Abstract
The ICP0 protein (bICP0) encoded by bovine herpesvirus 1 is the major viral regulatory protein because it stimulates all viral promoters and, consequently, productive infection. Like other ICP0 analogues encoded by Alphaherpesvirinae subfamily members, bICP0 contains a zinc RING finger near its amino terminus that is necessary for activating transcription, regulating subcellular localization, and inhibiting interferon-dependent transcription. In this study, we discovered that sequences near the C terminus, and the zinc RING finger, are necessary for inhibiting the human beta interferon (IFN-β) promoter. In contrast to herpes simplex virus type 1-encoded ICP0, bICP0 reduces interferon response factor 3 (IRF3), but not IRF7, protein levels in transiently transfected cells. The zinc RING finger and sequences near the C terminus are necessary for bICP0-induced degradation of IRF3. A proteasome inhibitor, lactacystin, interfered with bICP0-induced degradation of IRF3, suggesting that bICP0, directly or indirectly, targets IRF3 for proteasome-dependent degradation. IRF3, but not IRF7, is not readily detectable in the nuclei of productively infected bovine cells during the late stages of infection. In the context of productive infection, IRF3 and IRF7 are detected in the nucleus at early times after infection. At late times after infection, IRF7, but not IRF3, is still detectable in the nuclei of infected cells. Collectively, these studies suggest that the ability of bICP0 to reduce IRF3 protein levels is important with respect to disarming the IFN response during productive infection.
Bovine herpesvirus 1 (BHV-1) is a significant bovine pathogen because infection leads to conjunctivitis, pneumonia, genital disorders, abortions, and “shipping fever,” an upper respiratory infection (50). Infection of bovine cells (10) or calves (53) leads to rapid cell death and an increase in apoptosis. As with other Alphaherpesvirinae subfamily members, viral gene expression is temporally regulated in three distinct phases: immediate-early (IE), early, or late (29).
The BHV-1 ICP0 protein (bICP0) is encoded by IE transcription unit 1 (54) and activates expression of all three classes of viral promoters (14). During productive infection, bICP0 protein expression is constitutive because the gene has an IE promoter and an early promoter (Fig. 1A), and both promoters are activated by bICP0 (21). The ICP0 homologues encoded by BHV-1 and herpes simplex virus type 1 (HSV-1) contain a well-conserved C3HC4 zinc RING finger near their respective N termini. Mutational analysis has demonstrated the importance of the C3HC4 zinc RING finger domains of bICP0 and ICP0 (12, 13, 15, 28). ICP0 (16-18, 35, 36) and bICP0 (28, 41) colocalize with and disrupt the proto-oncogene promyelocytic leukemia protein-containing nuclear domains. bICP0 associates with chromatin-remodeling enzymes, histone deacetylase 1 (57) plus p300 (57), and stimulates plaque formation when BHV-1 DNA is transfected into bovine cells (22, 28). A panel of bICP0 transposon insertion mutations that span the entire protein-coding domain was generated (58) (Fig. 1A). These bICP0 mutant proteins are expressed at similar levels in transfected cells (58). Sequences located between the zinc RING finger and the acidic domain are necessary for efficient transactivation of a simple viral promoter (58). Although mutations within the acidic domain do not apparently play an important role in transactivation, the nuclear localization signal (NLS) at the C terminus is necessary for wild-type (wt) levels of transactivation. Collectively, these studies suggest that bICP0 contains multiple functional domains that activate productive infection in differentiated cell types and reactivation from latency.
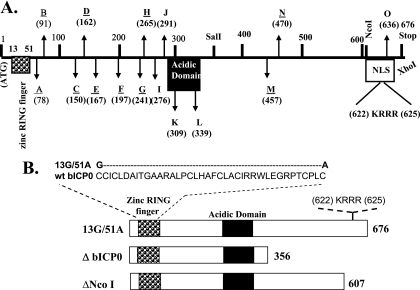
FIG. 1.
Schematic of bICP0 mutants used to localize the domains necessary for inhibiting IFN-dependent transcription. (A) Construction and identification of the transposon insertion mutants were previously described (50). The transposon insertion sites were first mapped by restriction endonuclease digestion, and then precise insertion sites were identified by DNA sequencing. The mutants were designated A to O, and the numbers in parentheses denote the amino acid number that was disrupted by transposon insertion. The positions of the zinc RING finger, acidic domain, consensus NLS (KRRR), ATG, and bICP0 stop codon are shown. The transposon mutants that had an effect on activating the thymidine kinase promoter are underlined. (B) Schematic of the 13G/51A and bICP0 deletion mutants. Two amino acid substitutions were inserted into conserved C's of the zinc RING finger of bICP0. The ΔbICP0 mutant construct was prepared by digestion of the wt construct with SalI and XhoI, which deleted sequences from amino acids 357 to 676. The details of the 13G/51A and ΔbICP0 mutants were described previously . The ΔNcoI mutant was previously described (50). Except for mutants B and L, the remainder of transposon mutants and deletion mutants express similar levels of bICP0 protein (25, 50).
Infection of cultured human cells with HSV-1 leads to production and secretion of interferon (IFN). The ICP0, ICP34.5, and Us11 genes are the known viral genes that inhibit IFN activation after infection (33, 38-40, 43). The HSV-1 glycoprotein gD induces IFN-α production in mononuclear cells (30), in part, because HSV-1 activates interferon response factor 3 (IRF3) in certain cell types (44). Mice lacking type I and type II interferon receptors in combination with having RAG-2 gene deletions die within a few days following BHV-1 infection (2). In contrast, BHV-1 infection of wt mice does not lead to clinical symptoms or extensive viral replication, highlighting the importance that IFN plays in controlling BHV-1 replication and pathogenesis. To date, the bICP0 gene is the only BHV-1 gene known to inhibit interferon signaling (25).
