Abstract
Members of the mitochondrial carrier family have been reported in eukaryotes only, where they transport metabolites and cofactors across the mitochondrial inner membrane to link the metabolic pathways of the cytosol and the matrix. The genome of the giant virus Mimiviridae mimivirus encodes a member of the mitochondrial carrier family of transport proteins. This viral protein has been expressed in Lactococcus lactis and is shown to transport dATP and dTTP. As the 1.2-Mb double-stranded DNA mimivirus genome is rich in A and T residues, we speculate that the virus is using this protein to target the host mitochondria as a source of deoxynucleotides for its replication.
Mimiviridae mimivirus (25) is the largest known virus, with a diameter of 400 nm, and was first identified as a pathogen of Acanthamoeba polyphaga living in the water of a hospital cooling tower following a pneumonia outbreak (19). Subsequently, mimivirus has been detected in the respiratory tracts of patients suffering from hospital-acquired pneumonia (5) and a laboratory technician, who developed acute pneumonia that did not respond to antibiotics after working with samples containing the virus (26). In the latter case, serologic studies were negative for the usual pneumonia-causing agents, but high levels of mimivirus seroconversion were found, as were proteins in the patient's sera that are unique to mimivirus (26). The mimivirus genome is 1.2 Mb long, it contains 911 genes, and its nucleotide composition is 72% A and T residues (25). It is larger than any other viral genome and exceeds the length of some bacterial genomes. Phylogenetic analysis of its proteins places mimivirus among the nucleocytoplasmic large DNA viruses (NCLDV), which includes the poxviruses, asfarviruses, iridoviruses, and phycodnaviruses (16). It has been speculated that mimivirus is the descendant of a more-complex entity that existed before the emergence of the three domains of life (25) and, furthermore, that mimivirus and its ancestors may represent a new domain of life and that they contributed to the genesis of eukaryotes (24). However, an alternative proposition is that the NCLDV are relatively new viruses with a simple ancestor, which developed after the evolution of eukaryotes and that they acquired many genes subsequently by horizontal gene transfer and gene duplication (16).
Among the NCLDV, the best-matching homologues of mimivirus proteins are found most often in phycodnaviruses (25). Putative RNA polymerase subunits are present in the Emiliania huxleyi phycodnavirus (33), the poxviruses, and the iridoviruses (16). Mimivirus also possesses subunits of RNA polymerase, suggesting that it can replicate independently of the machinery of the host cell. NCLDV often possess enzymes for nucleotide biosynthesis because their DNA synthesis requires more deoxynucleotides than can be provided by the available cytosolic pool or from degradation of host DNA by viral endonucleases (31). The mimivirus genome has genes for enzymes of nucleotide metabolism, including (deoxy)cytidine deaminase (R197), thymidine kinase (L258), large and small subunits of ribonucleotide reductase (L312 and R313), nucleoside diphosphate kinase (R418), thymidylate synthase (R497), deoxynucleotide kinase (R512), and GMP synthase (L716). Genes for thymidylate kinase and dUTPase, which are found in other NCLDV, have not been detected in the mimivirus genome. This absence is curious given its AT-rich genome (25). However, the mimivirus nucleoside diphosphate kinase has been reported as having a preferential affinity for deoxypyrimidine nucleotides, which may reflect an evolutionary adaptation to provide dTTP from the limited dTDP pool (17).
In the genomic sequence of mimivirus we have identified a member of the mitochondrial carrier family of transport proteins. Thus far, members of this family have been reported in eukaryotes only, where they transport metabolites and cofactors across the mitochondrial inner membrane to link the metabolic pathways of the cytosol and the matrix. Here we show that the viral mitochondrial carrier (VMC1) transports dATP and, to a lesser extent, dTTP, TTP, UTP, and ADP. VMC1 may support the replication of the large mimivirus genome by acquiring additional nucleotide triphosphates from the mitochondrial pool in exchange for cytosolic ADP.
MATERIALS AND METHODS
Enzymes and chemicals.
Restriction enzymes were purchased from New England Biolabs. Piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) was obtained from Fluka (United Kingdom). The substrate dATP was obtained from Sigma (United Kingdom), and the [3H]dATP was obtained from Amersham Biosciences (United Kingdom). The radiolabeled chemicals used for the initial substrate screen were [14C]ADP, [2-14C]oxoglutarate, [14C]glutamine, [14C]ATP, [14C]glutamate, [3H]glutathione, [14C]putrecine, and (S)-[adenosyl-14C]methionine from PerkinElmer. The radiolabeled compounds [14C]malate, [14C]malonate, [14C]aminolevulinic acid, [14C]GDP, [14C]cysteine, [14C]methionine, [14C]tryptophan, [3H]pyridoxine, [14C]pantothenate, and [14C]pyridoxal phosphate were from American Radiolabeled Chemicals. The substrates [14C]lactate, [14C]pyruvate, [14C]oxoisocaproate, [14C]citrate, [3H]cAMP, [3H]CTP, [14C]histidine, [14C]lysine, [14C]alanine, [14C]glycine, [14C]ornithine, [14C]leucine, [14C]tyrosine, [14C]proline, [3H]biotin, [3H]folate, [14C]choline, and [14C]NAD+ were from Amersham Pharmacia, whereas [14C]GTP, [3H]riboflavin, [14C]nicotinate, and [14C]nicotinamide were from Moravek.
DNA techniques.
The L276 gene from the mimivirus genome was amplified by PCR using the KOD HiFi DNA polymerase (Novagen) to introduce NcoI and XbaI sites at the 5′ and 3′ ends of the gene, respectively. The DNA fragments were restricted and ligated into the pNZ8048 vector (9, 10), previously restricted by NcoI and XbaI. The ligation mixtures were electroporated into electrocompetent Lactococcus lactis strain NZ9000 (18). All constructs were confirmed by DNA sequencing.
