Abstract
During adenovirus virion assembly, the packaging sequence mediates the encapsidation of the viral genome. This sequence is composed of seven functional units, termed A repeats. Recent evidence suggests that the adenovirus IVa2 protein binds the packaging sequence and is involved in packaging of the genome. Study of the IVa2-packaging sequence interaction has been hindered by difficulty in purifying the protein produced in virus-infected cells or by recombinant techniques. We report the first purification of a recombinant untagged version of the adenovirus IVa2 protein and characterize its binding to the packaging sequence in vitro. Our data indicate that there is more than one IVa2 binding site within the packaging sequence and that IVa2 binding to DNA requires the A-repeat consensus, 5′-TTTG-(N8)-CG-3′. Furthermore, we present evidence that IVa2 forms a multimeric complex on the packaging sequence. These data support a model in which adenovirus DNA packaging occurs via the formation of a IVa2 multiprotein complex on the packaging sequence.
Assembly of adenovirus (Ad) virions is a step-wise process, occurring through an ordered series of intermediates that is thought to resemble the assembly of double-stranded DNA bacteriophages, such as Φ29, T4, and λ, in which the virus genome is packaged into preformed capsids (reviewed in references 2, 5, and 27). The assembly of adenovirus virions initiates with the formation of empty, precursor capsids (4, 16). Formation of immature virions, the next intermediate in the assembly pathway, is generated by the simultaneous insertion of core proteins and the Ad DNA (4, 20). The virus genome is selectively packaged in a polar fashion starting at the left end of the genome (3, 31). Subsequent to the insertion of the genome and core proteins, an Ad-encoded endoprotease processes a subset of virion proteins to transform the immature particle into a mature virion, the final infectious form (1).
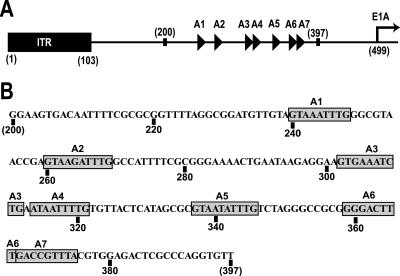
The packaging sequence, which mediates the specific packaging of the Ad genome, consists of a series of repeats of adenosine/thymidine-rich sequences, referred to as A repeats (7, 14). In human adenovirus type 5 (Ad5) the packaging sequence is a 192-bp segment of the genome, positioned between the left-end inverted terminal repeat (ITR) and the start site of the E1A early transcript (Fig. 1) (15). The Ad5 packaging sequence contains seven A repeats, designated A1 through A7 (reviewed in reference 23). Genetic analyses, based on deletions and mutations of nucleotides within the packaging sequence, demonstrated that the seven A repeats are functionally redundant for packaging (7, 8, 29). However, there is evidence to suggest that the A repeats are not functionally identical and that the A1, A2, A5, and A6 repeats are the most crucial for genome encapsidation (7, 8, 29, 30). Studies using site-directed mutagenesis of the A repeats most important for packaging sequence function have further defined a minimal functional domain, 5′-TTTG-(N8)-CG-3′, designated the bipartite consensus, which is vital for the packaging of the adenovirus genome (29, 30).
FIG. 1.
(A) Arrangement of the left end of the Ad5 genome. The A repeats, represented as triangles, are located between nucleotides 200 and 397, upstream of the transcription start site of the E1A promoter, shown as a right-facing arrow. Modified with permission from P. Hearing. (B) Ad5 sequence from nucleotide 200 to 397. The A repeats are highlighted with gray boxes. Specific nucleotide numbers are located below the sequence.
Recent attempts to characterize packaging sequence function in Ad assembly have concentrated on identifying factors that interact with the packaging sequence. The results of electrophoretic mobility shift assays (EMSAs) using minimal packaging sequence probes and nuclear extracts from Ad-infected cells implicate the binding of proteins that are specific to virus-infected cells (21, 33). The adenovirus-encoded IVa2 protein has been identified as one of the proteins involved in protein-DNA complex formation with DNA probes containing A repeats that are the most crucial to genome encapsidation (33). Early studies classified IVa2 as a minor component of the mature capsid that exhibited the ability to bind viral DNA (28). Later studies identified IVa2 as an enhancer of transcription, initiating from the adenovirus major late promoter (MLP), that acts on the MLP by binding to the proximal downstream sequence elements (DE) (32). Based on the observation that the sequences of the bipartite motifs within the packaging sequence were similar to the DE of the MLP, it was further demonstrated that IVa2 in nuclear extracts of virus-infected cells interacts with probes containing the A1-A2 and the A4-A5 repeats (33). The ability of IVa2 to associate with the packaging sequence during Ad infection was later confirmed by chromatin immunoprecipitation analysis (24, 26). IVa2 interaction with the L1 52/55-kDa protein, an Ad-encoded nuclear protein that is required for DNA packaging, further suggested that IVa2 functions in encapsidation of the genome (11, 12). In addition to the putative role of IVa2 in Ad DNA packaging, the inability of a IVa2-null mutant virus to form capsids suggests that IVa2 is required for capsid assembly (34). The functions of IVa2 in both of the early stages of Ad capsid assembly as well as in genome encapsidation emphasize its importance in multiple processes during virus morphogenesis. Moreover, the dual functions of IVa2 also indicate an association between the packaging sequence and the nucleation of the procapsid assembly and suggest that interaction between IVa2 and the packaging sequence plays a major role in the initiation of the Ad assembly process.
