Abstract
Coronavirus envelope (E) proteins are small (∼75- to 110-amino-acid) membrane proteins that have a short hydrophilic amino terminus, a relatively long hydrophobic membrane domain, and a long hydrophilic carboxy-terminal domain. The protein is a minor virion structural component that plays an important, not fully understood role in virus production. It was recently demonstrated that the protein forms ion channels. We investigated the importance of the hydrophobic domain of the mouse hepatitis coronavirus (MHV) A59 E protein. Alanine scanning insertion mutagenesis was used to examine the effect of disruption of the domain on virus production in the context of the virus genome by using a MHV A59 infectious clone. Mutant viruses exhibited smaller plaque phenotypes, and virus production was significantly crippled. Analysis of recovered viruses suggested that the structure of the presumed α-helical structure and positioning of polar hydrophilic residues within the predicted transmembrane domain are important for virus production. Generation of viruses with restored wild-type helical pitch resulted in increased virus production, but some exhibited decreased virus release. Viruses with the restored helical pitch were more sensitive to treatment with the ion channel inhibitor hexamethylene amiloride than were the more crippled parental viruses with the single alanine insertions, suggesting that disruption of the transmembrane domain affects the functional activity of the protein. Overall the results indicate that the transmembrane domain plays a crucial role during biogenesis of virions.
Coronaviruses are enveloped positive-stranded RNA viruses that belong to the Coronaviridae family in the Nidovirales order. The viruses cause primarily respiratory and enteric infections in humans and a broad range of animals. Recently several new human coronaviruses, including severe acute respiratory syndrome coronavirus (SARS-CoV), were identified, which significantly increased the interest in understanding this family of viruses and identification of antiviral targets for development of therapeutic treatments.
The coronavirus virion envelope contains at least three integral membrane proteins. All members of the family contain the membrane (M), spike (S), and envelope (E) proteins. Some members of the family have an additional envelope protein, the hemagglutinin esterase (HE) (5). The genomic RNA is encapsidated by the nucleocapsid (N) phosphoprotein (22). The S glycoprotein is the receptor binding protein that facilitates infection through fusion of viral and cellular membranes and is the major target of neutralizing antibodies (13). The M glycoprotein is a major component of the envelope that plays an important role in virus assembly (10, 18, 31, 37). The E protein is a minor component of the viral envelope. Assembly of these components into virions occurs at intracellular membranes in the region of the endoplasmic reticulum Golgi complex (ERGIC) (19, 35).
The focus of this paper is the E protein. Coronavirus E proteins are small (76- to 109-amino-acid) integral membrane proteins with rather long hydrophobic domains. The protein plays an important, not yet fully defined role in virus production (7, 11, 21, 32). Coexpression of the E and M proteins alone is sufficient for virus-like particle (VLP) assembly (3, 6, 37). E protein-containing vesicles are released from cells when E is expressed alone (6, 27). Deletion of the E gene from mouse hepatitis coronavirus (MHV) results in severely crippled virus (21), whereas removal of the protein from porcine transmissible gastroenteritis coronavirus blocks virus production (7, 32). The SARS-CoV E protein is important for virus production, but it is not absolutely required, since deletion of the gene results in virus yields that are 20- to 200-fold lower than those of the wild-type virus, depending on the cell type (9). Recently, it was demonstrated that E proteins of several coronaviruses, including MHV, are viroporins that exhibit ion channel activity (23, 26, 39, 40).
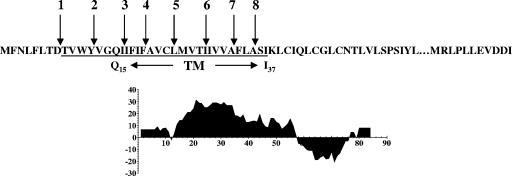
The MHV A59 E protein consists of 83 amino acids with a stretch of 29 hydrophobic residues located toward the amino end of the protein (Fig. 1). Although coronavirus E proteins share little homology at the sequence level, a long hydrophobic domain is a conserved feature. Two topologies, one transmembrane domain or a hairpin conformation, have been proposed for coronavirus E proteins (1, 6, 17, 28, 43). It remains to be determined if the proteins from different viruses adopt different topologies or if they assume two membrane conformations during the virus life cycle. For the purposes of our study we assumed that the hydrophobic domain can adopt a transmembrane topology. We hypothesized that the long hydrophobic domain must be important for the functional role(s) of the protein. To test this idea, alanine scanning insertion mutagenesis (4, 29) was used to examine the importance of the predicted α-helical structure of the domain. Insertion of an alanine residue into a transmembrane α-helix causes all amino acids on its carboxy side to be rotated by ∼100 degrees, which disrupts the potential helix-helix packing interface of residues on both sides of the insertion. Eight alanine insertion mutants were constructed by positioning the residues at various places across the hydrophobic domain (Fig. 1). The mutations were studied in the context of an MHV A59 infectious clone. Preliminary analysis of one of the mutant viruses suggested that shifting the relative positions of polar hydrophilic residues in the domain could be important for the function of the E protein (41). In the study reported here, we examined the impact of the single alanine insertions across the membrane on virus production and release. Viruses with the restored wild-type helical pitch and positions of polar hydrophilic residues were constructed to demonstrate the importance of the structural integrity of the domain for virus production and release. The ability of the ion channel inhibitor hexamethylene amiloride (HMA) to decrease virus yields correlated with the modification that restored the transmembrane integrity. Altogether, the results clearly illustrate the importance of the E protein transmembrane domain for virus production and release. The data strongly suggest that the integrity of the helix and positioning of residues along the α-helical structure are important for the function of the protein.
FIG. 1.
MHV A59 E protein hydrophobic domain alanine insertions. (A) Amino acid sequence of the E protein hydrophobic domain and positions of alanine insertions, with the hydrophobic domain underlined and the putative transmembrane (TM) domain spanning amino acids Q15 to I37 indicated by arrows. (B) Kyte-Doolittle hydropathy plot of the E protein. Hydrophobic and hydrophilic domains are plotted above and below the line, respectively.
MATERIALS AND METHODS
Cells and viruses.
Mouse L2 cells and 17 clone 1 (17Cl1) cells were maintained in Dulbecco's modified Eagle's medium containing 5% heat-inactivated fetal calf serum supplemented with glutamine and the antibiotics penicillin and streptomycin. Baby hamster kidney (BHK) cells expressing the MHV Bgp1a receptor (BHK-MHVR cells) were kindly provided by Ralph Baric, University of North Carolina at Chapel Hill (42). BHK and BHK-MHVR cells were grown in Glasgow minimal essential medium containing 5% fetal calf serum, supplemented with 10% tryptose phosphate broth and as described above. BHK-MHVR cells were maintained under selection with 800 μg/ml of Geneticin (G418) for selection of cells expressing the receptor. Stocks of wild-type MHV A59 and infectious cloned viruses were grown in mouse 17Cl1 and L2 cells. Virus titers were determined in L2 cells.
