Abstract
Curcumin (diferuloylmethane), a natural polyphenolic compound extracted from the spice turmeric, has been reported to have anti-inflammatory, antioxidant, and antiproliferative properties by modulating multiple cellular machineries. It inhibits several intracellular signaling pathways, including the mitogen-activated protein kinases (MAPKs), casein kinase II (CKII), and the COP9 signalosome (CSN), in various cell types. It has also been recently demonstrated that exposure to curcumin leads to the dysregulation of the ubiquitin-proteasome system (UPS). Coxsackievirus infection is associated with various diseases, including myocarditis and dilated cardiomyopathy. In searching for new antiviral agents against coxsackievirus, we found that treatment with curcumin significantly reduced viral RNA expression, protein synthesis, and virus titer and protected cells from virus-induced cytopathic effect and apoptosis. We further demonstrated that reduction of viral infection by curcumin was unlikely due to inhibition of CVB3 binding to its receptors or CVB3-induced activation of MAPKs. Moreover, gene silencing of CKII and Jab1, a component of CSN, by small interfering RNAs did not inhibit the replication of coxsackievirus, suggesting that the antiviral action of curcumin is independent of these pathways. Finally, we showed that curcumin treatment reduced both the 20S proteasome proteolytic activities and the cellular deubiquitinating activities, leading to increased accumulation of ubiquitinated proteins and decreased protein levels of free ubiquitin. We have recently demonstrated that the UPS-mediated protein degradation and/or modification plays a critical role in the regulation of coxsackievirus replication. Thus, our results suggest an important antiviral effect of curcumin wherein it potently inhibits coxsackievirus replication through dysregulation of the UPS.
Group B3 coxsackievirus (CVB3) is a major human pathogen that causes meningitis and myocarditis (10, 14). Despite extensive efforts, no specific and approved treatment has been developed that is effective against CVB3-induced diseases. New therapeutic options and antiviral drugs need to be explored.
We and others have previously demonstrated that CVB3 employs strategies similar to those of other viruses, such as host signaling manipulation and host protein regulation, to facilitate its own replication. Upon CVB3 infection, several intracellular signaling pathways are activated, including the extracellular signal-regulated kinases 1 and 2 (ERK1/2) (15, 19), c-Jun N-terminal kinase (JNK) (12, 25), p38 mitogen-activated protein kinase (MAPK) (25), and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (PKB) pathways (8, 38). Activation of these pathways is required for CVB3 infectivity. We have also demonstrated that CVB3 infection promotes host protein degradation and that proteasome inhibition reduces CVB3 replication, suggesting a critical role for the ubiquitin-proteasome system (UPS) in the viral life cycle (16).
The UPS is a major intracellular pathway for extralysosomal protein degradation (1, 7, 22, 35). There are two coupled steps involved in protein degradation: (i) covalent attachment of ubiquitin to the target protein substrate and (ii) degradation of the polyubiquitinated protein by the proteasome with the release of recyclable ubiquitin. Ubiquitin is a highly conserved 76-amino-acid protein that is activated in an ATP-dependent process by the ubiquitin-activating enzyme (E1) and subsequently transferred to a ubiquitin-conjugating enzyme (E2). Final transfer of ubiquitin to the target protein requires ubiquitin-protein ligase (E3). After several rounds of ubiquitination, a polyubiquitin chain is formed (21, 32). The ubiquitinated substrate is recognized and subsequently degraded by the 26S proteasome, and ubiquitin is recycled via the action of deubiquitinating enzymes (DUBs). The 26S proteasome consists of a central catalytic core, the 20S proteasome, and two regulatory 19S complexes. Three distinct proteolytic activities of the 20S proteasome have been reported: trypsin-like, chymotrypsin-like, and peptidylglutamyl-peptide hydrolase (PDGH) activities.
Curcumin (diferuloylmethane) is a natural polyphenolic compound extracted from the spice turmeric (Curcuma longa). Recent studies have shown a promising potential for curcumin in the treatment of various forms of cancer and cardiovascular diseases (1, 7, 22, 35). Curcumin has been reported to have profound anti-inflammatory, antioxidant, and antiproliferative properties, acting through modulation of multiple cellular machineries. It inhibits several intracellular signaling pathways, including the MAPKs, PI3K/PKB, and nuclear factor kappa B (NF-κB) (4, 6, 28). It has also been well documented that curcumin is an inhibitor of the COP9 signalosome (CSN) through inhibition of its associated kinases, such as casein kinase II (CKII) and protein kinase D (PKD) (6, 31). In addition, recent evidence has demonstrated that exposure to curcumin leads to dysregulation of the UPS (9, 18).
Based on our previous knowledge of the regulation of CVB3 replication and the known biological properties of curcumin, we propose that curcumin may be an attractive agent for the treatment of CVB3 infection. In this study, we investigated the effects of curcumin treatment on CVB3 infectivity and explored the potential mechanisms of its action. We found that curcumin treatment effectively reduced CVB3 replication. Further, we demonstrated that curcumin inhibition of CVB3 replication is independent of its ability to interfere with intracellular signaling pathways, including MAPKs, CKII, and Jab1, and instead is likely mediated by curcumin-induced dysregulation of the UPS.
MATERIALS AND METHODS
Cell culture and materials.
HeLa cells were obtained from the American Type Culture Collection and grown in complete medium (Dulbecco's modified Eagle's medium [DMEM] supplemented with 10% heat-inactivated newborn calf serum).
The monoclonal anti-β-actin antibody was purchased from Sigma-Aldrich. The monoclonal anti-VP1 antibody was obtained from DakoCytomation. The Jab1 small interfering RNA (siRNA); the CKIIβ siRNA; scramble control siRNA; the polyclonal anti-JNK antibody; the monoclonal anti-phospho-JNK, anti-caspase 3, anti-CKIIβ, and anti-Jab1 antibodies; and the horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology. The polyclonal anti-ubiquitin antibody and curcumin were obtained from Calbiochem. The CKIIα siRNA and the polyclonal anti-CKIIα antibody were from Upstate. The polyclonal anti-p38 and anti-ERK1/2 antibodies and the monoclonal anti-phospho-p38 and anti-phospho-ERK1/2 antibodies were purchased from Cell Signaling. The purified 20S proteasome (human) was from Biomol.
