Abstract
Unlike other retrovirus Gag proteins, the prototype foamy virus (PFV) p71gag protein is not processed into mature matrix (MA), capsid (CA), and nucleocapsid (NC) subunits. Little information about sequence motifs involved in FV capsid assembly and release is available. The recent analysis of candidate L-domain motifs in PFV Gag identified an evolutionarily conserved YXXL sequence motif with a potential function in capsid assembly. Here we provide support for the hypothesis that this motif does not function like a conventional L domain, by demonstrating that, unlike the PFV Gag PSAP L-domain motif, it cannot be functionally replaced by heterologous L-domain sequences. Furthermore, mutation of individual amino acids Y464, I466, L467, and L469, but not E465, to alanine led to reduced particle release and production of noninfectious, aberrant capsid structures, although relative structural protein incorporation and processing were not affected. In contrast, mutation of G468 to alanine resulted in an intermediate, temperature-sensitive phenotype characterized by reduced particle release and reduced infectivity. Despite similar relative RNA genome incorporation for all mutants, analysis and quantification of particle-associated viral nucleic acids demonstrated defects in genomic reverse transcription for all the noninfectious mutants, a process that, unlike that of orthoretroviruses, in the case of FVs takes place in the virus-producing cell. In correlation with the reduced infectivity, the G468A mutant displayed an intermediate level of genomic reverse transcription. Taken together, these results demonstrate that the conserved YXXLGL motif in PFV Gag is involved in correct capsid assembly, which in turn is essential for reverse transcription of the FV genome.
The orthoretrovirus Gag polyproteins are the driving force for viral particle assembly and release from infected host cells (reviewed in references 5 and 30). Typically, these polyproteins are processed in the newly formed viral particles during (or shortly after) budding into at least three mature subunits: MA, CA, and NC. Several functional domains within the polyprotein or the mature subunits have been identified. These include membrane-targeting (M) domains, located at or near the N terminus in the MA subunit, as well as interaction (I) domains, which are important for Gag oligomerization, and nucleic acid binding domains, which are required for viral genome packaging. The I domains and the nucleic acid binding domains are often located in the NC subunit. Furthermore, late assembly (L) domains have been characterized that are essential for late events in retrovirus budding, i.e., the pinching off and release of virions from infected host cells (reviewed in reference 3). At least three different late domain sequence motifs found in different virus families have been demonstrated to link the viral budding process to the cellular ubiquitylation machinery and the vacuolar protein sorting pathway. These are normally responsible for the sorting of cargo to the multivesicular body, a process that is topologically reminiscent of viral budding at cellular membranes.
Foamy viruses (FVs) use a replication strategy distinct from that of orthoretroviruses in several of its features and similar in some aspects to that of hepadnaviruses (reviewed in references 9 and 36). The FV particle release process, in particular, following a B/D-type assembly strategy with preassembled capsid structures in the cytoplasm, is unique among retroviruses in that coexpression of FV Gag and Env is essential for this process (1, 14). This is due to a very specific interaction, which has been mapped to the N-terminal, cytoplasmic region of the FV glycoprotein leader peptide (LP). An evolutionarily conserved W10XXW13 sequence motif interacts with the N-terminal region of FV Gag, which is essential for membrane association and budding to occur (6, 26, 48). This suggests that both the Gag and Env proteins of FVs contain structural information essential for particle formation and release. However, some functions might be redundant in the two proteins. This possibility is supported by the observation that FV glycoprotein expression is sufficient for the release of subviral particles into the supernatant (41), the extent of which is regulated by posttranslational ubiquitylation of FV Env (43). Furthermore, artificial membrane targeting of FV Gag by N-terminal addition of a myristoylation signal sequence results in release of virus-like particles, similar to what is observed for orthoretroviruses upon Gag expression (12).
The prototype FV (PFV) Gag protein displays a highly unusual structural organization and remarkable properties for a retrovirus capsid protein (reviewed in reference 28). Unlike orthoretrovirus Gag proteins, it is not processed into MA, CA, and NC subunits. Only a single proteolytic cleavage event, resulting in the removal of a C-terminal 3-kDa peptide from the p71gag precursor protein, takes place during particle assembly (reviewed in reference 16). Whereas p71gag expression alone leads to incompletely closed capsids that are noninfectious, the morphology of particles derived from expression of the processed p68gag protein is indistinguishable from that of wild-type particles, but their specific infectivity is reduced by 2 orders of magnitude (13, 14, 44, 51). No further Gag maturation upon particle release is observed. However, secondary cleavage sites within PFV Gag have been characterized by in vitro assays and have been demonstrated to be essential for infectivity (24, 33). This suggests an unusual uncoating process involving essential PFV protease-mediated Gag processing at secondary cleavage sites.
So far, no high-resolution structure of the PFV Gag protein or any of its subdomains is available, and very few functional domains and sequence motifs have been characterized. Close to the N terminus, a sequence motif with strong homology to the cytoplasmic targeting and retention signal (CTRS) of Mason-Pfizer monkey virus (MPMV), which is essential for the intracytoplasmic assembly of MPMV capsids, has been identified (7, 12). Interestingly, while mutation of a conserved arginine (R50) residue of the PFV CTRS motif led to complete abolishment of PFV particle assembly and release, mutation of the corresponding residue in MPMV Gag (R55) resulted in a switch of the particle assembly strategy from B/D type to C type. Furthermore, two coiled-coil domains located in the N-terminal third of the PFV Gag protein have been characterized. The first, which spans amino acids (aa) 130 to 160, was implicated in Gag-Gag protein interactions, whereas the second, encompassing aa 160 to 180, seemed to play a role in microtubule-mediated intracellular trafficking of intact, incoming PFV particles to the microtubule organizing center by an interaction with the dynein light chain 8 protein (32, 38, 46). Primate FVs contain three glycine/arginine-rich basic sequences (GR boxes) in the C-terminal third of the p68gag protein, instead of the canonical cysteine-histidine motifs usually found in the NC domain of orthoretrovirus Gag proteins (40). GR box I has binding activities for both RNA and DNA; GR box II contains a nuclear localization signal that leads to a transient nuclear Gag staining pattern during PFV infection; and GR box III has no assigned function yet (40, 49). In addition, analysis of the particle density of Rous sarcoma virus (RSV) Gag chimeras that had their NC domain replaced by PFV sequences spanning various parts of the C-terminal domain (CTD) of p68gag from aa 414 to 613, harboring the GR boxes, suggested the presence of two I domains in this region of the PFV Gag protein (4).
Recently, the analysis of candidate L-domain sequence motifs in PFV Gag led to the characterization of a PSAP (aa 284 to 287) motif with classical L-domain activity that is important for particle release and can be functionally replaced by heterologous L-domain sequence motifs (31, 42). In these studies a linkage of PFV egress in a tumor susceptibility gene (Tsg-101)-dependent manner to the ESCRT (endosomal sorting complex required for transport) machinery of the vacuolar protein sorting pathway by the PFV Gag PSAP motif was demonstrated (31, 42). In addition, an evolutionarily conserved YXXL motif (aa 464 to 467) with no conventional L-domain function was identified (42). Mutation of this motif led to a moderate reduction in PFV particle release and a complete loss of particle infectivity as a result of aberrant capsid morphogenesis (42). In this study we further characterize and analyze the function of this sequence motif for PFV replication.
