Abstract
The AIDS epidemic continues to spread at an alarming rate worldwide, especially in developing countries. One approach to solving this problem is the generation of anti-human immunodeficiency virus (HIV) compounds with inhibition spectra broad enough to include globally prevailing forms of the virus. We have examined the HIV type 1 (HIV-1) envelope specificity of a recently identified entry inhibitor candidate, HNG-105, using surface plasmon resonance spectroscopy and pseudovirus inhibition assays. The combined results suggest that the HNG-105 molecule may be effective across the HIV-1 subtypes, and they highlight its potential as a lead for developing therapeutic and microbicidal agents to help combat the spread of AIDS.
Human immunodeficiency virus type 1 (HIV-1), the major pathogen responsible for the AIDS pandemic, is among the most genetically diverse viral pathogens described to date. Isolates can be divided into groups, subtypes, and circulating recombinant forms (CRFs). The group M viruses, which are the most widespread and account for approximately 99% of infections worldwide, can be further subdivided into nine distinct genetic subtypes, or clades (1, 15). These clades show characteristic geographic localization, with clade B viruses dominating Europe, the Americas, and Australia, while clade C, which presently infects more people worldwide than any other clade, is most prevalent in southern Africa, China, and India (5, 15, 20, 25). Despite the prevalence of subtype C, most of the antiretroviral drugs available to treat HIV-1 have been developed in the Western world using in vitro studies of subtype B isolates. However, there is a growing body of evidence that the different subtypes, and in particular the subtype C viruses, have unique antigenic, infectivity, and replicative characteristics (1, 15, 25). Therefore, in the development of prophylactics and topical microbicides, and eventually in the generation of a viable vaccine, the genetic diversity of HIV-1 and its potential consequences for naturally occurring and acquired drug resistance must be considered.
During the past several years, a new class of antiretroviral drugs, often referred to as entry inhibitors, has emerged (5, 7, 10, 12, 21). This class of antiretroviral agents disrupts one or more steps involved in the initial docking, coreceptor binding, or fusion events that are crucial to the HIV infection process by targeting components of the envelope proteins (2, 6, 22, 26, 29-31, 34). However, the therapeutic targeting of the envelope proteins, gp120 and gp41, is not without its potential pitfalls. First, the Env gene is the most variable HIV-1 gene, with up to 35% sequence diversity between clades, 20% sequence diversity within a clade, and up to 10% sequence diversity within a single infected person (3). Second, by comparison of the recently determined unliganded structure of simian immunodeficiency virus (SIV) gp120 (8) to the known liganded structures of HIV-1 gp120 (16, 19), and also indirectly via thermodynamic methods (28), gp120 is thought to be extremely flexible and to undergo extensive structural rearrangement upon binding of its ligands, particularly CD4.
HNG-105 (Fig. 1A) is an entry inhibitor generated by our group by the click conjugation of the 12p1 peptide and has been shown to work by inhibiting key interactions of gp120 (4, 11, 14). HNG-105 inhibits the interactions of both monomeric and trimeric soluble gp120 with soluble CD4 (sCD4), and this molecular inhibition translates to viral inhibition (H. N. Gopi et al., unpublished data). Mechanistic studies of the inhibitory action of HNG-105 reveal that it works by a novel allosteric mechanism, interacting with a site other than that of the CD4 or coreceptor binding sites, and dramatically lowering the affinity of gp120 for either of its receptors (Gopi et al., unpublished). Given the unique inhibitory mechanism, we wanted to study the molecular consequences of HIV-1 envelope variation on the inhibitory action of HNG-105.
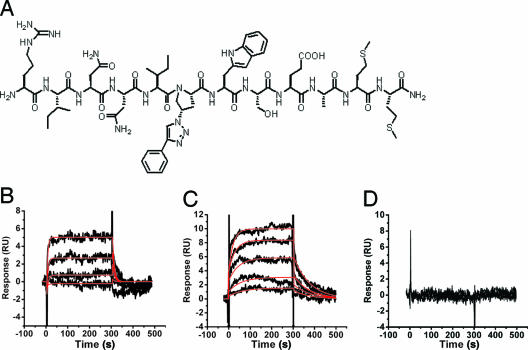
FIG. 1.
(A) Diagram of the HNG-105 conjugate peptide. (B through D) Sensorgrams depicting the interaction of HNG-105 with 92UG037-08 (clade A) (B), 96ZM651 (clade C) (C), and 90CM243 (CRF01_AE) (D). HNG-105 was used at concentrations of 0.125, 0.25, 0.5, 1, 2, and 4 μM. Experimental data are shown in black. Red lines indicate fitting to a 1:1 Langmuir binding model with a parameter included for mass transport.