Although previous studies have defined specific functional elements within the packaging sequence (14, 22, 29, 30), very little is known about how IVa2 binds to the complete packaging sequence. Current understanding of this is based on in vitro binding studies with nuclear extracts (21, 24, 26, 33). These assays were valuable in the identification of multiprotein complexes that are important for Ad packaging, but they did not allow the characterization of IVa2 interaction with DNA in the absence of other packaging sequence binding proteins. Furthermore, the location of the packaging sequence within the E1A promoter has made the study of IVa2 interaction with the complete packaging sequence DNA using nuclear extracts difficult, due to the contamination of E1A promoter binding proteins. Finally, IVa2 functional studies have also been hindered by the difficulty in purifying IVa2 synthesized in virus-infected cells or by recombinant techniques (19, 32).
To extend the results of the previous studies of IVa2 function, we wanted to quantify the binding of IVa2 to the complete packaging sequence in a purified system. To this end, we purified the first recombinant untagged version of the Ad5 IVa2 protein from a prokaryotic expression system and established an in vitro system to characterize its binding to the adenovirus packaging sequence. We compare IVa2 binding to the A-repeat probes and to the complete packaging sequence probe. The results indicate that the IVa2 protein forms a multisubunit complex on the packaging sequence.
MATERIALS AND METHODS
Plasmid construction.
The full-length IVa2 protein coding sequence was amplified by PCR from plasmid pBK-IVa2 (35) using primers 5′NAT4a2 (5′-CATGCCATGGAAACCAGAGGGCGAAGACCGGCA-3′) and 3′Ad5IVa2-Xho1 FUS (5′-CGCTCGAGTTTAGGGGTTTTGCGCGCGCGGT-3′). The PCR product was digested with NcoI and XhoI (New England Biolabs) and then inserted into pTYB4 (New England Biolabs) to create pTYB4-IVa2.6. The Ad5 nucleotide sequence 200 to 397, encompassing the packaging sequence, was amplified by PCR using primers 5′-PS(200) (5′-GGAAGTGACAATTTTCGCG-3′) and 3′-PS(397) (5′-AACACCTGGGCGAGTCTC-3′) and cloned into the vector pCR-Blunt II-TOPO (Invitrogen) according to the manufacturer's protocol to create vector pCRBT-FL.1. The integrity of the inserts in the constructed plasmids was verified by DNA sequencing (University of Michigan Sequencing Core).
Cells, viruses, and bacterial strains.
The Escherichia coli strain BL21-CodonPlus RIL (Stratagene) was used for recombinant protein expression. 293 cells, a human embryonic kidney cell line expressing the adenovirus E1A and E1B proteins, were grown and maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (10). The growth and manipulation of wild-type Ad5 were performed as previously described (9).
Preparation of adenovirus-infected whole cell and nuclear extracts.
293 cells were infected with 5 PFU per cell of Ad5. Twenty-four hours postinfection, whole cell lysates or nuclear extracts were prepared as previously described (13, 33).
Western blotting analysis.
Approximately 15 μg of total cell lysates were boiled in 3× sodium dodecyl sulfate (SDS) sample buffer, separated by 9% SDS-polyacrylamide gel electrophoresis (PAGE) (18), and transferred to a nitrocellulose membrane. The IVa2 protein was detected as previously described (26).
Expression and purification of recombinant IVa2 protein.
Recombinant IVa2 protein fused to an intein tag at the carboxy terminus was isolated from cultures of E. coli using the New England Biolabs IMPACT-CN system protocol. Briefly, the expression strain was grown in 3 liters of Luria broth in the presence of 100 μg/ml ampicillin and 20 μg/ml chloramphenicol to an optical density at 600 nm of 0.65 to 0.80. The cultures were then induced with isopropyl-β-d-thiogalactopyranoside at a final concentration of 0.5 mM for 16 h at 4°C. The cells were harvested, frozen, and stored at −80°C until lysis. The bacterial pellet was lysed in 75 ml lysis buffer (20 mM HEPES, pH 7.9, 500 mM NaCl, 0.5 mM EDTA, 0.1% Triton X-100, 20 μM phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 5 μg/ml leupeptin) by passage through a French press. The cell debris was separated from the soluble protein by centrifugation at 14,000 × g for 10 min at 4°C, and the cleared lysate was then added to 25 ml of a 50% slurry of chitin beads (New England Biolabs) equilibrated in lysis buffer. The protein was bound to the beads by batch method for 1 h at 4°C with shaking. The protein-bound beads were added to a column, allowing the unbound proteins to flow through, and then the beads were washed with 10 column volumes (CVs) of lysis buffer containing 1 M NaCl, followed by the addition of 3 CVs of lysis buffer containing 150 mM NaCl. The protein was then cleaved off the column via the addition of lysis buffer containing 150 mM NaCl and 50 mM dithiothreitol (DTT).
The eluted protein was concentrated in Amicon ultra-15 concentrators (Millipore) and further purified on a Mono S 4.6/100 PE column (GE Healthcare) equilibrated with buffer containing lysis buffer with 150 mM NaCl. The protein was eluted with a linear salt gradient from 150 mM to 1 M NaCl over 20 CVs. The fractions containing the recombinant IVa2 protein were pooled and concentrated before the addition of glycerol to 10% and storage at −80°C. Protein purity was assessed by 9% SDS-PAGE followed by detection with ProtoBlue (National Diagnostics) stain. The protein concentration was calculated by using a Bio-Rad protein assay using bovine serum albumin (BSA; Sigma) as a standard.
Preparation of mobility shift probes.
