcriptional activity of the IRF3 promoter and does not affect the stability of IRF3 mRNA. In fact, CSFV Npro induces proteasomal degradation of IRF3, as demonstrated by proteasome inhibition studies. Furthermore, Npro coprecipitates with IRF3, suggesting that the proteasomal degradation of IRF3 is induced by a direct or indirect interaction with Npro. Finally, we show that Npro does not downregulate IRF7 expression.
Alpha/beta interferon (IFN-α/β) represents one of the first lines of defense of the innate immune system and is produced rapidly when viral factors are recognized by pattern recognition receptors (41, 49). Viruses have evolved a multitude of strategies to subvert the IFN-α/β system (for selected reviews, see references 25, 27, 29, and 79). It is well established that the pestiviruses enhance the replication of other viruses by suppressing IFN-α/β induction (4, 9, 14, 21, 23, 30, 34, 36, 44, 46, 61, 63, 66, 74). The interference of pestiviruses with the IFN system is apparently restricted to the IFN-α/β induction pathway. There is good evidence that they do not counteract IFN-α/β signaling in the cell systems studied (4, 65). A unique feature of the pestiviruses compared with the other genera of the family Flaviviridae is the presence of the Npro gene at the 5′ end of the single large open reading frame. For the two pestiviruses classical swine fever virus (CSFV) and bovine viral diarrhea virus (BVDV), Npro is dispensable for virus replication in cell culture (23, 45, 75). CSFV lacking Npro (ΔNpro CSFV) replicates with comparable efficiency in cells devoid of a functional IFN-α/β induction system but is impaired in IFN-competent cells (63) and attenuated in animals (51). In fact, pestiviruses in which Npro was deleted have lost the capacity to suppress double-stranded RNA (dsRNA)- and virus-induced IFN production, and they induce IFN-α/β in porcine cell lines, macrophages, and monocyte-derived dendritic cells (DC) (8, 23, 63). Recent data demonstrated that the Npro protein of pestiviruses functions as an antagonist of IFN-α/β induction independently of other viral elements (30, 34, 46, 61).
Central players of the signaling cascade leading to IFN-α/β induction are interferon regulatory factor 3 (IRF3) and IRF7 (41, 42, 55). After virus infection of a cell, pattern recognition receptors recognize viral elements, such as dsRNA, single-stranded RNA, DNA, or viral glycoproteins, depending on the virus (for selected reviews, see references 41 and 49). For pestiviruses, there is evidence that dsRNA from secondary structures and from replicative forms of the viral RNA represents a trigger for IFN-α/β induction in primary target cells (8). Typically, infected cells sense viral dsRNA via Toll-like receptor 3 (50, 67) and/or via the helicases RIG-I and MDA-5 (1, 17, 38, 40, 80). Recently however, it was shown that RIG-I can sense uncapped viral single-stranded RNA bearing a 5′ triphosphate (33, 57). All these signals result in phosphorylation of IRF3 via the two IκB kinase (IKK)-related kinases IKKɛ and TANK-binding kinase 1 (22, 41, 52, 69). The phosphorylated IRF3 then dimerizes and translocates to the nucleus, where it associates with transcriptional coactivators and binds to the DNA elements of the IFN-α/β promoters to upregulate IFN-α/β mRNA transcription (31, 72). For CSFV, La Rocca and coworkers demonstrated that the virus did not provide a signal for nuclear translocation of IRF3 but in fact induced the loss of IRF3 prior to translocation to the nucleus (46). Expression of Npro was sufficient to downregulate IRF3 expression. It was suggested that the loss of IRF3 was due to the inhibition of transcription of the IRF3 gene (46). For BVDV, it was shown that the virus inhibits dsRNA-mediated IFN-α/β induction by preventing the activated IRF3 from binding to DNA (4). Very recently, Hilton et al. demonstrated that this mechanism is mediated by Npro and that Npro of BVDV targets IRF3 for proteasomal degradation (30).
In the present study, we demonstrate that CSFV does not inhibit the transcription of IRF3, contrary to the observations of La Rocca et al. mentioned above. Furthermore, IRF3 mRNA remains continuously present in CSFV-infected cells that have lost the IRF3 protein concomitant with Npro expression. Also, CSFV does not inhibit the phosphorylation and translocation of IRF3. Pull-down experiments showed that Npro interacts directly or indirectly with IRF3. Importantly, in the presence of the proteasome inhibitor MG132, CSFV does not induce the loss of IRF3, demonstrating that CSFV induces the degradation of IRF3 via a proteasome-dependent mechanism. IRF7 expression, however, is not affected by the presence of Npro.
MATERIALS AND METHODS
Cells.
The porcine kidney cell lines SK-6 (39) and PK-15 (ATCC, Manassas, VA) were propagated in Earl's minimal essential medium containing 7% horse serum and in Dulbecco's modified Eagle medium supplemented with nonessential amino acids, 1 mM Na-pyruvate and 5% horse serum (Gibco-BRL), respectively. HEK 293T cells were cultured in Earl's minimal essential medium supplemented with 7% fetal bovine serum (Biochrom AG, Switzerland). Porcine DC were differentiated from peripheral blood mononuclear cell (PBMC)-derived monocytes isolated from specific-pathogen-free pigs by incubation in the presence of porcine interleukin-4 and granulocyte/monocyte colony-stimulating factor as described elsewhere (13). The clonal cell lines PK15-EGFP-Npro and PK15-EGFP, constitutively expressing the enhanced green fluorescent protein (EGFP)-Npro fusion protein or EGFP alone, were previously described (61).
Viruses.
CSFV vA187-1 and vA187-ΔNpro (ΔNpro CSFV) were rescued by electroporation of SK-6 cells with RNA transcribed in vitro from the plasmid cDNA clones pA187-1 (62) and pA187-ΔNpro (63), respectively, as previously described (53). All the virus titers were determined on SK-6 cells by standard endpoint dilution and were expressed as 50% tissue culture infectious doses (TCID50)/ml. The multiplicity of infection (MOI) used with SK-6 and PK-15 cells was calculated from the titer obtained on the respective cell line. Titration on DC was not performed, and the MOI was calculated based on the SK-6 titer.
Plasmids.
Plasmid pEGFP-IRF3 was constructed for cytomegalovirus (CMV) promoter-driven expression of EGFP fused to the amino terminus of porcine IRF3. For this purpose, the coding sequence of the porcine IRF3 mRNA was amplified from total RNA extracted from porcine PBMCs using standard reverse transcription (RT)-PCR and primers designed from the published sequence with GenBank accession number AB116563. The 5′ and 3′ halves of the IRF3-coding sequence were amplified and cloned separately using the TOPO TA cloning system (Invitrogen). DNA sequencing was performed with the Thermo Sequenase DYEnamic direct cycle sequencing kit (GE Healthcare) and the Global IR2 Sequencer with e-Seq software (LI-COR). For the N-terminal fusion of EGFP with IRF3, the IRF3 gene was cloned downstream of the EGFP gene and of a Gly-Gly-Ser-Gly-Gly-Ser linker in the pEGFP-N1 expression vector (Clontech). Plasmid pEGFP-IRF3 was also used for the construction of a phosphomimetic mutant of IRF3 that lacked the nuclear export signal (NES). The phosphomimetic mutations were introduced into IRF3 by changing the serine residues at positions 394 and 396 to aspartic acid, in analogy to a phosphomimetic mutant described for human IRF3 (47). In addition, the NES was knocked out in the phosphomimetic IRF3 by mutating the serine at position 139; the leucine residues at positions 140, 143, 144, and 149; the methionine at position 147; and the aspartic acid at position 148 to alanine residues, based on mutants described by Yoneyama and coworkers (81). Finally, the EGFP gene and the linker were replaced by the FLAG tag in both the mutant construct and pEGFP-IRF3, resulting in pFLAG-IRF3-S394,396D-ΔNES and pFLAG-IRF3, respectively. The plasmid pIRF3 expressing untagged porcine IRF3 was obtained by deleting the FLAG tag from plasmid pFLAG-IRF3. The coding sequence of the porcine IRF7 gene was amplified from total RNA extracted from porcine PBMCs using standard RT-PCR and rapid amplification of cDNA ends techniques. Overlapping cDNA fragments were amplified using internal oligonucleotide primers designed from the consensus nucleotide sequence of human, mouse, and rat IRF7 mRNA (GenBank accession numbers AF076494, NM_016850, and NM_001033691). The PCR fragments were cloned and sequenced as described above. Plasmid pFLAG-IRF7 for CMV-driven IRF7 expression was constructed in analogy to pFLAG-IRF3 by replacing the EGFP gene in the pEGFP-N1 backbone with the FLAG tag sequence fused upstream of the IRF7 sequence. The details of the constructions can be obtained on request. Plasmid pIRF3(−779)Luc carrying the full-length human IRF3 promoter upstream of the firefly luciferase reporter gene (48) was a kind gift of Paula Pitha-Rowe (Johns Hopkins University School of Medicine, Baltimore, MD). Plasmid pFLAG-Npro was constructed by inserting the FLAG tag coding sequence upstream of the Npro gene derived from pA187-1 (62) in the pCI expression vector (Promega). The pEAK-Npro construct for eukaryotic expression of Npro was described earlier (61).