It has been demonstrated previously that HNG-105 binds directly to the gp120 derived from the primary isolate HIV-1YU-2. However, no other direct binding or molecular inhibition data exist for different clade B envelopes or for envelopes derived from viruses of other subtypes. We therefore measured the binding of HNG-105 to gp120 envelope proteins from several strains of HIV-1 from differing clades (Table 1, column 1) by using surface plasmon resonance interaction analysis (Biacore 3000 instrument). The respective gp120 proteins from HIV-1 clades A, B, C, and D and from two major circulating recombinant forms, CRF01_AE and CRF07_BC, were immobilized on a CM5 sensor chip and exposed to different concentrations of HNG-105 (0.125 to 4 μM). An additional gp120 from the SIV strain PBj was also included. Nonspecific binding and instrument artifacts were accounted for by subtraction of the response from a control surface (anti-IL5Rα antibody 2B6R). HNG-105 was found to bind to all HIV-1 gp120s tested, except for that of CRF01_AE, with equilibrium dissociation constants (KD) in the range of 0.04 to 7 μM (Table 1, column 2; Fig. 1B to D). This indicates a degree of conservation of the binding epitope across the subtypes studied. Like its parent peptide, 12p1, HNG-105 did not bind to the gp120 derived from SIV-PBj (9) (Table 1, column 2).
TABLE 1.
Direct binding affinity to HNG-105 and sCD4, along with IC50s for HNG-105-mediated inhibition of the sCD4 interaction with gp120 variants
| Variant (subtype) | Affinity (KD)a for:
|
IC50b of HNG-105 for sCD4-gp120 interaction | |
|---|---|---|---|
| HNG-105 | sCD4 | ||
| 92UG037-08 (A) | 1.9 μM | 15.8 nM | 666.1 nM |
| 92TH014-12 (B) | 83.2 nM | 19.0 nM | 88.4 nM |
| 92US715-06 (B) | 6.8 μM | 63.3 nM | 5.1 μM |
| BORI-15 (B) | 291.0 nM | 5.4 nM | 446.0 nM |
| BORI (B) | 381 nM | 14.4 nM | 457 nM |
| BaL (B) | 156.0 nM | 5.7 nM | 312.0 nM |
| YU-2 (B) | 38.6 nM | 14.2 nM | 187.5 nM |
| SF162 (B) | 70.4 nM | 30.3 nM | 253.0 nM |
| IIIB-8X (B) | 176.0 nM | 9.0 nM | 230.8 nM |
| 93MW959 (C) | 39.8 nM | 5.3 nM | 104.6 nM |
| 96ZM651 (C) | 136.7 nM | 97.2 nM | 230.4 nM |
| 92UG021-9 (D) | 254.0 nM | 53.5 nM | 865.0 nM |
| 97CN001-CN54 (BC) | 454 nM | 15.0 nM | 441.5 nM |
| 90CM243 (AE) | — | 21.8 nM | NA |
| SIV-PBj | — | 150.0 nM | NA |
The average kinetic parameters (association and dissociation rates) generated from a minimum of four data sets were used to define KD. —, no binding.
Derived from a minimum of two data sets. NA, not applicable.
Similarly, we assessed whether or not HNG-105 could inhibit the pivotal interaction of the non-subtype B gp120s with sCD4. Prior to the assessment of inhibitory effect, we characterized the interaction of sCD4 with a given gp120 under the conditions of the inhibition assay (Table 1, column 3). Then sCD4, in the absence or presence of increasing amounts of the peptide, was injected over the immobilized variant gp120s. Increasing concentrations of HNG-105 significantly inhibited the interaction of sCD4 with the gp120 variants that bound HNG-105, whereas those gp120s that did not interact with HNG-105 showed no effect. The inhibitory effect of HNG-105 on the gp120-sCD4 interactions was quantified from the corresponding decrease in the response at equilibrium (Req) at a given peptide concentration and was used to derive the respective 50% inhibitory concentrations (IC50s) (Table 1, column 4). The derived IC50s showed good correlation with the affinity (KD) of HNG-105 for a given variant.