MATERIALS AND METHODS
Cells.
The human kidney cell line 293T (11) and the human fibrosarcoma cell line HT1080 (34) were cultivated in Dulbecco′s modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics.
Expression constructs.
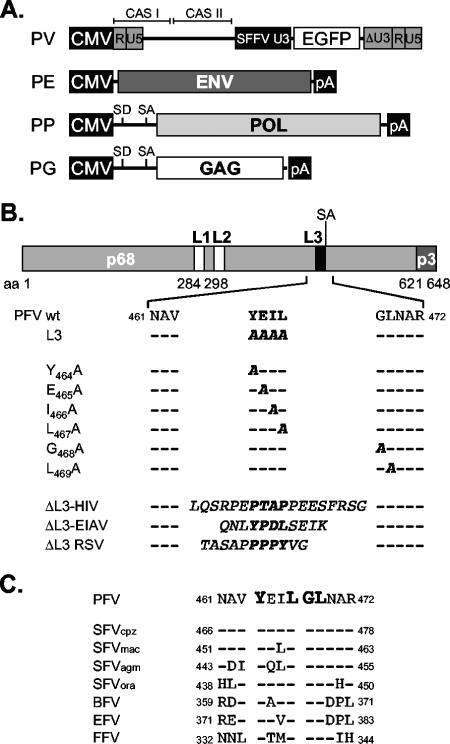
A schematic outline of the constructs used in this study is shown in Fig. 1. The four-plasmid PFV vector system (Fig. 1A), consisting of the PFV Gag (PG) expression vector pcziPG2 (pcziGag2), the PFV Pol (PP) expression vector pcziPol, the PFV Env (PE) expression construct pczHFVenvEM002, and the enhanced green fluorescent protein (EGFP)-expressing PFV transfer vector (PV) pMD9, has been described previously (19). For some experiments a variant PFV Pol expression construct (p6iPolΔRT), expressing a catalytically inactive Pol protein containing the M69 (YVDD312-315GAAA) mutation described previously (29), was used. The original PFV Gag L3 mutant, with the YEIL motif starting at aa 464 changed to AAAA, has been described previously (42). All other mutants were generated using standard PCR cloning techniques and mutagenesis primers. Details are available upon request. The PFV Gag point mutants harbor the following mutations: Y464A, E465A, I466A, L467A, G468A, and L469A (Fig. 1B). The ΔL3 L-domain replacement constructs contain heterologous L-domain sequence motifs replacing the YEIL motif in PFV Gag (Fig. 1B). The ΔL3-HIV construct has an 18-amino-acid sequence (LQSRPEPTAPPEESFRSG) of human immunodeficiency virus type 1 (HIV-1) Gag p6 inserted, ΔL3-EIAV harbors a peptide (QNLYPDLSEIK) with the L domain of equine infectious anemia virus (EIAV) Gag p9, and ΔL3-RSV contains the complete RSV Gag p2b peptide (TASAPPPPYVG).
FIG. 1.
Schematic illustration of the expression constructs and PFV Gag mutants. (A) Schematic outline of the four-plasmid PFV vector system consisting of a gene transfer vector (PV) as well as PFV Env (PE), Pol (PP), and Gag (PG) expression vectors. CAS I or II, cis-acting sequence I or II; CMV, cytomegalovirus virus promoter; R, repeat region of the long terminal repeat; U5, long-terminal-repeat unique 5′ region; U3, long-terminal-repeat unique 3′ region; ΔU3, enhancer-deleted U3 region; SFFV U3, spleen focus-forming virus U3 promoter; SD, splice donor; SA, splice acceptor; pA, polyadenylation signal sequence. (B) Schematic organization of the PFV Gag precursor protein and processing products p68 and p3. The locations of the putative L-domain motifs are indicated as boxes. Below, the amino acid sequences of L3 motif regions of wild-type and mutant PFV Gag proteins are shown. Putative L-domain sequence motifs are boldfaced, and amino acids altered in the mutant constructs are italicized. (C) Sequence alignment of the L3 motif regions of FV isolates from different species. Evolutionarily conserved amino acids are boldfaced. GenBank accession numbers for the cited viral genomes are NC001736 for PFV, U04327for simian FV-chimpanzee (SFVcpz), X54482 for simian FV-macaque (SFVmac), M74895 for simian FV-African green monkey (SFVagm), AJ544579 for simian FV-orangutan (SFVora), U94514 for bovine FV (BFV), AF201902 for equine FV (EFV), and U85043 for feline FV (FFV).
Generation of viral supernatants and analysis of transduction efficiency.
FV supernatants containing recombinant viral particles were generated essentially as described previously (25, 27). Briefly, FV supernatants were produced by cotransfection of 293T cells with equal amounts of pMD9, pczHFVenvEM002, pcziPol, and pcziPG2 or mutants thereof as indicated using polyethyleneimine or Polyfect transfection reagents. Twenty-four hours posttransfection, sodium butyrate (final concentration, 10 mM) was added to the growth medium for 8 h. Subsequently, the medium was replaced, and viral supernatants were harvested an additional 16 h later. Extra- and intracellular viral particles were harvested as described previously (26). Transductions of recombinant EGFP-expressing PFV vector particles were performed by infection of 2 × 104 HT1080 cells, plated 24 h in advance in 12-well plates, for 4 to 6 h using 1 ml of the viral supernatant or dilutions thereof. The percentage of EGFP-positive cells was determined by fluorescence-activated cell sorter (FACS) analysis 48 to 72 h after infection. All transduction experiments were performed at least three times, and in each independent experiment the values obtained with the wild-type pcziPG2 construct were arbitrarily set to 100%.
Analysis of temperature sensitivity.
For assaying the potential temperature sensitivity of specific mutants, virus was produced either at 37°C or at 30°C, by shifting one plate per construct of duplicate transfections to 30°C during the sodium butyrate induction step. Subsequently, the HT1080 target cells were incubated with viral supernatants or dilutions thereof for 6 h at 37°C or 30°C. After removal of the viral supernatant, the target cells were incubated for an additional 4 h at the corresponding temperature before they were all shifted to 37°C and assayed by FACS 48 h after addition of the viral supernatants to the target cells.
Antisera, Western blot expression analysis, and quantification of particle release.
Western blot expression analysis of cell- and particle-associated viral proteins was performed essentially as described previously (26). Polyclonal antisera used were specific for PFV Gag (2) or the LP of PFV Env, aa 1 to 86 (26). Furthermore, hybridoma supernatants specific for PFV reverse transcriptase (RT) (clone 15E10) or PFV integrase (IN) (clone 3E11) were employed in some experiments (21). The chemiluminescence signal was digitally recorded using a LAS-3000 imager and quantified using the Image Gauge software package (both from Fujifilm). Particle release levels relative to that for the wild-type PFV Gag control were determined in independent experiments by quantification of Gag proteins in purified viral particle samples, and values were corrected for different intracellular Gag expression levels in the individual samples.
