Abstract
Treating chronic hepatitis C virus (HCV) infection using pegylated alpha interferon and ribavirin leads to sustained clearance of virus and clinical improvement in approximately 50% of patients. Response rates are lower among patients with genotype 1 than with genotypes 2 and 3 and among African-American (AA) patients compared to Caucasian (CA) patients. Using DNA microarrays, gene expression was assessed for a group of 33 African-American and 36 Caucasian American patients with chronic HCV genotype 1 infection during the first 28 days of treatment. Results were examined with respect to treatment responses and to race. Patients showed a response to treatment at the gene expression level in RNA isolated from peripheral blood mononuclear cells irrespective of degree of decrease in HCV RNA levels. However, gene expression responses were relatively blunted in patients with poor viral response (<1.5 log10-IU/ml decrease at 28 days) compared to those in patients with a marked (>3.5 log10-IU/ml decrease) or intermediate (1.5 to 3.5 log10-IU/ml decrease) response. The number of genes that were up- or down-regulated by pegylated interferon and ribavirin treatment was fewer in patients with a poor response than in those with an intermediate or marked viral response. However AA patients had a stronger interferon response than CA patients in general. The induced levels of known interferon-stimulated genes such as the 2′5′-oligoadenylate synthetase, MX1, IRF-7, and toll-like receptor TLR-7 genes was lower in poor-response patients than in marked- or intermediate-response patients. Thus, the relative lack of viral response to interferon therapy of hepatitis C virus infection is associated with blunted interferon cell signaling. No specific regulatory gene could be identified as responsible for this global blunting or the racial differences.
Chronic hepatitis C virus (HCV) infection is a major cause of chronic liver disease and liver cancer (1, 2). An estimated 170 million persons worldwide, including 3 million in the United States, are actively infected with HCV. The prevalence of HCV infection varies by race and ethnicity, HCV infection being two times more common in African-Americans (AA) than Caucasian Americans (CA). The incidence of hepatocellular carcinoma is also greater in AA (9). The current recommended treatment for chronic HCV infection is the combination of pegylated alpha interferon (peginterferon) and the oral antiviral drug ribavirin given for 24 or 48 weeks (11, 15, 23, 31). This regimen is effective in eradicating virus in 70 to 80% of patients with genotype 2 or 3 HCV infection, but in only 40 to 50% of individuals infected with genotype 1, the most common genotype in the United States. For unknown reasons AA are less likely to respond to interferon-based therapy of HCV infection than CA (18, 20, 25, 27).
Alpha interferon mediates its antiviral and pharmacological effects by binding to type I interferon receptors on the cell surface membrane, which leads to transcription of up to 1,000 interferon-stimulated genes, presumably via the Janus-activated kinase 1 (JAK1)-STAT (signaling transducers of activation and transcription) signaling pathway (28, 30). A potential explanation for a lack of response to interferon therapy of HCV infection is an underlying deficient cellular response to interferon with a blunted response to interferon signaling, this being more common among AA patients than CA patients. To test this hypothesis, global gene expression in peripheral blood mononuclear cells (PBMC) before and during the first 28 days of therapy with peginterferon and ribavirin was analyzed in a cohort of AA and CA patients with genotype 1 HCV infection. These patients were undergoing therapy in the Study of Viral Resistance to Antiviral Therapy for Hepatitis C (Virahep-C), a large, prospective, multicenter study designed to define the differences in response rates among AA and CA patients and to determine clinical, immunological, host genetic, viral genetic, and interferon cell signaling factors that were associated with lack of response to treatment (5). The current analysis summarizes results of global gene expression in PBMC during the first 28 days of therapy, comparing patients with a marked (decrease of more than 3.5 log10 at 28 days following treatment initiation), intermediate (1.4 log10 to 3.5 log10 decrease), or poor (<1.4 log10 decrease) viral response.
MATERIALS AND METHODS
Patient treatment and samples.
The Virahep-C study enrolled a cohort of 196 AA and 205 CA participants from eight U.S. clinical centers, who were recruited between July 2002 and December 2003. The institutional review boards of participating centers approved the protocol, and all patients gave informed, written consent for both the therapy and investigations of viral, immunological, and host cell-signaling responses. Eligible patients were naive to interferon and ribavirin treatment and had detectable HCV RNA in serum, and nearly all had a liver biopsy performed within the previous 18 months showing chronic hepatitis. Only patients who were born in the United States and designated themselves as “African-American/black” or “Caucasian/white” were eligible. The clinical protocol called for participants to be treated for up to 48 weeks with peginterferon-2a (Pegasys; Roche Pharmaceuticals, Nutley, NJ) in a dose of 180 μg weekly by self-administered subcutaneous injection and ribavirin (Copegus; Roche) orally in a dose of 1,000 or 1,200 mg daily based on body weight of less than 75 kg or equal to or greater than 75 kg. Serum samples were tested for HCV RNA levels using a quantitative PCR-based assay (Cobas Amplicor HCV monitor test, version 2.0; Roche) on days 0 (pretreatment), 1, 2, 7, 14, and 28 and at weeks 12, 24, 48, and 72. PBMC were collected from patients before treatment (day 0) and on days 1 (after the initial supervised injection of peginterferon), 2, 7, 14, and 28. Treatment was discontinued at week 24 in participants whose serum was still HCV RNA positive by a sensitive qualitative PCR-based assay (Roche Cobas Amplicor HCV test, v2.0) that had a lower limit of sensitivity of 50 IU/ml. The primary end point of the study was a sustained virologic response, defined as lack of detectable serum HCV RNA in serum drawn 24 weeks after completing treatment. The overall results of this study have been published and demonstrated that the sustained virological response rate was higher among CA patients (52%) than AA patients (28%) and that the racial difference was not explained by clinical features such as age, gender, weight, severity of the underlying hepatitis, pretreatment viral levels, or amount of drug taken (6).
From the Virahep-C cohort, 72 patients who did not have dose reductions of either peginterferon or ribavirin in the first 28 days of treatment were selected such that 12 patients of each race (CA and AA) were included by virological-response category. The three categories of response were marked, defined as a decrease in HCV RNA levels of more than 3.5 log10 IU/ml or to an undetectable level on day 28; intermediate, defined as a decrease of 1.4 to 3.5 log10 IU/ml on day 28; and poor, defined as less than a 1.4 log10 IU/ml decline on day 28 relative to baseline. These definitions were made a priori in an attempt to analyze the biological basis for virological responses. Of these, RNA adequate to provide gene expression information was not obtained from three patients.
PBMC preparation.
PBMC were collected in sodium-heparin cell preparation tubes at day 0 (before treatment) and days 1, 2, 7, 14, and 28 after initiation of treatment. Whole blood was diluted with an equal volume (8 ml) of phosphate-buffered saline (PBS), carefully layered over a 10-ml Ficoll-Hypaque gradient (Amersham/Pharmacia, Piscataway, NJ), and centrifuged at 800 rpm for 20 min at room temperature. The buffy coat layer was transferred to a 15-ml RNase-free tube, diluted with PBS, and centrifuged at 100 × g for 15 min at room temperature. The supernatants were discarded, and the PBMC were retained.
RNA extraction.
Samples were shipped overnight by express courier at 4°C to a central repository, where RNA was isolated on arrival. The PBMC were lysed in 1 ml of TRI reagent (Molecular Research Center Inc., Cincinnati, OH). The PBMC lysate was mixed with 1-bromo-3-chloropropane phase separation agent for 1 min and incubated at room temperature for 15 min. After centrifugation for 15 min at 12,000 rpm and 4°C, RNA was precipitated from the supernatant overnight at −20°C with an equal volume of isopropanol and 1/10 volume of 7.5 M ammonium acetate. The precipitate was washed twice with 75% ethanol and then with 95% ethanol. RNA was briefly air dried and then further purified using RNeasy columns (QIAGEN, Valencia, CA). The amount and quality of RNA were determined by spectrophotometry and by electrophoresis through 1% agarose with ethidium bromide, and RNA quality was analyzed by the Agilent Bioanalyzer according to the manufacturer's instructions. Samples that did not show two clear bands of rRNA were discarded.
RNA labeling and hybridization.
Preparation of cDNA and cRNA and labeling were carried out according to the protocols recommended by Affymetrix (Santa Clara, CA) in the GeneChip expression analysis technical manual, as previously described (34).
Array analysis and data processing.