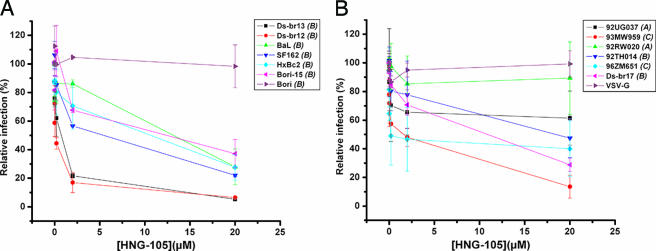
In addition to establishing molecular inhibition of the variant gp120s by HNG-105, we characterized the inhibitory action of HNG-105 on HIV-1 entry by using a pseudovirus single-round infection assay (23). Given the variability of the env gene, we sought to characterize the efficacy of HNG-105 against a panel of viruses that embodied this diversity. We therefore chose HIV-1 variants isolated from diverse body compartments (periphery, central nervous system, and brain) that embodied significant genetic diversity and included members of the globally prevalent subtypes A, B, and C (Fig. 2A and B). HNG-105 showed specific inhibition of viruses from all of the clades tested, albeit with differing efficacies. Interestingly, the subtype B peripheral isolate HIV-1BORI (33) was not sensitive to the inhibitory effects of HNG-105 over the concentration range studied. This is in stark contrast to the sensitivity displayed by the highly related HIV-1BORI-15 isolate (33) (Fig. 2A) and the biochemical data indicating that the monomeric forms of the gp120s from both of these strains bound and were affected by HNG-105 (Table 1). The major differences between these isolates (23, 24, 32), coupled with the localization of the potential binding site for HNG-105 (4; Gopi et al., unpublished), suggest that one mechanism by which an isolate can be resistant to HNG-105 may be conformational masking of the binding site in the context of the viral spike (17, 18). This concept of differential access to the HNG-105 binding site, which is thought to reside within the gp120 inner domain, is currently under investigation in our laboratory.
FIG. 2.
Inhibition of pseudotype infection by HNG-105. Recombinant luciferase-containing viruses pseudotyped with the envelope proteins from different HIV-1 isolates were used in single-round infection assays in the presence of increasing concentrations of HNG-105. HOS cells stably expressing CD4 and CCR5 were used as target cells for R5 viruses, whereas U87 cells stably expressing CD4 and CXCR4 were used for X4 viruses. For the sake of clarity, the data have been distributed into two panels, A and B. The subtype of each HIV-1 strain is given in parentheses after the variant name.
From this study and others, it is clear that variability in the component proteins of the viral envelope (gp41 and g120) has a modulatory effect on the susceptibility of a given isolate to drugs that target viral entry. Subtype sequence variability can result in either a reduction in an entry inhibitor's effectiveness (27) or an increase in efficacy (13). Moreover, this study highlights the need to consider the effect of sequence variation on the conformation of the viral spike and how that, in turn, affects susceptibility to entry inhibitors. Therefore, the correlation of biochemical binding analyses with virally derived inhibition parameters for an extended panel of isolates not only should become routine in the clinical development of this class of antivirals but may also yield important information regarding the molecular mechanism of cell entry by HIV-1.
Acknowledgments
This research was supported by National Institutes of Health (NIH) grants P01 GM 056550-08/C210JC and R21 AI071265-01 (to I.M.C.) and R21 NS047970-03 (to J.M.-G.).
Footnotes
Published ahead of print on 24 January 2007.
REFERENCES
- 1.Arien, K. K., A. Abraha, M. E. Quinones-Mateu, L. Kestens, G. Vanham, and E. J. Arts. 2005. The replicative fitness of primary human immunodeficiency virus type 1 (HIV-1) group M, HIV-1 group O, and HIV-2 isolates. J. Virol. 79:8979-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin, C. E., R. W. Sanders, and B. Berkhout. 2003. Inhibiting HIV-1 entry with fusion inhibitors. Curr. Med. Chem. 10:1633-1642. [DOI] [PubMed] [Google Scholar]
- 3.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biorn, A. C., S. Cocklin, N. Madani, Z. Si, T. Ivanovic, J. Samanen, D. I. Van Ryk, R. Pantophlet, D. R. Burton, E. Freire, J. Sodroski, and I. M. Chaiken. 2004. Mode of action for linear peptide inhibitors of HIV-1 gp120 interactions. Biochemistry 43:1928-1938. [DOI] [PubMed] [Google Scholar]
- 5.Borkow, G., and A. Lapidot. 2005. Multi-targeting the entrance door to block HIV-1. Curr. Drug Targets Infect. Disord. 5:3-15. [DOI] [PubMed] [Google Scholar]
- 6.Brelot, A., and M. Alizon. 2001. HIV-1 entry and how to block it. AIDS 15(Suppl. 5):S3-S11. [DOI] [PubMed] [Google Scholar]
- 7.Castagna, A., P. Biswas, A. Beretta, and A. Lazzarin. 2005. The appealing story of HIV entry inhibitors: from discovery of biological mechanisms to drug development. Drugs 65:879-904. [DOI] [PubMed] [Google Scholar]