Real-time PCR analysis.
Viral supernatants for real-time PCR analysis were generated as described above, with the modification that during the sodium butyrate induction step and the subsequent viral supernatant production step, DNase I (>3,000 U/mg; Applichem) was added to the medium to a final concentration of 500 μg/ml. Forty-eight hours posttransfection, the cell-free viral supernatant was harvested by sterile filtration (pore size, 0.45 μm) and centrifuged at 4°C and 25,000 rpm for 2 h in an SW40 or SW28 rotor through a 20% sucrose cushion. The supernatant was discarded, and the viral pellet was gently resuspended in phosphate-buffered saline (PBS) and again DNase I digested in a total volume of 150 μl using 150 μg DNase I (>3,000 U/mg) for 1 h at 37°C. Seventy-five microliters of the sample was added to an equal volume of 2× sodium dodecyl sulfate protein sample buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed by Western blotting as described above. Thirteen microliters was used for serial dilutions to determine infectivity as described above. The residual 62 μl was adjusted to 140 μl with PBS, used for isolation of viral nucleic acids using the QIAmp viral RNA minikit (QIAGEN) according to the manufacturer's instructions without the optional DNase I digest, and eluted in 60 μl. For analysis of particle-associated viral RNA and DNA, three separate reactions were set up for every sample. For analysis of RNA content, two parallel reactions (reactions 1 and 2) were set up using 5 μl of viral nucleic acid sample each; these were treated in parallel for 30 min at 37°C with DNase I (DNA-free; Ambion) using 2 U DNase I in a total volume of 18 μl each. The third sample (reaction 3) analyzed for viral DNA was mock incubated, omitting only the DNase I. DNase I digestion or mock incubation was terminated according to the manufacturer's protocol using DNase inactivation reagent (Ambion), resulting in a total final reaction sample volume of 10 μl each. Subsequently, all three reaction samples were reverse transcribed (reaction 1) or mock incubated (reactions 2 and 3) in a total volume of 20 μl using the Superscript II reverse transcription kit (Invitrogen) and oligo(dT)30 as the primer according to the manufacturer's instructions. Finally, 5 μl of each reverse transcription reaction product was analyzed by real-time PCR in duplicate in a total volume of 25 μl using the Brilliant SYBR Green QPCR kit (Stratagene) and an MX4000 multiplex quantitative PCR system (Stratagene). Primers (sense, 5′-GCAGTGCTTCAGCCGCTAC-3′; antisense, 5′-AAGAAGATGGTGCGCTCCTG-3′) were specific for the EGFP open reading frame and were used at 400 nM (22). Samples were initially denatured for 10 min at 95°C and subsequently amplified in 45 cycles of 30 s at 95°C, 1 min at 59°C, and 45 s at 72°C. Finally, the melting curve of each sample was determined. Quantification was determined in reference to a standard curve prepared by serial dilution of a pUC19 plasmid harboring the complete EGFP open reading frame. The viral genome (DNA or RNA) contents of the individual samples were determined in independent experiments at relative levels compared to that of the wild-type PFV Gag control, and values were corrected for differential particle release of the individual samples, determined by quantitative Western blot analysis as described above.
Reverse transcriptase assay.
For analysis of particle-associated reverse transcriptase activity, foamy virus particles were isolated and purified from transiently transfected 293T cell culture supernatants (six 10-cm dishes per sample) by a first ultracentrifugation step as described above in an SW28 rotor. Resuspended viral particles were subsequently proteolytically digested for 2 h at 37°C in a total volume of 200 μl, using 1 μl (5 mg/ml) subtilisin. Digestion was terminated by addition of 2 μl phenylmethylsulfonyl fluoride (20 mg/ml) and incubation at room temperature for 30 min, followed by addition of 10 ml complete cell culture medium and a second ultracentrifugation through 20% sucrose at 25,000 rpm and 4°C for 2 h in an SW40 rotor. Supernatants were discarded and viral pellets gently resuspended in 60 μl PBS. Fifty microliters of each sample was used for Western blot analysis as described above to quantitate PFV particle release for the individual samples. The residual 10 μl and three to five fivefold dilutions thereof were analyzed for PFV RT activity using a RetroSys C-type RT activity assay kit according to the manufacturer's instructions and using the provided murine leukemia virusRT standard. The RT activities of the individual samples were determined in independent experiments at relative levels compared to the RT activity of the wild-type PFV Gag control, and values were corrected for differential particle release of the individual samples, determined by Western blot analysis as described above. As a negative control, a PFV Pol expression construct harboring mutations in the active site (Y312G, V313A, D314A, and D315A) abolishing RT activity (29) was used in combination with the wild-type PFV Gag, Env, and vector constructs.
Electron microscopy analysis.
At 48 h posttransfection, the 293T cells were harvested and processed for electron microscopy analysis as described previously (23).
RESULTS
PFV Gag YXXL domain function cannot be complemented by heterologous L domains.