Antibodies.
The viral Npro protein was detected using the rabbit anti-Npro serum previously described (61). The antisera against the porcine IRF3 and against the C protein of CSFV vA187-1 were produced in rabbits by using recombinant proteins expressed in Escherichia coli M15 with the pQE31 vector (QIAGEN) and purified by nickel chelate affinity chromatography as previously described for Npro (61). The monoclonal antibody (MAb) 34/1 used for the detection of the unphosphorylated and phosphorylated doublet of IRF3 was produced from hybridoma cells derived from Sp2/0-Ag14 myeloma cells and spleen cells of BALB/c mice (Japan SLC) immunized with recombinant porcine IRF3. For this purpose, the porcine IRF3 gene was cloned into the expression vector pET-32b (Novagen) and the protein was expressed in E. coli BL21(DE3). The recombinant IRF3 was solubilized with urea lysis solution, purified with the HisTrap HP Kit (GE Healthcare), and dialyzed against phosphate-buffered saline (PBS). The immunoglobulin G1 isotype of MAb 34/1 was identified with mouse monoclonal antibody isotyping reagents (Sigma). EGFP was detected in Western blots with MAb JL-8 (Clontech) and FLAG-tagged proteins with anti-FLAG M2 MAb (Sigma). The MAb HC/TC26 directed against the viral envelope protein E2 (26) was a kind gift of Irene Greiser-Wilke (Hannover Veterinary School, Hannover, Germany). Alexa Fluor 680 goat anti-mouse and goat anti-rabbit immunoglobulin G were obtained from Molecular Probes Inc.
Plasmid transfection and reporter gene assay.
For transient protein expression, cells were transfected with eukaryotic expression plasmids by electroporation with a Gene Pulser device (Bio-Rad). Briefly, the cells were washed twice with PBS and resuspended in ice-cold PBS at a density of 2 × 107 cells/ml. Electroporation was carried out in 0.2-cm-gap cuvettes containing 0.4 ml of cell suspension mixed with 5 μg of DNA. The cell-DNA mixture was pulsed twice at 200 V and 500 μF. The cells were then transferred into cell culture vessels at an appropriate density and incubated in complete growth medium. Alternatively, cells were transfected with FuGENE 6 transfection reagent (Roche) according to the manufacturer's protocol using a 6-to-1 (vol/wt) FuGENE 6-to-DNA ratio. For the reporter gene assays, PK-15 cells were seeded in a 24-well plate at a densitiy of 1.5 × 105 cells/well and infected with the respective CSFV at an MOI of 2 TCID50/cell or mock treated. After 24 h of incubation at 37°C, the cells were transfected with FuGENE 6 with a mixture of 1 μg of reporter plasmid pIRF3(−779)Luc and 20 ng of plasmid phRL-SV40 for internal normalization. Extraction and quantification of firefly and Renilla luciferases were performed using the Dual Luciferase Reporter assay system (Promega) and a Lumat LB 9507 luminometer with two automated injectors (Berthold Technologies).
Immunoprecipitation.
Protein coimmunoprecipitation was performed essentially as described by Rottenberg and coworkers (60). Cells were lysed for 20 min on ice with a hypotonic buffer (20 mM Tris-HCl, pH 7.5, 75 mM NaCl, 2.5 mM MgCl2, 0.1% Nonidet P40) containing 5 μl/ml protease inhibitor cocktail (Sigma) and 10 μl/ml phosphatase inhibitor (Sigma). The lysate was cleared by centrifugation and incubated for 2 h at 4°C with 20 μl anti-FLAG M2-agarose (Sigma) in Tris-buffered saline (20 mM Tris-HCl, 75 mM NaCl, pH 7.6). The agarose was then washed three times with Tris-buffered saline, and the protein complexes were eluted from the beads with an equal volume of 2× sample buffer (125 mM Tris-HCl, pH 6.8, 20% glycerol, 1% bromophenol blue, 4% sodium dodecyl sulfate [SDS]) and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting.
Isolation of total cellular RNA.
Total RNA was extracted from cells using the NucleoSpin RNA II extraction kit (Macherey-Nagel). Typically, monolayers of 106 cells were lysed according to the manufacturer's protocol, including the DNase digestion step. The purified RNA was eluted with 60 μl RNase-free water. Prior to RT-PCR, an additional DNase digestion was performed. For this purpose, 1 μg of RNA was treated with 1 unit of DNase I (Fermentas) for 30 min at 37°C prior to heat inactivation of the DNase I at 65°C for 10 min in the presence of 2.5 mM EDTA.
Quantitative real-time RT-PCR for porcine IRF3 mRNA.
Oligonucleotides for the quantification of porcine IRF3 mRNA by TaqMan real-time RT-PCR were designed based on the porcine IRF3 coding sequence described above using Primer Express version 1.5 software (Applied Biosystems). The sense primer porcIRF3-F (5′-GCCCACCTGGAAGAGGAATT-3′) and the antisense primer porcIRF3-R (5′-CCTCTGCTAAACGCAATGCTT-3′) were used in combination with the 5′ 6-carboxyfluorescein/3′ 6-carboxytetramethylrhodamine-labeled probe porcIRF3-P (5′-CCGGTCTGCCCTGAACCGGAA-3′). The two-step RT-PCR was performed as previously described (61). The individual samples were normalized to each other using the respective cycle threshold (CT) value obtained for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA (61). The relative IRF3 mRNA content was calculated by subtracting the mean CT value obtained for the RT-PCR from the mean CT value of the “no-RT” control PCR (residual DNA).
Confocal microscopy.
Cells were seeded and infected in eight-well LabTek II culture slides (Becton Dickinson). Prior to being immunostained, the cultures were washed with PBS and fixed with 4% (wt/vol) paraformaldehyde for 10 min. The cells were washed again and incubated for 1 h at room temperature with the respective antibody diluted in PBS-0.3% (wt/vol) saponin (Sigma). Between treatments, the cells were washed with PBS containing 0.1% (wt/vol) saponin. The IRF3 signal was amplified with a biotin-conjugated anti-rabbit antibody and the TSA amplification kit (Molecular Probes) according to the manufacturer's protocol. Detection was performed with Alexa fluorochrome-labeled secondary antibodies or streptavidin (Molecular Probes). The image was acquired with a Leica TCS-SL spectral confocal microscope and Leica LCS software.