In this study, we identified bICP0 sequences that are necessary for inhibiting transactivation of the human IFN-β promoter by IRF3 or IRF7. The mechanisms by which bICP0 inhibits IRF3 versus IRF7 transactivation of the IFN-β promoter are different because bICP0 reduces steady-state IRF3 protein levels but not IRF7 protein levels. The zinc RING finger and sequences near the C terminus are necessary for inhibiting IFN-β promoter activity and reducing IRF3 protein levels. In contrast, an expression plasmid containing HSV-1 ICP0 had little effect on IRF3 protein levels in transfected cells. Following infection of bovine cells, the levels of IRF3 protein, but not IRF7, are reduced. These studies suggest that the ability of bICP0 to repress IRF3 protein levels promotes productive infection by suppressing IFN signaling.
MATERIALS AND METHODS
Cell culture and viruses.
Human epithelial 293 cells and Madin-Darby bovine kidney (MDBK) cells were grown in monolayer cultures in Earle's modified Eagle's medium supplemented with 5% fetal bovine serum, penicillin (10 U/ml), and streptomycin (100 μg/ml) in a humidified 5% CO2 atmosphere at 37°C. Bovine testicle cells (9.1.3 cells) were grown in the same medium with 10% fetal bovine serum.
The BHV-1 wt Cooper virus strain was propagated and titrated in MDBK cells. For viral infections, cells were plated onto 100-mm2 culture dishes 24 h prior to virus infection to obtain 90% confluence at the time of infection. Cells were infected at a multiplicity of infection (MOI) of 1. After 1 h of adsorption at 37°C, cells were rinsed with phosphate-buffered saline (PBS) and overlaid with Earle's modified Eagle's medium containing 5% or 10% fetal bovine serum.
Expression plasmids.
The IFN-β chloramphenicol acetyltransferase (CAT) plasmid was obtained from Stavros Lomvardas (Columbia University, NY). This construct contains a minimal human IFN-β promoter (positions −110 to +20) upstream of the bacterial CAT gene. The plasmid pCMV2C-bICP0 expresses Flag-tagged wt bICP0 under the control of the human cytomegalovirus (CMV) promoter. Generation of bICP0 transposon mutants was described previously (58). The zinc RING finger mutant 13G/51A contains point mutations within two conserved amino acids of the C3HC4 zinc RING finger. bICP0 C-terminus deletion mutants (ΔbICP0 and ΔNcoI) were generated by deleting the SalI-XhoI fragment (amino acids [aa] 356 to 676) and NcoI-XhoI fragment (aa 607 to 676), respectively, from the Flag-tagged bICP0 construct (58). A plasmid expressing HSV-1 ICP0 was obtained from Saul Silverstein (Columbia University, NY). ICP0-coding sequences were cloned in frame with the Flag epitope in pCMV2C, and this plasmid is designated Flag-ICP0.
IRF3 and IRF7 expression constructs were obtained from Luwen Zhang (University of Nebraska, Lincoln, NE).
CAT assays.
The IFN-β CAT reporter plasmid (2 μg) was cotransfected with bICP0 (1 μg) wild-type or transposon plasmids into 293 cells by using TransIT transfection reagents (Mirus) as described by the manufacturer. Cells were incubated with the transfection mix for 5 h and then replaced with fresh medium. After 40 h, cells were lysed by three freeze-thaw cycles in 250 mM Tris-HCl (pH 8.0). CAT assays was performed with 0.2 μCi (7.4 kBq) [14C]chloramphenicol (Amersham Biosciences, catalog no. CFA754) and 0.5 mM acetyl coenzyme A (Sigma, catalog no. A2181). Chloramphenicol and its acetylated forms were separated by thin-layer chromatography and CAT activity measured with a PhosphorImager (Molecular Dynamics, CA). CAT activity is expressed as fold induction relative to the vector control. Transfection experiments for CAT assays were repeated at least three times to confirm the results.
Western blot analysis.
293 or 9.1.3 cells were cotransfected with plasmids expressing IRF3 or IRF7 and the designated bICP0 plasmids. At 40 h after transfection, cells were washed with phosphate-buffered saline and suspended in lysis buffer (100 mM Tris [pH 8.0], 1 mM EDTA, 100 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride, and one tablet of complete protease inhibitor [Roche Molecular Biochemicals] per 10 ml). Cell lysate was incubated on ice for 10 min and then at 4°C with rotation for 10 min and then was clarified by centrifugation at 10,000 × g at 4°C for 15 min. Protein concentrations were quantified by the Bradford assay. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were mixed with an equal amount of 1× sample loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate, 50 mM dithiothreitol, 0.1% bromophenol blue, 10% glycerol) and boiled for 5 min. Proteins were separated in a 10% bis-cross-linked polyacrylamide gel. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore). Membranes were blocked in Tris-buffered saline that contained 5% milk. Primary antibodies to IRF3 (C-20 and SC-15991; 1:500 dilution) or IRF7 (Y-19 and SC-15993; 1:500 dilution) were purchased from Santa Cruz Biotechnology. Membranes were then incubated overnight with the indicated primary antibody in 5% milk-containing 0.1% Tween 20-Tris-buffered saline. After washing with 0.1% Tween 20-Tris-buffered saline, membranes were incubated with donkey anti-goat immunoglobulin G (IgG)-horseradish peroxidase (SC-2020; Santa Cruz Biotechnology) secondary antibody. Immunodetection was performed with enhanced chemiluminescence Western blotting detection reagents (Perkin-Elmer, MA) in accordance with the manufacturer's protocol.
Immunofluorescence.