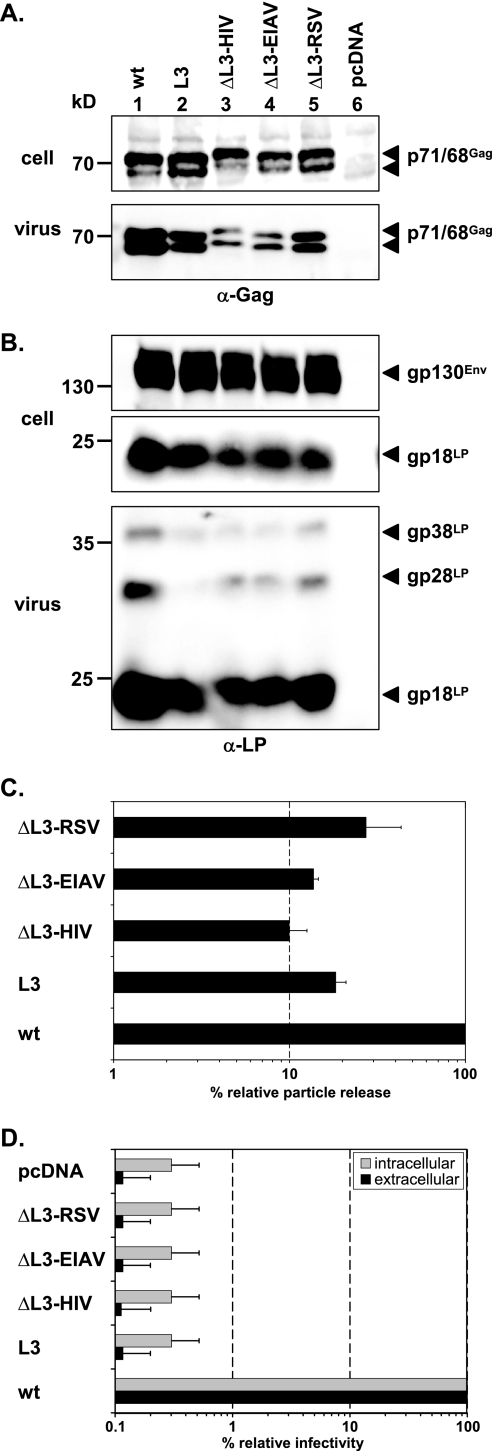
Our previous analysis of a role for the PFV YEIL motif in particle egress suggested that it does not function as a classical L domain. Mutagenesis of a classical L domain typically results in a strong reduction in particle release without affecting capsid morphology (42). In contrast, mutation of the PFV YEIL motif led to only a moderate reduction in PFV particle release, although it was associated with a complete loss in particle infectivity. We therefore tested whether removal of the PFV YEIL motif could be complemented by heterologous L-domain sequence motifs. As with the previously described PFV Gag L1 (PSAP) replacement mutants (42), we generated PFV Gag expression constructs that had the PFV Gag L3 (YEIL) motif replaced by the L-domain motif of HIV-1 (PTAP), EIAV (YPDL), or RSV (PPPY) (Fig. 1B). All constructs were expressed intracellularly at comparable levels upon transient cotransfection with a PFV retrovirus vector construct and wild-type PFV Pol and Env expression constructs (Fig. 2A). Quantification of PFV particle release supported by these mutant Gag proteins revealed for the ΔL3 RSV construct a reduction in particle egress in comparison to that for the wild type, at a level analogous to that of the original L3 mutant (Fig. 2A, lanes 1, 2, and 5; Fig. 2C). The ΔL3 HIV and ΔL3 EIAV mutants showed even greater reductions in particle release than the original L3 mutant (Fig. 2A, lanes 2, 3, and 4; Fig. 2C). All mutant particles showed normal levels of Gag processing in secreted particulate material (Fig. 2A), which is also an indirect indication of correct viral genome and PFV Pol incorporation, as well as of normal levels of Env incorporation (Fig. 2B). Furthermore, none of the mutant proteins showed any indication of restoration of particle infectivity, which was abolished upon mutation of the YEIL motif in PFV Gag (Fig. 2D). This apparent lack of functional complementation of the PFV Gag YEIL motif by heterologous L-domain sequence motifs further supports our previous view that this sequence motif does not function as a classical L domain.
FIG. 2.
Analysis of PFV Gag ΔL3 L-domain replacement mutants. Mutant PFV particles as indicated were generated by transient transfection of 293T cells using the four-plasmid PFV vector system. (A and B) Representative Western blot analysis of 293T cell lysates (cell) and viral particles (virus) purified by ultracentrifugation through 20% sucrose using either a polyclonal anti-PFV Gag (α-Gag) (A) or an anti-PFV Env LP (α-LP) (B) specific antiserum. (C) Quantification of PFV particle release. Means and standard deviations of particle-associated PFV Gag protein expression corrected for intracellular expression levels (n = 3) are shown. (D) Relative infectivities of cell supernatants (extracellular) and freeze-thaw cell lysates (intracellular) using the EGFP marker gene transfer assay. The values obtained using the wild-type PFV Gag expression plasmid (wt) were arbitrarily set to 100%. Means and standard deviations from three independent experiments are shown. 293T cells were cotransfected with pMD9, pcziPol, pczHFVenvEM002, and either pcziPG2 (wt), pcziPG L3 (L3), pcziPG ΔL3-HIV (ΔL3-HIV), pcziPG ΔL3-EIAV (ΔL3-EIAV), or pcziPG ΔL3-RSV (ΔL3-RSV). As controls, cells were transfected with only pcDNA3.1+zeo (pcDNA).
Characterization of YEILGL point mutants.
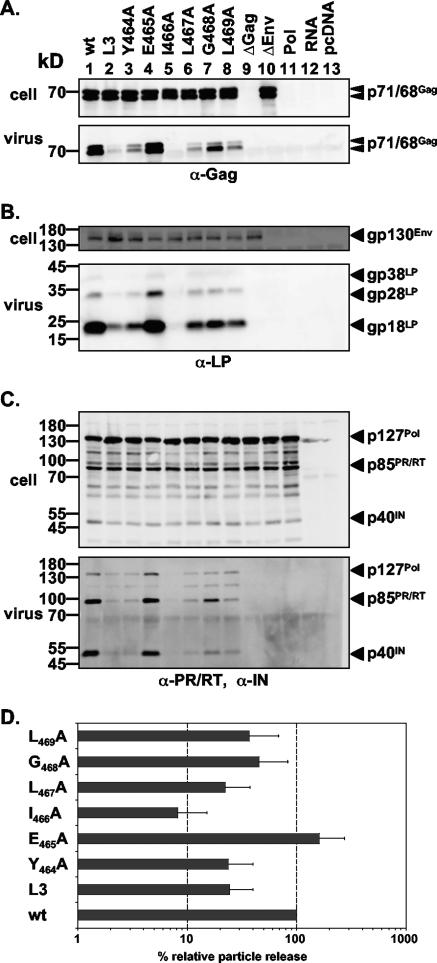
As illustrated in Fig. 1C, alignment of the PFV Gag YEIL motif and flanking sequences with their counterparts in different FV isolates revealed the presence of an evolutionarily conserved YXXLGL consensus sequence motif. This suggests an important function of this motif for FV replication and, in particular, for infectivity. In order to determine its potential function in more detail, we performed alanine-scanning mutagenesis of individual residues of this consensus motif in PFV Gag (Fig. 1B). All mutant Gag proteins were expressed at levels comparable to that of the wild type upon cotransfection with a PFV retrovirus vector and wild-type PFV Pol and Env expression constructs into 293T cells (Fig. 3A). However, the effects of the individual mutations could be distinguished based on the analysis of particle release (Fig. 3). Mutants containing the Y464A, L467A, G468A, or L469A substitution showed particle release levels that were lower than that for the wild type (Fig. 3A to C, lanes 1, 3, 6, 7, and 8; Fig. 3D) and similar to that for the original L3 mutant (Fig. 3A to C, lanes 2; Fig. 3D). The I466A mutant showed an even greater reduction in particle egress than the original L3 mutant (Fig. 3A to C, lanes 2 and 5; Fig. 3D). The E465A mutant supported levels of PFV particle release similar to that of the wild type (Fig. 3A to C, lanes 1 and 4; Fig. 3D). For all mutant particles, Gag processing (Fig. 3A, lower panel), Pol incorporation and processing (Fig. 3C, lower panel), and Env content (Fig. 3B, lower panel) were similar to those for the wild type.
FIG. 3.