Construction of amino acid substitution mutants.
pScript-E, a pPCR-Script Amp SK(+) vector (Stratagene), and pcDNA-E, a pcDNA3.1/Zeo(−) vector (Invtrogen), each containing the MHV A59 E gene, were used for mutagenesis. Site-directed alanine insertions were made in the hydrophobic domain of the E gene by using the primers shown in Table 1. Mutants were constructed by whole-plasmid PCR using high-fidelity Pfu polymerase (Stratagene). Following an initial incubation at 95°C for 3 min, 18 cycles of 95°C for 15 s, 74°C for 60 s, and 68°C for 12 min were applied. The PCR products were incubated at 37°C for 2 h with DpnI to destroy methylated template DNA before transformation into Escherichia coli DH5α. Mutations were confirmed by sequencing the entire insert from pScript-E before subcloning into the MHV G clone at the SbfI and EcoRV restriction sites.
TABLE 1.
Sequences of primers used for mutagenesis
| Primer | Sequence (5′→3′) |
|---|---|
| MHVE_Ala1 (forward) | GTTTAATTTATTCCTTACAGACGCCACAGTATGGTATGTGGGGCA |
| MHVE_Ala1 (reverse) | TAAAAATAATCTGCCCCACATACCATACTGTGGCGTCTGTAAGG |
| MHVE_Ala2 (forward) | CCTTACAGACACAGTATGGTATGCCGTGGGGCAGATTATTTTTAT |
| MHVE_Ala2 (reverse) | CACACTGCGAATATAAAAATAATCTGCCCCACGGCATACCATACT |
| MHVE_Ala3 (forward) | CAGTATGGTATGTGGGGCAGATTGCCATTTTTATATTCGCAGTG |
| MHVE_Ala3 (reverse) | CCATCAAACACACTGCGAATATAAAAATGGCAATCTGCCCCA |
| MHVE_Ala4 (forward) | GGGCAGATTATTTTTATATTCGCCGCAGTGTGTTTGATGGTC |
| MHVE_Ala4 (reverse) | CACAATTATGGTGACCATCAAACACACTGCGGCGAATATAAAA |
| MHVE_Ala5 (forward) | ATTTTTATATTCGCAGTGTGTTTGGCCATGGTCACCATAATTGTG |
| MHVE_Ala5 (reverse) | GCAACCACAATTATGGTGACCATGGCCAAACACACTGCG |
| MHVE_Ala6 (forward) | CAGTGTGTTTGATGGTCACCATAGCCATTGTGGTTGCCTTCCTTG |
| MHVE_Ala6 (reverse) | TGATAGACGCAAGGAAGGCAACCACAATGGCTATGGTGACCATC |
| MHVE_Ala7 (forward) | CCATAATTGTGGTTGCCGCCTTCCTTGCGTCTATCAAAC |
| MHVE_Ala7 (reverse) | GAATACAAAGTTTGATAGACGCAAGGAAGGCGGCAACC |
| MHVE_Ala8 (forward) | GTGGTTGCCTTCCTTGCGGCCTCTATCAAACTTTGTATTC |
| MHVE_Ala8 (reverse) | CCGCAAAGTTGAATACAAAGTTTGATAGAGGCCGCAAGG |
Generation of alanine insertion mutant viruses.
Viruses containing the alanine insertions in the E gene hydrophobic domain were generated by using an MHV A59 full-length infectious clone (42). Full-length cDNA clones were assembled, transcribed, and electroporated into BHK-MHVR cells as previously described (38).
At 24 to 48 h after electroporation, the media were harvested and an aliquot was used to infect L2 cells. Total RNA was extracted from cells remaining on the flasks by using an RNAqueous-4PCR extraction kit (Ambion). The extracted RNA was treated with DNase prior to being used as the template for reverse transcription (RT) with an oligo(dT) primer. The RT product was subjected to 30 cycles of PCR amplification using Ambion's SuperTaq Plus with forward (5′-CAGAACTGTCCAACAGGCCGTTAGCAAG-3′) and reverse (5′-GCAACCCAGAAGACACCTCAATGC-3′) primers to obtain E and M cDNA gene products. PCR products were cleaned up using QIAGEN′s MiniElute columns and sequenced directly.
Viruses were plaque purified from the electroporated media and passaged six times in L2 cells. RNA was extracted from the infected cells at passages 1 and 6 for RT-PCR. The E and M gene cDNA products were sequenced each time to determine the stability of the mutations and to identify any potential compensating changes.
Growth kinetics.
Growth kinetic experiments were carried out in L2 cells infected with passage 6 virus stocks at a multiplicity of infection (MOI) of 0.01. Cell culture supernatants were collected at various times after infection. Titers were determined by plaque assay on L2 cells. At approximately 48 h postinfection (p.i.), agarose/medium overlays were removed and the cells were fixed and stained with crystal violet in ethanol.
Indirect immunofluorescence analysis.
Mouse 17Cl1 cells were infected with mutant viruses and analyzed in parallel with wild-type MHV to determine the localization of the E and M proteins. Cells were plated on two-well glass slides 1 day before infection at a MOI of 0.5. At 10 h p.i. cells were washed with phosphate-buffered saline (PBS) and fixed witl 100% methanol for 15 min at −20°C. Fixed cells were washed with PBS and then blocked with 2% gelatin in PBS for 2 h. Slides were incubated with a mixture of anti-MHV E 9410 polyclonal (L. Lopez and B. G. Hogue, unpublished data) and anti-MHV M J1.3/2.7 monoclonal (12) primary antibodies for 1 h at room temperature. Cells were washed multiple times with 2% gelatin in PBS before incubation with fluorescein isothiocyanate-labeled anti-mouse and AlexaFluor-labeled anti-rabbit secondary antibodies. Cells were washed extensively with PBS containing 2% gelatin and then once with PBS alone. Slides were mounted in ProLong Gold antifade reagent (Molecular Probes) plus 4,6-diamino-2-phenylindole (DAPI) to stain nuclei. Images were viewed using an epifluorescence Nikon inverted microscope (Nikon Inc., Melville, NY) with MetaMorph imaging software (Universal Imaging Corporation, Downingtown, PA). Images were processed using Adobe Photoshop.
Titration of intracellular and extracellular virus.