Virus infection.
HeLa cells were infected at a multiplicity of infection of 10 with CVB3 (Kandolf strain) or sham infected with phosphate-buffered saline (PBS) for 1 h in serum-free DMEM. The cells were then washed with PBS and cultured in serum-free medium for various times until harvest. For inhibition experiments, HeLa cells were infected with CVB3 in the presence or absence of various inhibitors or vehicle (dimethyl sulfoxide [DMSO]) for 1 h, washed with PBS, and then incubated with fresh DMEM containing different concentrations of inhibitors unless otherwise specified in the figure legend.
Plaque assay.
The virus titer in the cell supernatant was determined by an agar overlay plaque assay in triplicate as described previously (16). In brief, the supernatant from CVB3-infected HeLa cells was serially 10-fold diluted and overlaid on a 90 to 95% confluent monolayer of HeLa cells. After 1 h of incubation, the HeLa cells were washed with PBS, followed by overlaying them with 2 ml of complete medium containing 0.75% agar in each well. The cells were incubated at 37°C for 72 h and then fixed with Carnoy's fixative (75% ethanol-25% acetic acid) and stained with 1% crystal violet. Plaques were counted, and the viral titer was calculated as PFU per milliliter.
In situ hybridization.
HeLa cells were grown on two-well chamber slides. Subconfluent HeLa cells were infected with CVB3 in the presence or absence of curcumin for 6 h. After two washes with PBS, the cells were fixed with 10% formalin buffer for 15 min and then air dried at room temperature. The slides were subjected to in situ hybridization for the detection of the sense strand coxsackievirus RNA as previously described (16). In brief, cells were hybridized using digoxigenin-labeled CVB3 antisense riboprobes, which were prepared from the full-length CVB3 cDNA by in vitro transcription. Hybridized riboprobes were detected using an alkaline phosphatase-conjugated antidigoxigenin antibody (Roche) and a color substrate, Vector Red (Vector Laboratories). Cell nuclei were counterstained with hematoxylin.
Western blot analysis.
After treatment, the cells were incubated with lysis buffer (50 mM pyrophosphate, 50 mM NaF, 50 mM NaCl, 5 mM EDTA, 5 mM EGTA, 100 μM Na3VO4, 10 mM HEPES, pH 7.4, 0.1% Triton X-100, 10 μg of leupeptin/ml, and 1 mM phenylmethylsulfonyl fluoride) on ice for 10 min. Cell lysates were then harvested by scraping and were centrifuged at 12,000 × g for 10 min at 4°C. The protein concentration was determined by the Bradford assay (Bio-Rad). Equal amounts of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (Amersham). The membranes were blocked for 1 h with 5% nonfat dry milk solution containing 0.1% Tween 20. The blots were then incubated for 1 h with the primary antibody, followed by incubation for another hour with a secondary antibody. Immunoreactive bands were visualized by enhanced chemiluminescence (Pierce). For examination of protein-ubiquitin conjugates and free ubiquitin, membranes were heat activated by autoclaving them at 120°C for 30 min prior to blocking them with nonfat dry milk solution in order to increase antigenic-site recognition.
20S proteasome activity assay.
The cell lysates were prepared as described above but in the absence of protease inhibitors. Fresh cellular extracts and purified human 20S proteasomes were used to measure 20S proteasome activities as described previously (16). In brief, 20 micrograms of cytoplasmic protein was added to an assay buffer (20 mM Tris-HCl [pH 8.0], 1 mM ATP, and 2 mM MgCl2) in the presence of 75 μM synthetic fluorogenic substrate to a final volume of 100 μl. Alternatively, 100 nanograms of purified 20S proteasome was preincubated with DMSO, curcumin, or MG132 in the assay buffer at 37°C for 30 min before synthetic fluorogenic substrates were added. The fluorogenic substrates Bz-Val-Gly-Arg-AMC (Biomol), Suc-Leu-Leu-Val-Tyr-AMC (Calbiochem), and Z-Leu-Leu-Glu-AMC (Calbiochem) were used to determine the 20S proteasome trypsin-like, chymotrypsin-like, and PDGH activities, respectively. The tubes were incubated at 30°C for 1 h, and the fluorescence product AMC in the supernatant was measured at a 465-nm emission wavelength using a fluorometer.
Deubiquitinating enzyme activity assay.
Cell lysates were prepared as described previously (2). Briefly, HeLa cells were harvested in suspension buffer (20 mM HEPES-K, pH 7.5, 10 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride, 0.5% NP-40, DNase I [50 μg/ml], 4% glycerol) and incubated on ice for 20 min. The suspension was centrifuged at 100,000 × g for 10 min at 4°C, and the supernatant was collected. Ten micrograms of supernatant was added to an assay buffer (50 mM HEPES-NaOH, pH 7.8, 0.5 mM EDTA, 1 mM dithiothreitol, 0.1 mg/ml ovalbumin) and incubated for 1 h at 37°C in the presence of 1 μM ubiquitin-AMC substrate (Biomol). The fluorescence of the liberated AMC was measured at a 465-nm emission wavelength using a fluorometer.
siRNA transfection.
HeLa cells were grown to 50% confluence and then transiently transfected with 200 nM Jab1, CKIIα, or CKIIβ siRNA using oligofectamine according to the manufacturer's instructions (Invitrogen). A scrambled siRNA (200 nM) was used as a control. The silencing efficiency was detected by Western blot analyses using the anti-Jab1, anti-CKIIα, or anti-CKIIβ antibody, respectively. Twenty-four hours after transfection, cells were infected with CVB3 as described above.
Virus binding assay.