Biochemical analysis of PFV Gag L3 domain motif point mutants. Mutant PFV particles as indicated were generated by transient transfection of 293T cells using the four-plasmid PFV vector system. (A to C) Representative Western blot analysis of 293T cell lysates (cell) and viral particles (virus) purified by ultracentrifugation through 20% sucrose using either a polyclonal anti-PFV Gag (α-Gag) antiserum (A), a polyclonal anti-PFV Env LP (α-LP) specific antiserum (B), or a mixture of anti-PFV PR/RT (α-PR/RT) and anti-PFV IN (α-IN) monoclonal antibodies (C). (D) Quantification of PFV particle release. Means and standard deviations of particle-associated PFV Gag protein expression, corrected for intracellular expression levels (n = 6), are shown. 293T cells were cotransfected with pMD9, pcziPol, pczHFVenvEM002, and either pcziPG2 (wt), pcziPG L3 (L3), pcziPG Y464A (Y464A), pcziPG E465A (E465A), pcziPG I466A (I466A), pcziPG L467A (L467A), pcziPG G468A (G468A), pcziPG L469A (L469A), no pcziPG (ΔGag), or no pczHFVenvEM002 (ΔEnv). As controls, cells were transfected with either pcziPol only (Pol), pMD9 only (RNA), or pcDNA3.1+zeo only (pcDNA).
The PFV Gag G468A point mutant displays a temperature-sensitive phenotype.
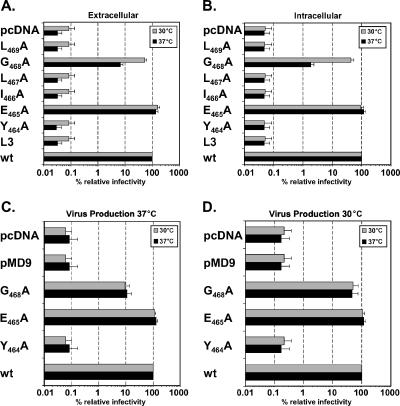
Next, we analyzed the infectivities of the individual mutants using a flow cytometric EGFP gene transfer assay. This analysis revealed comparable supernatant and cell-associated infectivities for the wild type and the E465A mutant (Fig. 4A and B). In contrast, analysis of the other mutants characterized by reduced PFV particle release revealed an interesting result. Whereas for most of these mutants (Y464A, I466A, L467A, L469A, and L3) no infectivity (at least 1,000-fold lower than that of the wild type) could be measured, the G468A mutant displayed extracellular and cell-associated infectivities that were only reduced to about 8% and 2% of those of the wild type, respectively (Fig. 4A and B). Furthermore, in contrast to those of the other mutants, the extracellular and cell-bound infectivities of the G468A mutant were increased to 53% and 43% of those of the wild type upon virus production and transduction at 30°C, indicating a temperature-sensitive phenotype (Fig. 4A and B). Further experiments with selected mutants demonstrated that the temperature-sensitive phenotype of the G468A mutant was established during particle production rather than at the stages of entry into the target cell, since extracellular infectivities were dependent on the incubation temperature used during virus production, regardless of the incubation temperature during subsequent infection of target cells (Fig. 4C and D).
FIG. 4.
Infectivity analysis of PFV Gag L3 domain motif point mutants. Mutant PFV particles as indicated were generated by transient transfection of 293T cells using the four-plasmid PFV vector system in duplicate. (A and B) During the sodium butyrate induction step, one plate per construct was shifted to 30°C (shaded bars) whereas the other plate was incubated at 37°C (solid bars) until virus was harvested from the culture supernatant (extracellular) (A) or freeze-thaw cell lysates (intracellular) (B). Subsequently, HT1080 target cells were incubated for 6 h with viral supernatants or dilutions thereof; after removal of the viral supernatants, target cells were incubated for an additional 4 h at the same temperature as before. Then all target cells were shifted to 37°C, and they were analyzed by FACS 48 h after addition of the viral supernatants to the target cells. (C and D) Reciprocal incubation at 37°C (C) or 30°C (D) during extracellular virus production and subsequent infection of HT1080 target cells using selected PFV Gag L3 motif mutants. Mean relative infectivities and standard deviations (n = 3) of cell supernatants (extracellular) and freeze-thaw cell lysates (intracellular) by the EGFP marker gene transfer assay are shown. The values obtained using the wild-type PFV Gag expression plasmid (wt) were arbitrarily set to 100%. 293T cells were cotransfected with pMD9, pcziPol, pczHFVenvEM002, and either pcziPG2 (wt), pcziPG L3 (L3), pcziPG Y464A (Y464A), pcziPG E465A (E465A), pcziPG I466A (I466A), pcziPG L467A (L467A), pcziPG G468A (G468A), or pcziPG L469A (L469A). As controls, cells were transfected either with pMD9 only (pMD9) or with pcDNA3.1+zeo only (pcDNA).
Analysis of RNA packaging and genome reverse transcription.
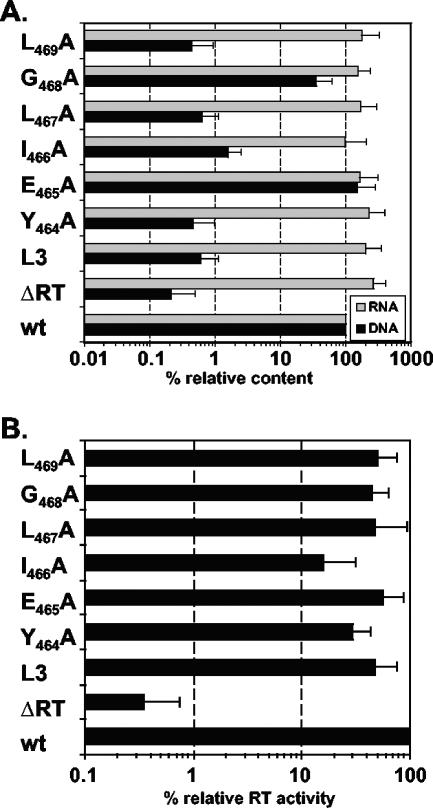
Although FVs initially package viral RNA genomes during intracytoplasmic assembly of the viral capsid, one peculiarity of the FV replication strategy is the generation of particle-associated reverse genome transcripts in the virus-producing cell (1, 29, 37). Particle-associated foamy virus DNA, and not RNA, has been demonstrated to be the infectious genome (37, 50). To characterize the replication defect of the different PFV Gag L3-motif mutants in further detail, we analyzed vector RNA packaging and reverse transcription in extracellular virions by real-time PCR. The results shown in Fig. 5A demonstrate that all PFV Gag mutants, including the wild type cotransfected with a Pol expression construct harboring an inactive RT mutant (ΔRT), packaged vector RNA efficiently (within a threefold range of the wild type). In contrast, all PFV Gag mutants, which showed no detectable infectivities in the GFP transfer assay (L3, Y464A, I466A, L467A, and L469A), contained 70- to 200-fold fewer genomic DNA copies per particle than the wild type, at levels comparable to that of the ΔRT sample (Fig. 5A). E465A mutant particles had amounts of reverse-transcribed vector genomes similar to that of the wild type, whereas the G468A mutant showed a threefold reduction in particle-associated reverse transcript levels (Fig. 5A). The residual amounts of reverse transcripts detected for the ΔRT sample, in the range of 0.3% of the wild-type amount, reflect the detection limit of the real-time DNA PCR and are residual copies of the input plasmid DNA used to generate the particles by transient transfection. These were present in the samples despite extensive DNase I digestion of intact viral particles prior to isolation of particle-associated nucleic acids (Fig. 5A). A similar background signal was also observed in analysis of supernatant samples of 239T cells transfected with the pMD9 gene transfer vector alone or a nonexpressing pUC19 EGFP control plasmid (data not shown).