Confluent mouse 17Cl1 cells were infected with viruses at an MOI of 0.1. At 16 h p.i the supernatant and cells were harvested separately. The supernatant containing extracellular virus was clarified by centrifugation to remove cell debris. Medium was added to the cell monolayer, followed by three freeze-thaw cycles. Virus titers for both fractions were determined by plaque assay as described above.
HMA inhibition of plaque formation.
Confluent mouse L2 cells were infected with viruses as described above with ∼50 to 150 PFU After infection, cells were overlaid for plaque assay with agarose/medium as described above. The overlay contained 20 μM of HMA dissolved in a 1:1 mix of 50% dimethyl sulfoxide and 50% methanol or an equivalent amount of dimethyl sulfoxide-methanol without drug. At 3 days after infection, cells were stained with crystal violet as described above.
Transmembrane α-helix and helical wheel analysis.
Prediction of E protein transmembrane helices was performed using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/). The transmembrane domain was modeled by helical wheel analysis using Gene Runner (Hastings Software, Inc.).
RESULTS
Alanine insertions in the hydrophobic domain of the E protein affect virus growth and output.
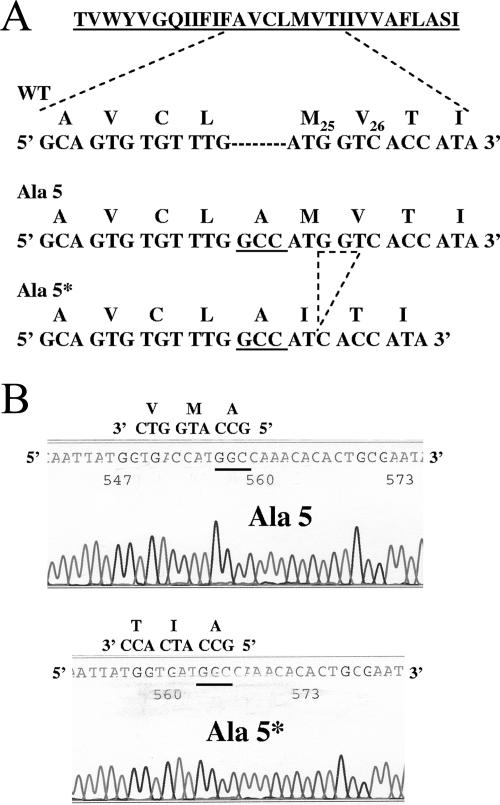
To begin understanding the specific requirements of the E protein hydrophobic domain for virus assembly, we generated a series of alanine scanning insertion mutations within the domain of the MHV A59 protein. Alanine residues were introduced singly across the hydrophobic domain by site-directed mutagenesis at eight positions (Fig. 1). The effect of the insertions on virus production was studied by reverse genetics using a full-length MHV A59 infectious clone. All full-length mutant RNAs produced cytopathic effects characterized by centers of fusion after electroporation into BHK-MHVR cells. Viruses, named Ala 1 to Ala 8, were subsequently recovered for all of the mutants after passage of the medium from the electroporated cells onto mouse L2 cells. RT-PCR and sequence analysis of the E and M genes were confirmed for each of the mutant viruses after passage 1 in L2 cells. The presence of the alanine insertion and no other mutations in either the E or M genes was confirmed for all mutants, with one exception. When the Ala 5 mutant was initially made, the recovered virus, designated Ala 5*, retained the inserted alanine codon insertion (GCC), but three nucleotides had been lost, i.e., one nucleotide (G) from the methionine codon and two nucleotides (GT) from the valine codon (Fig. 2). This resulted in the loss of the methionine and valine residues at positions 25 and 26, respectively, and replacement of these residues with an isoleucine (codon ATC) (Fig. 2). A second independent Ala 5 mutant full-length clone was assembled. The recovered Ala 5 virus had the insertion and no additional changes (Fig. 2).
FIG. 2.
Sequence confirmation of alanine 5 insertions. (A) Partial nucleotide and corresponding amino acid sequences of the E protein for wild-type (A21 to I28), Ala5, and Ala5* viruses. Codons for alanine insertions are underlined. The nucleotides (GGT) that were lost in the Ala 5* recovered virus that resulted in loss of M25 and V26 and addition of isoleucine (codon ATC) are indicated by the dashed-line triangle. (B) Sequences of the region surrounding the Ala insertions in the Ala 5 and Ala 5* E proteins, illustrating the loss of the M and V residues and addition of an I residue. The sequence shown is the complement of the coding strand. Key codons for the coding strand are shown above each sequence.
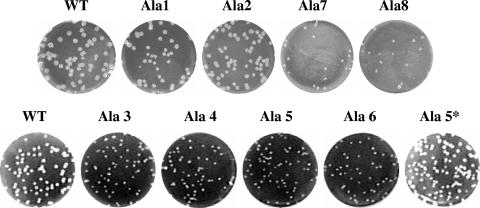
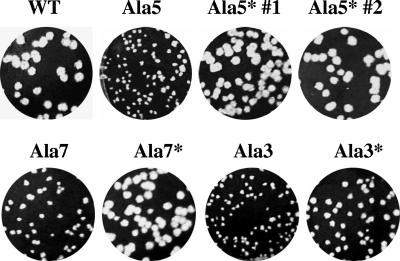
All of the mutant viruses, including Ala 5*, were plaque purified on mouse L2 cells, and multiple isolated plaques were passaged six times on L2 cells. RT-PCR and sequence analysis of each mutant virus confirmed the genetic stability of the introduced mutations and that no additional changes were present in the remainder of the E gene or within the M gene. After the final passage, the viruses were analyzed for their growth characteristics. The viruses with insertions at the amino end of the hydrophobic domain (Ala 1 and Ala 2) gave rise to plaques that were similar in size to those of the wild-type virus, whereas the other mutant viruses exhibited smaller plaques (Fig. 3).
FIG. 3.
Plaque morphologies of alanine insertion mutant viruses. Mouse L2 cells were infected with wild-type (WT) infectious cloned virus or alanine insertion mutant viruses and analyzed by plaque assay.