Virus was radiolabeled and purified as previously described (34). Briefly, HeLa cells were infected with CVB3 in methionine-free medium for 3 h and then incubated with medium containing 19.4 MBq/ml (525 μCi/ml) of Tran35S-label (MP Biomedicals) for an additional 8 h. The HeLa cells and medium were harvested and then frozen and thawed three times. After centrifugation, the pellet containing 35S-labeled CVB3 was resuspended in serum-free DMEM, the virus titer was measured by plaque assay, and aliquots were stored at −80°C.
For the virus binding assay, HeLa cells were incubated with 35S-labeled CVB3 for 1 h. The cells were washed thoroughly with PBS and lysed with 2% sodium dodecyl sulfate. The amounts of 35S-labeled CVB3 in the samples were measured using a 1217 Rackbeta liquid scintillation counter (LKB Wallac).
RESULTS
Curcumin inhibits replication of CVB3.
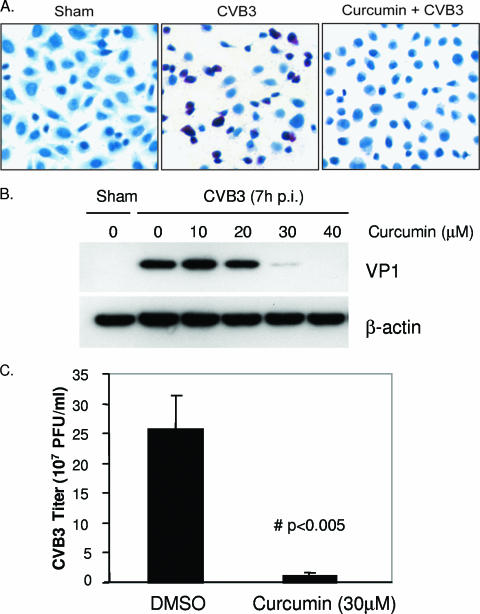
Curcumin is a natural compound that has been reported to have antioxidant, anti-inflammatory, and antiproliferative properties (1, 7, 22). In this study, we found that curcumin was a potent inhibitor of CVB3 replication. Curcumin treatment reduced expression of CVB3 viral RNA in infected HeLa cells, as measured by in situ hybridization (Fig. 1A). Curcumin treatment also decreased synthesis of the viral capsid protein VP1 in a dose-dependent manner (Fig. 1B). In addition, the CVB3 viral titer in the medium was reduced by more than 20-fold upon curcumin treatment (Fig. 1C).
FIG. 1.
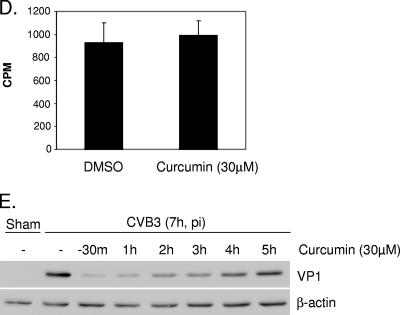
Curcumin decreases replication of CVB3. (A) HeLa cells were sham infected with PBS or infected with CVB3 in the absence or presence of curcumin (30 μM). Six hours postinfection, viral RNA was detected by in situ hybridization using antisense riboprobes for CVB3 (red). Cell nuclei were counterstained with hematoxylin (blue). The data represent the results from two independent experiments. (B) HeLa cells were infected and treated with various concentrations of curcumin as for panel A. Seven hours postinfection, CVB3 viral-protein synthesis was examined using a monoclonal antibody that recognizes the CVB3 capsid protein VP1. To verify equal loading, the same blots were stripped and reprobed for β-actin. The data represent results from three independent experiments. (C) HeLa cells were infected and treated with curcumin as for panel A. Nine hours postinfection (p.i.), medium was collected and CVB3 progeny were measured by plaque assay. The data are means plus standard deviations (SD); n = 3; #, P < 0.005 compared to DMSO treatment. (D) HeLa cells were infected with 35S-labeled CVB3 for 1 h. Cell lysates were collected and measured for radioactivity using a scintillation counter (mean ± SD; n = 6). CPM, counts per minute. (E) HeLa cells were infected with CVB3, and curcumin (30 μM) was added to the cells at different times as indicated. Seven hours postinfection, cell lysates were collected and expression levels of VP1 and β-actin were examined.
In trying to understand which step(s) of the CVB3 life cycle was disrupted by curcumin, we performed a virus binding assay using 35S-labeled CVB3. As shown in Fig. 1D, treatment with curcumin did not reduce virus binding to its receptors. We also examined the expression of viral protein in infected cells that were treated with curcumin at various times, either pre- or postinfection. We found that the reduction of viral-protein expression was more profound when curcumin was added earlier in the infectious cycle than when it was added later (Fig. 1E), suggesting that curcumin interfered with early steps in the CVB3 life cycle. This finding is consistent with our other observation that curcumin treatment significantly reduced the replication of the CVB3 genome (Fig. 1A), an early step in the CVB3 life cycle.
Selection of 10 to 40 μM of curcumin for these studies was based on the observed low toxicity, but also the dose-dependent antiviral effects, of the compound under our experimental conditions. Treatment with higher concentrations of curcumin (>40 μM) led to cell toxicity, as determined by cell viability assays and morphological assessment (data not shown).