For transfection studies, 9.1.3 cells were plated in four-well Lab-Tek culture slides (Nunc Life Science Products, catalog no. 154526) at 16 h before transfection. The designated plasmids were cotransfected by using Lipofectamine 2000 (Invitrogen, catalog no. 116668-019) according to the manufacturer's protocol. For infection studies, MDBK or 9.1.3 cells were split into four-well Lab-Tek slides, infected with BHV-1 at an MOI of 1, and then processed at different time intervals. At 24 h after transfection or the indicated infection times, cells were fixed in 4% formaldehyde for 10 min, followed by three washes with phosphate-buffered saline. Cells were permeabilized by incubating with 100% ethanol (−20°C) for 2 min. Slides were then washed three times and blocked in 4% donkey serum (Sigma, catalog no. D9663) in PBS for 1 h. The designated primary antibodies were used at a 1:50 dilution in PBS and incubated for 2 h. After three washes with 0.05% Tween 20 in PBS, slides were incubated with the secondary antibody for 1 h in the dark. Secondary antibodies used were as follows: Cy2-conjugated donkey anti-goat (catalog no. 705-226-147), Cy5-conjugated donkey anti-goat (catalog no. 705-176-147), Cy2-conjugated donkey anti-rabbit (catalog no. 711-225-152), and Cy5-conjugated donkey anti-mouse (catalog no. 715-176-150) (Jackson ImmunoResearch Laboratories Inc.). After slides were washed with 0.05% Tween 20 in PBS, coverslips were mounted on slides using Gelmount aqueous mounting medium (Sigma, catalog no. G0918). To visualize the nucleus, DAPI (4′,6′-diamidino-2-phenylindole) staining was performed. Images were obtained with a Bio-Rad confocal laser-scanning microscope (MRC-1024ES) with excitation/emission at 488/520 nm.
RNA extraction and RT-PCR.
Total RNA was extracted from 293 cells cotransfected with IRF3, bICP0, or transposon mutant O by using TRIzol reagent (Invitrogen, catalog no. 15596-018) as described by the manufacturer. Samples were digested with DNase I and subjected to reverse transcription-PCR (RT-PCR). RNA was reverse transcribed using oligo(dT) primers. A mock reaction was carried out with no reverse transcriptase added. Ten percent of the resulting cDNA was used as a template for PCR using specific primers for human IRF3 and β-actin. PCR products were analyzed on a 1% agarose gel. The following primer sequences were used: for IRF3, forward primer 5′TGGGAGTTCGAGGTGAC3′ and reverse primer 5′GGGCTCAGCTCTCCCCAG3′; for β-actin, forward primer 5′GTGGGG CGCCCCAGGCACCA3′ and reverse primer 5′CTCCTTAATGTCACGCACGATTTC3′.
RESULTS
Identification of bICP0 domains necessary for inhibiting IFN-β promoter activity.
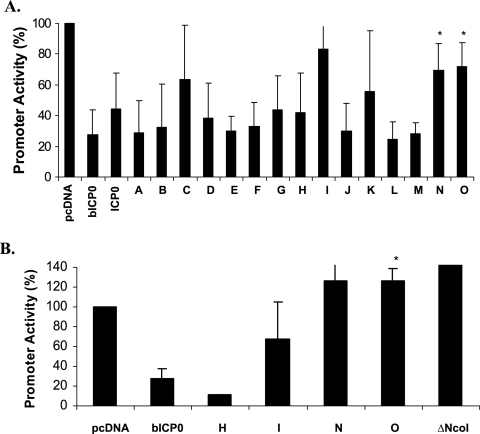
A previous study demonstrated that bICP0 repressed IFN-β promoter activity and interferon-stimulated response element-dependent transcription (25). Furthermore, the zinc RING finger is necessary for inhibiting IFN-dependent transcription in bovine cells (25). To test whether other functional domains within bICP0 are necessary for inhibiting IFN-β promoter activity, a series of bICP0 transposon mutants (58) were used (Fig. 1A). These mutants were previously used to identify functional domains within bICP0 that are necessary for activating a minimal HSV-1 thymidine kinase promoter (58). The transposon mutants used for this study express similar levels of bICP0 protein in transfected cells (58). wt bICP0 or each transposon mutant (mutants A to O) was cotransfected with either the IRF3 or IRF7 expression plasmid and the human IFN-β CAT construct in human 293 cells. After 40 h of transfection, cell lysate was collected and CAT assays performed. As expected, IFN-β promoter activity induced by IRF3 or IRF7 was suppressed four- to fivefold in the presence of wt bICP0 (Fig. 2A and B). In general, the N-terminus mutants (mutants A, B, D, E, F, G, and H) suppressed IFN-β promoter activation with similar efficiency relative to wt bICP0. Transposon mutants N and O were unable to suppress IFN-β promoter activity induced by IRF7 as efficiently as wt bICP0 or mutant H (Fig. 2A). Transposon mutants I, N, and O also did not inhibit IRF3 induction of the IFN-β promoter (Fig. 2B). Mutant I, however, was not as efficient as mutant N or O. With respect to mutant N, there was more variability in the degree of repression compared to mutant O. The deletion mutant ΔNcoI (which lacks aa 607 to 676 at the C terminus and the nuclear localization signal of bICP0 [Fig. 1A]) (58) slightly increased IFN-β promoter activity (Fig. 2B). In addition to the zinc RING finger, these results suggested that disruption of bICP0 sequences at site N or O and deletion of the C terminus (mutant ΔNcoI) eliminated the ability of bICP0 to suppress IFN-β promoter activity.
FIG. 2.