FIG. 5.
Nucleic acid composition and reverse transcriptase activities of mutant PFV particles. Mutant PFV particles as indicated were generated by transient transfection of 293T cells using the four-plasmid PFV vector system. (A) Relative nucleic acid composition of mutant particles. Following DNase I digestion of intact, purified particles, nucleic acids were isolated, and the relative amounts of vector RNA and DNA copies (expressed as percentages) were determined in comparison to that for the wild type by real-time PCR. Mean relative RNA and DNA contents and standard deviations (n = 3), normalized for the differential particle releases of the individual mutants, are shown. (B) Relative reverse transcriptase activities of mutant particles. Following subtilisin digestion of intact, purified particles, particle lysates were generated after an additional ultracentrifugation step and analyzed for PFV reverse transcriptase activity using a RetroSys C-type RT enzyme-linked immunosorbent assay. Mean relative reverse transcriptase activities (n = 3) and standard deviations, normalized for the differential particle releases of the individual mutants and expressed as percentages, are shown. 293T cells were cotransfected with pMD9, pcziPol, pczHFVenvEM002, and either pcziPG2 (wt), pcziPG L3 (L3), pcziPG Y464A (Y464A), pcziPG E465A (E465A), pcziPG I466A (I466A), pcziPG L467A (L467A), pcziPG G468A (G468A), or pcziPG L469A (L469A). As a control, cells were transfected with pMD9, pcziPG2, pczHFVenvEM002, and an RT-deficient Pol expression vector, pGiPolΔRT (ΔRT).
The biochemical characterization of the different mutant particles shown in Fig. 3 revealed normal Gag processing (Fig. 3A), which is an indirect indication of viral genome and Pol incorporation. This is in line with the quantification of particle-associated nucleic acids, which indicated normal RNA genome packaging of all Gag mutants analyzed (Fig. 5A). Furthermore, no defects in PFV Pol incorporation or Pol processing were observed by using Pol-specific antibodies in Western blot analysis (Fig. 3C). Therefore, the failure of most of the mutants to reverse transcribe their genomes was not the result of an absence of RT. Alternatively, either the incorporated RT could be enzymatically inactive or it was incapable of reverse transcribing the viral genome because of the lack of a proper environment for this process. To address the former possibility, we analyzed particle-associated RT activity by using a commercially available enzyme-linked immunosorbent assay-based C-type RT assay that has previously been shown to work for PFV RT (20). All RT activity values were normalized for physical particles by quantification of particle-associated Gag protein by Western blotting. The relative RT activities of the particles of the individual mutants in comparison to that for wild-type particles are shown in Fig. 5B. Except for I466A mutant particles, which displayed fivefold lower relative RT activity, the RT activities of the mutants were only two- to threefold lower than that of the wild type. In contrast, particles generated by cotransfection of wild-type PFV Gag and the RT-deficient Pol packaging constructs resulted in background RT activity signals similar to those for mock controls (300-fold lower than that of the wild type).
Taken together, these results demonstrate a good correlation between the lack of infectiousness of individual mutants and their failure to generate particle-associated reverse transcripts, although they packaged normal amounts of the viral RNA genome and showed only marginally reduced particle-associated RT activities when analyzed in vitro.
Ultrastructural analysis of particle morphogenesis.
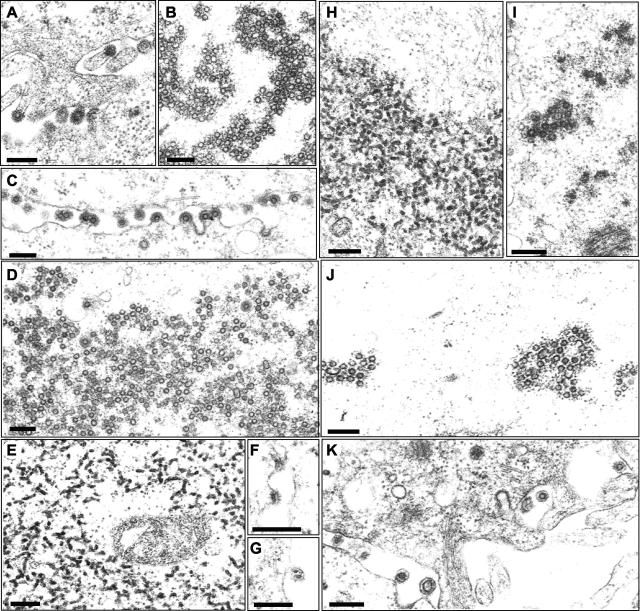
The previous analysis of the original PFV Gag L3 mutant revealed aberrant particle morphogenesis (42). To determine the role of the YEILGL motif in FV particle morphogenesis in further detail, 293T cells transiently cotransfected with expression vectors for PFV Env, PFV Pol, a packageable vector RNA, and selected PFV Gag point mutants were examined by electron microscopy. The results shown in Fig. 6 demonstrate PFV particle-release from cellular membranes and accumulation of naked capsids in the cytoplasm of cells expressing the wild-type PFV Gag protein (Fig. 6A and B). The E465A mutant showed a wild-type phenotype, including budding from cellular membranes (Fig. 6C) and naked capsids in the cytoplasm (Fig. 6D). Figure 6E to G show the Y464A mutant as an example of the group of mutants that showed no infectivity and completely failed to reverse transcribe their genomes. Similarly to the L3 mutant, electron-dense aggregates that probably represent aggregated Gag proteins were detected in the cytoplasm of cells expressing the Gag Y464A mutant (Fig. 6E). In addition, aberrant budding structures at cellular membranes were observed associated with the typical prominent PFV Env spike structures (Fig. 6F and G). In contrast, the G468A mutant showed an intermediate phenotype, with electron-dense material and only very few normally shaped capsids being detected in the cytoplasm when samples were incubated at 37°C prior to fixation (Fig. 6H and I). In samples incubated overnight at a lower temperature prior to fixation, more capsids with nearly wild type morphology were detectable in the cytoplasm, reflecting the temperature-sensitive phenotype of this mutant (Fig. 6J and K). However, the capsids in the cytoplasm seemed to contain incompletely closed capsids, and both larger and smaller capsids were present at greater frequencies (Fig. 6J and K).
FIG. 6.
Electron microscopy analysis. Electron micrographs show representative thin sections of 293T cells transiently cotransfected with the four-plasmid PFV vector system using either wild-type PFV Gag (A and B), PFV Gag E465A (C and D), PFV Gag Y464A (E to G), or PFV Gag G468A (H to K) expression constructs. Magnifications: ×61,000 for panels A, B, E, H, I, J, and K; ×45,000 for panels C and D; ×96,000 for panel F; ×77,000 for panel G. Bars, 200 nm.