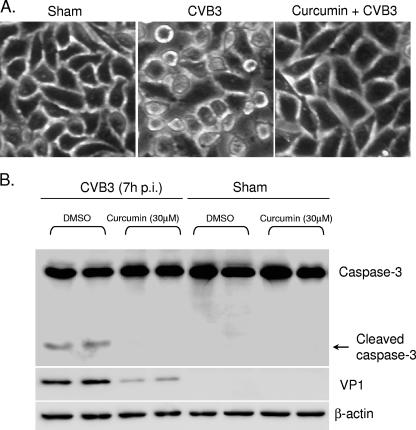
CVB3 infection leads to cytopathic effect (CPE) and apoptosis of infected cells (5, 16, 37). To investigate whether the antiviral activities of curcumin are associated with a reduction of CPE and apoptotic events, we compared the morphologies of infected cells in the presence or absence of curcumin. We found that curcumin treatment attenuated CVB3-induced CPE in infected cells (Fig. 2A). We further demonstrated that exposure to curcumin prevented the cleavage of caspase 3, a hallmark of apoptosis, in infected cells, which was accompanied by a reduction of viral protein synthesis (Fig. 2B). It was noted that, without viral infection, curcumin (30 μM) treatment did not induce caspase 3 cleavage under the same experimental conditions described in the presence of viral infection (Fig. 2B). Several previous studies have reported that curcumin treatment can activate caspase 3 (9, 30, 39). One explanation for the disparity is that in the present study we treated the cells with curcumin for only 7 h, whereas cells were treated for at least 16 h in previous reports. Indeed, we found that prolonged exposure to 30 μM curcumin did induce apoptosis in HeLa cells (data not shown). Thus, it is unlikely that curcumin has a direct inhibitory effect on apoptosis. We therefore conclude that curcumin inhibits replication of CVB3 and subsequently reduces CVB3-induced CPE and apoptosis.
FIG. 2.
Curcumin reduces CVB3-induced cytopathic effect and apoptosis. HeLa cells were sham infected with PBS or infected with CVB3 in the absence or presence of curcumin (30 μM). (A) Seven hours postinfection, the morphologies of infected cells were examined under a phase-contrast microscope. Representative images from three independent experiments are presented. (B) Cell lysates were prepared 7 h postinfection (p.i.), and cleavage of caspase 3 was examined using a monoclonal anti-capase 3 antibody. β-Actin was examined to show equal protein loading. The data represent the results from two independent experiments.
Curcumin inhibits CVB3 replication independently of the activation of MAPKs.
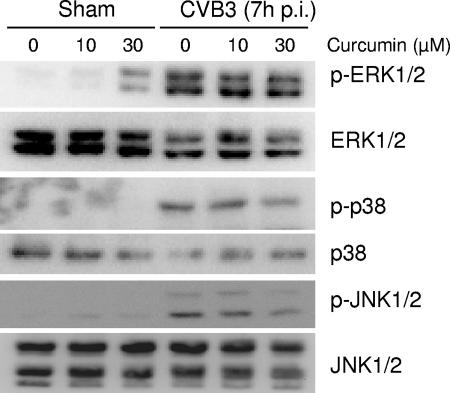
CVB3 infection results in activation of MAPKs, including ERK1/2, p38 MAPK, and JNK1/2, and activation of these kinases plays critical roles in various steps of the CVB3 life cycle (15, 19, 25). Curcumin has been reported to be a potent MAPK inhibitor in various models (27, 28). To investigate whether the antiviral activity of curcumin depends on its role in the modulation of MAPK activation, we examined the phosphorylation status of MAPKs. As shown in Fig. 3, in the absence of viral infection, treatment with curcumin did not induce MAPK phosphorylation. Following CVB3 infection, phosphorylation of ERK1/2, p38, and JNK1/2 was increased at 7 h postinfection. However, no apparent changes in the phosphorylation levels of MAPKs were observed after the addition of curcumin to infected cells, suggesting that it is unlikely that curcumin inhibits replication of CVB3 through the regulation of MAPK activation.
FIG. 3.
Curcumin does not inhibit replication of CVB3 through MAPK pathways. HeLa cells were infected with CVB3 in the absence or presence of curcumin (10 and 30 μM). Seven hours postinfection (p.i.), the phosphorylation status of MAPKs was examined using anti-phospho-ERK1/2, anti-phospho-p38, and anti-phospho-JNK1/2 antibodies, respectively. Expression levels of total ERK1/2, p38, and JNK1/2 were examined as the loading controls. The data represent the results from two independent experiments.
We also found that curcumin treatment did not inhibit CVB3-induced PKB phosphorylation in our model (data not shown), indicating that curcumin suppression of CVB3 is also not via the PI3K/PKB pathway.
Suppression of CVB3 replication by curcumin is independent of its inhibitory effect on CSN and associated CKII.
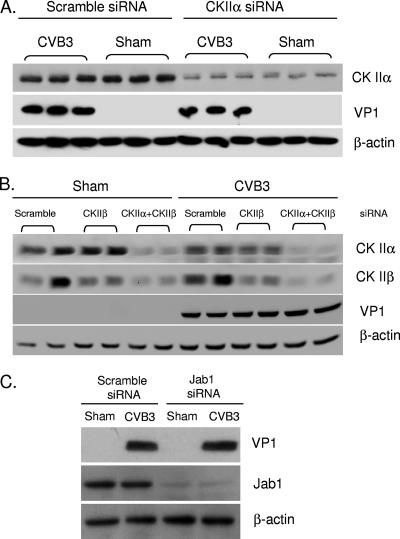
It has been well documented that curcumin is an inhibitor of the COP9 signalosome through the inhibition of its associated kinases, such as CKII and PKD (6, 31). To determine whether curcumin suppresses the replication of CVB3 by inhibiting CSN, we used various siRNAs to gene silence the expression of CSN and CKII. We first examined whether CKII was necessary for the replication of CVB3. CKII, a serine/threonine kinase that phosphorylates acidic protein, such as casein, has a tetrameric α2β2 structure with two α subunits, α and α′, and one common β subunit. We found that knockdown of the α subunit (Fig. 4A) or both the α and β subunits (Fig. 4B) of CKII did not reduce synthesis of viral protein in infected cells, suggesting that the activity of CKII was not essential for the replication of CVB3 and thus was not likely the cellular target of curcumin responsible for viral inhibition.
FIG. 4.
Curcumin probably does not inhibit replication of CVB3 through CKII and CSN. HeLa cells were transiently transfected with specific siRNAs to knock down the expression of CKIIα (A), CKIIα/β (B), or Jab1 (C), respectively, for 24 h. Seven hours after CVB3 infection, viral protein expression (VP1) and levels of CKIIα, CKIIβ, and Jab1 were determined by Western blotting. β-Actin was examined to verify equal loading. The data represent the results from three independent experiments.