Western blotting.
Cells were lysed with a hypotonic buffer (20 mM morpholinepropanesulfonic acid, 10 mM NaCl, 1.5 mM MgCl2, 1% Triton X-100, pH 6.5). Proteins were separated by SDS-PAGE under reducing conditions and analyzed by Western blotting using the Odyssey Infrared Imaging system (LI-COR) as previously described (61).
Nucleotide sequence accession number.
The consensus coding sequence of porcine IRF7 was deposited in GenBank under accession number EF195267.
RESULTS
CSFV Npro induces loss of IRF3 expressed from a CMV promoter.
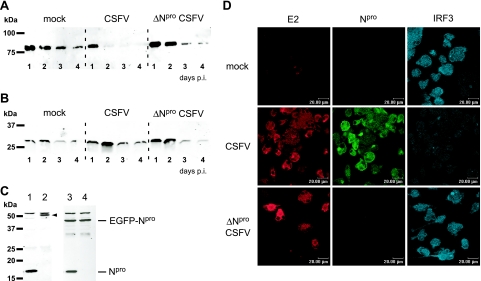
It was recently reported that IRF3 is lost from CSFV-infected cells and that this loss is related to an inhibition of transcription of the IRF3 gene by the viral Npro protein (46). Here, we show that CSFV induces the depletion of EGFP-IRF3 expressed from a CMV promoter (Fig. 1A), although expression from this promoter is not affected by CSFV, as demonstrated with the expression of EGFP (Fig. 1B). To this end, we transfected PK-15 cells with plasmids for CMV promoter-driven expression of the EGFP-IRF3 fusion protein or of EGFP alone and subsequently mock infected the cells or infected them with either CSFV or ΔNpro CSFV. Whereas the EGFP expression levels were not influenced by the infection (Fig. 1B), the amount of EGFP-IRF3 was clearly reduced in cells infected with CSFV compared to those infected with ΔNpro CSFV (Fig. 1A). To verify whether the loss of IRF3 is dependent on the presence of Npro alone, we analyzed IRF3 expression in a stable clonal cell line constitutively expressing Npro fused to EGFP. In the control PK15-EGFP cell line expressing EGFP alone, IRF3 was present as expected (Fig. 1C, lane 2) and was lost after infection with CSFV (Fig. 1C, lane 1). Note that the rabbit anti-IRF3 serum detected two nonspecific bands above and below IRF3. The IRF3 protein, however, could not be detected in the PK15-EGFP-Npro cell line expressing the 46-kDa EGFP-Npro fusion protein, whether the cells were infected or not (Fig. 1C, lanes 3 and 4). This demonstrates that Npro alone can mediate the loss of IRF3 and suggests that this process does not require an activation signal for IRF3. In order to exclude the possibility that the IRF3 downregulation by CSFV was a cell line-specific observation, we also analyzed the effect of CSFV on endogenous IRF3 expression in porcine DC. IRF3 expression was strongly reduced in the presence of CSFV (Fig. 1D). In contrast, infection of DC with ΔNpro CSFV did not affect IRF3 expression but rather induced nuclear translocation of IRF3 in a small percentage of infected cells (Fig. 1D). These data show that CSFV induces loss of IRF3 by means of Npro in a porcine cell line and in porcine DC and that the downregulation of IRF3 by Npro is independent of the promoter from which IRF3 is expressed.
FIG. 1.
CSFV infection induces loss of CMV-driven IRF3, a mechanism dependent on the presence of Npro. PK-15 cells were transfected with pEGFP-IRF3 (A) or with pEGFP-N1 (B). Twenty-four hours after transfection, the cells were mock treated or infected with CSFV or ΔNpro CSFV at an MOI of 0.2 TCID50/cell. On the indicated days p.i., the cells were lysed and extracts were analyzed for EGFP-IRF3 (A) and EGFP (B) expression by Western blotting with the anti-EGFP MAb JL-8. (C) IRF3 expression was analyzed in the clonal PK15-EGFP (lanes 1 and 2) and PK15-EGFP-Npro (lanes 3 and 4) cell lines. Cells were infected with CSFV at an MOI of 2 TCID50/cell (lanes 1 and 3) or mock treated (lanes 2 and 4). Twenty-four hours p.i., cell extracts were analyzed for IRF3, EGFP-Npro, and Npro expression by Western blotting using the rabbit anti-IRF3 and anti-Npro sera, respectively. The IRF3 protein in the PK15-EGFP extract (lane 2) is indicated with an arrowhead. (D) Endogenous IRF3 was analyzed in DC after mock, CSFV, and ΔNpro CSFV infection. After 4 days of differentiation, DC were transferred into chamber slides and mock infected or infected with CSFV or ΔNpro CSFV at an MOI of 20 TCID50/cell (based on the virus titer on SK-6 cells). Confocal microscopy was performed 48 h p.i. For this purpose, the cells were fixed and stained for the viral proteins E2 and Npro with MAb HC/TC26 and with the rabbit anti-Npro serum, respectively, and for IRF3 with the rabbit anti-IRF3 serum as indicated.
Npro does not inhibit IRF3 promoter activity.
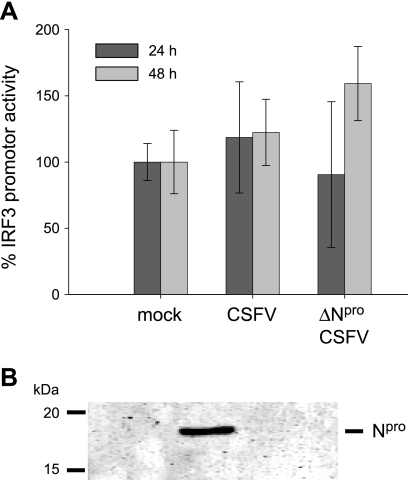
In view of the above-mentioned results, it was considered important to analyze whether Npro inhibits IRF3 promoter activity. For this purpose, we performed an IRF3 promoter-dependent luciferase reporter assay in CSFV-, ΔNpro CSFV-, and mock-infected PK-15 cells. To this end, we used the reporter plasmid pIRF3(−779)Luc for the expression of firefly luciferase under the control of the human IRF3 promoter (48). Each individual transfection was normalized using a plasmid constitutively expressing Renilla luciferase under the control of a simian virus 40 (SV40) promoter. The normalized IRF3 promoter activity was lowest in mock-infected cells (Fig. 2A). In CSFV- and ΔNpro CSFV-infected cells, the activity was slightly increased. The differences in luciferase expression between mock, CSFV-, and ΔNpro CSFV-infected cells were not significant (P > 0.113). However, when we analyzed the raw data for the IRF3- and the SV40 promoter-driven luciferase expression separately, considerable variations were observed that were dependent on the presence of virus and on the time postinfection (p.i.) but independent of the presence of Npro. Both CSFV and ΔNpro CSFV reduced the expression of firefly and Renilla luciferases at 24 h p.i. and induced their expression after 48 h (data not shown). Whether this was due to reduced transfection efficiency or to a downstream effect of the virus was not further investigated. Parallel Western blot analysis of protein extracts confirmed the expression of Npro in CSFV-infected cells (Fig. 2B). SV40 promoter-normalized IRF3 promoter activity was also assayed in the presence of Npro expression in HEK 293T cells. No significant reduction of the IRF3 promoter activity was observed, confirming the data obtained with CSFV in PK-15 cells (data not shown). These experiments demonstrated that Npro does not specifically influence IRF3 promoter activity.
FIG. 2.