Forty-two- to 45-bp double-stranded probes of sections of the packaging sequence were created by the annealing of single-stranded oligonucleotides. The A1-A2, A2, A3, A4-A5, and A5-A7 probes correspond to Ad5 nucleotides 238 to 279, 260 to 300, 280 to 320, 315 to 358, and 335 to 380, respectively. The double-stranded probe for the hexon coding region contained nucleotides 19981 to 20021. The full-length packaging sequence probe for the mobility shift assays was generated by PCR amplification. The blunt-ended amplicon was generated with Pfx polymerase (Invitrogen) using primers 5′-PS(200) and 3′-PS(397) and purified from a 2% agarose gel using a QIAGEN gel extraction kit. The probes were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs) in buffer containing 50 mM Tris-HCl (pH 9.0), 10 mM MgCl2, 5 mM DTT, 45 μCi [γ-32P]ATP (6,000 Ci/mmol; GE Healthcare), and 1 mM spermidine and then purified on a Sephadex G-25 spin column (Roche). The double-stranded oligonucleotides were consistently labeled with an estimated specific activity of 2,000 to 3,000 cpm/fmole DNA, while the complete packaging sequence probe had a consistent label of 1,000 cpm/fmole DNA.
Electrophoretic mobility shift assays.
Purified protein was combined with 200 pM of radiolabeled probe in a total volume of 10 μl for 30 min at 23°C in buffer containing 20 mM HEPES (pH 8.0), 10 mM Mg2(CH3O2), 5 mM KCl, 0.5 mM EDTA, 1 mM DTT, a 40-fold excess (mass/mass) of poly(dI-dC) (Roche), 500 ng of BSA, and 12% glycerol. The complexes were electrophoretically resolved at 200 V at 4°C on either a 5% (complete packaging sequence probe) or 6% (42- to 45-bp probes) native polyacrylamide (37.5:1, acrylamide/bisacrylamide) gel in 0.5× Tris-borate-EDTA buffer for 90 min or 45 min, respectively. Control experiments demonstrated that a 30-min incubation allowed for steady-state binding. For competition assays, the unlabeled oligonucleotides were mixed with the radiolabeled probe prior to being added to the binding reaction mixtures. The dried gels were analyzed either by autoradiography or scanning with a Molecular Dynamics STORM PhosphorImager. The data used to calculate the percentage of bound probe in the gel retardation assays was collected by exposing the dried gels to PhosphorImager plates for 12 to 15 h. The percentage of total DNA was calculated as [counts bound/(counts bound + counts free)] × 100%. Nonlinear regression analysis of the DNA binding data was performed using SigmaPlot 8.0. The data from the calculation of the percentage of DNA in complex with the IVa2 protein were fit into the sigmoidal logistic, 3 parameter equation.
To compare the binding of purified protein to nuclear extracts, reactions were performed essentially as described previously (26), with the following modifications: all reactions were performed in a 20-μl volume containing 1 μg BSA, 5 ng probe, and 250 ng poly(dI-dC). The products were analyzed on a 4.5% polyacrylamide gel as previously reported (33).
Mass spectrometric analysis.
Trypsin digestion and tandem mass spectrometry (MS/MS) analysis were performed by the Michigan Proteome Consortium.
RESULTS
Expression and purification of a full-length recombinant IVa2 protein.
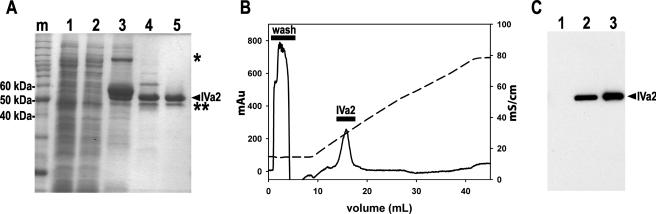
We used the IMPACT system from New England Biolabs for the initial capture step in the purification of the recombinant IVa2 protein expressed in E. coli. In this system, the recombinant IVa2 protein was expressed as a preprotein species fused with a carboxy-terminus tag comprised of a chitin binding domain and an internal intein protease domain (Fig. 2A, lane 3). After binding to a chitin matrix, the recombinant IVa2 protein was liberated from the tag by the addition of buffer containing DTT (Fig. 2A, lane 4), leaving four amino acids connected to the carboxy-terminal residue (amino acid 449) of the IVa2 protein. Western blot analysis of the eluted proteins from this purification step using an anti-IVa2 polyclonal antibody that was made against an amino-terminal peptide confirmed that the major protein band at 50 kDa was IVa2 (Fig. 2C). The similarity of the mobility of the recombinant IVa2 protein to that of the native protein on an SDS-PAGE gel suggested that no undesirable modification of IVa2 occurred during its expression and purification (Fig. 2C). Although the IVa2 protein from this purification step was nearly 70 to 80% pure, there were two contaminating proteins of approximately 45 kDa and 60 kDa (Fig. 2A, lane 4) that were not detected by Western blotting. The IVa2 protein was therefore further purified by cation exchange chromatography. After injection and binding, the protein eluted as a major peak around 400 mM NaCl, which corresponded to a buffer conductivity of about 25 to 30 mS/cm (Fig. 2B, lane 5). Analysis of the recombinant protein by staining of an SDS-PAGE gel showed that the IVa2 protein was further purified away from the 60-kDa protein contaminant (Fig. 2A, lane 5); however, the 45-kDa protein band was still present in the sample. Mass spectrometric analysis of this minor 45-kDa species indicated that it is a recombinant IVa2 protein missing a small portion of the amino terminus that was most likely generated by degradation or through the internal initiation of translation near the beginning of the IVa2 coding sequence. In addition, the results from the MS analysis of the purified protein explain the previously observed lack of recognition by the anti-IVa2 serum made against the amino terminus.
FIG. 2.