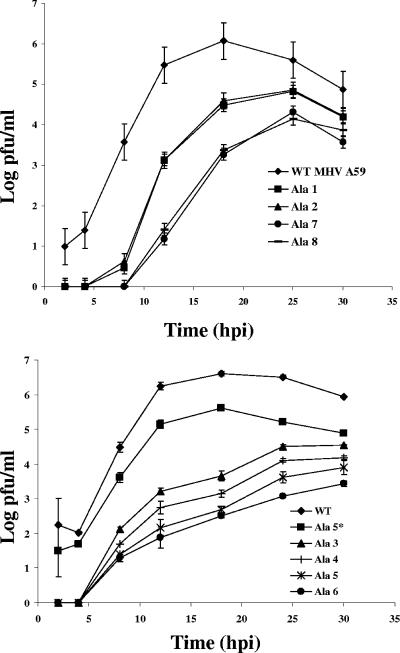
Growth kinetic analysis was carried out in mouse L2 cells. Cells were infected with the wild-type and mutant viruses at a MOI of 0.01, and extracellular virus was assayed at between 2 and 30 h p.i. None of the mutant viruses grew as well as the wild-type virus (Fig. 4). The Ala 1, Ala 2, Ala 7, and Ala 8 viruses reached peak titers that were approximately 1.5 to 2 logs lower than that of the wild-type virus. The mutants reached their peak titers at 25 h p.i., compared with 18 h p.i. for the wild-type virus. The viruses with alanine insertions at positions 3 to 6 yielded titers at 30 h p.i. that were 2 to 3 logs lower than the peak titer of the wild-type virus at 18 h p.i. Together, these results indicated that the hydrophobic domain of the E protein is sensitive to disruption by insertion of alanine residues.
FIG. 4.
Growth kinetics of alanine insertion viruses. Mouse L2 cells were infected with wild-type (WT) or alanine insertion mutant viruses at an MOI of 0.01. Titers were determined by plaque assay on L2 cells at the indicated times. Error bar represent the standard deviations from the means for three independent growth kinetic experiments.
The recovered Ala 5* was also analyzed for its growth properties. Interestingly, the virus yielded plaques that were similar in size to those of the wild-type virus (Fig. 3). Significantly, the growth properties of the virus were more like those of the wild type than like those of Ala 5 or any of the other mutant viruses (Fig. 4). Ala 5* reached its peak titer like the wild-type virus at ∼18 h p.i., even though the virus yield was about 1 log lower. Since Ala 5* clearly grew better than Ala 5, this strongly suggested that the difference in the hydrophobic domains of these viruses was responsible for their phenotype difference.
Disruption of E and M interaction is not due to the protein mislocalization.
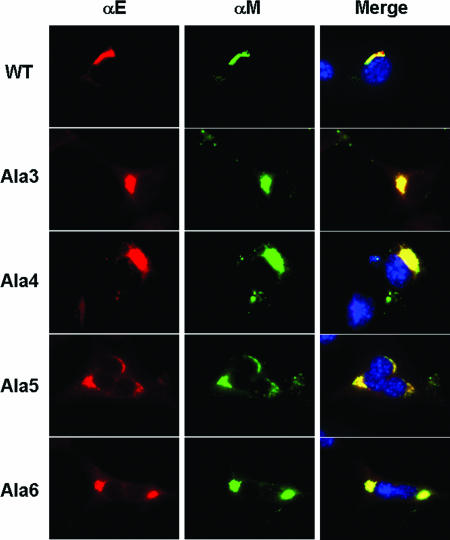
MHV E protein was previously shown to accumulate in the ER and ERGIC membranes (34). We have determined that wild-type E localization overlaps with ER markers, but the protein colocalizes in the ERGIC/Golgi when coexpressed with the M protein or in virus-infected cells (Lopez and Hogue, unpublished data). To rule out the possibility that the mutant E proteins were mislocalized in cells, which could contribute to the significant reduction in virus output, we examined the cellular localization of the E and M proteins in cells infected with the most crippled mutant viruses, Ala 3, Ala 4, Ala 5, and Ala 6 (Fig. 5) and Ala 5* (data not shown). In each case the E and M proteins colocalized like the wild-type virus, indicating that mislocalization does not account for the crippled phenotype of the viruses.
FIG. 5.
Colocalization of MHV E and M proteins. Mouse 17Cl1 cells were infected with wild-type (WT), Ala 3, Ala 4, Ala 5, and Ala 6 viruses. Cells were fixed and analyzed by immunofluorescence at 10 h p.i. using mouse and rabbit antibodies against the M and E proteins, respectively. Fluorescein isothiocyanate-conjugated mouse and AlexaFluor 594-conjugated rabbit secondary antibodies were used to visualize the localization of the proteins. Colocalization of M and E proteins is represented in the merged images by yellow. Nuclei were stained with DAPI.
Analysis of the E protein hydrophobic domain.
Since the Ala 1 and Ala 2 viruses exhibited phenotypes more like that of the wild-type virus than like those of the other mutant viruses, this suggested that the insertions within the amino end of the hydrophobic domain were less disruptive. We reasoned that the positions of the first and second insertions might lie outside of the transmembrane α-helix, considering the length of the hydrophobic domain. Thus, we used the TMHMM server at the Technical University of Denmark Center for Biological Sequence Analysis, which predicts α-helices in protein, to analyze the MHV E protein hydrophobic domain. Helices are predicted based on the hidden Markov model (20). TMHMM predicted that the α-helix spans residues Q15 to I37 in the protein (Fig. 1). This places the Ala 1 and Ala 2 insertions within the hydrophobic domain but outside the predicted α-helix, consistent with the lesser effect of insertions at these positions on virus production.
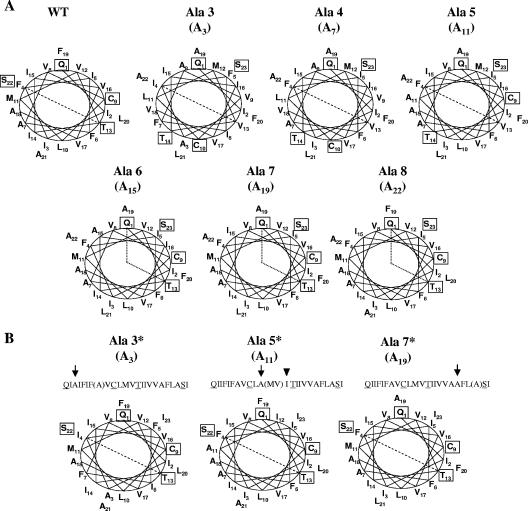
To further analyze possible effects of disruption of the predicted α-helix, helical wheel analysis was used to provide insight into the effect on positioning of residues along the helix. Four hydrophilic polar residues, Q1, C9, T13, and S22 (corresponding to Q15, C23, T27, and S36 in the full-length protein [Fig. 1]), are predicted to align along the same face of the wild-type E α-helix (Fig. 6A). The residues are predicted to be evenly distributed along the length of the helix, with C9 and T13 positioned two turns on the helix below and above Q1 and S22, respectively. The residues are positioned such that they would cover the entire 180o face of the helix. However, in the case of the Ala 3 and Ala 4 mutants, two of the residues, C9 and T14, are predicted to be positioned on the opposite face of the α-helix. Insertions of Ala 6, Ala 7, and Ala 8 are all predicted to change the separating distance of the hydrophilic residues, such that only roughly 120o of the helix face would be covered. The positioning of the four hydrophilic resides is also predicted to be disrupted in the Ala 5 mutant transmembrane; however, the residues would be returned to their wild-type positions in the partially compensated Ala 5* mutant. These results suggested that the integrity of the helix and possibly positions of the polar hydrophilic residues may be functionally important.