Npro does not inhibit transcription of IRF3. (A) PK-15 cells were transfected with a plasmid encoding firefly luciferase under the control of the human IRF3 promoter and a second plasmid expressing Renilla luciferase under the SV40 promoter. Twenty-four hours after transfection, the cells were mock treated or infected with CSFV or ΔNpro CSFV. Cells were lysed 24 and 48 h p.i. and analyzed for the relative firefly and Renilla luciferase activities using the dual-luciferase assay system. The normalized relative induction is represented as a percentage of the promoter activity of mock-infected cells (mock = 100%). The error bars represent the standard deviations. (B) In parallel, protein extracts were analyzed for the presence of Npro by Western blotting using the rabbit anti-Npro serum.
Npro does not destabilize IRF3 mRNA.
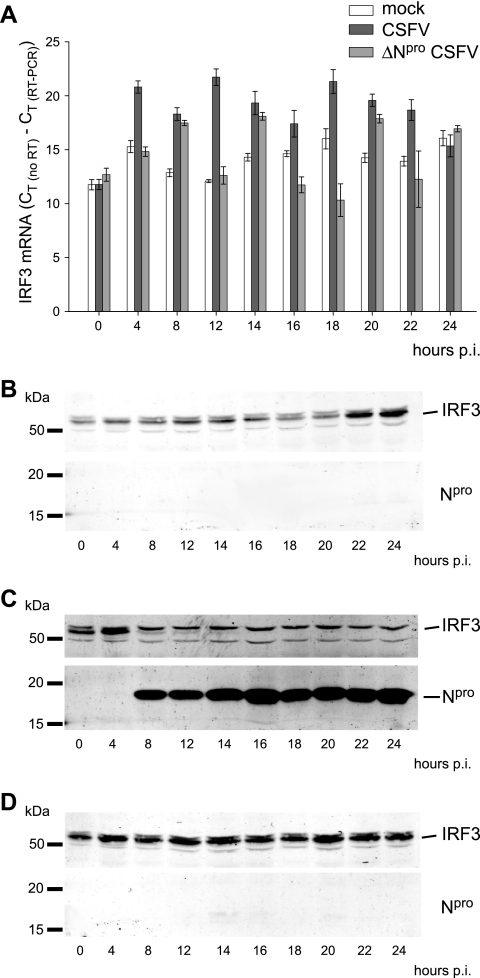
Since Npro did not affect the transcriptional activity of the IRF3 promoter, we asked whether Npro would destabilize IRF3 mRNA. To investigate this, we measured IRF3 mRNA by real-time RT-PCR in RNA extracts from either mock-, CSFV-, or ΔNpro CSFV-infected PK-15 cells. At any time between 4 and 24 h p.i., the IRF3 mRNA content in CSFV-infected cells was higher than or equal to the IRF3 mRNA levels detected in cells infected with ΔNpro CSFV and mock-infected cells (Fig. 3A). Interestingly, in CSFV-infected cells, the endogenous IRF3 protein disappeared 8 h p.i. when Npro became detectable, despite the fact that the levels of IRF3 mRNA increased (Fig. 3A and C). In mock- and ΔNpro CSFV-infected cells, the IRF3 protein was not affected throughout the experiment (Fig. 3B and D). Taken together, the IRF3 promoter reporter assay (Fig. 2A) and the quantitative IRF3 mRNA analysis (Fig. 3) demonstrated that Npro regulates the IRF3 protein turnover rather than mRNA transcription and/or stability.
FIG. 3.
Npro does not destabilize IRF3 mRNA. (A) The IRF3 mRNA contents of PK-15 cells at different times after mock treatment or infection with either CSFV or ΔNpro CSFV were measured by real-time RT-PCR. The amount of IRF3 mRNA is shown as CT (no RT) − CT (RT-PCR), with error bars representing the 95% confidence interval. In parallel with the RNA extraction, cells were lysed at the indicated times, and the IRF3 and Npro proteins were analyzed by Western blotting in mock- (B), CSFV- (C), and ΔNpro CSFV-infected (D) cells using the rabbit anti-IRF3 and anti-Npro sera.
Npro does not interfere with the phosphorylation of IRF3 or with the nuclear translocation of a constitutively active form of IRF3.
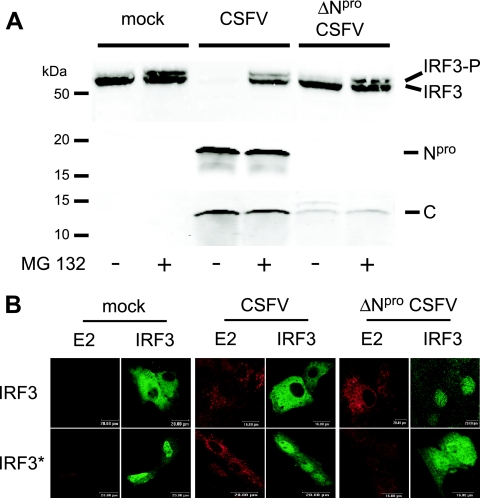
Phosphorylation of IRF3 at distinct amino acid positions represents an activation signal for its dimerization and nuclear translocation, as well as for its proteasomal degradation (47, 68). Consequently, it was of interest to investigate the influence of Npro on the general IRF3 phosphorylation and on translocation to the nucleus. For this purpose, we used a monoclonal antibody capable of detecting a doublet of the unphosphorylated and phosphorylated forms of IRF3 independently of the sites of phosphorylation. As expected from the results shown in Fig. 1 and 3, endogenous IRF3 was barely detectable in CSFV-infected PK-15 cells (Fig. 4A, third lane from left), whereas a single band representing the unphosphorylated form of IRF3 was present in the lysate of mock-infected cells (Fig. 4A, first lane on left). In the presence of the proteasome inhibitor MG132, however, the IRF3 doublet was detected in both the mock- and the CSFV-infected cells (Fig. 4A, second and fourth lanes), indicating that CSFV cannot prevent IRF3 phosphorylation and that it targets IRF3 for proteasomal degradation. In PK-15 cells infected with ΔNpro CSFV, the phosphorylated form of IRF3 was detected as a faint band in the untreated cells and as a stronger band in the MG132-treated cells. This is consistent with the fact that ΔNpro CSFV induces IFN-α/β, as opposed to CSFV. To analyze whether CSFV interferes with the nuclear translocation of a constitutively active IRF3, we constructed the plasmid pFLAG-IRF3-S394,396D-ΔNES for the expression of a phosphomimetic form of IRF3 carrying in addition a deletion of the NES to prevent IRF3 from shuttling back to the cytoplasm. The localization of IRF3 was then analyzed by confocal microscopy in mock-infected PK-15 cells and cells infected with CSFV or ΔNpro CSFV and transfected with the plasmids expressing the respective FLAG-tagged authentic or mutant form of IRF3. With FLAG-IRF3, no nuclear translocation was detectable in mock- and CSFV-infected cells, whereas ΔNpro CSFV induced translocation as expected, showing that ΔNpro CSFV activates IRF3 in contrast to the parent CSFV (Fig. 4B, top row). The constitutively active IRF3 accumulated in the nucleus with comparable efficiencies in mock-, CSFV-, and ΔNpro CSFV-infected cells (Fig. 4B, bottom row). These results indicate that Npro does not inhibit the general phosphorylation of IRF3 and suggest that, at least for a constitutively active form of IRF3, Npro does not interfere with nuclear translocation.
FIG. 4.