Expression and purification of recombinant, untagged IVa2 protein. (A) ProtoBlue-stained 9% SDS-PAGE. Intein-tagged IVa2 protein was affinity purified from soluble E. coli extract with a chitin agarose matrix as described in Materials and Methods. Lanes: m, protein standard marker; 1, soluble extract from uninduced E. coli; 2, soluble extract from isopropyl-β-d-thiogalactopyranoside-induced E. coli; 3, boiled intein-tagged IVa2 protein-bound chitin matrix beads; 4, 5 μg of IVa2 protein eluted from the chitin matrix; 5, 5 μg of IVa2 protein eluted from the Mono S PE 4.5/100 column. The purified full-length IVa2 protein is indicated with a labeled arrowhead. *, intein-tagged fusion IVa2 protein; **, IVa2 protein degradation product. (B) Typical chromatogram of the elution profile of the IVa2 protein from a Mono S column. The optical density at 280 nm (mAU) and ionic strength (mS/cm) of the buffer eluted from the column are represented as a solid and a dashed line, respectively. (C) Western blot comparing the IVa2 protein and IVa2 protein from infected cell lysates. Lanes: 1, 15 μg of 293 whole cell lysate; 2, 15 μg of Ad5-infected 293 whole cell lysate; 3, 150 ng of IVa2.
Interaction of IVa2 with A-repeat DNA probes.
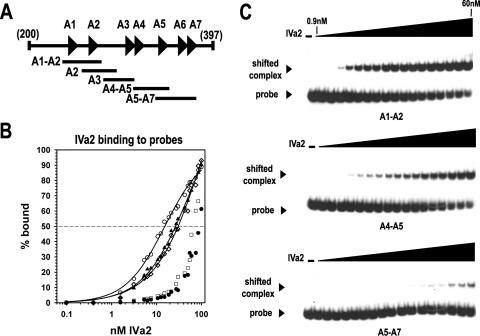
To extend the results of previous studies on IVa2 binding to packaging probes using nuclear extracts from virus-infected cells, we performed EMSA experiments with the purified IVa2 protein and small, 42- to 45-bp overlapping probes containing DNA sequences from the packaging sequence (Fig. 3A). The results from the EMSA analysis demonstrated that the IVa2 protein exhibited the highest affinity for a probe containing the A1 and A2 repeats (A1-A2) (Fig. 3). The plot of the steady-state binding data as a function of IVa2 protein concentration produced a sigmoidal curve (Fig. 3B). Nonlinear regression analysis of the IVa2 protein-DNA binding data produced an estimated half-maximal binding value of 15.4 ± 1.5 nM. The IVa2 protein exhibited a similar affinity for probes containing either the A2 repeat (A2) or the A4 and A5 repeats (A4-A5), albeit at a slightly lower affinity than the A1-A2 probe (Fig. 3B and C). The IVa2 protein affinity for these two probes was threefold lower (55 nM) than for the A1-A2-repeat probe. The IVa2 protein had an equally low affinity for probes containing either the A3 repeat (A3) or the A5, A6, and A7 repeats (A5-A7) (Fig. 3B and C). The data from IVa2 interaction with the A3 and A5-A7 probes did not produce a sigmoidal binding curve, making the accurate calculation of a half-maximal binding constant difficult, but we estimated that IVa2 affinity for each of these probes was at least fivefold lower than that for the A1-A2 probe (Fig. 3C). These data indicate that the five probes fall into three IVa2 binding categories, with the A1-A2 probe representing the high affinity probe, the A2 and the A4-A5 probes representing the medium affinity probes, and the A3 and the A5-A7 probes representing the low affinity probes.
FIG. 3.
IVa2 binding to packaging sequence probes. (A) Diagram of the packaging sequence region from nucleotide 200 to 397. The specific overlapping probes are indicated below the complete packaging sequence. (B) Binding of IVa2 to the specific probes. The percentages of the DNA probes bound by IVa2 (% bound) were calculated using PhosphorImager scanning data from the averages of three to four experiments. Symbols: ○, A1-A2 probe; ▴, A2 probe; ⋄, A4-A5 probe; □, A5-A7 probe; •, A3 probe. The fitting of a sigmoidal function to the data is shown as a solid line (see Material and Methods). (C) Representative autoradiograms from EMSAs using the A1-A2, A4-A5, and A5-A7 radiolabeled probes. The dashes above the first lanes indicate that the reaction mixture lacks IVa2. The triangles above the lanes represent increasing concentrations of IVa2 (0.1, 0.4, 1.6, 3.1, 5.3, 6.3, 7.5, 10.6, 12.5, 15, 18.8, 21.3, 27.5, 30, 42.5, 50, and 60 nM). The IVa2-DNA complex and the free probe are indicated with labeled arrowheads.
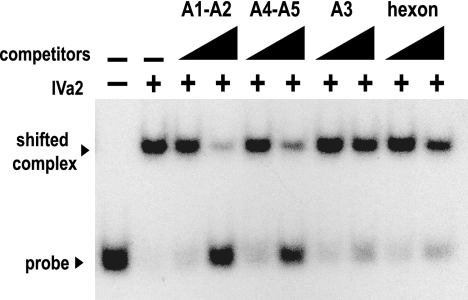
Competition assays with unlabeled competitor DNA from each of the three binding categories using the A1-A2 radiolabeled probe were performed to determine the specificity of the IVa2 binding to DNA. As shown in Fig. 4, IVa2 interaction with the A1-A2 probe was not easily disrupted by a 200-fold excess of the A3 probe or a nonspecific probe of similar length corresponding to a sequence from the hexon coding region. However, this interaction was competed by unlabeled A1-A2 or A4-A5 probe, though the A4-A5 competitor was slightly less efficient.
FIG. 4.
Competition assay of IVa2 binding to the A1-A2 probe. EMSAs were performed as described in the legend to Fig. 3 with 40 nM IVa2 and a radiolabeled A1-A2 probe. EMSA binding reaction mixtures were incubated with or without a 20- or 200-fold molar excess of unlabeled DNA encoding the A1-A2 repeats, the A4-A5 repeats, the A3 repeat, or an equal length of DNA from the hexon coding region. The presence or absence of IVa2 in the binding reaction mixture is indicated with a + or −, respectively. The first two lanes lack probes. The triangles above the lanes indicate increasing concentrations of the competitors.