Thus, ultrastructural analysis of the mutants correlated well with relative infectivity and with genetic analysis, suggesting that proper FV capsid morphogenesis is required for intraparticle reverse transcription of the foamy virus RNA genome.
DISCUSSION
In the present study we characterized the role of the conserved YXXLGL motif of PFV Gag in particle assembly, release, and infectivity. This motif was shown to be important for FV replication in previous studies aimed at characterization of the PFV L domains. The YXXL motif, unlike the PSAP L domain, is conserved in FV isolates of different species. The previous analysis suggested that the YXXL motif has a function in PFV capsid assembly rather than in particle release. This notion is further supported by the current study and indicates that the PFV Gag YEIL motif does not function like a classical retrovirus L domain. Mutant Gag proteins in which the YEIL motif was replaced with heterologous L-domain motifs failed to restore the defect in particle release and infectivity, in contrast to similar replacement mutants of the PFV Gag PSAP L domain (42).
In this study, the further characterization of the YEILGL motif by alanine-scanning mutagenesis revealed the importance of conserved amino acids Y464, I466, L467, and L469 for proper PFV capsid morphology and reverse transcription of the PFV RNA genome. In contrast, mutation of the E465 residue was well tolerated, a finding consistent with this amino acid being the least conserved in the sequence motif (Fig. 1C). Of particular interest was the G468A Gag mutant, which displayed an intermediate, temperature-sensitive phenotype. Biochemical characterization of the individual point mutants revealed normal relative levels of Env and Pol incorporation into particles. This excluded a lack of viral glycoprotein or enzyme incorporation as the cause of the lack of infectiousness of the released mutant particles.
FVs, unlike orthoretroviruses, reverse transcribe their RNA genome during, or shortly after, capsid assembly in the virus-producing cell (reviewed in reference 28). Previously it has been reported that PFV Gag p71/p68 cleavage is required for viral infectivity (13). Inactivation of the p71/p68 cleavage site by mutagenesis resulted in the generation of PFV particles with incompletely closed capsids at higher frequencies. Furthermore, these mutants were noninfectious and failed to accumulate linear PFV cDNA in transfected cells. However, neither particle-associated PFV cDNA content nor incorporation of Pol into particles was analyzed for the p71/p68 cleavage site mutants (13). These mutants are particularly interesting in light of the current study, in which all noninfectious Gag mutants displayed aberrant particle morphologies and failed to reverse transcribe the viral RNA genome. Altogether, this is reminiscent of certain HIV-1 Gag mutants that display aberrant capsid morphologies, are associated with defects in early steps of the viral replication cycle, and lead to incomplete reverse transcription of the viral genome or failure to initiate reverse transcription (10, 15, 35, 47).
Orthoretrovirus Gag proteins are processed by the viral protease into several mature subunits, including MA, CA, and NC. Structural analysis of HIV-1 Gag has revealed that the CA subunit contains two separately folding domains connected by a short 5-residue linker (17, 18). In general, the N-terminal domain (NTD) is involved in shaping the virion morphology, whereas the CTD is essential for capsid assembly. Unlike orthoretrovirus Gag proteins, FV Gag proteins are not processed into MA, CA, and NC subunits and lack the major homology domain found in all other retrovirus CA CTDs. Furthermore, due to a lack of sequence homology between FV Gag proteins and orthoretrovirus Gag proteins, no clear MA, CA, and NC subdomains can be assigned (reviewed in reference 28). When the phenotypes of the noninfectious PFV Gag mutants identified in this study are compared to those of Gag mutants of other retroviruses, they resemble most closely those of HIV-1 with mutations in the CA NTD. Like the PFV Gag mutants, these HIV Gag mutants show aberrant capsid morphologies, and in some studies they have been shown to harbor a defect in reverse transcription (15, 35, 45). This suggests that the YEILGL motif is part of a Gag subdomain with a role analogous to that of the HIV-1 CA NTD in influencing capsid morphology.
A short distance from the C terminus of the YEILGL motif (aa 464 to 469), another functional domain has been characterized: GR box I (aa 485 to 510) (49). GR box I, analogous to orthoretrovirus NC domain zinc finger motifs, displays nucleic acid binding activities and is essential for FV RNA genome packaging (49). Furthermore, the region spanning aa 414 to 540, containing both the YEILGL motif and GR box I, has previously been reported to contain an I domain, characterized by analysis of RSV Gag chimeras (4). The reduced particle release of YEILGL motif mutants may indicate that this motif contributes to the I-domain activity of the region spanning PFV Gag aa 414 to 540. However, all noninfectious PFV Gag mutants analyzed in this study package normal relative amounts of viral RNA, and in contrast to orthoretrovirus NC I domains, the YEILGL motif is surrounded by very few positively charged amino acids (8, 39). Therefore, a function of the YEILGL motif as an I domain seems unlikely, although a defect in Gag-Gag interactions due to mutation of the YEILGL motif (and resulting in the moderately decreased particle release observed) cannot be excluded and has not been directly addressed in this study.
Taken together, the results of this study demonstrate that the conserved PFV Gag YXXLGL motif is essential for correct particle assembly and capsid morphology. Furthermore, to our knowledge, our report of the G468A PFV Gag mutant is the first description of a temperature-sensitive assembly defect in PFV Gag, and this mutant should be a useful tool for further analysis of PFV capsid assembly and the genome reverse transcription process. In addition, this study suggests that proper PFV core assembly is required for reverse transcription of the viral genome. However, it is not a prerequisite for incorporation of the other structural proteins, nor does it lead to incorporation of inactive polymerase or an enzymatic defect of the viral reverse transcriptase per se. This suggests that PFV requires a proper microenvironment within the assembled viral capsid in order for reverse transcription of the viral genome to occur.
Acknowledgments
We thank Uta von Schwedler, Axel Rethwilm, and Welkin Johnson for critical reading of the manuscript.