Npro does not prevent IRF3 phosphorylation or nuclear translocation of a constitutively active form of IRF3. (A) PK-15 cells were treated with 0.2 μM of the proteasome inhibitor MG132 (+) or left untreated (−) and infected with CSFV or ΔNpro CSFV at an MOI of 0.2 TCID50/cell or mock treated as indicated. Sixteen hours p.i., the cells were lysed and analyzed by Western blotting for endogenous IRF3 using MAb 34/1 and for the viral proteins Npro and capsid C using the respective rabbit antisera as described in Materials and Methods. (B) PK-15 cells were mock treated or infected with CSFV or ΔNpro CSFV at an MOI of 2 TCID50/cell and transfected with pFLAG-IRF3 (top row) or pFLAG-IRF3-S394,396D-ΔNES (IRF3*; bottom row). Sixteen hours after transfection, the cells were fixed and stained for the viral E2 protein and for the FLAG tag of IRF3 as indicated and analyzed by confocal microscopy.
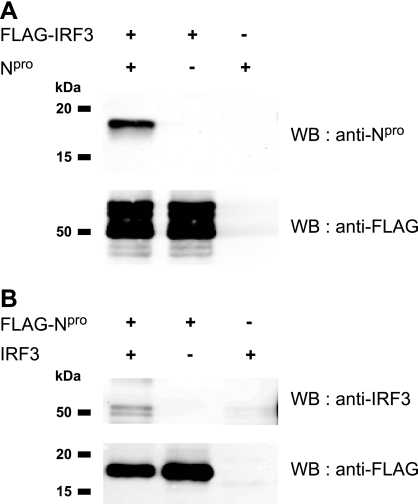
Npro interacts with IRF3.
The observation that the CSFV-mediated IRF3 protein degradation is linked to the presence of Npro (Fig. 1, 3, and 4) pointed to a putative interaction of Npro with IRF3. To address this question, HEK 293T cells were cotransfected with plasmids expressing FLAG-tagged IRF3 and untagged Npro (Fig. 5A) or FLAG-tagged Npro and untagged IRF3 (Fig. 5B) and analyzed for protein interaction by coimmunoprecipitation with anti-FLAG agarose. Npro was detected in Western blots after immunoprecipitation of FLAG-IRF3 from lysates containing both FLAG-IRF3 and Npro, but not from cells expressing Npro alone (Fig. 5A, first and third lanes). Conversely, IRF3 was detected in Western blots after immunoprecipitation of FLAG-Npro from cells expressing FLAG-Npro and IRF3, but not from cells expressing IRF3 alone (Fig. 5B, first and third lanes). However, Npro pulled down only a small fraction of IRF3, and untagged IRF3 was nonspecifically precipitated in trace amounts with the FLAG agarose (Fig. 5B, third lane from left). Although Npro did not pull down IRF3 with the same efficiency and specificity as IRF3 precipitated Npro, these data show that Npro interacts with IRF3 either directly or indirectly.
FIG. 5.
IRF3 and Npro interact with each other. HEK 293T cells were transfected with plasmids pFLAG-IRF3 and pEAK-Npro for the expression of FLAG-tagged IRF3 and untagged Npro (A) or with pFLAG-Npro and pIRF3 for the expression of FLAG-tagged Npro and untagged IRF3 (B), either individually or mixed at a 1:1 ratio. The presence and absence of the expression plasmid are indicated by + and −, respectively. Twenty-four hours after transfection, the cells were lysed and the proteins were immunoprecipitated with anti-FLAG M2 agarose. The precipitated proteins were eluted from the agarose, separated by SDS-PAGE, and analyzed by Western blotting (WB) with rabbit anti-Npro serum, anti-IRF3 MAb 34/1, and anti-FLAG MAb as indicated.
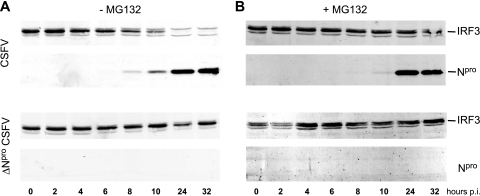
Npro induces degradation of IRF3 via the proteasome pathway.
We have shown above that Npro had no effect on IRF3 mRNA synthesis and turnover (Fig. 2 and 3). In addition, we observed that the proteasome inhibitor MG132 had a preventive effect on the loss of IRF3 in CSFV-infected cells (Fig. 4A). On the other hand, Npro is a protease and therefore might directly cleave IRF3 by trans activity and thus contribute to its degradation. Therefore, we analyzed how the proteasome inhibitor MG132 influenced the relationship between Npro expression and cellular IRF3 content. For this purpose, we determined the temporal IRF3 content in CFSV-infected PK-15 cells in the absence or presence of the proteasome inhibitor MG132. As seen previously (Fig. 3C), the appearance of the viral Npro protein approximately 8 h p.i. with CSFV was concomitant with the decrease of the IRF3 content in the cell (Fig. 6A, top). Inhibition of the proteasome activity with MG132 completely prevented the CSFV-dependent degradation of IRF3 (Fig. 6B, top). In cells infected with ΔNpro CSFV (Fig. 6A and B, bottom), the treatment with proteasome inhibitor had no influence on the cellular IRF3 content (Fig. 6B, bottom). These data indicate that Npro does not directly cleave IRF3 but indeed induces proteasome-dependent degradation of IRF3. Taken together, our data demonstrate that Npro does not interfere with the synthesis or with the phosphorylation of IRF3 but actively induces proteasome-mediated depletion of IRF3 by interacting with IRF3 either directly or via adaptor proteins.
FIG. 6.
Npro induces proteasomal degradation of IRF3. PK-15 cells were infected with CSFV (top) or with ΔNpro CSFV (bottom) at an MOI of 0.2 TCID50/cell in the absence (A) or in the presence (B) of 0.2 μM proteasome inhibitor MG132. At the indicated times p.i., the cells were lysed and the Npro and IRF3 contents were analyzed by Western blotting using the rabbit anti-Npro and anti-IRF3 sera, respectively.
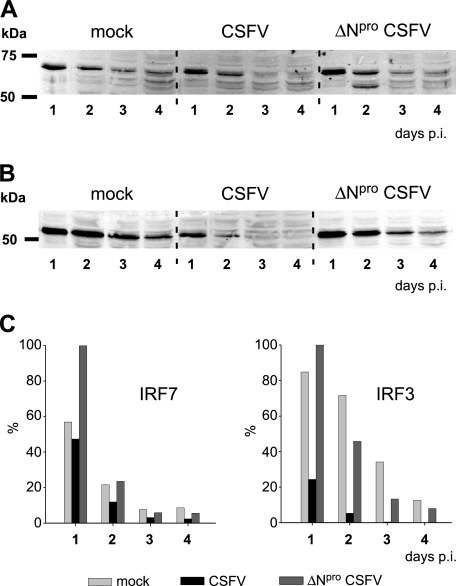
CSFV does not downregulate IRF7 expression.
In certain cell types, IFN-α/β induction is essentially IRF7 dependent (32). Therefore, we analyzed the effect of CSFV on IRF7 expression. The CMV promoter-driven IRF7 expression levels over time were similar in mock-, CSFV-, and ΔNpro CSFV-infected cells (Fig. 7A and C), while IRF3 levels decreased faster in CSFV-infected cells than in mock- or ΔNpro CSFV-infected cells (Fig. 7B and C). These data indicate that Npro does not downregulate IRF7 expression, as opposed to IRF3 expression.
FIG. 7.
CSFV Npro does not downregulate IRF7 expression. PK-15 cells were transfected with plasmid pFLAG-IRF7 (A) or pFLAG-IRF3 (B) for CMV promoter-driven expression of FLAG-tagged IRF7 or FLAG-tagged IRF3, respectively. Twenty-four hours after transfection, the cells were mock treated or infected with CSFV or ΔNpro CSFV at an MOI of 2 TCID50/cell. On the indicated days p.i., cells were lysed and extracts were analyzed for IRF7 (A) and IRF3 (B) expression by Western blotting using the rabbit anti-FLAG MAb. (C) The IRF7- and IRF3-specific signals from the Western blots shown in panels A and B were quantified with the Odyssey Imaging system and expressed as percentages of the strongest signal.