Identification of bICP0 domains that are necessary to inhibit activation of the human IFN-β promoter. 293 cells (1 × 105) were cotransfected with the IFN-β CAT reporter plasmid (1.0 μg DNA), 1.0 μg of IRF7 (A) or 1.0 μg or IRF3 (B) expression plasmid, and the designated bICP0 expression plasmids (1.0 μg DNA). An empty vector (pcDNA3.1) was used as a control. Cell extracts were collected after 40 h of transfection and analyzed for CAT expression as described in Material and Methods. The value for the IFN-β promoter and IRF3 or IRF7 in the presence of the blank expression vector was set at 100%. Data represent the means from at least three experiments. Error bars show the standard errors for triplicate transfections. *, P < 0.05.
bICP0 reduces IRF3 protein levels.
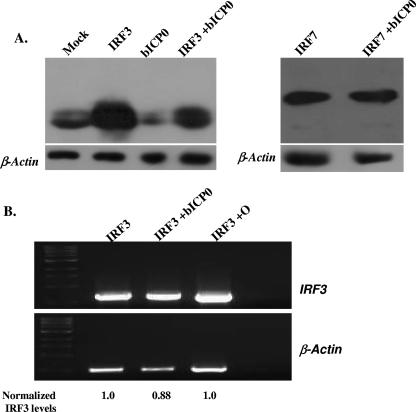
The ability of bICP0 to inhibit IRF3- or IRF7-induced IFN-β promoter activation suggested that bICP0 altered expression or activity of these transcription factors. To begin to understand the molecular basis of how bICP0 inhibits IFN-β promoter activity, we examined IRF3 or IRF7 protein levels in transfected cells. When 293 cells (Fig. 3 and 4A) or bovine cells (9.1.3) (Fig. 4B) were cotransfected with plasmids expressing IRF3 and bICP0, IRF3 protein levels were consistently reduced. In contrast, bICP0 did not have a dramatic effect on IRF7 protein levels in the cell lines examined (Fig. 3A and 4B). RT-PCR was performed to test whether bICP0 reduced IRF3 mRNA levels. IRF3 mRNA levels were not reduced dramatically by bICP0 (Fig. 3B), suggesting that bICP0 reduced IRF3 protein levels, not RNA levels.
FIG. 3.
bICP0 expression correlates with reduced IRF3 protein levels. (A) 293 cells (1 × 105) were transfected with a plasmid expressing IRF3 (2.5 μg) or IRF7 (2.5 μg) and Flag-tagged bICP0 (2.5 μg) expression vector as described for Fig. 2. Whole-cell lysate was collected at 40 h after transfection and IRF3 or IRF7 protein levels measured by Western blot analysis using 100 μg of total cell lysate. (B) 293 cells were transiently transfected with IRF3 alone, IRF3 and bICP0, or IRF3 and mutant O. Total RNA was isolated 40 h after transfection. For RT-PCR, 1 μg of total RNA was used, and 10% of the resulting cDNA was used as the template for PCR. Primers specific for IRF3 and β-actin (control) were used. The β-actin and IRF3 bands were quantified using BioRad Molecular Images. The ratio between IRF3 and β-actin was calculated for each lane. The IRF3 lane was normalized to 1.
FIG. 4.
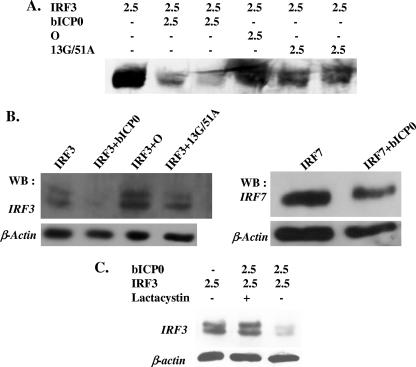
bICP0 reduces IRF3 levels in transiently transfected cells. (A) 293 cells were transfected with the designated amounts of plasmids (μg DNA). (B) IRF3 protein expression was analyzed in 9.1.3 cells. IRF3 plasmid (2.5 μg) was cotransfected with bICP0 plasmid (2.5 μg). At 40 h after transfection, cell lysate was collected as described in Materials and Methods. (C) 9.1.3 cells were transfected with the designated plasmids (2.5 μg DNA). At 24 h after transfection, the designated cultures were treated with lactacystin (15 μM; Calbiochem catalog no. 426100) or dimethyl sulfoxide, which was used to suspend lactacystin. IRF3 protein levels were detected by Western blot analysis. For each lane, 100 μg protein was used. The level of DNA in each lane was the same because an empty expression vector (pcDNA3.1−) was added to the transfection mixture to make the total DNA equal to 5 μg.
The zinc RING finger and C-terminal domains of bICP0 are important for reducing IRF3 protein levels.
Since the zinc RING finger, mutant O, and the ΔNcoI deletion construct consistently suppressed IRF3-induced IFN-β promoter activity (Fig. 2), we tested whether these mutants reduced IRF3 protein levels. For these studies, IRF3 was cotransfected with the zinc RING finger mutant (13G/51A) or the C-terminal transposon mutant O into 293 or 9.1.3 cells. The 13G/51A mutant or mutant O did not reduce IRF3 protein levels to the extent that wt bICP0 did in 293 cells (Fig. 4A) or 9.1.3 cells (Fig. 4B). To test whether a functional proteasome was necessary for bICP0-induced IRF3 degradation, cultures were treated with lactacystin, a cell-permeative and irreversible inhibitor of the proteasome (19). Treatment with lactacystin inhibited the ability of bICP0 to reduce IRF3 protein levels (Fig. 4C).