FIG. 6.
Helical wheel analysis of the E protein predicted transmembrane domains of alanine insertion mutant viruses. (A) Schematic top views from the amino ends of wild-type (WT) and alanine insertion mutant E protein transmembrane domains. Hydrophilic polar residues Q1, C9, T13, and S22, corresponding to Q15, C23, T27, and S36 in the full-length wild-type protein, are boxed. Residues toward the carboxy end of the helix are shown adjacent to the corresponding positions relative to the first 18 amino acids in the transmembrane domain. Numbers below the names indicate the position of the inserted alanine residue in the helical wheel. Lines across the wheels are added to emphasize the positioning on one face of the α-helix for wild-type, Ala 5*, Ala 3*, and Ala 7* viruses and the disruption of the positions of these residues in the Ala 3 to Ala 8 mutant proteins. Helical wheels plots were prepared using Gene Runner version 3.05. (B) Schematic tops views of Ala 3*, Ala 5*, and Ala 7* E proteins, illustrating positioning of the hydrophilic residues with removal of one amino acid on the carboxy-terminal side of the alanine insertion. Alanine insertions are indicated by arrows in the sequences above each wheel. Amino acids that were removed are enclosed in parentheses. The arrowhead notes the isoleucine insertion resulting from codon rearrangement in the recovered Ala 5* virus.
Resetting the pitch of the polar hydrophilic residues partially compensates for the alanine insertions.
To confirm that the changes in the recovered Ala 5* virus were indeed responsible for its apparent growth advantage compared with the Ala 5 virus, an independent Ala 5* virus was constructed. The alanine insertion in combination with deletion of the three nucleotides that resulted in replacement of M25V26 with isoleucine were all introduced into a new infectious cloned virus to ensure that no other incidental changes elsewhere in the genome were providing a growth advantage to the virus. The second Ala 5* virus recovered from the newly constructed clone was genetically stable and exhibited growth and plaque size/morphology like those of the originally recovered Ala 5* virus (Fig. 7), thus confirming that the new changes were indeed responsible for the growth advantage of the virus.
FIG. 7.
Plaque morphologies of reconstructed and modified alanine mutant viruses. Mouse L2 cells were infected with wild-type (WT) virus or alanine insertion mutant viruses and analyzed by plaque assay. Ala 5* #1 and Ala 5* #2 designate the originally recovered and reconstructed viruses, respectively. Ala 3* and Ala 7* were constructed with the originally inserted alanine residues (Ala 3 and Ala 7) plus removal of a upstream amino acid to restore the wild-type transmembrane helical pitch.
To further explore the idea that restoration of the pitch of the transmembrane α-helix provides an advantage for the virus, we constructed two additional viruses. Ala 3* and Ala 7* were constructed. The original alanine insertion clones were modified by site-directed deletion of one amino acid carboxy-terminal of the insertions to restore the pitch of the helix (Fig. 6B). Residues A21 and A35 were deleted from Ala 3 and Ala 7, respectively (Fig. 1). The infectious clones for both viruses were assembled in parallel with the second Ala 5* virus described above and wild-type virus, thus controlling for the possibility that additional changes within the genome likely account for the observed phenotype of the recovered viruses. Both viruses were genetically stable. The recovered Ala 7* virus exhibited a plaque phenotype like that of the wild-type virus (Fig. 7). The recovered Ala 3* virus had plaques larger than those of the Ala 3 virus but smaller than those of the wild-type virus that was constructed in parallel (Fig. 7). Altogether, these results strongly suggest that the integrity of the helix is important.
Disruption of the E transmembrane domain affects virus assembly and possibly virus release.
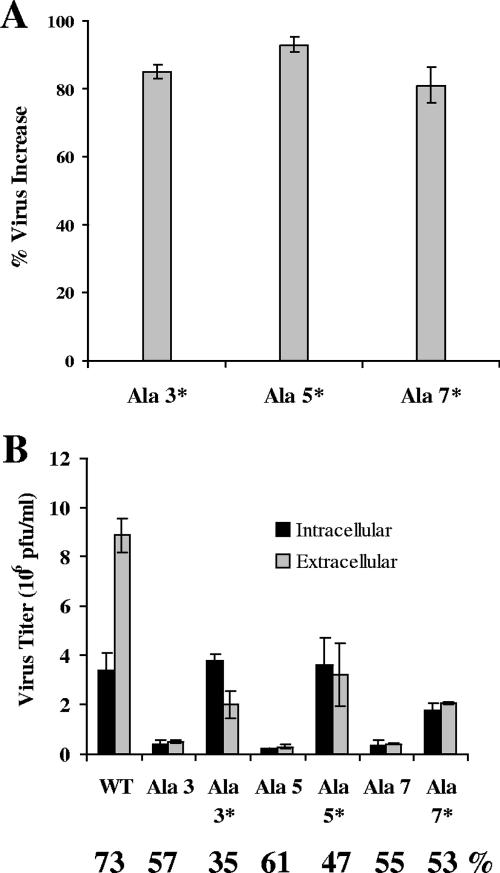
To gain insight into how the alanine insertions might be affecting the function of the E protein, multiple functional analyses were done. Initially we examined the effect on VLP output from coexpression of the mutant E and wild-type M proteins. The amount of VLPs in the medium was decreased with all of the E proteins with the single alanine insertions (data not shown). Second, the amount of total infectious virus produced by each mutant virus was also measured. The medium and cells were harvested separately. Both intracellular and extracellular virus were assayed by plaque titration. At 16 h p.i. Ala 3*, Ala 5*, and Ala 7* each yielded 81 to 93% more total (intracellular plus extracellular) virus than their respective Ala 3, Ala 5, and Ala 7 virus counterparts (Fig. 8A). Thus, the viruses with the restored pitch produced significantly more total virus than their parental virus counterpart with the disrupted helix, even though the virus yields were still 2 to 3 times lower than that of the wild-type virus. This strongly suggests that the transmembrane domain and its integrity are important for virus assembly.
FIG. 8.