DISCUSSION
Pestiviruses subvert the innate immune system by suppressing IFN-α/β induction (4, 9, 63, 66). This property is mediated by the viral protein Npro independently of other viral elements (23, 30, 34, 46, 61). A recent study revealed that the inhibition of IFN-α/β induction by CSFV is due to an Npro-dependent loss of IRF3 (46). These authors proposed that the loss of IRF3 was due to an inhibition of IRF3 transcription. In the present study, we demonstrated that CSFV does not affect the transcriptional activity of the IRF3 promoter but rather induces IRF3 degradation by a proteasome-dependent mechanism. The degradation of IRF3 is mediated by Npro alone, in accordance with our previous data on the Npro-mediated prevention of IFN-α/β induction (61) and with the very recent results obtained for BVDV by Hilton and coworkers (30). The reason for the discrepancy with the data of La Rocca et al. showing inhibition of IRF3 transcription (46) could result from the different experimental setup used for the analysis of the IRF3 promoter activity. In the latter report, the effects of CSFV and mock infection on IRF3 promoter-driven luciferase activity were analyzed using a single reporter plasmid (46). Using the same cell line and the same reporter plasmid for IRF3 promoter activity (46, 48), we performed a dual-luciferase reporter assay, including an internal control plasmid driving the expression of Renilla luciferase under the control of an SV40 promoter (phRL-SV40). We have previously used this plasmid in reporter assays to eliminate variations between individual transfections and nonspecific effects due to foreign protein expression and viral infection (61). In addition, the comparison of CSFV with ΔNpro CSFV infections permitted us to account for virus-mediated Npro-independent effects on the IRF3 promoter. Although the nonnormalized data of the two reporter plasmids indicated a slight reduction of transcription in the presence of CSFV (data not shown), the comparison with ΔNpro CSFV and the normalization with SV40-mediated transcription demonstrated that Npro had no specific effect on the IRF3 promoter activity. We further confirmed our reporter assay data by showing that CSFV-infected cells had a higher IRF3 mRNA content than uninfected cells. These data clearly show that neither the transcriptional activity of the IRF3 promoter nor IRF3 mRNA stability is influenced by Npro. For BVDV also, IRF3 mRNA levels remained stable during infection (30). We did not investigate the influence of Npro on IRF3 translation, considering that any inhibition of translation is likely to result in a general shutdown of host protein synthesis, which is not observed with CSFV infections.
Viruses have evolved many different strategies to counteract the IFN system. Virtually all components of the IFN system are targeted by the multitude of IFN-antagonistic proteins that have been described (for recent reviews, see references 18, 27, 29, and 79). One of these components is IRF3, which can be targeted by viruses at the levels of activation and phosphorylation (7, 10, 12, 43, 58, 70), dimerization (37, 56), nuclear translocation (19, 28), and downstream IRF3 functions (4, 71). There are only a few reports of viruses that target IRF3 for proteasomal degradation. Besides the very recent data with BVDV (30), targeting of IRF3 to the proteasome was reported for Sendai virus (47) and for rotavirus (6), for which degradation of IRF3 is mediated by nonstructural protein 1 (NSP1). NSP1 possesses an IRF3 binding domain. Sequence comparison of Npro and rotavirus NSP1 did not reveal any similarity. The induction of proteasomal degradation of components of the IFN system by viruses has been extensively described for the signal transducer and activator of transcription (STAT) proteins. Simian virus 5 targets STAT1, human parainfluenza virus 2 targets STAT2, and mumps virus targets both STAT1 and STAT3 to the proteasome pathway (reviewed in reference 35). For all these viruses, the V protein mediates the assembly of a ubiquitin ligase complex involving the target and nontarget STATs, the 127-kDa subunit of the UV-damaged DNA-binding protein (DDB1), Cullin 4a, and the regulator of cullins 1 (Roc1) (2, 20, 59, 76, 77, 78). Whether Npro associates with IRF3 to a ubiquitin ligase complex is under investigation. Alternatively, Npro might mimic another component of the ubiquitin-conjugating machinery. However, using an anti-ubiquitin antibody, we were not able to pull down ubiquitinated Npro, whereas ubiquitin-conjugated IRF3 could be precipitated as a control (data not shown). This indicates that Npro does not carry ubiquitin and is therefore unlikely to serve functions similar to those of the E1 and E2 protein family (for selected reviews, see references 24 and 54). Attempts to pull down ubiquitinated IRF3 in the presence of Npro failed, probably due to the rapid elimination of IRF3 by Npro. In a very recent report, Saitoh and coworkers demonstrated that IRF3 is negatively regulated by interaction with peptidylprolyl isomerase Pin1 (64). Here also, amino acid comparison did not reveal any similarity with Npro. It is unknown whether this mechanism is of any significance for Npro-dependent IRF3 degradation. Nevertheless, the involvement of the proteasome pathway in Npro-mediated IRF3 degradation is consistent with the observation that the protease activity of Npro is not required for the inhibition of IFN-α/β induction (23).
The finding that IRF3 can be phosphorylated in the presence of Npro suggests that the upstream kinase complex inducing IRF3 phosphorylation is not affected by Npro. At the same time, CSFV does not inhibit the nuclear translocation of constitutively active IRF3, an observation that is consistent with previous reports showing that both BVDV and CSFV do not interfere with IRF3 translocation to the nucleus (3, 4, 30, 46). Interestingly, it was shown for BVDV that the nuclear IRF3 is resistant to degradation and that Npro prevents IRF3 from binding to DNA (4, 30). Due to the rapid elimination of IRF3 after CSFV infection, we did not investigate whether CSFV Npro prevents the binding of IRF3 to DNA. Future work will focus on the detailed characterization of the mechanisms employed by Npro to induce IRF3 degradation by the proteasome system.
The inhibition of IFN-α/β induction by CSFV in epitheloidal cell lines, endothelial cells, macrophages, and conventional DC is in apparent contradiction to the in vivo observation showing strong IFN-α activity in sera of animals suffering from CSF (73). A similar apparent discrepancy between inhibition and induction of IFN-α/β has been described for BVDV (15, 16). In fact, whereas pestiviruses inhibit IFN-α/β induction in most cells, there are specialized cell populations that respond to a pestivirus infection with high IFN-α production (5, 11). For CSFV, Balmelli and coworkers identified these cells as natural interferon producing cells (NIPC), also known as plasmacytoid DC (5). In these cells, we were not able to identify an IFN-α/β-antagonistic effect of Npro (unpublished data), which is in agreement with our results indicating that IRF7 is not downregulated by CSFV, considering the fact that in mouse NIPC, viruses use an IRF3-independent, IRF7-dependent IFN-α induction pathway (32). In light of CSFV pathogenesis, the differential modulation of IFN-α/β induction by CSFV let us postulate the following model. At the early stages of viral entry into the host, CSFV counteracts IFN-α/β induction by depleting IRF3 in the primary target cells through proteasomal degradation. This allows the virus to establish a productive infection at initial sites of replication and to spread within the host. Once the virus enters the circulation and colonizes lymphoid tissue, it will infect NIPC, resulting in IRF7-dependent overproduction of IFN-α and other cytokines, mediating the immunopathological effects typical of CSF (73).
Acknowledgments
This work was funded by the Swiss Federal Veterinary Office (grant 1.03.04).
We are grateful to Markus Gerber, Luzia Liu, and Heidi Gerber for excellent technical assistance, and we thank Christian Griot for continuous support. The SK-6 cell line was kindly provided by M. Pensaert. We also thank Shigeki Inumaru for the porcine granulocyte/monocyte colony-stimulating factor, Irene Greiser-Wilke for MAb HC/TC26, and Paula Pitha-Rowe for plasmid pIRF3(−779)Luc.