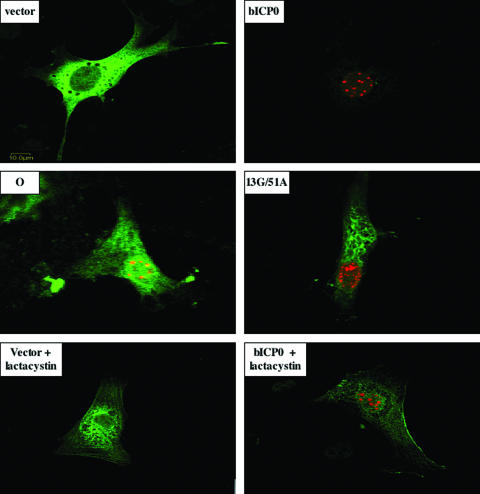
Immunofluorescent staining of IRF3 and bICP0 was performed to confirm the reduction of IRF3 protein levels in transfected cells (Fig. 5). bICP0 localizes to promyelocytic leukemia protein-containing nuclear domains (28), and IRF3 shuttles between the cytoplasm and nucleus. In general, the IRF3 protein is detected primarily in the cytoplasm of uninfected cells (55) or cells transfected with an empty vector (Fig. 5, vector panel). As expected, punctate staining of bICP0 was detected in the nucleus. High levels of IRF3 were detected in cells cotransfected with IRF3 and pcDNA3.1 (empty vector). Cells expressing both Flag-tagged bICP0 (red, anti-Flag antibody) and IRF3 plasmid contained low levels of IRF3 (green, anti-IRF3 antibody) (Fig. 5, panel bICP0). 9.1.3 cells expressing either Flag-tagged 13G/51A or the Flag-tagged O mutant contained levels of IRF3 similar to those in cells transfected with an empty vector. However, many cells cotransfected with the 13G/51A construct had an altered cytoplasmic IRF3 distribution. With respect to mutant O, a subset of the total IRF3 appeared to be present in the nucleus because of the yellowish appearance of bICP0 spots and reduction of bICP0 fluorescence.
FIG. 5.
Effect of bICP0 expression on IRF3 protein expression and subcellular localization. Equivalent amounts of IRF3 and Flag-tagged bICP0 plasmids (wt, 13G/51A mutant, or the mutant O construct) were transfected into 9.1.3 cells. After 24 h of transfection, immunostaining was performed using the anti-IRF3 and anti-Flag antibodies as described in Materials and Methods. Cultures were then stained with Cy2-conjugated anti-goat IgG antibody (IRF3, green) and Cy5-conjugated anti-mouse IgG antibody (bICP0, red). Stained cells were visualized by confocal microscopy. In the cultures containing lactacystin, cultures were transfected and then treated with 15 μM lactacystin. The images are representative of three different experiments (at least 100 cells were examined for each sample in each experiment).
To confirm that a functional proteasome was necessary for bICP0-induced IRF3 degradation, cultures were treated with lactacystin. Lactacystin treatment yielded numerous bICP0-positive cells with high levels of IRF3 (Fig. 5, bICP0 + lactacystin panel). As expected, when cells were cotransfected with the empty expression vector plus IRF3 and then treated with lactacystin (vector + lactacystin panel), many IRF3-expressing cells were detected. In conclusion, the studies shown in Fig. 3 to 5 demonstrated that wt bICP0 reduced IRF3 protein levels and that a functional proteasome was necessary for bICP0-induced IRF3 degradation.
The HSV-1-encoded ICP0 does not reduce IRF3 protein levels in transiently transfected cells.
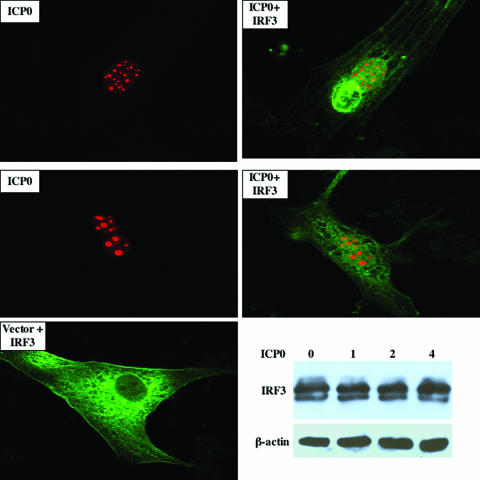
Although bICP0 and ICP0 activate productive infection and repress IFN-dependent transcription, only the C3HC4 zinc RING finger domains of bICP0 and ICP0 are well conserved (28, 34). To test whether ICP0, in the absence of other viral genes, reduced IRF3 protein levels, 9.1.3 cells were transfected with a plasmid expressing Flag-tagged ICP0 and the IRF3 expression plasmid. Confocal microscopy revealed that all ICP0-positive cells contained IRF3 (Fig. 6, ICP0+IRF3 panels), which was in stark contrast to the results obtained with bICP0. Although most ICP0-positive cells contained IRF3 distributed throughout the cell, there were ICP0-positive cells that had IRF3 localized to the nucleus (Fig. 6, middle panel). As expected, ICP0 was localized primarily in the nucleus and yielded punctate staining. In cells transfected with an empty expression vector, IRF3 was consistently localized in the cytoplasm (Fig. 6, Vector + IRF3 panel). Consistent with the confocal microscopy results, Western blot analysis demonstrated that ICP0 did not have a dramatic effect on IRF3 protein levels (Fig. 6, lower right panel).
FIG. 6.