Since CKII is not the only kinase associated with CSN, we also examined the role of CSN in CVB3 replication by gene silencing Jab1, a subunit of CSN, using siRNA (Fig. 4C). The finding that VP1 expression after knockdown of Jab1 was as robust as that in control cells suggested that CSN may not be the critical target for curcumin to execute its potent antiviral activities.
Curcumin likely inhibits replication of CVB3 through the dysregulation of the UPS.
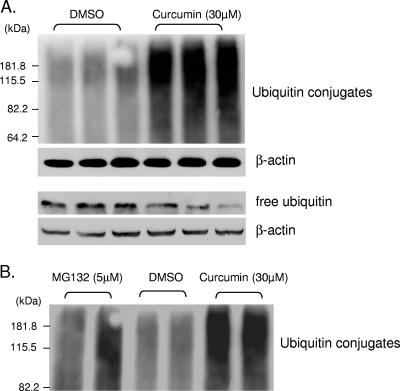
Several recent studies have shown that curcumin is capable of modulating the function of the ubiquitin-proteasome system (9, 18). We have previously demonstrated that the UPS plays a critical role in the regulation of coxsackievirus replication (16, 26). To explore the potential roles of the UPS in mediating the antiviral activities of curcumin, we examined the effect of curcumin on the UPS. As shown in Fig. 5A, curcumin treatment resulted in an increased accumulation of protein-ubiquitin conjugates, which was accompanied by a decrease in the free ubiquitin level. Interestingly, we found that accumulation of ubiquitin conjugates in curcumin-treated cells far exceeded that in MG132-incubated cells (Fig. 5B).
FIG. 5.
Curcumin promotes protein polyubiquitination and decreases free ubiquitin levels. (A) HeLa cells were treated with DMSO or curcumin (30 μM) for 5 h. Protein polyubiquitination and free ubiquitin levels were examined by Western blot analysis using a polyclonal anti-ubiquitin antibody. β-Actin was examined to verify equal loading. (B) HeLa cells were incubated with either DMSO, MG132 (5 μM), or curcumin (30 μM) for 5 h. Ubiquitin conjugates were determined as for panel A. Multiple lanes represent the results from independent experiments.
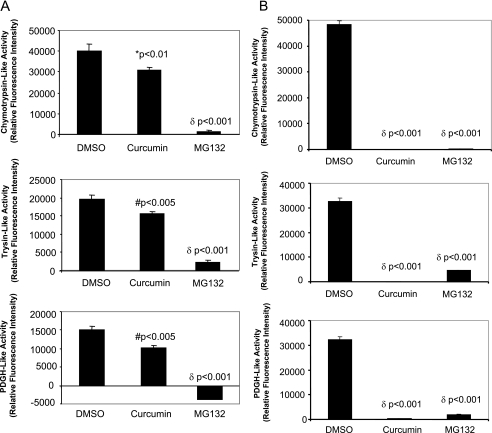
To understand how curcumin manipulates the UPS, we examined proteasome activities in curcumin-treated cells. We have previously demonstrated that proteasome activities were unchanged after CVB3 infection of HeLa cells and murine cardiomyocytes (16, 17). Thus, it is unlikely that virus infection affects the role of curcumin in the regulation of UPS. Here, we measured the proteasome activities after curcumin treatment in the absence of viral infection. As shown in Fig. 6A, curcumin treatment reduced chymotrypsin-like, trypsin-like, and PDGH-like activities of the 20S proteasome by nearly 30%. However the inhibition was not as profound as that with MG132 treatment (5 μM). Considering that synthetic fluorogenic substrates can be cleaved not only by the 20S proteasome, but also by other proteases present in the cell lysates, we then investigated the direct effects of curcumin on proteasome proteolytic activities using purified 20S proteasome. As shown in Fig. 6B, curcumin dramatically inhibited the chymotrypsin-like, trypsin-like, and PDGH-like hydrolytic activities of purified 20S proteasome by more than 90%. Interestingly, the inhibitory effect of curcumin (30 μM) on proteasome activities was found to be greater than that by MG132 (5 μM). This result is consistent with our observation in Fig. 5B, which shows that curcumin treatment induced a larger accumulation of ubiquitin conjugates than MG132 treatment.
FIG. 6.
Curcumin inhibits 20S proteasome activities. (A) HeLa cells were treated with DMSO, curcumin (30 μM), or MG132 (5 μM) for 5 h. The activities of 20S proteasome, including trypsin-like, chymotrypsin-like, and PDGH activities, in the cell lysates were measured as described in Materials and Methods. (B) Purified 20S proteasome (100 ng) was used to examine the effects of curcumin (30 μM) and MG132 (5 μM) on proteasome hydrolytic activities. The data are means plus standard deviations; n = 3. *, P < 0.01; #, P < 0.005; δ, P < 0.001 compared to DMSO treatment.
MG132 has been reported to be a potent yet nonspecific inhibitor of the 20S proteasome. It also targets other cellular proteases, such as cathepsins and calpains (13). The greater reduction of 20S proteasome activities by MG132 (5 μM) than by curcumin (30 μM) in cellular extracts (Fig. 6A) is likely due to the nonspecific inhibitory effects of MG132 on other proteases.