The microarrays were scanned using a dedicated model 3000 scanner controlled by GCOS software. The average intensity on each array was normalized by global scaling to a target intensity of 1,000. Data were extracted using the Affymetrix Microarray Suite 5 (MAS5) algorithm and exported into a custom-designed database (MicroArray Data Portal) in the Center for Medical Genomics (Indiana University-Purdue University Indianapolis, Indianapolis). All DNA microarray chips were analyzed for unequal distribution or artifacts as described previously (4). Any chip shown to be defective was corrected or dropped from the analysis.
The MicroArray Data Portal, in addition to its role as a database and analytical tool, is an informatics platform with active links from each sequence to several public databases. Sequence information for each gene on the HG-U133A GeneChip was obtained by parsing the HG-U133 target file obtained from the Affymetrix informatics website, http://www.affymetrix.com/analysis/download_center.affx). A GenBank accession number and a Unigene cluster were used to match sequences to their corresponding LocusLink number, gene symbol, and map position and to link with Gene Ontology (GO) terms and Enzyme Nomenclature (19) EC numbers. EC numbers were then used in conjunction with the Ligand database to link genes to KEGG pathways (www.genome.ad.jp/kegg/).
Statistical analysis.
The MAS5 data were filtered to eliminate any gene that was not called present in at least 50% of the samples in any one group (fraction present ≥ 0.5) (24). Changes (n-fold) for each gene were calculated using the ratio of the MAS5 signals of the baseline and the posttreatment time. If the signal for the posttreatment time point was greater than the baseline the change was calculated as +averageposttreatment/averagebaseline; otherwise, the change was calculated as −averagebaseline/averageposttreatment. The asymptotic standard errors (ASE) were estimated using the delta method, and 95% confidence intervals were calculated by multiplying the ASE by 1.96, with the product added to and subtracted from the change.
Welch's t test using the MAS5 signals was used to test for differences in gene expression between CA and AA, and one-way analysis of variance was used to test for differences among the three response groups.
For each gene, the expression levels of posttreatment time points were compared to the baseline (pretreatment) expression levels using a paired t test of the MAS5 signals. Genes whose P value ≤0.001 and for which the absolute value of the change was at least 1.5-fold were selected as significant. Because of the filtering and differences in power, the numbers of genes considered to be significant in mutually exclusive and exhaustive subgroups will not necessarily add up to the number of genes considered to be significant in the entire sample. Genes that are significant in both racial groups contribute twice to the sum of genes but only once to the number of significant genes in the entire sample. On the other hand, gene expression differences that meet the change criterion may not meet the criterion of a P value <0.001 in either racial group but, due to the increased number of observations for the entire sample compared to each racial group, do have a P of <0.001 for the entire sample; such genes do not contribute to the sum of significant genes across racial groups but do contribute to the number of significant genes in the entire sample.
All analyses were performed using the R statistical language and environment.
Microarray data accession number.
Microarray data presented in this paper have been deposited with NCBI/GEO under accession no. GSE7123.
RESULTS
The baseline features of the three response groups are shown in Table 1. The participants who were included in this analysis of gene expression were not representative of the total Virahep-C cohort since they were selected for nearly equal representation in the various race and response groups early in the study and the total cohort did not divide equally into these categories at the end of the study. Baseline features were similar across response groups, though marked responders had somewhat lower HCV RNA levels than the other response groups. However, this difference was not statistically significant (P = 0.11). Ultimately, sustained virological responses occurred in 81% of the marked responders, 35% of the intermediate responders, and 8% of the poor responders. Characteristics of AA and CA were also similar (Table 2), although AA tended to have lower alanine aminotransferase (ALT) values than did CA (P = 0.01), a difference that was also found in the total Virahep-C cohort (6). Racial differences in sustained virologic response were not apparent (39% for versus 47% for CA; P = 0.51), though they were in the total cohort, due to the selection criteria within each race whereby the three early-response groups were nearly equally represented in each race.
TABLE 1.
Baseline participant characteristics by response group
| Feature | Value for patients with:
|
P | ||
|---|---|---|---|---|
| Marked response | Intermediate response | Poor response | ||
| No. | ||||
| Total | 27 | 17 | 25 | |
| AA | 13 (48%) | 9 (53%) | 11 (44%) | 0.85a |
| Male | 17 (63%) | 14 (82%) | 19 (76%) | 0.33a |
| Body wt (kg) | ||||
| Mean (SD) | 84.7 (22.5) | 92.1 (23.4) | 93.1 (14.6) | 0.28b |
| Median (25th, 75th) | 79.8 (70.8, 97.1) | 94.3 (81.2, 99.8) | 93.9 (79.8, 104.3) | |
| HCV RNA level (log10 IU/ml) | ||||
| Mean (SD) | 6.0 (0.8) | 6.5 (0.6) | 6.4 (0.5) | |
| Median (25th, 75th) | 5.7 (5.3, 6.8) | 6.7 (6.2, 6.9) | 6.4 (6.2, 6.6) | 0.11c |
| ALT (mg/dl) | ||||
| Mean (SD) | 101.7 (94.1) | 69.5 (28.7) | 129.8 (106.3) | |
| Median (25th, 75th) | 68.0 (42.0, 123.0) | 65 (52.0, 81.0) | 83.0 (58.0, 164.0) | 0.13c |
| Liver histology | ||||
| Ishak necroinflammatory score (0-18) | ||||
| Mean (SD) | 8.2 (2.6) | 7.2 (2.4) | 7.8 (3.3) | |
| Median (25th, 75th) | 8 (6, 9) | 8 (6, 9) | 7 (5, 9) | 0.48c |
| Ishak fibrosis score (0-6) | ||||
| Mean (SD) | 2.1 (1.0) | 1.7 (1.3) | 2.4 (1.4) | |
| Median (25th, 75th) | 2 (1, 3) | 1 (1, 2) | 2 (2, 3) | 0.08c |
Chi-square test.
Analysis of variance.
Kruskal-Wallis test.
TABLE 2.
Baseline patient characteristics by race
| Feature | Value for:
|
P | |
|---|---|---|---|
| AA | CA | ||
| No. | |||
| Total | 33 | 36 | |
| Male | 24 (73%) | 26 (72%) | 0.96a |
| Body wt (kg) | |||
| Mean (SD) | 91.7 (23.3) | 87.6 (17.2) | 0.41b |
| Median (25th, 75th) | 84.4 (78.9, 104.3) | 91.2 (75.8, 100.2) | |
| HCV RNA level (log10 IU/ml) | |||
| Mean (SD) | 6.2 (0.7) | 6.3 (0.7) | |
| Median (25th, 75th) | 6.4 (5.5, 6.8) | 6.5 (5.7, 6.7) | 0.54c |
| ALT (mg/dl) | |||
| Mean (SD) | 79.2 (63.2) | 126.6 (104.9) | |
| Median (25th, 75th) | 63.0 (47.0, 83.0) | 91.5 (56.0, 169.0) | 0.01c |
| Liver histology | |||
| Ishak necroinflammatory score (0-18) | |||
| Mean (SD) | 7.8 (2.3) | 7.8 (3.2) | |
| Median (25th, 75th) | 7.0 (6.0, 9.0) | 7.5 (5.5, 9.0) | 0.60c |
| Ishak fibrosis score (0-6) | |||
| Mean (SD) | 1.9 (1.3) | 2.3 (1.1) | |
| Median (25th, 75th) | 2.0 (1.0, 2.0) | 2.0 (1.5, 3.0) | 0.16c |
Chi-square test.
Analysis of variance.
Wilcoxon rank sum test.
Oligonucleotide microarray analysis of gene expression in PBMC demonstrated less difference among poor, intermediate, and marked viral responders before therapy than would be expected by chance. For example, of the 10,910 genes whose expression was detected at baseline in at least one patient in each response category (and whose fraction present was at least 0.5 in any one group) only 73 differed across response groups at a P of ≤0.01 and 2 at a P of ≤0.001, whereas 109 and 11, respectively, would be expected by chance at these significance levels.
Global gene expression response is greater in marked responders than in poor responders.
Gene expression in PBMC changed substantially during peginterferon and ribavirin therapy, with major changes being evident by days 1 and 2 after the initial injection of peginterferon and administration of ribavirin. The numbers of genes that were significantly modified (absolute value of change greater than 1.5-fold and P ≤ 0.001) at each time point for each response group and for each racial group within the response group are shown in Table 3. Many genes were altered in expression at the early time points in patients in all three response groups. The number of differentially expressed genes dropped between day 2 and day 7 and increased again slightly between days 7 and 28. Postbaseline PBMC samples were generally taken before administration of interferon. For only one subject at two time points, samples were taken 4 h after administration of interferon; this did not appear to affect the results for this patient.
TABLE 3.