Virus production from wild-type and alanine insertion mutant viruses. Mouse 17Cl1 cells were infected with the indicated viruses at an MOI of 0.1. At 16 h p.i., titers of both intracellular and extracellular virus were determined by plaque assay. (A) The percent increase of total (intracellular plus extracellular) virus was determined for Ala 3*, Ala 5*, and Ala 7* in comparison with the original parental virus counterpart (Ala 3, Ala 5, and Ala 7) for each virus. (B) The titers of intracellular and extracellular virus for the wild-type and mutant viruses are shown expressed as 106 PFU/ml. The numbers below the graph indicate the percentage of virus released. The percentage of virus released was calculated by dividing the extracellular virus by the total (intracellular plus extracellular) virus. The error bars represent the standard deviations of three and two independent measurements for the wild-type and mutant viruses, respectively.
We also compared the amounts of infectious intracellular and extracellular virus to determine if release of any of the mutant viruses might be affected. Less total infectious virus was released for all mutant viruses than 73% release for the wild-type virus (Fig. 8B). At 16 h p.i., 57% of the total infectious Ala 3 was present in the medium, whereas only 35% of the Ala 3* virus was released. At the same time point, 61% and 47% of the total infectious Ala 5 and Ala 5*, respectively, were present in the media (Fig. 8B). Comparable amounts, i.e., 55% and 53% of Ala 7 and Ala 7*, respectively, were present in the media. Thus, it appears that while Ala 3* and Ala 5* produced more total virus, less was released into the medium.
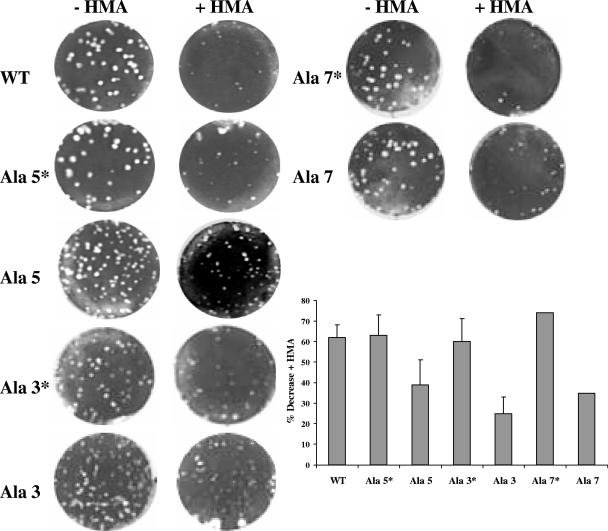
HMA inhibits growth of Ala 3*, Ala 5*, and Ala 7* more than that of their mutant virus counterparts.
MHV E protein was recently shown to form cation-selective ion channels (39). Activity of the channels is inhibited by the Na+ ion channel inhibitor drug HMA. Furthermore, virus growth was inhibited by the drug, whereas no antiviral affect was observed with a recombinant MHV lacking the E protein. Thus, as an additional approach to study the functional importance of the E protein transmembrane domain, we asked if disruption of the domain affects sensitivity to HMA. Cells were infected with wild-type MHV and the Ala 3, Ala 3*, Ala 5, Ala 5*, Ala 7, and Ala 7* mutant viruses in both the absence and presence of HMA. In the presence of HMA the numbers and plaque sizes of the wild-type, Ala 3*, Ala 5*, and Ala 7* viruses were reduced more than those of the parental mutant viruses Ala 3, Ala 5, and Ala 7 (Fig. 9). Based on measurement of the number of plaques, virus replication was reduced by ∼60 to 75%. Replication of the parental viruses was inhibited by only ∼25 to 40%. This suggests that disruption of the hydrophobic domain affects the ion channel activity of the protein, since the drug had a greater effect on the Ala 3*, Ala 5*, and Ala 7* viruses. These results are consistent with the HMA antiviral effect on the wild-type MHV compared with no effect on the MHVΔE virus (39).
FIG. 9.
HMA inhibition of replication of wild-type (WT) and alanine insertion mutant viruses. Mouse L2 cells were infected with wild-type, Ala 5, Ala 5*, Ala 3, ala 3*, Ala 7, and Ala 7* viruses before being overlaid with medium/agarose with and without 20 μM HMA for plaque assay. Three days after infection, cells were stained with crystal violet. Plaques were counted and expressed as as the percentage of virus decrease in the presence of HMA compared with the control without drug treatment. The error bars represent the standard deviations for four independent experiments for wild-type, Ala 5, and 5* viruses and two independent experiments for Ala 3 and Ala 3*. The percent decreases for Ala 7 and Ala 7* are representative of only one measurement.
DISCUSSION
Coronavirus assembly at internal cellular membranes and release of virions from cells are areas of significant interest. The role of the minor virion structural protein E is not understood, but it is clear that the protein plays an important role in the virus life cycle (7, 9, 11, 21, 32). The protein is not universally required for virus assembly. However, in the cases where the protein could be deleted, crippled viruses with low yields were produced. Here, we demonstrate that the MHV A59 E protein hydrophobic domain is important for virus production. Alteration of the MHV E putative transmembrane domain by insertion of single alanine residues results in smaller virus plaques and reduced virus production. Examination of the recovered virus Ala 5* virus, which exhibited a partially restored wild-type-like phenotype, suggested that the predicted transmembrane α-helix structure and sequestration of polar hydrophilic residues on one face of the helix are important for the function of the protein. We tested this idea by building additional viruses with the restored wild-type helical pitch and positioning of the polar residues. These viruses exhibit larger plaques and significantly increased virus production compared with their original parental single-alanine-insertion viruses. This strongly supports that structural integrity of the hydrophobic, presumably transmembrane, domain is important for virus assembly. Interestingly, an additional phenotype was observed with two of these viruses, Ala 3* and Ala 5*. Both viruses exhibited reduced efficiency of virus release even though the virus yields were increased. This suggests that the transmembrane domain plays a role in virus release and that minor changes affect this role. Disruption of the domain also appears to be important, as would be expected, for the presumed ion channel activity of the E protein, since the HMA channel inhibitor reduced infectivity of the wild-type and “star” (Ala 3*, Ala 5*, and Ala 7*) viruses to a greater degree than that of the parental viruses with the more disrupted helix. Our findings significantly expand earlier preliminary observations in our lab, as well as results with infectious bronchitis virus (IBV) reported by Machamer and Youn, which indicated that the E protein transmembrane domain is important for virus biogenesis (25, 41).