HSV-1-encoded ICP0 does not reduce IRF3 protein levels in transfected cells. IRF3 (0.5 μg DNA) and the Flag-tagged ICP0 construct (0.5 μg DNA) were transfected into 9.1.3 cells. As designated, certain cultures were transfected with an empty vector (pCMV2C; 0.5 μg DNA) and the IRF3 expression construct (0.5 μg DNA). After 24 h of transfection, immunostaining was performed using the anti-IRF3 and anti-Flag antibodies. Cultures were then stained with Cy2-conjugated anti-goat IgG antibody (IRF3, green) and Cy5-conjugated anti-mouse IgG antibody (ICP0, red). Stained cells were visualized by confocal microscopy. The ICP0 panels show just the Flag antibody staining, and the ICP0+IRF3 panels show the merge between ICP0 and IRF3 staining. Images are representative of three different experiments (at least 100 cells were examined for each sample). The lower right panel shows a Western blot study demonstrating that transfection of 9.1.3 cells with increasing concentrations of HSV-1 ICP0 does not reduce IRF3 protein levels. The amount of Flag-ICP0 construct (μg DNA) is shown. For all lanes, 2.5 μg IRF3 was used. As a loading control, β-actin levels were examined.
Analysis of IRF3 and IRF7 following infection.
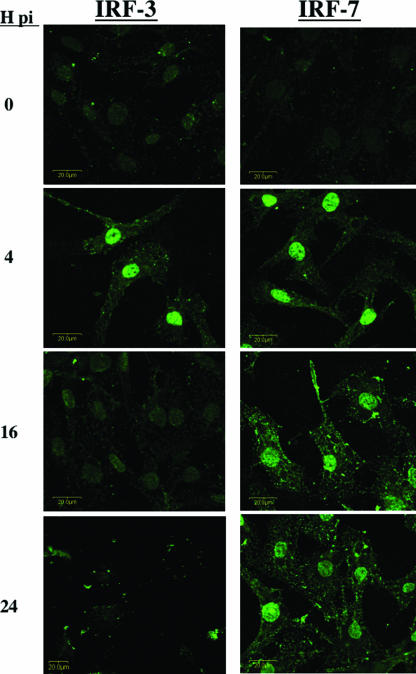
In general, virus infection stimulates transcription of IFN genes and transactivation of these genes requires IRF3 activation (30). Phosphorylation of IRF3 is necessary for translocation to the nucleus, where it is transcriptionally active (55). To assess the effect of BHV-1 infection on activation and nuclear translocation of IRF3 or IRF7, two bovine cell types (9.1.3 and MDBK) were infected with wt BHV-1 at an MOI of 1 for 4, 16, or 24 h. Using an MOI of 1, we detected viral proteins in more than 75% of the infected cells at 16 h after infection by confocal microscopy. After infection, IRF3 or IRF7 localization was examined by confocal immunofluorescence (Fig. 7). Mock-infected cultures were used as controls because IRF3 was localized to the cytoplasm. At 4 h after infection, IRF3 was located primarily in the nuclei of 9.1.3 cells, indicating that infection activated IRF3 (Fig. 7). At 16 or 24 h after infection, IRF3 protein levels appeared to be reduced relative to those at 4 h after infection. IRF7, like IRF3, was detected in the nucleus at 4 h after infection (Fig. 7). In contrast to the results obtained with IRF3, IRF7 was readily detected in the nucleus at 16 or 24 h after infection. Similar results were obtained with MDBK cells, and Western blot studies confirmed that IRF3 protein levels were reduced at late times after infection (unpublished data). In summary, these studies suggested that BHV-1 infection triggered IRF3 and IRF7 activation at early times after infection, but IRF3 was not detected in the nucleus at late times after infection.
FIG. 7.
Localization of IRF3 or IRF7 (green) in BHV-1-infected cells. 9.1.3 cells were infected with BHV-1 at an MOI of 1. Cells were then immunostained with the anti-IRF3 antibody or anti-IRF7 antibody at 0, 4, 16, or 24 h postinfection (pi). Images were visualized by confocal microscopy, and the results are representative of three different experiments.
DISCUSSION
A previous study demonstrated that bICP0 inhibited IFN-dependent transcription in the absence of other viral genes (25). In this study, evidence was provided that bICP0 induced proteasome-dependent degradation of IRF3 but not IRF7. The zinc RING finger and sequences near the C terminus (transposon mutant N and O or the ΔNcoI deletion construct) were necessary for inhibiting IRF7- or IRF3-induced IFN-β promoter activity and IRF3 degradation. Since IFN production and IFN signaling pathways restrict the host range of BHV-1 (2), identifying viral genes that inhibit IFN signaling and their mechanism of action is important. Although these studies indicated that bICP0 inhibited IRF3 proteins levels and IFN-β promoter activity, it is likely that other viral genes activate and then inhibit IFN activity during productive infection, because HSV-1 encodes several proteins that inhibit the IFN response to infection.
Following virus infection, two cellular protein kinases, IKK-ɛ and TBK-1, phosphorylate serine residues at the C terminus of IRF3, which induces IRF3 homodimerization and nuclear translocation (20, 47). Nuclear IRF3 associates with other transcriptional activators, resulting in direct binding and stimulation of IFN-β promoter activity (52). IRF3 also directly binds several consensus DNA binding sites, including the interferon-stimulated response element, and can consequently stimulate transcription of IFN-stimulated genes in the absence of IFN (24, 38). HSV-1-encoded ICP0 is an essential component of the IFN resistance to infection and is sufficient to inhibit induction of IFN-stimulated genes (38-40). Inhibition of IRF3- and IRF7-mediated activation of IFN-responsive genes requires an intact ICP0 RING finger domain (33). IRF3 and IRF7 protein levels do not change dramatically following infection of human embryonic lung cells for 10 h with specific ICP0 mutants or ICP0-expressing virus strains (33). To further examine the effects of HSV-1 on IRF3 activation, human endometrial adenocarcinoma cells (HEC-1-B) were infected with Sendai virus to induce IRF3 nuclear accumulation and IFN production (37). Under these experimental conditions, HSV-1-encoded ICP0 inhibits IRF3 nuclear accumulation and IFN-β production in HEC-1-B cells. Relative to Sendai virus-infected cells, IRF3 degradation was enhanced in HEC-1-B cells following infection with Sendai virus and then with HSV-1 (37). The ability of HSV-1 to induce IRF3 is cell type dependent (44), suggesting that in certain cell types HSV-1 productive infection can reduce IRF3 protein levels in an ICP0-dependent fashion. The finding that the bICP0 zinc RING finger domain and sequences near the C terminus, including the NLS, are important for inhibiting IFN-dependent transcription (25) is consistent with published findings for HSV-1-encoded ICP0.