Sequence specificity of the IVa2 protein binding to the A1-A2 repeats.
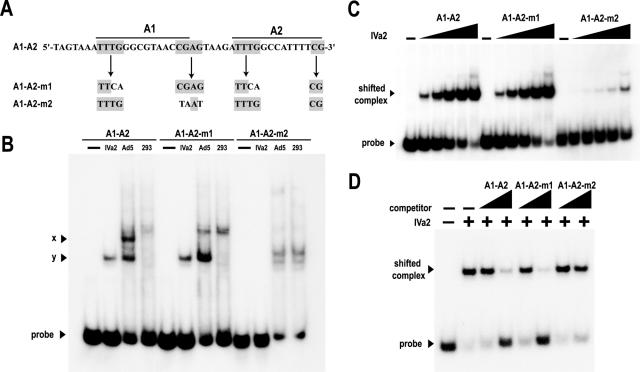
Previous data from EMSA analyses using adenovirus-infected nuclear extracts demonstrated that the IVa2 protein can form two complexes, designated x and y, on a radiolabeled A1-A2-repeat probe (33). Formation of the slower-migrating x complex requires the A-repeat TTTG motif, while both complexes require the CG motif. More recent evidence indicates that the x complex contains the L4 22-kDa protein in addition to IVa2, while the y complex contains only IVa2 (21, 26). To determine if mutation of these motifs also disrupts the binding of IVa2 to DNA in the absence of other nuclear proteins, we analyzed the binding of the purified IVa2 protein to radiolabeled mutant A1-A2 probes (Fig. 5A). As seen in the results of previous studies with virus-infected nuclear extracts, mutation of the TTTG sequence to TTAC in both the A1 and A2 repeat (probe A1-A2-m1) did not affect binding, while mutation of the CG sequence to AT in the A1 repeat (probe A1-A2-m2) disrupted the IVa2 interaction with the probe (Fig. 5C). This binding pattern was corroborated by the results of competition assays with unlabeled DNA competitors (Fig. 5D). Competitors possessing mutations of the two TTTG motifs competed for IVa2 protein interaction with the A1-A2 probe as well as the wild-type competitor did, and the A1-A2-m2 probe, in which the CG motif is mutated, did not efficiently compete for IVa2 binding to the A1-A2 probe. These data demonstrate that the CG motif from the A1 repeat is critical for IVa2 interaction with DNA. We also performed an EMSA experiment in which we directly compared the binding of the purified protein to the IVa2 protein in nuclear extracts (Fig. 5B). The complex formed by the purified protein corresponds to the y complex, confirming that the y complex does not contain additional protein components and that formation of the y complex requires the CG motif.
FIG. 5.
Interaction of IVa2 with mutant A1-A2 probes. (A) Sequences of the wild-type and mutant probes. The A1-repeat bipartite consensus, 5′-TTTG-(N8)-CG-3′, is indicated by bold lines over the sequence, with the conserved nucleotide motifs in gray boxes. (B) EMSAs comparing binding of A1-A2 wild-type and mutant radiolabeled probes to either 60 nM recombinant IVa2 (IVa2) or 4 μg nuclear extract from adenovirus type 5-infected (Ad5) and mock-infected (293) 293 cells. The dash above the first lane of each set indicates that the reaction mixture contains no IVa2 or extract. The labeled arrowheads indicate the formation of Ad5-specific complexes x and y on the A1-A2 wild-type probe. (C) EMSAs with the A1-A2 wild-type and mutant radiolabeled probes. The dash above the first lane of each set indicates that the reaction mixture contains no IVa2. The triangles above the lanes represent increasing concentrations of IVa2 (3.1, 6.3, 12.5, 25, and 50 nM). (D) Representative autoradiograms from competition assays of IVa2 protein binding to radiolabeled A1-A2. EMSAs were performed as described in the legend to Fig. 4, using the indicated competitors. The IVa2 protein-DNA-shifted complex and free probe are indicated with labeled arrowheads. The presence or absence of IVa2 in the binding reaction is indicated with a + or −, respectively. The first two lanes lack competitor. The triangles above the lanes indicate increasing concentrations of the competitors.
IVa2 protein interaction with the full-length packaging sequence.
The results from the EMSA analysis using probes with the A repeats suggested the presence of more than one IVa2 binding site. To analyze IVa2 interaction with the complete packaging sequence, we created a DNA probe by radiolabeling a PCR product corresponding to nucleotides 200 to 397 of the Ad5 genome.
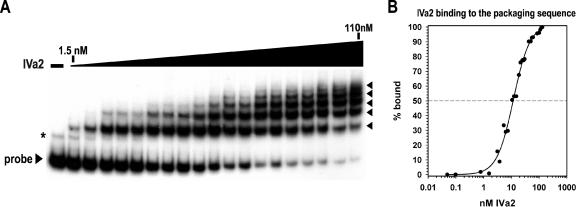
We observed IVa2 interaction with the packaging sequence probe at very low nanomolar concentrations and the appearance of more than one shifted complex with increasing amounts of IVa2 (Fig. 6A). Five complexes formed at 110 nM of IVa2, the concentration of IVa2 in which nearly all the free probe was bound. Additional complexes assembled with increasing amounts of IVa2 beyond 110 nM (data not shown). Nonlinear regression analysis of the binding data yielded a sigmoidal curve, with an R2 value greater than 0.99 and a half-maximal binding constant at around 11.5 ± 0.8 nM (Fig. 6B). These results suggest that the IVa2 protein has a very high affinity for the complete packaging sequence region, similar to that for the smaller A1-A2 probe (Fig. 3C).
FIG. 6.