- 8.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 9.Dewhurst, S., J. E. Embretson, D. C. Anderson, J. I. Mullins, and P. N. Fultz. 1990. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature 345:636-640. [DOI] [PubMed] [Google Scholar]
- 10.Dorr, P., M. Westby, S. Dobbs, P. Griffin, B. Irvine, M. Macartney, J. Mori, G. Rickett, C. Smith-Burchnell, C. Napier, R. Webster, D. Armour, D. Price, B. Stammen, A. Wood, and M. Perros. 2005. Maraviroc (UK-427,857), a potent, orally bioavailable, and selective small-molecule inhibitor of chemokine receptor CCR5 with broad-spectrum anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 49:4721-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrer, M., and S. C. Harrison. 1999. Peptide ligands to human immunodeficiency virus type 1 gp120 identified from phage display libraries. J. Virol. 73:5795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fung, H. B., and Y. Guo. 2004. Enfuvirtide: a fusion inhibitor for the treatment of HIV infection. Clin. Ther. 26:352-378. [DOI] [PubMed] [Google Scholar]
- 13.Geretti, A. M. 2006. HIV-1 subtypes: epidemiology and significance for HIV management. Curr. Opin. Infect. Dis. 19:1-7. [DOI] [PubMed] [Google Scholar]
- 14.Gopi, H. N., K. C. Tirupula, S. Baxter, S. Ajith, and I. M. Chaiken. 2006. Click chemistry on azidoproline: high-affinity dual antagonist for HIV-1 envelope glycoprotein gp120. ChemMedChem 1:54-57. [DOI] [PubMed] [Google Scholar]
- 15.Gray, E. S., T. Meyers, G. Gray, D. C. Montefiori, and L. Morris. 2006. Insensitivity of paediatric HIV-1 subtype C viruses to broadly neutralising monoclonal antibodies raised against subtype B. PLoS Med. 3:e255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong, P. D. 2005. Human immunodeficiency virus: refolding the envelope. Nature 433:815-816. [DOI] [PubMed] [Google Scholar]
- 18.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 19.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 20.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazzarin, A. 2005. Enfuvirtide: the first HIV fusion inhibitor. Expert Opin. Pharmacother. 6:453-464. [DOI] [PubMed] [Google Scholar]
- 22.Markovic, I., and K. A. Clouse. 2004. Recent advances in understanding the molecular mechanisms of HIV-1 entry and fusion: revisiting current targets and considering new options for therapeutic intervention. Curr. HIV Res. 2:223-234. [DOI] [PubMed] [Google Scholar]
- 23.Martin, J., C. C. LaBranche, and F. Gonzalez-Scarano. 2001. Differential CD4/CCR5 utilization, gp120 conformation, and neutralization sensitivity between envelopes from a microglia-adapted human immunodeficiency virus type 1 and its parental isolate. J. Virol. 75:3568-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Garcia, J., S. Cocklin, I. M. Chaiken, and F. Gonzalez-Scarano. 2005. Interaction with CD4 and antibodies to CD4-induced epitopes of the envelope gp120 from a microglial cell-adapted human immunodeficiency virus type 1 isolate. J. Virol. 79:6703-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, J. P., F. E. McCutchan, S. W. Poon, J. Mascola, J. Liu, Y. Cao, and D. D. Ho. 1994. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J. Virol. 68:8350-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore, J. P., A. Trkola, and T. Dragic. 1997. Co-receptors for HIV-1 entry. Curr. Opin. Immunol. 9:551-562. [DOI] [PubMed] [Google Scholar]
- 27.Moore, P. L., T. Cilliers, and L. Morris. 2004. Predicted genotypic resistance to the novel entry inhibitor, BMS-378806, among HIV-1 isolates of subtypes A to G. AIDS 18:2327-2330. [DOI] [PubMed] [Google Scholar]
- 28.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Brien, W. A. 1994. HIV-1 entry and reverse transcription in macrophages. J. Leukoc. Biol. 56:273-277. [DOI] [PubMed] [Google Scholar]
- 30.Pierson, T. C., and R. W. Doms. 2003. HIV-1 entry and its inhibition. Curr. Top. Microbiol. Immunol. 281:1-27. [DOI] [PubMed] [Google Scholar]
- 31.Ryser, H. J., and R. Fluckiger. 2005. Progress in targeting HIV-1 entry. Drug Discov. Today 10:1085-1094. [DOI] [PubMed] [Google Scholar]
- 32.Shieh, J. T., J. Martin, G. Baltuch, M. H. Malim, and F. Gonzalez-Scarano. 2000. Determinants of syncytium formation in microglia by human immunodeficiency virus type 1: role of the V1/V2 domains. J. Virol. 74:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strizki, J. M., A. V. Albright, H. Sheng, M. O'Connor, L. Perrin, and F. Gonzalez-Scarano. 1996. Infection of primary human microglia and monocyte-derived macrophages with human immunodeficiency virus type 1 isolates: evidence of differential tropism. J. Virol. 70:7654-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomkowicz, B., and R. G. Collman. 2004. HIV-1 entry inhibitors: closing the front door. Expert Opin. Ther. Targets 8:65-78. [DOI] [PubMed] [Google Scholar]