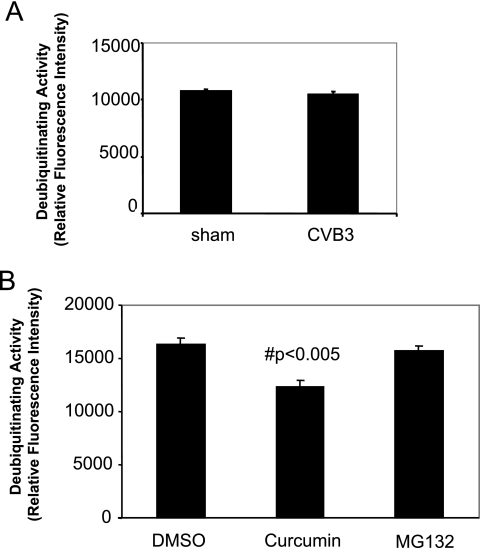
Protein ubiquitination can also be regulated by DUBs that specifically cleave ubiquitin from ubiquitin-conjugated protein substrates (33). We found that CVB3 infection did not change the DUB activities (Fig. 7A). However, treatment with curcumin significantly reduced the activities of DUBs by nearly 30% under our experimental conditions (Fig. 7B), which is consistent with a previous report that curcumin is an inhibitor of ubiquitin isopeptidases (18). The combined inhibitory effects on 20S proteasome activities and on DUB activities may explain the marked accumulation of protein-ubiquitin conjugates in curcumin-treated cells. The reduction of the available pool of free ubiquitin in infected cells upon curcumin treatment may thus limit replication of CVB3 in a fashion similar to what we have previously suggested using proteasome inhibitors.
FIG. 7.
Curcumin reduces cellular deubiquitinating activities. (A) HeLa cells were sham infected with PBS or infected with CVB3 for 6 h. Cell lysates were prepared, and cellular deubiquitinating activities were measured using a fluorogenic substrate, ubiquitin-AMC, as described in Materials and Methods. (B) HeLa cells were treated with DMSO, curcumin (30 μM), or MG132 (5 μM) for 5 h, and cellular deubiquitinating activities were measured. The data are means plus standard deviations; n = 3. #, P < 0.005 compared to DMSO treatment.
DISCUSSION
In the present study, we have provided evidence that curcumin is a novel inhibitor of CVB3 replication. We showed that curcumin strongly reduces viral-RNA expression, protein synthesis, and virus titer but does not interfere with CVB3 binding to its receptors. We further demonstrated that inhibition of CVB3 by curcumin is likely attributable to the impairment of UPS function. Other potential targets of curcumin, such as MAPKs, CKII, and CSN, are unlikely to be involved in its antiviral mechanisms against CVB3. Several novelties compared to previous research are presented in this paper. First, to our knowledge, this is the first report showing that curcumin can inhibit coxsackievirus replication. Although curcumin has been previously implicated in the treatment of cancer and cardiovascular diseases, the antiviral mechanism of this naturally occurring compound gives us a unique opportunity to design more specific molecular medicines against coxsackievirus infection. Second, many biological activities have been associated with the function of curcumin; however, we provide the first evidence that inhibition of 20S proteasome and isopeptidase activities contributes to its anticoxsackieviral properties. Third, the CSN is important in the regulation of several host proteins that are substrates of the 26S proteasome. Our findings that exclude a role of the CSN pathway in coxsackievirus replication further narrow the possible UPS substrates relevant to controlling viral replication. Finally, we demonstrate for the first time that curcumin inhibits 20S core proteasome activities directly and that such inhibition is at least as potent as that of MG132.
There is increasing evidence that the UPS, the major protein degradation pathway, plays critical roles in various steps of viral life cycles, including virus entry, viral replication, budding and progeny release, and latent-virus reactivation (20, 24, 29, 36, 40). We have previously shown that CVB3 infection results in degradation of several host proteins, such as cyclin D1, p53, and β-catenin, through the UPS (17, 26, 37). Specific inhibitors of the proteasome significantly reduce the replication of CVB3, possibly by decreasing free ubiquitin levels, thereby preventing CVB3 from utilizing the host protein ubiquitination machinery for its own benefit. Indeed, we have found that protein ubiquitination is increased after CVB3 infection and that depletion of the cellular pool of ubiquitin by siRNA significantly inhibits CVB3 replication in infected cells (unpublished data). In this study, we reported that curcumin dysregulates the UPS function through inhibition of 20S proteasome activities and isopeptidase activities, consistent with previous observations (9, 18). Dysregulation of the UPS, especially diminution of the free ubiquitin levels by curcumin, may largely account for its strong anticoxsackieviral activities.
The CSN is a multiprotein complex composed of eight subunits. There is a striking homology between the CSN and the 19S regulatory complex of the proteasome, suggesting that they have a common ancestor and possibly possess similar biochemical properties. The CSN has been demonstrated to regulate the degradation of several substrates of the 26S proteasome, such as p53 and c-Jun, through phosphorylation by its associated kinases, CKII and PKD (3, 31). Curcumin has been reported to be an inhibitor of the CSN, working through inhibition of the activities of its associated kinases (3). Treatment with curcumin significantly increases the expression levels of p53 (3). We have previously shown that CVB3 infection results in the degradation of p53 (17). Here, we found that inhibition of the CSN by curcumin or Jab1 siRNA partially restores the expression of p53 in infected cells (data not shown). However, only curcumin, but not Jab1 siRNA, shows strong antiviral activity, suggesting that curcumin inhibits CVB3 replication through mechanisms independent of its effects on CSN. The finding that siRNAs for α and β subunits of CKII have no effect on the expression of CVB3 protein further supports the view that the CSN is not the primary target for curcumin inhibition of CVB3 replication.
Several other biological actions of curcumin have been reported previously, such as inhibition of MAPKs and protein kinase C (PKC) (11, 23). We and others have shown that activation of MAPKs following CVB3 infection plays a critical role in the regulation of viral replication (15, 19, 25). In this study, we explored whether inhibition of MAPKs contributed to the antiviral activities of curcumin and found that MAPKs were activated in infected cells regardless of the presence or absence of curcumin, excluding such a possibility. We did not examine the correlation between PKC activation and curcumin inhibition of CVB3 in the present study, since we have previously found that PKC inhibitors did not reduce the replication of CVB3 (unpublished results). The importance of other physiological targets of curcumin, such as NF-κB, is under investigation in our laboratory.