Multiple complexes form as IVa2 interacts with the packaging sequence. (A) Representative autoradiogram from an EMSA using a radiolabeled probe to nucleotide 200 to 397. The first lane lacks IVa2. The triangle above the lanes represents increasing concentrations of IVa2 (1.5, 3.1, 5.3, 6.3, 7.5, 10.6, 12.5, 15, 18.8, 21.3, 27.5, 30, 42.5, 50, 55, 60, 85, 100, and 110 nM). The IVa2-packaging sequence complexes are indicated with arrowheads. A nonspecific band that was consistently observed in the preparation of the complete packaging sequence probe is indicated with an asterisk. (B) Binding of IVa2 to the packaging sequence probe. The percentage of probe bound by IVa2 (% bound) was calculated as described in the legend to Fig. 3.
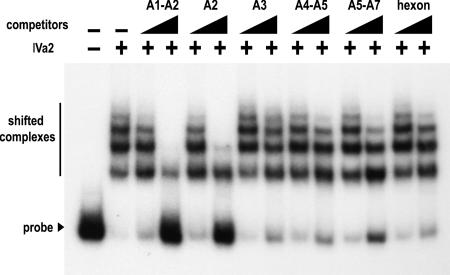
To assess the specificity of IVa2 for the probe containing the complete packaging sequence, competition analyses were performed using unlabeled competitor DNA corresponding to the A1-A2, A2, A3, A4-A5, and A5-A7 repeats (Fig. 3A). In these assays, a concentration of IVa2 protein was maintained at 40 nM, the concentration of protein that produced four to five IVa2-packaging sequence complexes when incubated with the full-length packaging sequence probe. Additionally, we used 100 and 1,000-fold molar excesses of unlabeled DNA for competition due to the length differences between the small A-repeat probes and the packaging sequence and because of the presence of multiple-binding IVa2 binding sites in the packaging sequence. The addition of a nonspecific competitor, such as a probe to a region of the hexon coding sequence (Fig. 7) or increasing amounts of poly(dI-dC) carrier DNA (data not shown), did not disrupt the interaction of IVa2 with the packaging sequence probe. However, the A1-A2 or A2 probes competed for IVa2 binding to the packaging sequence probe, although the A2 probe was slightly less efficient at competing than the A1-A2 probe (Fig. 7). In addition, the formation of the IVa2-packaging sequence complexes was not efficiently competed by the addition of unlabeled DNA competitors containing the A3, A4-A5, or A5-A7 repeats (Fig. 7). The inability of an A4-A5 unlabeled DNA competitor to efficiently compete IVa2 binding to the packaging sequence did not correlate with the results of the previous EMSA experiments using a radiolabeled A4-A5 probe (Fig. 3), which suggested that IVa2 affinity for the A4-A5 probe is similar to its affinity for the A2-repeat probe. In addition, the cold competition results indicated that the slower-migrating complexes that appeared with the addition of increasing amounts of IVa2 protein were also specific, only being disrupted by competition with A1-A2 and A2 probes.
FIG. 7.
Competition assay of IVa2 binding to the packaging sequence probe. EMSAs were performed as described in the legend to Fig. 4, using either a 100 or 1,000-fold molar excess of the indicated unlabeled A-repeat DNA competitors. The IVa2-packaging sequence probe complexes and the free probe are indicated with a bold line and an arrowhead, respectively. The presence or absence of IVa2 in the binding reaction is indicated with a + or −, respectively. The first two lanes lack competitor. The triangles above the lanes indicate increasing concentrations of the competitors.
DISCUSSION
We report the isolation of an untagged recombinant form of the IVa2 protein. The isolation of this protein allowed us to establish a system to analyze the steady-state binding characteristics between the IVa2 protein and the packaging sequence without the contamination of other DNA binding proteins from nuclear extracts. The results of our in vitro analysis indicate that IVa2 binds specifically to the packaging sequence through interactions at multiple binding sites. In addition, IVa2 protein binding to the DNA required the second part of the bipartite consensus sequence, 5′-TTTG-(N8)-CG-3′. The results of our EMSA analyses investigating the ability of the IVa2 protein to bind to DNA in vitro are consistent with results from previous genetic studies of the packaging sequence. In this study, we demonstrated that IVa2 binding to the A-repeat probes in vitro occurred in a hierarchy similar to the proposed functional hierarchy in vivo: IVa2 possessed the highest affinity for probes containing the A1, A2, A4, and A5 repeats, which were previously shown to be the most critical for packaging sequence function in vivo (7, 8, 29). The sequence requirements for IVa2 binding to the A1-A2-repeat probe were also consistent with the results of previous studies identifying the extended 5′-TTTG-(N8)-CG-3′ bipartite consensus as important for A1-A2 function (29). In addition, the results from EMSA analyses using A-repeat probes indicate that there are multiple IVa2 binding sites within the adenovirus packaging sequence. There are at least two IVa2 binding sites within the packaging sequence, with at least one high-affinity binding site in the A1- and A2-repeat region and one in the region encompassing the A4 and A5 repeats. Although the results from the EMSA analyses indicate that IVa2 has a very low affinity for a probe to the A5-A7 repeats, the higher affinity for the A4-A5-repeat probe suggests that the A4 repeat represents a IVa2 binding site or that IVa2 interaction with the A5 repeat requires the presence of the A4 repeat. In summary, the binding preference of IVa2 for regions and motifs within the packaging sequence that are the most crucial for Ad packaging in vivo suggests that IVa2 mediates the connection between the Ad genome and the rest of the virus packaging machinery.