Number of genes (proportion present > 0.5) modified (at least 1.5-fold change; P ≤ 0.001) during peginterferon and ribavirin treatment
| Day (n) | No. of genes modified for:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Marked responders
|
Intermediate responders
|
Poor responders
|
|||||||
| AA | CA | All | AA | CA | All | AA | CA | All | |
| 1 (69) | 1,373 | 553 | 1,250 | 233 | 198 | 760 | 228 | 195 | 418 |
| 2 (68) | 586 | 336 | 669 | 177 | 110 | 384 | 206 | 87 | 272 |
| 7 (68) | 190 | 94 | 293 | 40 | 59 | 180 | 9 | 4 | 58 |
| 14 (63) | 181 | 151 | 240 | 50 | 74 | 183 | 50 | 9 | 72 |
| 28 (61) | 229 | 117 | 307 | 21 | 45 | 133 | 40 | 56 | 149 |
In another study, similar experiments were run with RNA samples taken directly from patient PBMC and cultured in vitro for 6 or 24 h with interferon/ribavirin, and similar patterns of gene induction or down regulation were obtained (34). Likewise, samples processed immediately (without shipping) in an ongoing study show very little difference from the data in these experiments.
Within marked responders, there were more genes changed in AA than in CA at every time point (Table 3). Among poor responders, the same relationship held except at day 28. The relationship was more mixed in intermediate responders, where more genes changed expression in AA than CA in the first 2 days after treatment but more genes changed expression in CA than in AA after that.
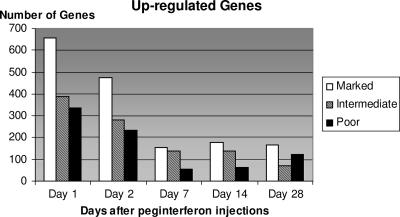
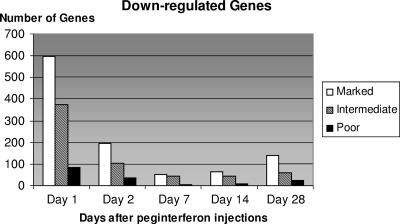
The number of genes that changed in expression was greater in the marked responders than in the intermediate or poor responders at all time points. There was a smaller difference in the numbers of genes that changed in expression between the intermediate and poor responders at most points. Figures 1 and 2 show the numbers of genes whose expressions increased or decreased, respectively, using the change filter of 1.5-fold and a P value of ≤0.001. The numbers of genes that were increased in gene expression were far higher in the marked responders than in either the intermediate or poor responders, and the intermediate responders had numbers intermediate between the other two groups. Slightly more genes were up- than down-regulated. Although the differences among the three response categories of patients held for both up- and down-regulation of genes, the decline over time in numbers of down-regulated genes was much sharper than the decline in up-regulated genes.
FIG. 1.
Number of genes up-regulated (P < 0.001; ≥1.5-fold change) at each time point compared to baseline in each response category of patient.
FIG. 2.
Number of genes down-regulated (P < 0.001; ≥1.5-fold change) at each time point compared to baseline in each category of patients.
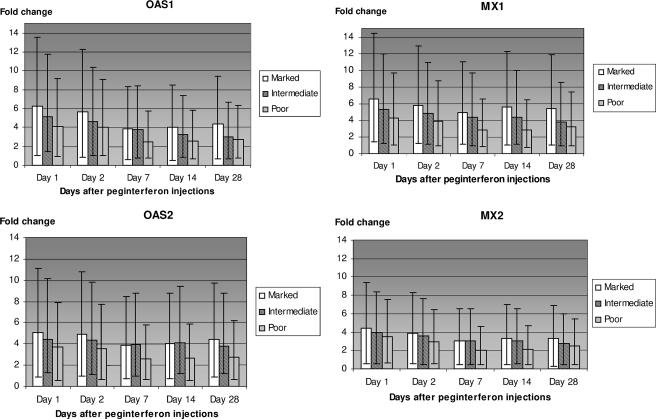
Compared to the baseline expression, there was an interferon response in all three categories of patients at each time point. The increases in four well-defined interferon-stimulated genes (2′5′-oligoadenylate synthetase 1 [OAS1] and OAS2, MX1, and MX2 genes) are shown in Fig. 3. At all time points, there was a significant difference in increases of these four genes between the marked and poor virological responders (P < 0.04 at each time point). There was less of a difference between marked and intermediate responders, and this difference did not reach statistical significance at every time point.
FIG. 3.
Increase in mRNA as detected by microarrays for oligo(A) synthetase 1 and 2 and MX1 and MX2 in all three response categories of patients at days 1, 2, 7, 14, and 28 after initiation of treatment.
Table 4 presents a selected group of genes that were up-regulated in marked and poor responders by function. These genes were chosen from the total list (See Table S1 in the supplemental material) because of previous association with interferon activity or because they form a functional group. The change for genes in poor responders was consistently lower than that found in marked responders. Supplemental Table S1 presents a list of individual induced genes, for patients of all three response groups at different time points after treatment initiation. Supplemental Table 2 presents a list of those genes that were down-regulated on different days. The majority of genes that were down-regulated encode products which are involved in translational regulation, such as eukaryotic elongation or translation factors and ribosomal proteins. In general, these were down-regulated substantially in marked-response patients but marginally in poor- and intermediate-response patients. As in the case of up-regulated genes in patients, the magnitude of the decrease in gene expression in poor responders was considerably less than in marked or intermediate responders. Table 5 compares the response between AA patients and CA patients on day 1. The responses in terms of change (n-fold) were very similar between the two groups.
TABLE 4.
Change in gene expressiona
| Function/description | Response | Symbol | Fold change in gene expression on day:
|
||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 7 | 14 | 28 | |||
| Antiviral response | |||||||
| Interferon-stimulated gene 20 kDa | Marked | ISG20 | 3.21 | 2.49 | 2.21 | 2.07 | 2.16 |
| Interferon-stimulated gene 20 kDa | Poor | ISG20 | 2.75 | 2.11 | 1.65 | 1.50 | 1.57 |
| 2′5′-Oligoadenylate synthetase 1 (40/46 kDa) | Marked | OAS1 | 7.03 | 6.78 | 4.18 | 3.80 | 4.29 |
| 2′5′-Oligoadenylate synthetase 1 (40/46 kDa) | Poor | OAS1 | 4.94 | 4.86 | 2.82 | 2.98 | 3.01 |
| 2′5′-Oligoadenylate synthetase 2 (69/71 kDa) | Marked | OAS2 | 5.10 | 4.91 | 3.88 | 4.04 | 4.44 |
| 2′5′-Oligoadenylate synthetase 2 (69/71 kDa) | Poor | OAS2 | 3.67 | 3.55 | 2.57 | 2.63 | 2.77 |
| 2′5′-Oligoadenylate synthetase-like | Marked | OASL | 7.83 | 6.72 | 3.57 | 4.09 | 4.16 |
| 2′5′-Oligoadenylate synthetase-like | Poor | OASL | 5.56 | 4.94 | 2.64 | 2.63 | 2.99 |
| Viperin, cig5 | Marked | RSAD2 | 18.64 | 17.94 | 10.3 | 14.28 | 15.58 |
| Viperin, cig 5 | Poor | RSAD2 | 12.11 | 12.1 | 6.70 | 7.13 | 7.66 |
| Apoptosis | |||||||
| XIAP associated factor 1 | Marked | HSXIAPAF1 | 3.48 | 4.00 | 3.42 | 3.74 | 3.52 |
| XIAP associated factor 1 | Poor | HSXIAPAF1 | 2.87 | 2.99 | 2.36 | 2.41 | 2.56 |
| TRAIL | Marked | TNFSF10 | 5.4 | 4.33 | 2.31 | 2.16 | 2.42 |
| TRAIL | Poor | TNFSF10 | 3.28 | 2.99 | 1.56 | 1.59 | 1.67 |
| Cell proliferation | |||||||
| Lysosomal-associated membrane protein 3 | Marked | LAMP3 | 11.52 | 6.61 | 5.5 | 5.13 | 6.19 |