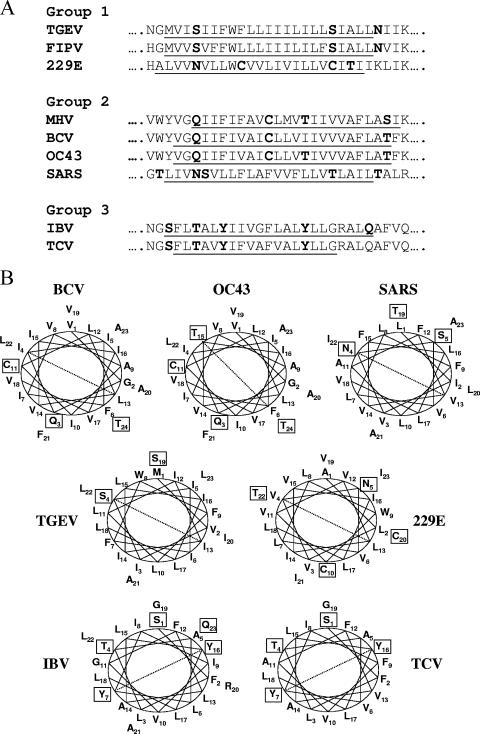
Clearly the most striking difference between the single-insertion mutants and the “star” viruses is the positioning of four hydrophilic polar residues (Q15, C23, T27, and S36) along one face of the helix. Positioning of hydrophilic polar residues on one face of the predicted α-helix of the E proteins of members of the three coronavirus subgroups is conserved, suggesting that this is a functionally relevant feature of the proteins (Fig. 10). Human coronavirus 229E is an exception, with one hydrophilic residue positioned on the opposite face. The structure of the transmembrane domain may be important for virus-virus or virus-host protein interactions which directly affect virus assembly and/or release of assembled virions as they mature through the exocytic pathway via transport vesicles.
FIG. 10.
Alignment and helical wheel analysis of predicted transmembrane domains for representative coronavirus E proteins. (A) Alignment of transmembrane domains for E proteins for viruses representative of the three subgroups. Transmembrane domains were predicted using TMHMM 2.0 (http://www.cbs.dtu.dk). Sequences for porcine transmissible gastroenteritis coronavirus (TGEV) (accession no. AY335549.1), feline infectious bronchitis virus (FIPV) (AY994055.1), human coronavirus 229E (AF304460.1)), MHV A59 (NC001846.1), bovine coronavirus (BCV) (U00735.2), human coronavirus OC43 (AY903460.1), SARS-CoV (AY278741.1), IBV (DQ001338.1), and turkey coronavirus (TCV) (AJ310640.1) were obtained from GenBank. Hydrophilic polar residues are in boldface. (B) Helical wheel plots for E proteins from the representative coronaviruses are shown as described for Fig. 6.
Specific positioning of polar hydrophilic residues on one face may contribute to the overall architecture of the E transmembrane α-helix, which in turn could play a role in the function(s) of the protein. Serine, threonine, and cysteine residues often form intrahelical hydrogen bonds with carbonyl oxygen atoms in the preceding turn of the α-helix (16). These residues exhibit a low turn propensity and thus remain α-helical in a stretch of otherwise hydrophobic residues (30). Certain conformational forms of Ser and Thr can also increase bending of an α-helix, and it has been suggested that changes in the rotamer configuration of these residues may cause conformational changes across the membrane that are important for protein function and signaling (2). Thus, the positioning of T13 and S22 on the same face (Fig. 6) may result in bending of the MHV E transmembrane α-helix, which could be mechanistically important for the protein's function.
Polar residues within transmembrane domains contribute to intermolecular interactions between proteins through interhelical hydrogen bonding (8, 15, 44, 45). A single Gln can mediate helix-helix association (8, 15). Single Ser or Thr residues do not promote oligomerization, but multiples of these residues, possibly in combination with other polar residues, can play a role in driving the process (8). Thus, the presence of polar residues on one side of the MHV E transmembrane helix may promote helix-helix interactions that contribute to oligomerization of E or its interaction with the M protein or possibly host proteins in the membrane.
SARS-CoV and MHV E proteins were recently shown to exhibit viroporin activity (23, 24, 26). Peptides corresponding to the E proteins of SARS-CoV, MHV, human coronavirus 229E, and IVB form cation-specific ion channels in planar lipid bilayers (39, 40). Thus, the coronavirus E proteins are now members of a group of interesting viral proteins that appear to be characteristic of many enveloped RNA viruses (for a review, see reference 14). Viroporins are small, highly hydrophobic proteins with transmembrane domains that generally form amphipathic α-helical structures in the membrane. The proteins assemble in the membrane as oligomers which form hydrophilic pores or channels, with hydrophilic and hydrophobic amino acids positioned inward toward the pore and outward toward the phospholipid bilayer, respectively (14). Viroporins are implicated in playing roles in virus assembly and release, as well as pathogenesis and cytotoxicity.
The significance of viroporin and ion channel activity in coronavirus-infected cells is not known. The E-dependent reduced MHV infectivity caused by drugs that block ion channel activity suggests that it may be important during infection (39). Interestingly, introduction of charged amino acid substitutions in the transmembrane domain of SARS-CoV E protein when express alone disrupted its membrane-permeabilizing activity (24). Our results in the context of the virus are consistent with the idea that the transmembrane domain is important for channel activity, since HMA treatment reduced infectivity of the MHV viruses with the restored wild-type helical pitch to a greater extent than for those with more disrupted helices that would possibly exhibit decreased functional activity. Positioning of the hydrophilic polar residues on one face of the E protein transmembrane α-helix is likely to be important for both channel formation and activity. We do not presently know the oligomeric status of the MHV E protein in virus-infected cells. Molecular dynamic simulations predict that SARS E forms pentamers (36). Intrahelical and/or interhelical interactions through hydrogen bonding as described above must be important for stabilization and organization of the protein, presumably as an oligomer with the polar hydrophilic residues positioned facing the pore. Placement of the polar residues along one face such that they cover ∼180o of the α-helix may be important for gating of the ion channel.
Overall, it appears that the E protein may provide more than one function for the virus. Our results are consistent with a role in virus assembly and possibly release. Recently published parallel studies also point to roles in assembly and release. Cells infected with a recombinant IBV that expresses a chimeric E protein where the transmembrane domain was replaced by that of vesicular stomatitis virus G protein are significantly defective in release of infectious virus (25). Cells infected with the IBV recombinant contain a larger number of virion-filled vacuoles than cells infected with the wild-type virus. A recombinant SARS-CoV lacking the E protein also suggests that assembly and possibly release are affected (9). Fewer mature SARS virions were observed at intracellular sites of assembly, and intracellular vacuoles appear to contain partially assembled viruses.
Recently, the structure of hepatitis C virus ion channel p7 protein was modeled (33). Various computational methods predict a hexameric oligomer with Ser and Thr residues facing the hydrophilic pore. Like coronavirus E proteins, no structural information and limited biochemical data are available for the protein. Understanding the structure/function of the E protein is important for insight into its role in virus assembly and release, as well as the potential role of viroporin and ion channel activity exhibited by the protein. The E protein is clearly an attractive target for antiviral therapy, and thus such studies are significant not only for better understanding of coronaviruses but also for design and development of reagents that target disruption of its function(s).