This work was supported by grants from the DFG (Li621/3-1, Li621/4-1) and the BMBF (01ZZ0102) to D.L.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Baldwin, D. N., and M. L. Linial. 1998. The roles of Pol and Env in the assembly pathway of human foamy virus. J. Virol. 72:3658-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baunach, G., B. Maurer, H. Hahn, M. Kranz, and A. Rethwilm. 1993. Functional analysis of human foamy virus accessory reading frames. J. Virol. 67:5411-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D. 2006. Late budding domains and host proteins in enveloped virus release. Virology 344:55-63. [DOI] [PubMed] [Google Scholar]
- 4.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukrinskaya, A. G. 2004. HIV-1 assembly and maturation. Arch. Virol. 149:1067-1082. [DOI] [PubMed] [Google Scholar]
- 6.Cartellieri, M., O. Herchenröder, W. Rudolph, M. Heinkelein, D. Lindemann, H. Zentgraf, and A. Rethwilm. 2005. N-terminal Gag domain required for foamy virus particle assembly and export. J. Virol. 79:12464-12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, G., S. Park, B. Choi, S. Hong, J. Lee, E. Hunter, and S. S. Rhee. 1999. Identification of a cytoplasmic targeting/retention signal in a retroviral Gag polyprotein. J. Virol. 73:5431-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delelis, O., J. Lehmann-Che, and A. Saib. 2004. Foamy viruses—a world apart. Curr. Opin. Microbiol. 7:400-406. [DOI] [PubMed] [Google Scholar]
- 10.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Göttlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DuBridge, R. B., P. Tang, H. C. Hsia, P. M. Leong, J. H. Miller, and M. P. Calos. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eastman, S. W., and M. L. Linial. 2001. Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J. Virol. 75:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enssle, J., N. Fischer, A. Moebes, B. Mauer, U. Smola, and A. Rethwilm. 1997. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J. Virol. 71:7312-7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, N., M. Heinkelein, D. Lindemann, J. Enssle, C. Baum, E. Werder, H. Zentgraf, J. G. Müller, and A. Rethwilm. 1998. Foamy virus particle formation. J. Virol. 72:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schröder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268:294-307. [DOI] [PubMed] [Google Scholar]
- 16.Flügel, R. M., and K. I. Pfrepper. 2003. Proteolytic processing of foamy virus Gag and Pol proteins. Curr. Top. Microbiol. Immunol. 277:63-88. [DOI] [PubMed] [Google Scholar]
- 17.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 18.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 19.Heinkelein, M., C. Leurs, M. Rammling, K. Peters, H. Hanenberg, and A. Rethwilm. 2002. Pregenomic RNA is required for efficient incorporation of Pol polyprotein into foamy virus capsids. J. Virol. 76:10069-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill, C. L., P. D. Bieniasz, and M. O. McClure. 1999. Properties of human foamy virus relevant to its development as a vector for gene therapy. J. Gen. Virol. 80:2003-2009. [DOI] [PubMed] [Google Scholar]
- 21.Imrich, H., M. Heinkelein, O. Herchenröder, and A. Rethwilm. 2000. Primate foamy virus Pol proteins are imported into the nucleus. J. Gen. Virol. 81:2941-2947. [DOI] [PubMed] [Google Scholar]
- 22.Klein, D., B. Bugl, W. H. Günzburg, and B. Salmons. 2000. Accurate estimation of transduction efficiency necessitates a multiplex real-time PCR. Gen. Ther. 7:458-463. [DOI] [PubMed] [Google Scholar]
- 23.Kräusslich, H. G., M. Fäcke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann-Che, J., M. L. Giron, O. Delelis, M. Löchelt, P. Bittoun, J. Tobaly-Tapiero, H. de The, and A. Saib. 2005. Protease-dependent uncoating of a complex retrovirus. J. Virol. 79:9244-9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindemann, D., M. Bock, M. Schweizer, and A. Rethwilm. 1997. Efficient pseudotyping of murine leukemia virus particles with chimeric human foamy virus envelope proteins. J. Virol. 71:4815-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M.Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindemann, D., and A. Rethwilm. 1998. Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing. J. Virol. 72:4088-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linial, M. L., and S. W. Eastman. 2003. Particle assembly and genome packaging. Curr. Top. Microbiol. Immunol. 277:89-110. [DOI] [PubMed] [Google Scholar]
- 29.Moebes, A., J. Enssle, P. D. Bieniasz, M. Heinkelein, D. Lindemann, M. Bock, M. O. McClure, and A. Rethwilm. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morikawa, Y. 2003. HIV capsid assembly. Curr. HIV Res. 1:1-14. [DOI] [PubMed] [Google Scholar]
- 31.Patton, G. S., S. A. Morris, W. Chung, P. D. Bieniasz, and M. O. McClure. 2005. Identification of domains in Gag important for prototypic foamy virus egress. J. Virol. 79:6392-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petit, C., M. L. Giron, J. Tobaly-Tapiero, P. Bittoun, E. Real, Y. Jacob, N. Tordo, H. De The, and A. Saib. 2003. Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J. Cell Sci. 116:3433-3442. [DOI] [PubMed] [Google Scholar]
- 33.Pfrepper, K. I., M. Löchelt, H. R. Rackwitz, M. Schnolzer, H. Heid, and R. M. Flügel. 1999. Molecular characterization of proteolytic processing of the Gag proteins of human spumavirus. J. Virol. 73:7907-7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasheed, S., W. A. Nelson-Rees, E. M. Toth, P. Arnstein, and M. B. Gardner. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027-1033. [DOI] [PubMed] [Google Scholar]
- 35.Reicin, A. S., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rethwilm, A. 2003. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277:1-26. [DOI] [PubMed] [Google Scholar]
- 37.Roy, J., W. Rudolph, T. Juretzek, K. Gärtner, M. Bock, O. Herchenröder, D. Lindemann, M. Heinkelein, and A. Rethwilm. 2003. Feline foamy virus genome and replication strategy. J. Virol. 77:11324-11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saib, A., F. Puvion Dutilleul, M. Schmid, J. Peries, and H. de The. 1997. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J. Virol. 71:1155-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55gag. J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schliephake, A. W., and A. Rethwilm. 1994. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 68:4946-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw, K. L., D. Lindemann, M. J. Mulligan, and P. A. Goepfert. 2003. Foamy virus envelope glycoprotein is sufficient for particle budding and release. J. Virol. 77:2338-2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stange, A., I. Mannigel, K. Peters, M. Heinkelein, N. Stanke, M. Cartellieri, H. Göttlinger, A. Rethwilm, H. Zentgraf, and D. Lindemann. 2005. Characterization of prototype foamy virus Gag late assembly domain motifs and their role in particle egress and infectivity. J. Virol. 79:5466-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanke, N., A. Stange, D. Lüftenegger, H. Zentgraf, and D. Lindemann. 2005. Ubiquitination of the prototype foamy virus envelope glycoprotein leader peptide regulates subviral particle release. J. Virol. 79:15074-15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stenbak, C. R., and M. L. Linial. 2004. Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. J. Virol. 78:9423-9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tobaly-Tapiero, J., P. Bittoun, M. L. Giron, M. Neves, M. Koken, A. Saib, and H. de The. 2001. Human foamy virus capsid formation requires an interaction domain in the N terminus of Gag. J. Virol. 75:4367-4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilk, T., V. Geiselhart, M. Frech, S. D. Fuller, R. M. Flügel, and M. Löchelt. 2001. Specific interaction of a novel foamy virus Env leader protein with the N-terminal Gag domain. J. Virol. 75:7995-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu, S. F., K. Edelmann, R. K. Strong, A. Moebes, A. Rethwilm, and M. L. Linial. 1996. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 70:8255-8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, S. F., M. D. Sullivan, and M. L. Linial. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zemba, M., T. Wilk, T. Rutten, A. Wagner, R. M. Flügel, and M. Löchelt. 1998. The carboxy-terminal p3Gag domain of the human foamy virus Gag precursor is required for efficient virus infectivity. Virology 247:7-13. [DOI] [PubMed] [Google Scholar]