Footnotes
Published ahead of print on 10 January 2007.
REFERENCES
- 1.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 101:17264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baigent, S. J., S. Goodbourn, and J. W. McCauley. 2004. Differential activation of interferon regulatory factors-3 and -7 by non-cytopathogenic and cytopathogenic bovine viral diarrhoea virus. Vet. Immunol. Immunopathol. 100:135-144. [DOI] [PubMed] [Google Scholar]
- 4.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balmelli, C., I. E. Vincent, H. Rau, L. Guzylack-Piriou, K. McCullough, and A. Summerfield. 2005. FcγRII-dependent sensitisation of natural interferon-producing cells for viral infection and interferon-alpha responses. Eur. J. Immunol. 35:2406-2415. [DOI] [PubMed] [Google Scholar]
- 6.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. USA 102:4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basler, C. F., A. Mikulasova, L. Martinez Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauhofer, O., A. Summerfield, K. C. McCullough, and N. Ruggli. 2005. Role of double-stranded RNA and Npro of classical swine fever virus in the activation of monocyte-derived dendritic cells. Virology 343:93-105. [DOI] [PubMed] [Google Scholar]
- 9.Bensaude, E., J. L. E. Turner, P. R. Wakeley, D. A. Sweetman, C. Pardieu, T. W. Drew, T. Wileman, and P. P. Powel. 2004. Classical swine fever virus induces proinflammatory cytokines and tissue factor expression and inhibits apoptosis and interferon synthesis during the establishment of long-term infection of porcine vascular endothelial cells. J. Gen. Virol. 85:1029-1037. [DOI] [PubMed] [Google Scholar]
- 10.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brackenbury, L. S., B. V. Carr, Z. Stamataki, H. Prentice, E. A. Lefevre, C. J. Howard, and B. Charleston. 2005. Identification of a cell population that produces alpha/beta interferon in vitro and in vivo in response to noncytopathic bovine viral diarrhea virus. J. Virol. 79:7738-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brzozka, K., S. Finke, and K. K. Conzelmann. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carrasco, C. P., R. C. Rigden, R. Schaffner, H. Gerber, V. Neuhaus, S. Inumaru, H. Takamatsu, G. Bertoni, K. C. McCullough, and A. Summerfield. 2001. Porcine dendritic cells generated in vitro: morphological, phenotypic and functional properties. Immunology 104:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrasco, C. P., R. C. Rigden, I. E. Vincent, C. Balmelli, M. Ceppi, O. Bauhofer, V. Tache, B. Hjertner, F. McNeilly, H. G. van Gennip, K. C. McCullough, and A. Summerfield. 2004. Interaction of classical swine fever virus with dendritic cells. J. Gen. Virol. 85:1633-1641. [DOI] [PubMed] [Google Scholar]
- 15.Charleston, B., L. S. Brackenbury, B. V. Carr, M. D. Fray, J. C. Hope, C. J. Howard, and W. I. Morrison. 2002. Alpha/beta and gamma interferons are induced by infection with noncytopathic bovine viral diarrhea virus in vivo. J. Virol. 76:923-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charleston, B., M. D. Fray, S. Baigent, B. V. Carr, and W. I. Morrison. 2001. Establishment of persistent infection with non-cytopathic bovine viral diarrhoea virus in cattle is associated with a failure to induce type 1 interferon. J. Gen. Virol. 82:1893-1897. [DOI] [PubMed] [Google Scholar]
- 17.Childs, K., N. Stock, C. Ross, J. Andrejeva, L. Hilton, M. Skinner, R. Randall, and S. Goodbourn. 16 October 2006. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology. doi: 10.1016/j.virol.2006.09.023. [DOI] [PubMed]
- 18.Conzelmann, K. K. 2005. Transcriptional activation of alpha/beta interferon genes: interference by nonsegmented negative-strand RNA viruses. J. Virol. 79:5241-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delhaye, S., V. van Pesch, and T. Michiels. 2004. The leader protein of Theiler's virus interferes with nucleocytoplasmic trafficking of cellular proteins. J. Virol. 78:4357-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diderholm, H., and Z. Dinter. 1966. Interference between strains of bovine virus diarrhea virus and their capacity to suppress interferon of a heterologous virus. Proc. Soc. Exp. Biol. Med. 121:976-980. [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 23.Gil, L. H., I. H. Ansari, V. Vassilev, D. Liang, V. C. Lai, W. Zhong, Z. Hong, E. J. Dubovi, and R. O. Donis. 2006. The amino-terminal domain of bovine viral diarrhea virus Npro protein is necessary for alpha/beta interferon antagonism. J. Virol. 80:900-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glickman, M. H., and A. Ciechanover. 2002. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol. Rev. 82:373-428. [DOI] [PubMed] [Google Scholar]
- 25.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 26.Greiser-Wilke, I., K. E. Dittmar, B. Liess, and V. Moennig. 1992. Heterogeneous expression of the non-structural protein p80/p125 in cells infected with different pestiviruses. J. Gen. Virol. 73:47-52. [DOI] [PubMed] [Google Scholar]
- 27.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He, B., R. G. Paterson, N. Stock, J. E. Durbin, R. K. Durbin, S. Goodbourn, R. E. Randall, and R. A. Lamb. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15-32. [DOI] [PubMed] [Google Scholar]
- 29.Hengel, H., U. H. Koszinowski, and K. K. Conzelmann. 2005. Viruses know it all: new insights into IFN networks. Trends Immunol. 26:396-401. [DOI] [PubMed] [Google Scholar]
- 30.Hilton, L., K. Moganeradj, G. Zhang, Y. H. Chen, R. E. Randall, J. W. McCauley, and S. Goodbourn. 2006. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J. Virol. 80:11723-11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiscott, J., P. Pitha, P. Genin, H. Nguyen, C. Heylbroeck, Y. Mamane, M. Algarte, and R. Lin. 1999. Triggering the interferon response: the role of IRF-3 transcription factor. J. Interferon Cytokine Res. 19:1-13. [DOI] [PubMed] [Google Scholar]
- 32.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Sato, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 33.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 34.Horscroft, N., D. Bellows, I. Ansari, V. C. Lai, S. Dempsey, D. Liang, R. Donis, W. Zhong, and Z. Hong. 2005. Establishment of a subgenomic replicon for bovine viral diarrhea virus in Huh-7 cells and modulation of interferon-regulated factor 3-mediated antiviral response. J. Virol. 79:2788-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horvath, C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 271:4621-4628. [DOI] [PubMed] [Google Scholar]
- 36.Inaba, Y., T. Omori, and T. Kumagai. 1963. Detection and measurement of non-cytopathogenic strains of virus diarrhea of cattle by the END method. Arch. Gesamte Virusforsch. 13:425-429. [DOI] [PubMed] [Google Scholar]
- 37.Jennings, S., L. Martinez Sobrido, A. Garcia Sastre, F. Weber, and G. Kochs. 2005. Thogoto virus ML protein suppresses IRF3 function. Virology 331:63-72. [DOI] [PubMed] [Google Scholar]
- 38.Kang, D. C., R. V. Gopalkrishnan, Q. Wu, E. Jankowsky, A. M. Pyle, and P. B. Fisher. 2002. mda-5: an interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl. Acad. Sci. USA 99:637-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kasza, L., J. A. Shadduck, and G. J. Christofinis. 1972. Establishment, viral susceptibility and biological characteristics of a swine kidney cell line SK-6. Res. Vet. Sci. 13:46-51. [PubMed] [Google Scholar]