In conclusion, we have shown for the first time that curcumin is a potent inhibitor of the replication of CVB3. Although many biological properties have been reported for curcumin, the dysregulation of the UPS likely contributes to its antiviral properties. Further, the integrity of CSN is unlikely to be necessary for the life cycle of CVB3, since gene silencing of either a CSN-associated kinase or subunits of CSN does not prevent CVB3 replication in infected HeLa cells.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) (to H.L.), the Heart and Stroke Foundation of Canada (HSFC) (to H.L. and B.M.M.), and the Canada Foundation for Innovation (to H.L.). X.S. is the recipient of a CIHR/HSFC IMPACT Post-Doctoral Fellowship, a CIHR Michael Smith Fellowship, and an HSFC Research Fellowship. Y.W. is supported by a fellowship from the China Scholarship Council. H.L. is the recipient of a New Investigator of the CIHR/St. Paul's Hospital Foundation Award and a Scholar of the Michael Smith Foundation for Health Research.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Ahsan, H., N. Parveen, N. U. Khan, and S. M. Hadi. 1999. Pro-oxidant, anti-oxidant and cleavage activities on DNA of curcumin and its derivatives demethoxycurcumin and bisdemethoxycurcumin. Chem. Biol Interact. 121:161-175. [DOI] [PubMed] [Google Scholar]
- 2.Balakirev, M. Y., M. Jaquinod, A. L. Haas, and J. Chroboczek. 2002. Deubiquitinating function of adenovirus proteinase. J. Virol. 76:6323-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bech-Otschir, D., R. Kraft, X. Huang, P. Henklein, B. Kapelari, C. Pollmann, and W. Dubiel. 2001. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 20:1630-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bharti, A. C., Y. Takada, and B. B. Aggarwal. 2004. Curcumin (diferuloylmethane) inhibits receptor activator of NF-kappa B ligand-induced NF-kappa B activation in osteoclast precursors and suppresses osteoclastogenesis. J. Immunol. 172:5940-5947. [DOI] [PubMed] [Google Scholar]
- 5.Carthy, C. M., B. Yanagawa, H. Luo, D. J. Granville, D. Yang, P. Cheung, C. Cheung, M. Esfandiarei, C. M. Rudin, C. B. Thompson, D. W. Hunt, and B. M. McManus. 2003. Bcl-2 and Bcl-xL overexpression inhibits cytochrome c release, activation of multiple caspases, and virus release following coxsackievirus B3 infection. Virology 313:147-157. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhary, L. R., and K. A. Hruska. 2003. Inhibition of cell survival signal protein kinase B/Akt by curcumin in human prostate cancer cells. J. Cell Biochem. 89:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Dutta, S., S. Padhye, K. I. Priyadarsini, and C. Newton. 2005. Antioxidant and antiproliferative activity of curcumin semicarbazone. Bioorg. Med. Chem. Lett. 15:2738-2744. [DOI] [PubMed] [Google Scholar]
- 8.Esfandiarei, M., H. Luo, B. Yanagawa, A. Suarez, D. Dabiri, J. Zhang, and B. M. McManus. 2004. Protein kinase B/Akt regulates coxsackievirus B3 replication through a mechanism which is not caspase dependent. J. Virol. 78:4289-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jana, N. R., P. Dikshit, A. Goswami, and N. Nukina. 2004. Inhibition of proteasomal function by curcumin induces apoptosis through mitochondrial pathway. J. Biol. Chem. 279:11680-11685. [DOI] [PubMed] [Google Scholar]
- 10.Kawai, C. 1999. From myocarditis to cardiomyopathy: mechanisms of inflammation and cell death: learning from the past for the future. Circulation 99:1091-1100. [DOI] [PubMed] [Google Scholar]
- 11.Kim, G. Y., K. H. Kim, S. H. Lee, M. S. Yoon, H. J. Lee, D. O. Moon, C. M. Lee, S. C. Ahn, Y. C. Park, and Y. M. Park. 2005. Curcumin inhibits immunostimulatory function of dendritic cells: MAPKs and translocation of NF-kappa B as potential targets. J. Immunol. 174:8116-8124. [DOI] [PubMed] [Google Scholar]
- 12.Kim, S. M., J. H. Park, S. K. Chung, J. Y. Kim, H. Y. Hwang, K. C. Chung, I. Jo, S. I. Park, and J. H. Nam. 2004. Coxsackievirus B3 infection induces cyr61 activation via JNK to mediate cell death. J. Virol. 78:13479-13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kisselev, A. F., and A. L. Goldberg. 2001. Proteasome inhibitors: from research tools to drug candidates. Chem. Biol. 8:739-758. [DOI] [PubMed] [Google Scholar]
- 14.Lee, C. K., K. Kono, E. Haas, K. S. Kim, K. M. Drescher, N. M. Chapman, and S. Tracy. 2005. Characterization of an infectious cDNA copy of the genome of a naturally occurring, avirulent coxsackievirus B3 clinical isolate. J. Gen. Virol. 86:197-210. [DOI] [PubMed] [Google Scholar]
- 15.Luo, H., B. Yanagawa, J. Zhang, Z. Luo, M. Zhang, M. Esfandiarei, C. Carthy, J. E. Wilson, D. Yang, and B. M. McManus. 2002. Coxsackievirus B3 replication is reduced by inhibition of the extracellular signal-regulated kinase (ERK) signaling pathway. J. Virol. 76:3365-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo, H., J. Zhang, C. Cheung, A. Suarez, B. M. McManus, and D. Yang. 2003. Proteasome inhibition reduces coxsackievirus B3 replication in murine cardiomyocytes. Am. J. Pathol. 163:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo, H., J. Zhang, F. Dastvan, B. Yanagawa, M. A. Reidy, H. M. Zhang, D. Yang, J. E. Wilson, and B. M. McManus. 2003. Ubiquitin-dependent proteolysis of cyclin D1 is associated with coxsackievirus-induced cell growth arrest. J. Virol. 77:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullally, J. E., and F. A. Fitzpatrick. 2002. Pharmacophore model for novel inhibitors of ubiquitin isopeptidases that induce p53-independent cell death. Mol. Pharmacol. 62:351-358. [DOI] [PubMed] [Google Scholar]