In the absence of other viral genes, bICP0 (directly or indirectly) reduced IRF3 protein levels in human or bovine cells. In contrast to bICP0, HSV-1-encoded ICP0 was unable to reduce IRF3 protein levels in transiently transfected 9.1.3 cells as judged by confocal microscopy or Western blot analysis (Fig. 6). The RING finger of HSV-1-encoded ICP0 is an E3 ubiquitin ligase (7, 8, 51). The bICP0 RING finger has also been reported to possess E3 ubiquitin ligase (11), suggesting that the bICP0 RING finger is involved with IRF3 degradation. The finding that a functional proteasome was necessary for bICP0-induced IRF3 degradation supported the contention that the bICP0 RING finger mediated IRF3 degradation by directly or indirectly inducing proteasome-dependent IRF3 degradation. At this time, it is not clear how the C terminus of bICP0 inhibited IRF3-dependent transcription and degradation. Although one could argue that mutations within the C terminus of bICP0 altered the zinc RING finger conformation, previous studies indicated that mutant O activated a simple HSV-1 thymidine kinase promoter with an efficiency similar to that of wt bICP0 (58). Since the zinc RING finger is necessary for activating transcription, the transposon insertion at O does not appear to disrupt the zinc RING finger.
As discussed above, IRF3 is a central component of innate immune responses following virus infection. Thus, it is not surprising that several viruses, i.e., hepatitis C virus (32), classical swine fever virus (45), bovine (6) or human (48) respiratory syncytial virus, Bunyamwera virus (31), bovine diarrhea virus (3), Ebola virus (5), influenza virus (49), human cytomegalovirus (1), human rhinovirus (42), rabies virus (9), and rotavirus (23), encode proteins that inhibit IRF3. A variety of mechanisms are utilized to disrupt IRF3 functions, and these include inhibiting IRF3 phosphorylation, inhibiting translocation to the nucleus, preventing DNA binding, degrading factors necessary for activating IRF3, inhibiting IRF3 transcription, blocking IRF3 dimerization, or binding and sequestering IRF3. The NPro protein encoded by bovine diarrhea virus also induces the proteolytic degradation of IRF3 (26). Furthermore, the nonstructural protein 1 encoded by rotavirus binds IRF3 (23) and then degrades IRF3 (4). Although bICP0 reduced steady-state levels of the IRF3 protein, we have been unable to coimmunoprecipitate IRF3 and bICP0, suggesting that these two proteins do not stably interact.
IRF3 activation is considered to be an immediate-early phase of the IFN response, whereas IRF7 is a component of the early response (46, 55, 56). Recent studies have suggested that IRF7 is more important with respect to inhibiting viral infection when IRF3 versus IRF7 knockout mice are compared (27). Although it seems clear that IRF7 was activated and translocated to the nuclei of bovine cells during the early stages of BHV-1 infection, IRF7 protein levels were not dramatically reduced relative to IRF3 protein levels. This highlights two important points: the effect on IRF3 was specific, and bICP0 apparently inhibited IRF7 activity following translocation to the nucleus. For example, bICP0 may (directly or indirectly) inhibit IRF7 from binding DNA or associating with cellular factors necessary for transcriptional activation. It is also possible that when IRF3 protein levels are very low, IRF7 does not efficiently activate IFN-β promoter activity. Studies designed to understand the mechanism by which bICP0 interferes with the ability of IRF7 to activate transcription and how BHV-1 infection induces nuclear translocation of IRF3 and IRF7 will help us to understand the mechanism by which bICP0 regulates the IFN response following infection.
Acknowledgments
This work was supported by USDA grants 2005-01554 and 2006-01627 and, in part, by Public Health Service grant P20RR15635.
We thank Terri Fangman for assistance with confocal microscopy.
Footnotes
Published ahead of print on 10 January 2007.