| Lysosomal-associated membrane protein 3 | Poor | LAMP3 | 7.18 | 4.57 | 2.78 | 2.69 | 3.45 |
| Chemokines | |||||||
| Chemokine (C-C motif) ligand 2 | Marked | CCL2 | 58 | 41.07 | 16.24 | 23.59 | 15.65 |
| Chemokine (C-C motif) ligand 2 | Poor | CCL2 | 52.43 | 27.33 | 7.07 | 75.62 | 10.38 |
| Chemokine (C-C motif) receptor 1 | Marked | CCR1 | 4.22 | 3.88 | 2.21 | 2.86 | 2.82 |
| Chemokine (C-C motif) receptor 1 | Poor | CCR1 | 2.84 | 2.86 | 1.78 | 2.08 | 1.84 |
| Chemokine (C-C motif) ligand 8 | Marked | CCL8 | 11.18 | 5.28 | 2.01 | 2.47 | 2.85 |
| Chemokine (C-C motif) ligand 8 | Poor | CCL8 | 7.04 | 2.70 | 1.40 | 1.57 | 1.56 |
| Chemokine (C-X-C motif) ligand 11 | Marked | CXCL11 | 5.09 | 2.22 | −1.01 | 1.00 | −1.07 |
| Chemokine (C-X-C motif) ligand 11 | Poor | CXCL11 | 4.83 | 2.66 | 3.66 | 1.33 | 1.22 |
| Chemokine (C-X-C motif) ligand 10 | Marked | CXCL10 | 24.42 | 11.88 | 1.79 | 1.93 | 2.00 |
| Chemokine (C-X-C motif) ligand 10 | Poor | CXCL10 | 19.95 | 8.94 | 1.51 | 1.47 | 1.56 |
| Complement pathway | |||||||
| Complement component 3a receptor 1 | Marked | C3AR1 | 3.62 | 3.28 | 1.76 | 2.22 | 1.92 |
| Complement component 3a receptor 1 | Poor | C3AR1 | 2.38 | 2.32 | 1.35 | 1.49 | 1.33 |
| Serine (or cysteine) proteinase inhibitor | Marked | SERPING1 | 11.93 | 7.09 | 2.19 | 2.57 | 2.43 |
| Serine (or cysteine) proteinase inhibitor | Poor | SERPING1 | 8.88 | 5.65 | 1.94 | 2.29 | 2.24 |
| Endonuclease | |||||||
| Liver RNase (neurotoxin) | Marked | RNAse2 | 2.85 | 2.70 | 2.10 | 2.71 | 2.24 |
| Liver RNase (neurotoxin) | Poor | RNAse2 | 2.19 | 2.13 | 1.74 | 1.64 | 1.82 |
| Helicases | |||||||
| Hypothetical protein FLJ20035 | Marked | FLJ20035 | 4.56 | 4.38 | 4.10 | 4.06 | 4.27 |
| Hypothetical protein FLJ20035 | Poor | FLJ20035 | 3.36 | 2.85 | 2.42 | 2.16 | 2.66 |
| Interferon induced with helicase C domain 1 | Marked | IFIH1 | 5.59 | 4.99 | 3.52 | 3.70 | 4.21 |
| Interferon induced with helicase C domain 1 | Poor | IFIH1 | 3.59 | 3.25 | 2.24 | 2.20 | 2.48 |
| Immune response | |||||||
| Alpha interferon-inducible protein (clone IFI-15K) | Marked | G1P2 | 10.51 | 9.76 | 6.53 | 7.72 | 8.35 |
| Alpha interferon-inducible protein (clone IFI-15K) | Poor | G1P2 | 7.43 | 6.93 | 3.89 | 4.24 | 4.75 |
| Alpha interferon-inducible protein (clone IFI-6-16) | Marked | G1P3 | 4.11 | 4.09 | 3.02 | 3.22 | 3.07 |
| Alpha interferon-inducible protein (clone IFI-6-16) | Poor | G1P3 | 3.39 | 2.67 | 1.73 | 1.99 | 2.29 |
| Alpha interferon-inducible protein 27 | Marked | IFI27 | 27.98 | 41.54 | 61.26 | 100.47 | 121.84 |
| Alpha interferon-inducible protein 27 | Poor | IFI27 | 21.45 | 35.28 | 52.67 | 69.51 | 89.4 |
| Interferon-induced protein 35 | Marked | IFI35 | 4.75 | 4.18 | 2.77 | 2.58 | 2.71 |
| Interferon-induced protein 35 | Poor | IFI35 | 3.56 | 3.00 | 1.82 | 1.87 | 1.95 |
| Interferon-induced protein 44 | Marked | IFI44 | 6.62 | 6.31 | 6.03 | 6.59 | 6.77 |
| Interferon-induced protein 44 | Poor | IFI44 | 4.29 | 4.12 | 3.41 | 3.36 | 4.09 |
| Interferon-induced protein 44 | Marked | IFI44L | 9.09 | 9.36 | 9.13 | 10.68 | 10.67 |
| Interferon-induced protein 44 | Poor | IFI44L | 5.91 | 5.93 | 5.02 | 4.95 | 6.06 |
| Interferon-induced protein with tetratricopeptide repeats 1 | Marked | IFIT1 | 17.57 | 17.1 | 10.72 | 12.27 | 13.55 |
| Interferon-induced protein with tetratricopeptide repeats 1 | Poor | IFIT1 | 9.23 | 9.23 | 5.79 | 5.27 | 6.31 |
| Interferon-induced protein with tetratricopeptide repeats 3 | Marked | IFIT3 | 11.95 | 10.3 | 6.38 | 7.53 | 8.47 |
| Interferon-induced protein with tetratricopeptide repeats 3 | Poor | IFIT3 | 7.42 | 6.73 | 3.56 | 3.74 | 4.23 |
| Interferon-induced protein with tetratricopeptide repeats 5 | Marked | IFIT5 | 3.32 | 3.19 | 2.63 | 2.72 | 2.94 |
| Interferon-induced protein with tetratricopeptide repeats 5 | Poor | IFIT5 | 2.49 | 2.30 | 2.05 | 2.02 | 2.00 |
| Interferon-induced transmembrane protein 1 (9-27) | Marked | IFITM1 | 1.86 | 1.76 | 1.67 | 1.71 | 1.95 |
| Interferon induced transmembrane protein 1 (9-27) | Poor | IFITM1 | 1.71 | 1.63 | 1.42 | 1.41 | 1.62 |
| Interferon-induced transmembrane protein 3 (1-8U) | Marked | IFITM3 | 2.71 | 2.45 | 1.89 | 2.00 | 1.91 |
| Interferon-induced transmembrane protein 3 (1-8U) | Poor | IFITM3 | 2.28 | 2.20 | 1.60 | 1.65 | 1.82 |
| IL-1 receptor antagonist | Marked | IL1RN | 5.83 | 4.12 | 1.43 | 1.99 | 1.98 |
| IL-1 receptor antagonist | Poor | IL1RN | 4.21 | 2.93 | 1.44 | 2.05 | 1.75 |
| Myxovirus (influenza virus) resistance 1 | Marked | MX1 | 6.51 | 5.80 | 4.95 | 5.57 | 5.39 |
| Myxovirus (influenza virus) resistance 1 | Poor | MX1 | 4.29 | 3.90 | 2.83 | 2.87 | 3.26 |
| Myxovirus (influenza virus) resistance 2 | Marked | MX2 | 4.39 | 3.89 | 3.07 | 3.28 | 3.30 |
| Myxovirus (influenza virus) resistance 2 | Poor | MX2 | 3.46 | 2.94 | 2.04 | 2.12 | 2.49 |
| Inflammatory response | |||||||
| Heparanase | Marked | HPSE | 4.16 | 2.45 | 1.73 | 2.05 | 1.80 |
| Heparanase | Poor | HPSE | 2.97 | 1.92 | 1.39 | 1.50 | 1.59 |
| Sialoadhesin | Marked | SN | 44.54 | 39.65 | 23.14 | 33.58 | 26.4 |
| Sialoadhesin | Poor | SN | 18.17 | 16.45 | 9.37 | 11.92 | 12.41 |
| JAK-STAT cascade | |||||||
| N-myc (and STAT) interactor | Marked | NMI | 3.07 | 2.68 | 1.91 | 1.93 | 2.08 |
| N-myc (and STAT) interactor | Poor | NMI | 2.19 | 2.01 | 1.50 | 1.46 | 1.59 |
| Signal transducer and activator of transcription 1 (91 kDa) | Marked | STAT1 | 2.38 | 2.05 | 1.88 | 1.85 | 2.09 |
| Signal transducer and activator of transcription 1 (91 kDa) | Poor | STAT1 | 1.98 | 1.77 | 1.46 | 1.37 | 1.61 |
| Phospholipid scrambling | |||||||
| Phospholipid scramblase 1 | Marked | PLSCR1 | 4.61 | 4.22 | 2.92 | 3.32 | 3.50 |
| Phospholipid scramblase 1 | Poor | PLSCR1 | 3.24 | 2.97 | 2.17 | 2.01 | 2.50 |
| RNA editing | |||||||
| Apolipoprotein B mRNA editing enzyme | Marked | APOBEC3A | 6.03 | 4.66 | 2.63 | 2.93 | 2.94 |
| Apolipoprotein B mRNA editing enzyme | Poor | APOBEC3A | 4.29 | 3.53 | 2.08 | 2.15 | 2.22 |
| Signal transduction | |||||||
| Membrane-spanning 4 domains, subfamily A, member 4 | Marked | MS4A4A | 7.00 | 7.36 | 2.35 | 2.89 | 2.48 |
| Membrane-spanning 4 domains, subfamily A, member 4 | Poor | MS4A4A | 3.68 | 4.39 | 1.61 | 1.71 | 1.89 |
| Toll pathway | |||||||
| Toll-like receptor 7 | Marked | TLR7 | 7.17 | 5.12 | 2.90 | 3.19 | 2.79 |
| Toll-like receptor 7 | Poor | TLR7 | 3.93 | 3.07 | 1.70 | 1.66 | 1.80 |
| Transcription factor/repressor | |||||||