Recent data indicate that the L4 22-kDa protein, a protein of previously unknown function, interacts with the TTTG packaging sequence motifs together with the IVa2 protein (21). Viruses that lack expression of the L4 22-kDa protein are nonviable and fail to assemble capsids and/or package the viral genome (6, 17, 21). These data also suggest that L4 22-kDa protein interaction with the IVa2-DNA complex at the TTTG motif is critical for virus assembly. The results of previous DNA binding studies using infected-cell nuclear extracts suggest that IVa2 interaction with the DNA occurs initially at the CG motif of the bipartite consensus, with the ensuing binding of IVa2 to the TTTG motif occurring via augmentation by the L4 22-kDa protein (21, 24, 26, 33). We observed a similar pattern of binding by IVa2 alone in EMSAs using mutant A1-A2-repeat probes. Mutation of the CG motif significantly decreased IVa2-DNA binding, while mutation of the TTTG motifs had no effect on the IVa2 interaction with the A1-A2 probe, confirming the results seen with nuclear extracts from infected cells (33).
The results of our previous study using a purified 6-His-tagged IVa2 protein demonstrated that IVa2 can directly interact with the packaging sequence (26). However, we were concerned that the inability of the 6-His-tagged protein to bind to an A5-A7 probe, although the IVa2 from nuclear extracts could bind, was due to the presence of the recombinant tag and a bulky linker region. The results from our current study using a recombinant untagged protein confirm our previous results with the 6-His-tagged IVa2 fusion protein and suggest that the ability of IVa2 to bind to the A5-A7 region of the packaging sequence requires additional proteins.
The data from the EMSA analyses using the complete packaging sequence suggest that multiple IVa2 proteins bind to the packaging sequence, possibly through the sequential interaction of increasing numbers of IVa2 molecules with different binding sites within the packaging sequence. The assembly of these complexes occurred in a step-wise manner (Fig. 6A). The steady-state binding measurements from our EMSA analyses indicate that the IVa2 protein exhibits a slightly higher affinity for the complete packaging sequence than for the A1-A2 probe. These results, along with the results of the competition assays using unlabeled A1-A2 DNA, suggest that the initial IVa2 binding site on the packaging sequence is within the A1- and A2-repeat region. Furthermore, the failure of a 1,000-fold molar excess of a nonspecific DNA competitor to disrupt the formation of the multiple slower-migrating complexes observed by EMSA suggested that the complexes result from a specific interaction with IVa2 and are not due to a general protein-DNA association. The formation of the higher-order, slower-mobility IVa2 protein-DNA complexes was not efficiently competed by an excess of probe containing the A4 and A5 repeats (Fig. 7). This result was surprising, since the binding studies using the A-repeat probes indicated that IVa2 possesses an affinity for this probe that is similar to its affinity for the A2-repeat probe (Fig. 3B, C, and 4). These studies suggest that the complete packaging sequence probe is quite different from the small A-repeat probes that have been previously used to characterize the function of packaging sequence-interacting proteins from nuclear extracts. Moreover, these data underscore the fact that interactions between the adenovirus packaging sequence and cognate binding proteins that are unique to the complete packaging sequence may be missed using smaller probes. Unfortunately, it may not be possible to detect more complicated interactions in nuclear extracts using larger probes, because the packaging sequence overlaps the E1A enhancer and binding of transcription factors is likely to confound the analysis.
Although we do not know the precise structure of the IVa2 protein-packaging sequence complex, the inability of the A3-repeat oligonucleotide to efficiently compete the IVa2 interaction with the packaging sequence probe suggests that IVa2 may not directly interact with the A3-repeat region during complex formation on the packaging sequence. Aside from the A7 repeat, the A3 repeat possesses the least homology to the other A repeats and it was bound poorly by IVa2 in the EMSAs. Furthermore, mutational analyses of the packaging sequence suggest that the A3 repeat does not contribute significantly to DNA packaging (7, 14). Therefore, the A3 repeat may not represent an important IVa2 binding site, but rather, a region that could function in the critical spatial organization of the A repeats.
The data from the experiments using the full-length packaging sequence as the probe and the data indicating the existence of multiple IVa2 binding sequences within the packaging sequence region suggest that multiple IVa2 proteins bind to the packaging sequence. Furthermore, analysis of the steady-state binding characteristics of IVa2 to the full-length packaging sequence indicates that IVa2 binding to the packaging sequence may be cooperative, due to the sigmoidal shape of the binding curve and to the observation that complete IVa2-packaging sequence complex formation occurred over less than 1.5 logs of IVa2 concentration. This mode of binding is likely facilitated by the close proximity of multiple-binding IVa2 binding sequences within the packaging sequence. In addition, evidence for a cooperative mode of binding is further strengthened by the results of previous cross-linking studies demonstrating that IVa2 binds as a dimer to a DNA probe containing the DE from the MLP and by the observation that IVa2 is found as a multimer in virus-infected cells (19). Therefore, the next step in understanding the role of IVa2 during adenovirus assembly will be to define the specific IVa2 protein-protein and protein-DNA interaction domains that are required for IVa2 binding to the packaging sequence. It has also been proposed, based on bioinformatic analysis, that the IVa2 protein is an ATPase, possibly functioning as the motor protein for packaging of the viral genome. Mutations in a lysine residue that would be predicted to be involved in ATP binding result in nonviable virus, but a direct demonstration of enzymatic activity has not been reported (25). The availability of purified protein should facilitate such studies.
Acknowledgments
We thank members of the Imperiale laboratory for help with this work and their insightful discussions. We also thank David Friedman and David Miller for critical reading of the manuscript.
This work was supported by an award from the American Heart Association to R.E.T. and by grant R01 AI52150 from the National Institutes of Health to M.J.I.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Anderson, C. W., P. R. Baum, and R. F. Gesteland. 1973. Processing of adenovirus 2-induced proteins. J. Virol. 12:241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D., and S. Grimes. 2005. The Φ29 DNA packaging motor, p. 102-116. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structure, and mechanism. Landes Bioscience/Eurekah.com, Georgetown, TX.