- 40.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 41.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131-137. [DOI] [PubMed] [Google Scholar]
- 42.Kawai, T., and S. Akira. 2006. TLR signaling. Cell Death Differ. 13:816-825. [DOI] [PubMed] [Google Scholar]
- 43.Komatsu, T., K. Takeuchi, J. Yokoo, and B. Gotoh. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325:137-148. [DOI] [PubMed] [Google Scholar]
- 44.Kumagai, T., T. Shimizu, S. Ikeda, and M. Matumoto. 1961. A new in vitro method (END) for detection and measurement of hog cholera virus and its antibody by means of effect of HC virus on Newcastle disease virus in swine tissue culture. I. Establishment of standard procedure. J. Immunol. 87:245-256. [PubMed] [Google Scholar]
- 45.Lai, V. C. H., W. D. Zhong, A. Skelton, P. Ingravallo, V. Vassilev, R. O. Donis, Z. Hong, and J. Y. N. Lau. 2000. Generation and characterization of a hepatitis C virus NS3 protease-dependent bovine viral diarrhea virus. J. Virol. 74:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.La Rocca, S. A., R. J. Herbert, H. Crooke, T. W. Drew, T. E. Wileman, and P. P. Powell. 2005. Loss of interferon regulatory factor 3 in cells infected with classical swine fever virus involves the N-terminal protease, Npro. J. Virol. 79:7239-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lowther, W. J., P. A. Moore, K. C. Carter, and P. M. Pitha. 1999. Cloning and functional analysis of the human IRF-3 promoter. DNA Cell Biol. 18:685-692. [DOI] [PubMed] [Google Scholar]
- 49.Malmgaard, L. 2004. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. 24:439-454. [DOI] [PubMed] [Google Scholar]
- 50.Matsumoto, M., K. Funami, H. Oshiumi, and T. Seya. 2004. Toll-like receptor 3: a link between toll-like receptor, interferon and viruses. Microbiol. Immunol. 48:147-154. [DOI] [PubMed] [Google Scholar]
- 51.Mayer, D., M. A. Hofmann, and J. D. Tratschin. 2004. Attenuation of classical swine fever virus by deletion of the viral Npro gene. Vaccine 22:317-328. [DOI] [PubMed] [Google Scholar]
- 52.Meylan, E., and J. Tschopp. 2006. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol. Cell 22:561-569. [DOI] [PubMed] [Google Scholar]
- 53.Moser, C., P. Stettler, J. D. Tratschin, and M. A. Hofmann. 1999. Cytopathogenic and noncytopathogenic RNA replicons of classical swine fever virus. J. Virol. 73:7787-7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nandi, D., P. Tahiliani, A. Kumar, and D. Chandu. 2006. The ubiquitin-proteasome system. J. Biosci. 31:137-155. [DOI] [PubMed] [Google Scholar]
- 55.O'Neill, L. A. 2006. How Toll-like receptors signal: what we know and what we don't know. Curr. Opin. Immunol. 18:3-9. [DOI] [PubMed] [Google Scholar]
- 56.Peng, T., S. Kotla, R. E. Bumgarner, and K. E. Gustin. 2006. Human rhinovirus attenuates the type I interferon response by disrupting activation of interferon regulatory factor 3. J. Virol. 80:5021-5031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 58.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 59.Precious, B., K. Childs, V. Fitzpatrick-Swallow, S. Goodbourn, and R. E. Randall. 2005. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J. Virol. 79:13434-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rottenberg, S., J. Schmuckli-Maurer, S. Grimm, V. T. Heussler, and D. A. Dobbelaere. 2002. Characterization of the bovine IκB kinases (IKK)α and IKKβ, the regulatory subunit NEMO and their substrate IκBα. Gene 299:293-300. [DOI] [PubMed] [Google Scholar]
- 61.Ruggli, N., B. H. Bird, L. Liu, O. Bauhofer, J. D. Tratschin, and M. A. Hofmann. 2005. Npro of classical swine fever virus is an antagonist of double-stranded RNA-mediated apoptosis and IFN-alpha/beta induction. Virology 340:265-276. [DOI] [PubMed] [Google Scholar]
- 62.Ruggli, N., J. D. Tratschin, C. Mittelholzer, and M. A. Hofmann. 1996. Nucleotide sequence of classical swine fever virus strain Alfort/187 and transcription of infectious RNA from stably cloned full-length cDNA. J. Virol. 70:3478-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruggli, N., J. D. Tratschin, M. Schweizer, K. C. McCullough, M. A. Hofmann, and A. Summerfield. 2003. Classical swine fever virus interferes with cellular antiviral defense: evidence for a novel function of Npro. J. Virol. 77:7645-7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saitoh, T., A. Tun-Kyi, A. Ryo, M. Yamamoto, G. Finn, T. Fujita, S. Akira, N. Yamamoto, K. P. Lu, and S. Yamaoka. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598-605. [DOI] [PubMed] [Google Scholar]
- 65.Schweizer, M., P. Matzener, G. Pfaffen, H. Stalder, and E. Peterhans. 2006. “Self” and “nonself” manipulation of interferon defense during persistent infection: bovine viral diarrhea virus resists alpha/beta interferon without blocking antiviral activity against unrelated viruses replicating in its host cells. J. Virol. 80:6926-6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schweizer, M., and E. Peterhans. 2001. Noncytopathic bovine viral diarrhea virus inhibits double-stranded RNA-induced apoptosis and interferon synthesis. J. Virol. 75:4692-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sen, G. C., and S. N. Sarkar. 2005. Transcriptional signaling by double-stranded RNA: role of TLR3. Cytokine Growth Factor Rev. 16:1-14. [DOI] [PubMed] [Google Scholar]
- 68.Servant, M. J., B. ten Oever, C. LePage, L. Conti, S. Gessani, I. Julkunen, R. Lin, and J. Hiscott. 2001. Identification of distinct signaling pathways leading to the phosphorylation of interferon regulatory factor 3. J. Biol. Chem. 276:355-363. [DOI] [PubMed] [Google Scholar]
- 69.Sharma, S., B. R. ten Oever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 70.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 79:5353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suhara, W., A. Yoneyama, I. Kitabayashi, and T. Fujita. 2002. Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J. Biol. Chem. 277:22304-22313. [DOI] [PubMed] [Google Scholar]
- 73.Summerfield, A., M. Alves, N. Ruggli, M. G. de Bruin, and K. C. McCullough. 2006. High IFN-alpha responses associated with depletion of lymphocytes and natural IFN-producing cells during classical swine fever. J. Interferon Cytokine Res. 26:248-255. [DOI] [PubMed] [Google Scholar]
- 74.Toba, M., and M. Matumoto. 1969. Role of interferon in enhanced replication of Newcastle disease virus in swine cells infected with hog cholera virus. Jpn. J. Microbiol. 13:303-305. [DOI] [PubMed] [Google Scholar]
- 75.Tratschin, J. D., C. Moser, N. Ruggli, and M. A. Hofmann. 1998. Classical swine fever virus leader proteinase Npro is not required for viral replication in cell culture. J. Virol. 72:7681-7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 77.Ulane, C. M., A. Kentsis, C. D. Cruz, J. P. Parisien, K. L. Schneider, and C. M. Horvath. 2005. Composition and assembly of STAT-targeting ubiquitin ligase complexes: paramyxovirus V protein carboxyl terminus is an oligomerization domain. J. Virol. 79:10180-10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ulane, C. M., J. J. Rodriguez, J. P. Parisien, and C. M. Horvath. 2003. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J. Virol. 77:6385-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weber, F., G. Kochs, and O. Haller. 2004. Inverse interference: how viruses fight the interferon system. Viral Immunol. 17:498-515. [DOI] [PubMed] [Google Scholar]
- 80.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 81.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]