| CD38 antigen (p45) | Marked | CD38 | 2.61 | 2.15 | 1.89 | 1.86 | 1.90 |
| CD38 antigen (p45) | Poor | CD38 | 2.43 | 2.03 | 1.64 | 1.46 | 1.56 |
| Homeobox (expressed in ES cells) 1 | Marked | HESX1 | 23.01 | 17.4 | 9.44 | 10.46 | 9.53 |
| Homeobox (expressed in ES cells) 1 | Poor | HESX1 | 17.56 | 12.78 | 9.01 | 5.74 | 6.23 |
| Gamma interferon-inducible protein 16 | Marked | IFI16 | 2.28 | 2.05 | 1.94 | 1.80 | 1.80 |
| Gamma interferon-inducible protein 16 | Poor | IFI16 | 1.84 | 1.87 | 1.55 | 1.45 | 1.51 |
| Interferon regulatory factor 7 | Marked | IRF7 | 4.56 | 4.25 | 3.12 | 3.20 | 3.37 |
| Interferon regulatory factor 7 | Poor | IRF7 | 3.34 | 3.11 | 2.04 | 2.19 | 2.33 |
| Lectin, galactoside-binding, soluble, 9 (galectin 9) | Marked | LGALS9 | 3.04 | 2.86 | 1.86 | 1.81 | 2.07 |
| Lectin, galactoside-binding, soluble, 9 (galectin 9) | Poor | LGALS9 | 2.30 | 2.07 | 1.32 | 1.61 | 1.65 |
| Lymphocyte antigen 6 complex, locus E | Marked | LY6E | 4.59 | 4.93 | 3.68 | 3.31 | 3.85 |
| Lymphocyte antigen 6 complex, locus E | Poor | LY6E | 3.71 | 3.71 | 2.25 | 2.95 | 2.86 |
| Nuclear antigen Sp100 | Marked | SP100 | 2.04 | 1.90 | 1.89 | 1.82 | 1.75 |
| Nuclear antigen Sp100 | Poor | SP100 | 1.78 | 1.78 | 1.51 | 1.37 | 1.43 |
| SP110 nuclear body protein | Marked | SP110 | 1.98 | 2.03 | 1.76 | 1.80 | 1.85 |
| SP110 nuclear body protein | Poor | SP110 | 1.64 | 1.64 | 1.46 | 1.46 | 1.53 |
| T-box 3 (ulnar mammary syndrome) | Marked | TBX3 | 7.36 | 6.13 | 4.26 | 4.42 | 4.42 |
| T-box 3 (ulnar mammary syndrome) | Poor | TBX3 | 4.91 | 4.02 | 2.47 | 2.35 | 2.6 |
| Ubiquitin pathway | |||||||
| Hect domain and RLD 5 | Marked | HERC5 | 8.64 | 7.50 | 4.64 | 5.06 | 5.46 |
| Hect domain and RLD 5 | Poor | HERC5 | 5.92 | 5.09 | 2.88 | 2.85 | 3.18 |
| Hect domain and RLD 6 | Marked | HERC6 | 5.61 | 5.46 | 5.84 | 5.54 | 5.61 |
| Hect domain and RLD 6 | Poor | HERC6 | 4.19 | 4.07 | 3.59 | 3.35 | 3.76 |
| Promyelocytic leukemia | Marked | PML | 3.68 | 2.89 | 1.80 | 1.93 | 1.91 |
| Promyelocytic leukemia | Poor | PML | 3.07 | 2.26 | 1.49 | 1.60 | 1.60 |
| Ubiquitin-conjugating enzyme E2L 6 | Marked | UBE2L6 | 2.97 | 2.52 | 1.76 | 1.81 | 1.82 |
| Ubiquitin-conjugating enzyme E2L 6 | Poor | UBE2L6 | 2.54 | 2.19 | 1.42 | 1.45 | 1.53 |
| Ubiquitin-specific protease 18 | Marked | USP18 | 12.53 | 10.06 | 7.10 | 8.66 | 9.96 |
| Ubiquitin-specific protease 18 | Poor | USP18 | 6.89 | 5.80 | 4.00 | 3.95 | 4.89 |
| Unknown function | |||||||
| 28-kDa interferon-responsive protein | Marked | IFRG28 | 4.68 | 4.87 | 3.38 | 3.52 | 3.76 |
| 28-kDa interferon-responsive protein | Poor | IFRG28 | 3.43 | 3.09 | 2.16 | 2.10 | 2.11 |
Changes in gene expression for marked responders and poor responders at days 1, 2, 7, 14, and 28 after initiation of treatment. Data were derived from Table S1 in the supplemental material. Numbers in boldface represent values below the cutoff of a 1.5-fold change or a P value of >0.001.
TABLE 5.
Changes on day 1 for CA and AA and the combined groupa
| Function/description | Symbol | Fold change for:
|
P | ||
|---|---|---|---|---|---|
| All | CA | AA | |||
| Antiviral response | |||||
| Interferon-stimulated gene 20 kDa | ISG20 | 2.94 | 2.71 | 3.19 | 0.15 |
| 2′5′-Oligoadenylate synthetase 1, 40/46 kDa | OAS1 | 5.77 | 5.29 | 6.26 | 0.32 |
| 2′5′-Oligoadenylate synthetase 2, 69/71kDa | OAS2 | 4.37 | 4.16 | 4.61 | 0.38 |
| 2′5′-Oligoadenylate synthetase-like | OASL | 6.68 | 6.19 | 7.15 | 0.28 |
| Radical S-adenosyl methionine domain containing 2 | RSAD2 | 15.66 | 14.77 | 16.56 | 0.60 |
| Apoptosis | |||||
| XIAP associated factor-1 | HSXIAPAF1 | 3.40 | 3.17 | 3.61 | 0.50 |
| TRAIL | TNFSF10 | 4.45 | 4.05 | 4.88 | 0.25 |
| Cell proliferation | |||||
| Lysosomal-associated membrane protein 3 | LAMP3 | 9.17 | 8.91 | 9.42 | 0.77 |
| Chemokines | |||||
| Chemokine (C-C motif) ligand 2 | CCL2 | 46.88 | 39.68 | 53.27 | 0.36 |
| Chemokine (C-C motif) receptor 1 | CCR1 | 3.58 | 3.46 | 3.69 | 0.68 |
| Chemokine (C-C motif) ligand 8 | CCL8 | 9.58 | 7.85 | 10.91 | 0.29 |
| Chemokine (C-X-C motif) ligand 11 | CXCL11 | 5.62 | 5.17 | 6.19 | 0.48 |
| Chemokine (C-X-C motif) ligand 10 | CXCL10 | 21.81 | 19.51 | 24.34 | 0.25 |
| Complement pathway | |||||
| Complement component 3a receptor 1 | C3AR1 | 3.02 | 2.80 | 3.31 | 0.21 |
| Serine (or cysteine) proteinase inhibitor | SERPING1 | 10.64 | 9.51 | 11.94 | 0.28 |
| Endonuclease | |||||
| Liver RNase (neurotoxin) | RNAse2 | 2.54 | 2.49 | 2.60 | 0.73 |
| Helicase | |||||
| Hypothetical protein FLJ20035 | FLJ20035 | 3.92 | 4.10 | 3.73 | 0.42 |
| Interferon induced with helicase C domain 1 | IFIH1 | 3.91 | 3.69 | 4.09 | 0.64 |
| Immune response | |||||
| Alpha interferon-inducible protein (clone IFI-15K) | G1P2 | 8.78 | 7.80 | 9.89 | 0.22 |
| Alpha interferon-inducible protein (clone IFI-6-16) | G1P3 | 3.75 | 3.42 | 4.11 | 0.31 |
| Alpha interferon-inducible protein 27 | IFI27 | 25.98 | 21.91 | 29.71 | 0.39 |
| Interferon-induced protein 35 | IFI35 | 3.96 | 3.63 | 4.34 | 0.31 |
| Interferon-induced protein 44 | IFI44 | 5.32 | 5.62 | 5.02 | 0.66 |
| Interferon-induced protein 44 | IFI44L | 7.38 | 7.25 | 7.52 | 0.49 |
| Interferon-induced protein with tetratricopeptide repeats 1 | IFIT1 | 12.68 | 13.10 | 12.30 | 0.88 |
| Interferon-induced protein with tetratricopeptide repeats 3 | IFIT3 | 9.40 | 9.62 | 9.19 | 0.78 |
| Interferon-induced protein with tetratricopeptide repeats 5 | IFIT5 | 2.98 | 2.89 | 3.08 | 0.84 |
| Interferon induced transmembrane protein 1 (9-27) | IFITM1 | 1.82 | 1.77 | 1.86 | 0.55 |
| Interferon induced transmembrane protein 3 (1-8U) | IFITM3 | 2.49 | 2.35 | 2.63 | 0.49 |
| IL-1 receptor antagonist | IL1RN | 4.76 | 4.19 | 5.47 | 0.31 |
| Myxovirus (influenza virus) resistance 1 | MX1 | 5.29 | 5.47 | 5.12 | 0.11 |
| Myxovirus (influenza virus) resistance 2 | MX2 | 3.92 | 3.67 | 4.19 | 0.65 |
| Inflammatory response | |||||
| Heparanase | HPSE | 3.65 | 3.22 | 4.19 | 0.03 |
| Sialoadhesin | SN | 28.31 | 28.66 | 28.02 | 0.96 |
| JAK-STAT cascade | |||||
| N-myc (and STAT) interactor | NMI | 2.60 | 2.47 | 2.76 | 0.35 |
| Signal transducer and activator of transcription 1 (91 kDa) | STAT1 | 2.17 | 2.11 | 2.24 | 0.54 |
| Phospholipid scrambling | |||||
| Phospholipid scramblase 1 | PLSCR1 | 4.00 | 3.75 | 4.29 | 0.31 |
| RNA editing | |||||
| Apolipoprotein B mRNA editing enzyme | APOBEC3A | 4.88 | 4.26 | 5.70 | 0.06 |
| Signal transduction | |||||
| Membrane-spanning 4 domains, subfamily A, member 4 | MS4A4A | 5.51 | 4.68 | 6.40 | 0.07 |