Acknowledgments
This work was supported by Public Health Service grant AI53704 from the National Institute of Allergy and Infectious Diseases to B.G.H.
We thank members of the Hogue lab for helpful discussions throughout the study and especially Lisa Lopez for help with the immunofluoresence analysis.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Arbely, E., Z. Khattari, G. Brotons, M. Akkawi, T. Salditt, and I. T. Arkin. 2004. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 341:769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballesteros, J. A., X. Deupi, M. Olivella, E. E. Haaksma, and L. Pardo. 2000. Serine and threonine residues bend alpha-helices in the chi(1) = g(−) conformation. Biophys. J. 79:2754-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos, E. C., W. Luytjes, H. V. van der Meulen, H. K. Koerten, and W. J. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, P., B. Persson, H. R. Kaback, and G. von Heijne. 1997. Alanine insertion scanning mutagenesis of lactose permease transmembrane helices. J. Biol. Chem. 272:29566-29571. [DOI] [PubMed] [Google Scholar]
- 5.Brian, D. A., B. G. Hogue, and T. E. Kienzle. 1995. The coronavirus hemagglutinin esterase glycoprotein, p. 165-179. In S. G. Siddell (ed.), The Coronaviridae. Plenum, New York, NY.
- 6.Corse, E., and C. E. Machamer. 2000. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 74:4319-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawson, J. P., J. S. Weinger, and D. M. Engelman. 2002. Motifs of serine and threonine can drive association of transmembrane helices. J. Mol. Biol. 316:799-805. [DOI] [PubMed] [Google Scholar]
- 9.DeDiego, M. L., E. Alvarez, F. Almazan, M. T. Rejas, E. Lamirande, A. Roberts, W. J. Shieh, S. Zaki, K. Subbarao, and L. Enjuanes. 2007. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 81:1701-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A., H. Vennema, and P. J. Rottier. 2000. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 74:4967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleming, J. O., S. A. Stohlman, R. C. Harmon, M. M. Lai, J. A. Frelinger, and L. P. Weiner. 1983. Antigenic relationships of murine coronaviruses: analysis using monoclonal antibodies to JHM (MHV-4) virus. Virology 131:296-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher, T. M., and M. J. Buchmeier. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, M. E., and L. Carrasco. 2003. Viroporins. FEBS Lett. 552:28-34. [DOI] [PubMed] [Google Scholar]
- 15.Gratkowski, H., J. D. Lear, and W. F. Degrado. 2001. Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. USA 98:880-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gray, T. M., and B. W. Matthews. 1984. Intrahelical hydrogen bonding of serine, threonine and cysteine residues within alpha-helices and its relevance to membrane-bound proteins. J. Mol. Biol. 175:75-81. [DOI] [PubMed] [Google Scholar]
- 17.Khattari, Z., G. Brotons, M. Akkawi, E. Arbely, I. T. Arkin, and T. Salditt. 2006. SARS coronavirus E protein in phospholipid bilayers: an X-ray study. Biophys. J. 90:2038-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klumperman, J., J. K. Locker, A. Meijer, M. C. Horzinek, H. J. Geuze, and P. J. Rottier. 1994. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 68:6523-6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 21.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laude, H., and P. S. Masters. 1995. The coronavirus nucleocapsid protein, p. 141-163. In S. G. Siddell (ed.), The Coronaviridae. Plenum, New York, NY.
- 23.Liao, Y., J. Lescar, J. P. Tam, and D. X. Liu. 2004. Expression of SARS-coronavirus envelope protein in Escherichia coli cells alters membrane permeability. Biochem. Biophys. Res. Commun. 325:374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao, Y., Q. Yuan, J. Torres, J. P. Tam, and D. X. Liu. 2006. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 349:264-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machamer, C. E., and S. Youn. 2006. The transmembrane domain of the infectious bronchitis virus E protein is required for efficient virus release. Adv. Exp. Med. Biol. 581:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madan, V., M. J. Garcia, M. A. Sanz, and L. Carrasco. 2005. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 579:3607-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maeda, J., A. Maeda, and S. Makino. 1999. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology 263:265-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda, J., J. F. Repass, A. Maeda, and S. Makino. 2001. Membrane topology of coronavirus E protein. Virology 281:163-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mingarro, I., P. Whitley, M. A. Lemmon, and G. von Heijne. 1996. Ala-insertion scanning mutagenesis of the glycophorin A transmembrane helix: a rapid way to map helix-helix interactions in integral membrane proteins. Protein Sci. 5:1339-1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monne, M., M. Hermansson, and G. von Heijne. 1999. A turn propensity scale for transmembrane helices. J. Mol. Biol. 288:141-145. [DOI] [PubMed] [Google Scholar]
- 31.Opstelten, D. J., M. J. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patargias, G., N. Zitzmann, R. Dwek, and W. B. Fischer. 2006. Protein-protein interactions: modeling the hepatitis C virus ion channel p7. J. Med. Chem. 49:648-655. [DOI] [PubMed] [Google Scholar]
- 34.Raamsman, M. J., J. K. Locker, A. de Hooge, A. A. de Vries, G. Griffiths, H. Vennema, and P. J. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tooze, J., and S. A. Tooze. 1985. Infection of AtT20 murine pituitary tumour cells by mouse hepatitis virus strain A59: virus budding is restricted to the Golgi region. Eur. J. Cell Biol. 37:203-212. [PubMed] [Google Scholar]
- 36.Torres, J., K. Parthasarathy, X. Lin, R. Saravanan, A. Kukol, and D. X. Liu. 2006. Model of a putative pore: the pentameric alpha-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 91:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verma, S., V. Bednar, A. Blount, and B. G. Hogue. 2006. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 80:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, L., P. Gage, and G. Ewart. 2006. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353:294-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, L., C. McKinlay, P. Gage, and G. Ewart. 2004. SARS coronavirus E protein forms cation-selective ion channels. Virology 330:322-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye, Y., and B. G. Hogue. 2006. Role of mouse hepatitis coronavirus envelope protein transmembrane domain. Adv. Exp. Med. Biol. 581:187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan, Q., Y. Liao, J. Torres, J. P. Tam, and D. X. Liu. 2006. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 580:3192-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou, F. X., M. J. Cocco, W. P. Russ, A. T. Brunger, and D. M. Engelman. 2000. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 7:154-160. [DOI] [PubMed] [Google Scholar]
- 45.Zhou, F. X., H. J. Merianos, A. T. Brunger, and D. M. Engelman. 2001. Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. USA 98:2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]