- 19.Opavsky, M. A., T. Martino, M. Rabinovitch, J. Penninger, C. Richardson, M. Petric, C. Trinidad, L. Butcher, J. Chan, and P. P. Liu. 2002. Enhanced ERK-1/2 activation in mice susceptible to coxsackievirus-induced myocarditis. J. Clin. Investig. 109:1561-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 22.Quiles, J. L., M. D. Mesa, C. L. Ramirez-Tortosa, C. M. Aguilera, M. Battino, A. Gil, and M. C. Ramirez-Tortosa. 2002. Curcuma longa extract supplementation reduces oxidative stress and attenuates aortic fatty streak development in rabbits. Arterioscler. Thromb. Vasc. Biol. 22:1225-1231. [DOI] [PubMed] [Google Scholar]
- 23.Reddy, S., and B. B. Aggarwal. 1994. Curcumin is a non-competitive and selective inhibitor of phosphorylase kinase. FEBS Lett. 341:19-22. [DOI] [PubMed] [Google Scholar]
- 24.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Si, X., H. Luo, A. Morgan, J. Zhang, J. Wong, J. Yuan, M. Esfandiarei, G. Gao, C. Cheung, and B. M. McManus. 2005. Stress-activated protein kinases are involved in coxsackievirus B3 viral progeny release. J. Virol. 79:13875-13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Si, X., B. M. McManus, J. Zhang, J. Yuan, C. Cheung, M. Esfandiarei, A. Suarez, A. Morgan, and H. Luo. 2005. Pyrrolidine dithiocarbamate reduces coxsackievirus B3 replication through inhibition of the ubiquitin-proteasome pathway. J. Virol. 79:8014-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sowa, H., H. Kaji, T. Yamaguchi, T. Sugimoto, and K. Chihara. 2002. Activations of ERK1/2 and JNK by transforming growth factor beta negatively regulate Smad3-induced alkaline phosphatase activity and mineralization in mouse osteoblastic cells. J. Biol. Chem. 277:36024-36031. [DOI] [PubMed] [Google Scholar]
- 28.Squires, M. S., E. A. Hudson, L. Howells, S. Sale, C. E. Houghton, J. L. Jones, L. H. Fox, M. Dickens, S. A. Prigent, and M. M. Manson. 2003. Relevance of mitogen activated protein kinase (MAPK) and phosphatidylinositol-3-kinase/protein kinase B (PI3K/PKB) pathways to induction of apoptosis by curcumin in breast cells. Biochem. Pharmacol. 65:361-376. [DOI] [PubMed] [Google Scholar]
- 29.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uddin, S., A. R. Hussain, P. S. Manogaran, K. Al-Hussein, L. C. Platanias, M. I. Gutierrez, and K. G. Bhatia. 2005. Curcumin suppresses growth and induces apoptosis in primary effusion lymphoma. Oncogene 24:7022-7030. [DOI] [PubMed] [Google Scholar]
- 31.Uhle, S., O. Medalia, R. Waldron, R. Dumdey, P. Henklein, D. Bech-Otschir, X. Huang, M. Berse, J. Sperling, R. Schade, and W. Dubiel. 2003. Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J. 22:1302-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissman, A. M. 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol. 2:169-178. [DOI] [PubMed] [Google Scholar]
- 33.Wing, S. S. 2003. Deubiquitinating enzymes—the importance of driving in reverse along the ubiquitin-proteasome pathway. Int. J. Biochem. Cell. Biol. 35:590-605. [DOI] [PubMed] [Google Scholar]
- 34.Wong, J., J. Zhang, G. Gao, M. Esfandiarei, X. Si, Y. Wang, B. Yanagawa, A. Suarez, B. McManus, and H. Luo. 2006. Liposome-mediated transient transfection reduces cholesterol-dependent coxsackievirus infectivity. J. Virol. Methods 133:211-218. [DOI] [PubMed] [Google Scholar]
- 35.Yang, X., D. P. Thomas, X. Zhang, B. W. Culver, B. M. Alexander, W. J. Murdoch, M. N. Rao, D. A. Tulis, J. Ren, and N. Sreejayan. 2006. Curcumin inhibits platelet-derived growth factor-stimulated vascular smooth muscle cell function and injury-induced neointima formation. Arterioscler. Thromb. Vasc. Biol. 26:85-90. [DOI] [PubMed] [Google Scholar]
- 36.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan, J., J. Zhang, B. W. Wong, X. Si, J. Wong, D. Yang, and H. Luo. 2005. Inhibition of glycogen synthase kinase 3β suppresses coxsackievirus-induced cytopathic effect and apoptosis via stabilization of beta-catenin. Cell Death Differ. 12:1097-1106. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, H. M., J. Yuan, P. Cheung, H. Luo, B. Yanagawa, D. Chau, N. Stephan-Tozy, B. W. Wong, J. Zhang, J. E. Wilson, B. M. McManus, and D. Yang. 2003. Overexpression of interferon-gamma-inducible GTPase inhibits coxsackievirus B3-induced apoptosis through the activation of the phosphatidylinositol 3-kinase/Akt pathway and inhibition of viral replication. J. Biol. Chem. 278:33011-33019. [DOI] [PubMed] [Google Scholar]
- 39.Zheng, S., and A. Chen. 2004. Activation of PPARγ is required for curcumin to induce apoptosis and to inhibit the expression of extracellular matrix genes in hepatic stellate cells in vitro. Biochem. J. 384:149-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, Q., J. Yao, G. Wani, J. Chen, Q. E. Wang, and A. A. Wani. 2004. The ubiquitin-proteasome pathway is required for the function of the viral VP16 transcriptional activation domain. FEBS Lett. 556:19-25. [DOI] [PubMed] [Google Scholar]