| All | CA | AA | |||
| Toll pathway | |||||
| Toll-like receptor 7 | TLR7 | 5.66 | 5.37 | 5.98 | 0.51 |
| Transcription factor/repressor | |||||
| CD38 antigen (p45) | CD38 | 2.46 | 2.29 | 2.64 | 0.16 |
| Homeobox (expressed in embryonic stem cells) 1 | HESX1 | 22.41 | 19.63 | 25.53 | 0.24 |
| Gamma interferon-inducible protein 16 | IFI16 | 2.07 | 2.04 | 2.10 | 0.67 |
| Interferon regulatory factor 7 | IRF7 | 3.84 | 3.49 | 4.22 | 0.13 |
| Lectin, galactoside-binding, soluble, 9 (galectin 9) | LGALS9 | 2.56 | 2.49 | 2.63 | 0.74 |
| Lymphocyte antigen 6 complex, locus E | LY6E | 3.95 | 3.79 | 4.13 | 0.73 |
| Nuclear antigen Sp100 | SP100 | 1.92 | 1.96 | 1.88 | 0.69 |
| SP110 nuclear body protein | SP110 | 1.71 | 1.75 | 1.67 | 0.65 |
| T-box 3 (ulnar mammary syndrome) | TBX3 | 5.93 | 5.90 | 5.96 | 0.94 |
| Ubiquitin pathway | |||||
| Hect domain and RLD 5 | HERC5 | 7.30 | 7.13 | 7.47 | 0.75 |
| Hect domain and RLD 6 | HERC6 | 5.01 | 4.82 | 5.24 | 0.43 |
| Promyelocytic leukemia | PML | 3.31 | 3.18 | 3.44 | 0.48 |
| Ubiquitin-conjugating enzyme E2L 6 | UBE2L6 | 2.69 | 2.51 | 2.90 | 0.07 |
| Ubiquitin-specific protease 18 | USP18 | 9.62 | 8.37 | 11.10 | 0.12 |
| Unknown function | |||||
| 28-kDa interferon-responsive protein | IFRG28 | 4.31 | 3.69 | 5.01 | 0.01 |
Changes (n-fold) were defined as +averageday 1/averageday 0, if averageday 1 > averageday 0, or as −averageday 0/averageday 1, if averageday 1 < averageday 0. The standard error of a change (n = fold) was estimated by the delta method. A two-sample Z test was constructed to compare changes (n = fold) between CA and AA according to the equation Z = FCCA − FCAA/(std.FCCA2 + std.FCAA2)1/2, where Z is approximately standard normal, FCCA and FCAA are the changes (n-fold) for CA and AA, respectively, and std.FCCA and std.FCAA are corresponding standard errors.
Most of the genes that were up-regulated during therapy and that are presented in Table 4 are well-known interferon-induced genes that have been described as being increased in expression in human PBMC in vitro with interferon stimulation (75% concordance) (34). Many of the genes that were induced at days 1 and 2 were no longer increased in expression in PBMC collected at days 7, 14, and 28 (see Tables S1 and S2 in the supplemental material). Among the major genes with early transient expression were those encoding chemokines CXCL10 (IP10), CXCL11, CCL8, and TNFSF10 (TRAIL). Several of these genes are known to be strongly induced by gamma interferon (5, 14). Other gamma interferon-induced genes such as the WARS, INDO, and caspase genes were also induced transiently at a low level during the first 24 h after the initial injection of peginterferon (see Table S1 in the supplemental material). However, gamma interferon mRNA was not detectable, nor was gamma interferon detectable by enzyme-linked immunosorbent assay (data not shown). Other genes transiently induced include the interleukin-1Ra (IL-1Ra) gene, previously shown to be induced only during the first 48 h of treatment (7). A subgroup of genes showed no change with time in any of the patient categories. These included those encoding IFI44, IFI44L, IFIH1, MX1, OAS2, SP100, SP110, IFIT1, IFITM1, and HERC6, genes known to be important in the antiviral response. Other genes such as the IFI27 gene increased with time in all three classes of patients. The function of this gene is unknown, but it was induced to very high levels (approximately 100-fold) by day 28. Other genes such as the carbonic anhydrase 1 gene were induced late (days 14 and 28) after treatment initiation.
DISCUSSION
The major finding of this study was that patients who exhibited a vigorous early virological response to treatment with peginterferon and ribavirin had concurrent vigorous alterations in PBMC gene mRNA levels, including genes whose levels were induced and repressed. Recognizing that several of the genes examined have expressions that may not meet the assumptions of the t tests, the analyses were repeated using a nonparametric rank sum test. Though this affected the results for some genes, the main conclusion that the number of genes whose levels were induced or repressed was greater among marked responders than poor responders remained true. Included in these genes whose expression was strongly altered were the classic interferon-induced genes. Thus, among marked responders (whose HCV RNA levels decreased by 3.5 logs or more 1 month after starting therapy) 655 genes were increased and 595 genes decreased in expression within 24 h of the first injection of peginterferon. For comparison, among poor responders (whose HCV RNA levels decreased by less than 1.4 logs after 1 month of therapy) the number of up-regulated genes was only 336 and only 82 were down-regulated within 24 h of starting therapy (Fig. 1 and 2). This difference was not due to lack of compliance because the initial doses of peginterferon and ribavirin were administered under observation. These findings suggest that poor or nonresponse to interferon-based therapy in chronic HCV infection may be due to a blunted induction of interferon-responsive genes. The finding that large number of genes are “down-regulated” in marked responders and not in poor responders may be indicative of a lack of sufficient oligo(A) synthetase, resulting in lowered activation of RNase L (26, 28, 29). However, a core of genes did appear to be actively down-regulated, since genes involved in translation regulation such as translation elongation factor and ribosomal protein genes appeared to be down-regulated in both marked and poor responders. Such genes have previously been reported to be down-regulated by alpha interferon in PBMC in culture (34).
In the Virahep-C study, AA had lower rates of sustained response and higher levels of serum HCV RNA than CA at almost all time points (6). The current analyses show that differences in the number of genes whose expression changed at least 1.5-fold between marked and poor responders in both AA and CA patients occurred within 24 h of starting treatment (Tables 4 and 5; Fig. 1 and 2). The level of gene expression and number of genes induced or down-regulated were considerably higher among marked virological responders than among poor responders (see Tables S1 and S2 in the supplemental material). However, it is puzzling that AA had higher numbers of genes induced and slightly higher levels of gene expression at day 1 than others.