- 3.Daniell, E. 1976. Genome structure of incomplete particles of adenovirus. J. Virol. 19:685-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edvardsson, B., E. Everitt, H. Jornvall, L. Prage, and L. Philipson. 1976. Intermediates in adenovirus assembly. J. Virol. 19:533-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feiss, M., and C. E. Catalano. 2005. Bacteriophage lambda terminase and mechanisms of DNA packaging, p. 5-39. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structure and mechanism. Landes Bioscience/Eurekah.com, Georgetown, TX.
- 6.Fessler, S. P., and C. S. Young. 1999. The role of the L4 33K gene in adenovirus infection. Virology 263:507-516. [DOI] [PubMed] [Google Scholar]
- 7.Grable, M., and P. Hearing. 1990. Adenovirus type 5 packaging domain is composed of a repeated element that is functionally redundant. J. Virol. 64:2047-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grable, M., and P. Hearing. 1992. cis and trans requirements for the selective packaging of adenovirus type 5 DNA. J. Virol. 66:723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham, F. L., and L. Prevec. 1991. Manipulation of adenovirus vectors. Methods Mol. Biol. 7:109-128. [DOI] [PubMed] [Google Scholar]
- 10.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 11.Gustin, K. E., and M. J. Imperiale. 1998. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72:7860-7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gustin, K. E., P. Lutz, and M. J. Imperiale. 1996. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70:6463-6467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, K. F., J. B. Christensen, and M. J. Imperiale. 1996. BK virus large T antigen: interactions with the retinoblastoma family of tumor suppressor proteins and effects on cellular growth control. J. Virol. 70:2378-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hearing, P., R. J. Samulski, W. L. Wishart, and T. Shenk. 1987. Identification of a repeated sequence element required for efficient encapsidation of the adenovirus type 5 chromosome. J. Virol. 61:2555-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hearing, P., and T. Shenk. 1983. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell 33:695-703. [DOI] [PubMed] [Google Scholar]
- 16.Horwitz, M. S., M. D. Scharff, and J. V. J. Maizel. 1969. Synthesis and assembly of adenovirus 2. I. Polypeptide synthesis, assembly of capsomeres, and morphogenesis of the virion. Virology 39:682-694. [DOI] [PubMed] [Google Scholar]
- 17.Kulshreshtha, V., L. A. Babiuk, and S. K. Tikoo. 2004. Role of bovine adenovirus-3 33K protein in viral replication. Virology 323:59-69. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lutz, P., and C. Kedinger. 1996. Properties of the adenovirus IVa2 gene product, an effector of late-phase-dependent activation of the major late promoter. J. Virol. 70:1396-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology 36:126-136. [DOI] [PubMed] [Google Scholar]
- 21.Ostapchuk, P., M. E. Anderson, S. Chandrasekhar, and P. Hearing. 2006. The L4 22-kilodalton protein plays a role in packaging of the adenovirus genome. J. Virol. 80:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ostapchuk, P., and P. Hearing. 2003. Minimal cis-acting elements required for adenovirus genome packaging. J. Virol. 77:5127-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostapchuk, P., and P. Hearing. 2003. Regulation of adenovirus packaging. Curr. Top. Microbiol. Immunol. 272:165-185. [DOI] [PubMed] [Google Scholar]
- 24.Ostapchuk, P., J. Yang, E. Auffarth, and P. Hearing. 2005. Functional interaction of the adenovirus IVa2 protein with adenovirus type 5 packaging sequences. J. Virol. 79:2831-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pardo-Mateos, A., and C. S. Young. 2004. A 40 kDa isoform of the type 5 adenovirus IVa2 protein is sufficient for virus viability. Virology 324:151-164. [DOI] [PubMed] [Google Scholar]
- 26.Perez-Romero, P., R. E. Tyler, J. R. Abend, M. Dus, and M. J. Imperiale. 2005. Analysis of the interaction of the adenovirus L1 52/55-kilodalton and IVa2 proteins with the packaging sequence in vivo and in vitro. J. Virol. 79:2366-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao, V. B., and L. W. Black. 2005. DNA packaging in bacteriophage T4, p. 40-58. In C. E. Catalano (ed.), Viral genome packaging machine: genetics, structure, and mechanisms. Landes Bioscience/Eurekah.com, Georgetown, TX.
- 28.Russell, W. C., and B. Precious. 1982. Nucleic acid-binding properties of adenovirus structural polypeptides. J. Gen. Virol. 63:69-79. [DOI] [PubMed] [Google Scholar]
- 29.Schmid, S. I., and P. Hearing. 1997. Bipartite structure and functional independence of adenovirus type 5 packaging elements. J. Virol. 71:3375-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid, S. I., and P. Hearing. 1998. Cellular components interact with adenovirus type 5 minimal DNA packaging domains. J. Virol. 72:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tibbetts, C. 1977. Viral DNA sequences from incomplete particles of human adenovirus type 7. Cell 12:243-249. [DOI] [PubMed] [Google Scholar]
- 32.Tribouley, C., P. Lutz, A. Staub, and C. Kedinger. 1994. The product of the adenovirus intermediate gene IVa2 is a transcriptional activator of the major late promoter. J. Virol. 68:4450-4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, W., and M. J. Imperiale. 2000. Interaction of the adenovirus IVa2 protein with viral packaging sequences. J. Virol. 74:2687-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, W., and M. J. Imperiale. 2003. Requirement of the adenovirus IVa2 protein for virus assembly. J. Virol. 77:3586-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, W., J. A. Low, J. B. Christensen, and M. J. Imperiale. 2001. Role for the adenovirus IVa2 protein in packaging of viral DNA. J. Virol. 75:10446-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]