REFERENCES
- 1.Abate, D. A., S. Watanabe, and E. S. Mocarski. 2004. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J. Virol. 78:10995-11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abril, C., M. Engels, A. Limman, M. Hilbe, S. Albini, M. Franchini, M Suter, and M. Ackerman. 2004. Both viral and host factors contribute to neurovirulence of bovine herpesvirus 1 and 5 in interferon receptor-deficient mice. J. Virol. 78:3644-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interfeon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. USA 102:4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H.-D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bossert, B., S. Marozin, and K.-K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boutell, C., and R. D. Everett. 2003. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with an ubiquitinates p53. J. Biol. Chem. 278:36596-36602. [DOI] [PubMed] [Google Scholar]
- 8.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzozka, K., S. Finke, and K.-K. Conzelmann. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phospoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devireddy, L. R., and C. J. Jones. 1999. Activation of caspases and p53 by bovine herpesvirus 1 infection results in programmed cell death and efficient virus release. J. Virol. 73:3778-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dia, L., B. Zhang, J. Fan, X. Gao, S. Sun, K. Yang, D. Xin, N. Jin, Y. Geng, and C. Wang. 2005. Herpes virus proteins ICP0 and BICP0 can activate NF-κB by catalyzing IκBα ubiquitination. Cell. Signalling 17:217-229. [DOI] [PubMed] [Google Scholar]
- 12.Everett, R. D. 1988. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J. Mol. Biol. 202:87-96. [DOI] [PubMed] [Google Scholar]
- 13.Everett, R. D. 1987. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 6:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R. D., P. Barlow, A. Milner, B. Luisi, A. Orr, G. Hope, and D. Lyon. 1993. A novel arrangement of zinc-binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J. Mol. Biol. 234:1038-1047. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., P. Lomonte, T. Sternsdorf, R. van Driel, and A. Orr. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112:4581-4588. [DOI] [PubMed] [Google Scholar]
- 18.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. (Corrected and republished from EMBO J. 16:566-577.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fenteany, G. R. F. S., W. S. Lane, S. Choi, E. J. Corey, and S. L. Schreiber. 1995. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science 268:726-731. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald, K. A., S. M. McWhirter, K. L. Faja, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S.-M. Liao, and T. Maniatis. 2003. IKKe and TBKI are essential components of the IRF3 signaling pathway. Nature 4:491-496. [DOI] [PubMed] [Google Scholar]
- 21.Fraefel, C., J. Zeng, Y. Choffat, M. Engels, M. Schwyzer, and M. Ackermann. 1994. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J. Virol. 68:3154-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiser, V., and C. Jones. 2003. Stimulation of bovine herpesvirus 1 productive infection by the adneovirus E1A gene and a cell cycle regulatory gene, E2F-4. J. Gen. Virol. 84:929-938. [DOI] [PubMed] [Google Scholar]
- 23.Graff, J. W., D. N. Mitzel, C. M. Weisend, M. L. Flenniken, and M. E. Hardy. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 25.Henderson, G., Y. Zhang, C. Jones. 2005. The bovine herpesvirus 1 gene encoding infected cell protein 0 (bICP0) can inhibt interferon-dependent transcription in the absence of other viral genes. J. Gen. Virol. 86:2697-2702. [DOI] [PubMed] [Google Scholar]
- 26.Hilton, L., K. Moganeradji, G. Zhang, Y.-H. Chen, R. E. Randall, J. W. McCauley, and S. Goodburn. 2006. The NPro product of bovine viral diarrhea virus inhibits DNA binding interfereon regulatory factor-3 and targets it for proteasomal degradation. J. Virol. 80:11723-11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda, K., H. Yanai, H. Negishi, M. Asagiri, M. Saton, T. Mizutani, N. Shimada, Y. Ohba, A. Takaoka, N. Yoshida, and T. Taniguchi. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434:772-777. [DOI] [PubMed] [Google Scholar]
- 28.Inman, M., Y. Zhang, V. Geiser, and C. Jones. 2001. The zinc ring finger in the bICP0 protein encoded by bovine herpes virus-1 mediates toxicity and activates productive infection. J. Gen. Virol. 82:483-492. [DOI] [PubMed] [Google Scholar]
- 29.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katze, M. G., Y. Heng, and M. Gale. 2002. Viruses and interferon: fight for supremacy. Nat. Rev. Immunol. 2:675-686. [DOI] [PubMed] [Google Scholar]
- 31.Kohl, A., R. F. Clayton, F. Weber, A. Bridgen, R. E. Randall, and R. M. Elliot. 2003. Bunyamwera virus nonstructural protein NSs counteracts interferon regulatory factor 3-mediated induction of early cell death. J. Virol. 77:7999-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. M. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, R., R. S. Noyce, S. E. Collins, R. D. Everett, and K. L. Mossman. 2004. The herpes simplex virus ICP0 RING finger domain inhibits IRF3- and IRF7-mediated activation of interferon-stimulated genes. J. Virol. 78:1675-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lium, E. K., and S. Silverstein. 1997. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J. Virol. 71:8602-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 36.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 37.Melroe, G. T., N. A. DeLuca, and D. M. Knipe. 2004. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 78:8411-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 76:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng, T., S. Kotla, R. E. Bumgarner, and K. E. Gustin. 2006. Human rhinovirus attenuates the type I interferon response by dsrupting activaiton of interferon regulatory factor 3. J. Virol. 80:5021-5031. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Peters, G. A., D. Khoo, I. Mohr, and G. S. Sen. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex simplex virus type 1 Us11 protein. J. Virol. 75:11054-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rocca, S. A., R. J. Herbert, H. Crooke, T. W. Drew, T. E. Wileman, and P. P. Powell. 2005. Loss of interferon regulatory factor 3 in cells infected with classical swine fever virus involves the N-terminal protease, Npro. J. Virol. 79:7239-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sato, M., T. Taniguchi, and N. Tanaka. 2001. The interferon system and interferon regulatory factor transcription factors—studies from gene knockout mice. Cytokine Growth Factor Rev. 12:133-142. [DOI] [PubMed] [Google Scholar]
- 47.Sharma, S., B. R. tenOever, N. Grandvaux, G.-P. Zhou, R. Lin, and J. Hiscott. 2003. Trigerring the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 48.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 or human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 79:5353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Talon, J., C. M. Horvath, R. Polley, C. F. asler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191-223. [DOI] [PubMed] [Google Scholar]
- 51.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 53.Winkler, M. T., A. Doster, and C. Jones. 1999. Bovine herpesvirus 1 can infect CD4+ T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 73:8657-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wirth, U. V., C. Fraefel, B. Vogt, C. Vlcek, V. Paces, and M. Schwyzer. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 66:2763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoneyama, M., W. Suhara, M. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type 1 interferon system by virus infection: activation of a transcription factors containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuang, Y., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y., and C. Jones. 2001. The bovine herpesvirus 1 immediate-early protein (bICP0) associates with histone deacetylase 1 to activate transcription. J. Virol. 75:9571-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, Y., and C. Jones. 2005. Identification of functional domains within the bICP0 protein encoded by BHV-1. J Gen. Virol. 86:879-886. [DOI] [PubMed] [Google Scholar]