Alpha interferon is known to act through induction of a large number of genes, the exact number and pattern of which have only been partially identified (33, 34). In this study, 801 genes were found to be increased during peginterferon and ribavirin therapy; many, but not all, of these were known interferon-induced genes (see Tables S1 and S2 in the supplemental material). While global interferon-induced gene expression was less among poor responders than marked responders, no specific gene could be linked to the differences in responses or to racial differences. Thus, poor or nonresponse appeared to be a global blunting of interferon cell signaling, rather than the lack of induction or function of a specific antiviral gene product. These data are in agreement with recent findings in which the gene expression of nonresponders was lower than that of responders when PBMC were cultured from such patients (16). Despite differences in the microarray systems used and assessment of in vitro versus in vivo responses, the levels of induction (n-fold) for many genes were remarkably similar. However, He et al. (16) found that the levels of gene induction in white patients was higher than in black patients. However, we could not find any difference between the racial groups in this study in levels of gene expression. The present study demonstrates that, controlling for virological response, gene expression changes were actually more common, within 2 weeks of treatment initiation, in AA than CA patients who had a marked or poor response. The reason for this difference is not known.
Previous studies using cell culture systems have suggested that HCV replication or presence of HCV antigens may interfere with specific interferon-induced gene products, such as the well characterized antiviral enzymes OAS, protein kinase R, and adenosine deaminase (8, 10, 12). The present analysis, in contrast, suggests that lack of response to administered interferon was due to an ongoing physiological defect that causes blunted regulation of interferon responsiveness. The blunted response might be due to a prior inflammatory response, interferon receptor deficiency or dysfunction, or lack of afferent cell signaling through the JAK-STAT pathway. In this regard, several recent studies in vitro and in vivo have suggested that a deficiency in STAT1 activation or DNA binding occurs in patients with chronic HCV infection (22). Such findings are compatible with the findings in this study. In fact genes such as the IRF-7 gene (Tables 4 and 5), a key gene in induction of interferon, was induced compared to baseline at lower levels in poor responders than in marked-response patients, as was the cig 5 (viperin) gene, previously identified as being important in the interferon response to hepatitis C virus (17). Toll-like receptor 7 (TLR7) has been shown to be important in the recognition of single-stranded viral RNA and subsequent signaling of the interferon, IκB kinase α/β/γ, and mitogen-activated protein kinase cascades leading to NF-κB and AP-1 activation and to IRF-7 and interferon production (13). Expression of the IRF-7 gene was increased from baseline to levels almost twice as high in marked-response than in poor-response patients and was thus strongly induced by peginterferon/ribavirin combination therapy.
Several limitations of the present findings deserve mention. First and foremost, the analysis of gene expression was conducted on PBMC and not on hepatocytes that harbor replicating HCV. Analysis of hepatocytes, however, requires liver biopsy, an invasive procedure which cannot be done repeatedly in humans during interferon therapy. Furthermore, analyses on liver tissue include PBMC and other nonparenchymal cells, and changes in expression in liver tissue may not reflect effects on hepatocytes only. Responses in PBMC are more likely to reflect a global response and not be under the local control of replicating virus or disease activity, which may modulate interferon responses. The chimpanzee model of HCV infection offers a potential approach to analyzing intrahepatic gene expression during interferon therapy (3, 21, 32). However, chimpanzees respond minimally to human alpha interferon therapy, and interpretation of results has to take into consideration interspecies differences.
A final limitation to this study was that it was based upon viral kinetic analyses done during the first 28 days of therapy and was not based on results of sustained virological responses. This design was purposeful, in that early virological responses are highly predictive of ultimate responses and are not affected by nonbiologic factors, such as dose modification, compliance, and dropout. Only patients who took the full prescribed dose of peginterferon were selected. Furthermore, the differences between responders and nonresponders in the strength of gene induction were found even at day 1, which occurred after an observed administration of peginterferon and ribavirin at the initiation of treatment. Thus, by using early viral responses, purely biological factors associated with response and nonresponse could be assessed.
In this study, a poor virological response to peginterferon and ribavirin therapy of HCV infection was found to be associated with global, blunted changes in interferon-responsive gene expression. These results indicate that the blunted response is not specific to the liver or to virally infected cells. This hyporesponsiveness may be determined by host genetics, or it may be due to an environmentally induced lesser sensitivity to interferon. It is also possible that PBMC are exposed to viral proteins in circulation or to hepatocyte-associated HCV antigens which might alter the immune response of such cells to interferon treatment.
Supplementary Material
Acknowledgments
This study was funded as a cooperative agreement by the National Institute of Diabetes and Digestive and Kidney Diseases with cosupport from the Intramural Research Program of the National Cancer Institute and with further support under a Cooperative Research and Development Agreement with Roche Laboratories, Inc. Grant numbers are as follows: U01 DK60329, U01 DK60340, U01 DK60324, U01 DK60344, U01 DK60327, U01 DK60335, U01 DK60352, U01 DK60342, U01 DK60345, U01 DK60309, U01 DK60346, U01 DK60349, and U01 DK60341. Other support was received from the National Center for Research Resources General Clinical Research Centers Program, grants M01 RR00645 (New York Presbyterian), M02 RR000079 (University of California, San Francisco), M01 RR16500 (University of Maryland), M01 RR000042 (University of Michigan), and M01 RR00046 (University of North Carolina). The Center for Medical Genomics is supported in part by grants from the Indiana 21st Century Research and Technology Fund and the Indiana Genomics Initiative (supported in part by the Lilly Endowment, Inc.).
Members of Virahep-C contributing to the study include, from the Beth Israel Deaconess Medical Center, Boston, MA, Nezam Afdhal (principal investigator) and Tiffany Geahigan (research coordinator); from the New York-Presbyterian Medical Center, New York, NY, Robert S. Brown, Jr. (principal investigator), Lorna Dove (coinvestigator), Shana Stovel (study coordinator), and Maria Martin (study coordinator); from the University of California, San Francisco, San Francisco, Norah Terrault, (principal investigator), Stephanie Straley, Eliana Agudelo, Melissa Hinds (clinical research coordinator), and Jake Heberlein (clinical research coordinator); from Rush University, Chicago, IL, Thelma E. Wiley (principal investigator) and Monique Williams (study coordinator); from the University of Maryland, Baltimore, Charles D. Howell (principal investigator), Kelly Gibson (project coordinator), Karen Callison (study coordinator), and Jane Lewis (study coordinator); from the University of Miami, Miami, FL, Lennox J. Jeffers (principal investigator), Shvawn McPherson Baker (coinvestigator), Maria DeMedina (project manager), and Carol Hermitt (project coordinator); from the University of Michigan, Ann Arbor, Hari S. Conjeevaram (principal investigator), Robert J. Fontana (coinvestigator), and Donna Harsh (study coordinator); from the University of North Carolina, Chapel Hill, Michael W. Fried (principal investigator [K24 DK066144]), Scott R. Smith (coinvestigator), Dickens Theodore (coinvestigator), Steven Zacks (coinvestigator), Roshan Shrestha (coinvestigator), Karen Dougherty (coinvestigator), Paris Davis (study coordinator), and Shirley Brown (study coordinator); from St. Louis University, St. Louis, MO, John E. Tavis (principal investigator), Adrian Di Bisceglie (coinvestigator), Ermei Yao (coinvestigator), Maureen Donlin (coinvestigator), Nathan Cannon (graduate student), and Ping Wang (lab technician); from Cedars-Sinai Medical Center, Los Angeles, CA, Huiying Yang (principal investigator), George Tang (project scientist), and Dai Wang (project scientist); from the University of Colorado Health Sciences Center, Denver, Hugo R. Rosen (principal investigator), James R. Burton (coinvestigator), and Jared Klarquist (lab technician); from Veteran's Administration, Portland, OR, Scott Weston (lab technician); from Indiana University, Bloomington, Milton W. Taylor (principal investigator), Corneliu Sanda (postdoctoral associate), Takuma Tsukahara (statistician), and Mary Ferris (lab assistant); from the Data Coordinating Center, Graduate School of Public Health at the University of Pittsburgh, Pittsburgh, PA, Steven H. Belle (principal investigator), Richard A. Bilonick (statistician), Geoffrey Block (coinvestigator), Jennifer Cline (data manager), Marika Haritos (statistician), KyungAh Im (statistician), Stephanie Kelley (data manager), Sherry Kelsey (coinvestigator), Laurie Koozer (project coordinator), Sharon Lawlor (data coordinator), Stephen B. Thomas (coinvestigator), Abdus Wahed (statistician), Yuling Wei (project coordinator), Leland J. Yee (consultant), and Song Zhang (statistician); from the National Institute of Diabetes and Digestive and Kidney Diseases, Patricia Robuck (project scientist), James Everhart (scientific advisor), Jay H. Hoofnagle (scientific advisor), Edward Doo (scientific advisor), T. Jake Liang (scientific advisor), and Leonard B. Seeff (scientific advisor); and from the National Cancer Institute, David E. Kleiner (central pathologist).
We thank Mary Ferris for the excellent record keeping and entering of data into the portal at the Center for Medical Genetics. We thank Ron Jerome and Chunxiao Zhu for expert assistance with the microarray studies, which were carried out using the facilities of the Center for Medical Genomics at Indiana University School of Medicine. We also thank Song Zhang and Jia Li from the data coordinating center, Pittsburgh, for statistical support and Jay H. Hoofnagle for help in editing the manuscript.
Footnotes
Published ahead of print on 31 January 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Armstrong, G. L., A. Wasley, E. P. Simard, G. M. McQuillan, W. L. Kuhnert, and M. J. Alter. 2005. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705-714. [DOI] [PubMed] [Google Scholar]
- 2.Benvegnu, L., M. Gios, S. Boccato, and A. Alberti. 2004. Natural history of compensated viral cirrhosis: a prospective study on the incidence and hierarchy of major complications. Gut 53:744-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bigger, C. B., B. Guerra, K. M. Brasky, G. Hubbard, M. R. Beard, B. A. Luxon, S. M. Lemon, and R. E. Lanford. 2004. Intrahepatic gene expression during chronic hepatitis C virus infection in chimpanzees. J. Virol. 78:13779-13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodsky, L., A. Leontovich, M. Shtutman, and E. Feinstein. 2004. Identification and handling of artifactual gene expression profiles emerging in microarray hybridization experiments. Nucleic Acids Res. 32:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassatella, M. A., S. Gasperini, F. Calzetti, A. Bertagnin, A. D. Luster, and P. P. McDonald. 1997. Regulated production of the interferon-gamma-inducible protein-10 (IP-10) chemokine by human neutrophils. Eur. J. Immunol. 27:111-115. [DOI] [PubMed] [Google Scholar]
- 6.Conjeevaram, H. S., M. W. Fried, L. J. Jeffers, N. A. Terrault, T. E. Wiley-Lucas, N. Afdhal, R. S. Brown, S. H. Belle, J. H. Hoofnagle, D. E. Kleiner, and C. D. Howell. 2006. Peginterferon and ribavirin treatment in African Americans and Caucasian American patients with chronic hepatitis C genotype I. J. Gastroenterology 131:470-477. [DOI] [PubMed] [Google Scholar]
- 7.Cotler, S. J., T. Craft, M. Ferris, M. Morrisey, J. McCone, K. R. Reddy, A. Conrad, D. M. Jensen, J. Albrecht, and M. W. Taylor. 2002. Induction of IL-1Ra in resistant and responsive hepatitis C patients following treatment with IFN-con1. J. Interferon Cytokine Res. 22:549-554. [DOI] [PubMed] [Google Scholar]
- 8.Duong, F. H., V. Christen, J. M. Berke, S. H. Penna, D. Moradpour, and M. H. Heim. 2005. Upregulation of protein phosphatase 2Ac by hepatitis C virus modulates NS3 helicase activity through inhibition of protein arginine methyltransferase 1. J. Virol. 79:15342-15350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Serag, H. B. 2004. Hepatocellular carcinoma: recent trends in the United States. Gastroenterology 127:S27-S34. [DOI] [PubMed] [Google Scholar]
- 10.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 102:2986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 12.Gale, M., Jr., and E. M. Foy. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436:939-945. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Sastre, A., and C. A. Biron. 2006. Type 1 interferons and the virus-host relationship: a lesson in detente. Science 312:879-882. [DOI] [PubMed] [Google Scholar]
- 14.Gasperini, S., M. Marchi, F. Calzetti, C. Laudanna, L. Vicentini, H. Olsen, M. Murphy, F. Liao, J. Farber, and M. A. Cassatella. 1999. Gene expression and production of the monokine induced by IFN-gamma (MIG), IFN-inducible T cell alpha chemoattractant (I-TAC), and IFN-gamma-inducible protein-10 (IP-10) chemokines by human neutrophils. J. Immunol. 162:4928-4937. [PubMed] [Google Scholar]
- 15.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346-355. [DOI] [PubMed] [Google Scholar]
- 16.He, X. S., X. Ji, M. B. Hale, R. Cheung, A. Ahmed, Y. Guo, G. P. Nolan, L. M. Pfeffer, T. L. Wright, N. Risch, R. Tibshirani, and H. B. Greenberg. 2006. Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology 44:352-359. [DOI] [PubMed] [Google Scholar]
- 17.Helbig, K. J., D. T. Lau, L. Semendric, H. A. Harley, and M. R. Beard. 2005. Analysis of ISG expression in chronic hepatitis C identifies viperin as a potential antiviral effector. Hepatology 42:702-710. [DOI] [PubMed] [Google Scholar]
- 18.Howell, C., L. Jeffers, and J. H. Hoofnagle. 2000. Hepatitis C in African Americans: summary of a workshop. Gastroenterology 119:1385-1396. [DOI] [PubMed] [Google Scholar]
- 19.International Union of Biochemistry and Molecular Biology. 1992. Enzyme nomenclature. Academic Press, San Diego, CA.
- 20.Jeffers, L. J., W. Cassidy, C. D. Howell, S. Hu, and K. R. Reddy. 2004. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology 39:1702-1708. [DOI] [PubMed] [Google Scholar]
- 21.Lanford, R. E., B. Guerra, H. Lee, D. Chavez, K. M. Brasky, and C. B. Bigger. 2006. Genomic response to interferon-alpha in chimpanzees: implications of rapid downregulation for hepatitis C kinetics. Hepatology 43:961-972. [DOI] [PubMed] [Google Scholar]
- 22.Lin, W., W. H. Choe, Y. Hiasa, Y. Kamegaya, J. T. Blackard, E. V. Schmidt, and R. T. Chung. 2005. Hepatitis C virus expression suppresses interferon signaling by degrading STAT1. Gastroenterology 128:1034-1041. [DOI] [PubMed] [Google Scholar]
- 23.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 24.McClintick, J. N., and H. J. Edenberg. 2006. Effect of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics 7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muir, A. J., J. D. Bornstein, and P. G. Killenberg. 2004. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N. Engl. J. Med. 350:2265-2271. [DOI] [PubMed] [Google Scholar]
- 26.Nijs, J., and K. De Meirleir. 2005. Impairments of the 2-5A synthetase/RNase L pathway in chronic fatigue syndrome. In Vivo 19:1013-1021. [PubMed] [Google Scholar]
- 27.Reddy, K. R., J. H. Hoofnagle, M. J. Tong, W. M. Lee, P. Pockros, E. J. Heathcote, D. Albert, T. Joh, et al. 1999. Racial differences in responses to therapy with interferon in chronic hepatitis C. Hepatology 30:787-793. [DOI] [PubMed] [Google Scholar]
- 28.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silverman, R. H. 2003. Implications for RNase L in prostate cancer biology. Biochemistry 42:1805-1812. [DOI] [PubMed] [Google Scholar]
- 30.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 31.Strader, D. B., T. Wright, D. L. Thomas, and L. B. Seeff. 2004. Diagnosis, management, and treatment of hepatitis C. Hepatology 39:1147-1171. [DOI] [PubMed] [Google Scholar]
- 32.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan, H., J. Derrick, J. Hong, C. Sanda, W. M. Grosse, H. J. Edenberg, M. Taylor, S. Seiwert, and L. M. Blatt. 2005. Global transcriptional profiling combination of type I and type II demonstrates the interferon enhances antiviral and immune responses at clinically relevant doses. J. Interferon Cytokine Res. 25:632-649. [DOI] [PubMed] [Google Scholar]
- 34.Taylor, M. W., W. M. Grosse, J. E. Schaley, C. Sanda, X. Wu, S. C. Chien, F. Smith, T. G. Wu, M. Stephens, M. W. Ferris, J. N. McClintick, R. E. Jerome, and H. J. Edenberg. 2004. Global effect of PEG-IFN-alpha and ribavirin on gene expression in PBMC in vitro. J. Interferon Cytokine Res. 24:107-118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.