Protein expression in L. lactis.
L. lactis cells were grown at 30°C in M17 medium (Difco), supplemented with 1% glucose and 5 μg/ml of chloramphenicol, until the optical density at 600 nm (OD600) reached 0.5, after which expression was induced by nisin A by adding a 1,000-fold dilution of the spent medium of the nisin-producing strain NZ9700. Cells were washed in PIPES buffer (10 mM PIPES, 50 mM NaCl, pH 7.0), and membrane vesicles were prepared by mechanical disruption at 30 kpsi using a 2.2-kW Z Plus Series cell disruptor (Constant Systems Ltd.). Whole cells were removed by centrifugation at 9,700 × g for 2 × 10 min at 4°C (Sorvall), and membranes were collected by ultracentrifugation at 140,000 × g at 4°C for 30 min (Beckman), washed once in PIPES buffer, and resuspended at a final protein concentration of approximately 5 mg/ml. The membrane proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. For quantification of expression levels, the intensities of the protein bands were determined in the linear range by using the Image Master 2D Elite software.
Transport assays.
Prior to transport, the lactococcal membrane vesicles were fused with liposomes, essentially as described previously (21). Liposomes were prepared by mixing E. coli total lipid extracts and egg yolk phosphatidylcholine (both from Avanti Polar Lipids, Inc.) in a 3:1 ratio (wt/wt) in PIPES buffer to a final concentration of 20 mg/ml. To prepare membrane vesicles loaded with substrate, membranes (1 mg protein) were mixed with liposomes in a ratio of 1:5 protein to lipid (wt/wt) in 1 ml of PIPES buffer with 5 mM substrate at pH 7. The mixture was frozen in liquid nitrogen and thawed slowly seven times before being stored in liquid nitrogen. This procedure leads to fusion of the liposomes with the vesicles but creates multilaminar fused membranes that are variable in size. Therefore, prior to the transport assays, the liposome-vesicle fusions were extruded through a 1-μm membrane (Whatman) and collected by centrifugation at 300,000 × g for 30 min at 4°C. As a consequence of this procedure, the fused membranes become unilaminar and uniform in size. The pellet was resuspended in 200 μl of PIPES-buffer with 5 mM substrate at pH 7. To remove the external substrate, the suspension was applied to a 3.5-ml bed volume Sephadex G-75 gel filtration column, previously equilibrated with PIPES buffer at 4°C. The fused membranes (1 ml) were collected in PIPES buffer and kept on ice for immediate use in transport assays.
Transport into fused membrane vesicles was initiated by diluting 100 μl of membrane vesicles into 300 μl of PIPES buffer in the presence of cold (final concentration of 1 μM) and radioactively labeled (concentration of about 1.8 nM) substrate at 30°C with constant stirring. At regular time intervals, the transport was quenched by adding 4 ml of ice-cold PIPES buffer, immediately followed by filtration through cellulose nitrate filters (0.45-μm pore size). The filters were washed once with 2 ml of ice-cold PIPES buffer and transferred to a scintillation vial, after which 2 ml of Ultima Gold AB scintillation liquid (Packard Bioscience) was added for counting in a Packard TriCarb 2100 TR-liquid scintillation analyzer.
Protein identification by mass spectrometry.
Gel bands were excised and digested with trypsin according to the method of Shevchenko et al. (30). Samples of the tryptic peptide mixtures were mixed with alpha-cyano-4-hydroxy-transcinnamic acid matrix and analyzed with an ABI 4700 Proteomics Analyzer with time of flight/time of flight optics (Applied Biosystems). For peptide mass fingerprints, mass calibration was performed with features internal to the spectrum, specifically the matrix-related ion peak at 1,060.048 Da and the trypsin autolysis peaks at 2,163.057 Da and 2,273.160 Da. For peptide fragment spectra, an external mass calibration was generated from the 2,163.057-Da trypsin autolysis peak, using the y10, y14, and y16 fragment ions. Spectra were interpreted by using the Mascot sequence search engine (22a) configured with a mass tolerance of 40 ppm and the gel-derived variable modifications “propionamide cysteine” and “methionine sulfoxide” on the nonredundant protein sequence data set of the National Center for Biotechnology Information (32).
Computational sequence analysis.
The BLASTP application of BLAST (1) was used to identify homologues in the nonredundant protein sequence data set of the National Center for Biotechnology Information. The searches used the BLOSUM62 substitution matrix (15) with BLAST parameters set to a word size of 3, a gap opening penalty of 11, and a gap extension penalty of 1. In addition, potential homologues were checked to see if they both had equivalent Pfam domains (14).
The TargetP application (13) was used to predict the subcellular localization of viral proteins by detecting potential N-terminal mitochondrial targeting sequences. To reduce false positives, accepted predictions were limited to those with a reliability class of either 1 or 2.
Jalview (8) was used for sequence editing and TeXshade (4) for sequence display.
RESULTS
The mimivirus genome encodes a eukaryotic mitochondrial carrier.
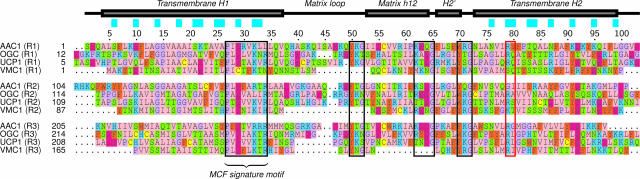
Members of the mitochondrial carrier family consist of three tandem sequence repeats of approximately 100 amino acids in length (29), each containing two transmembrane α-helices and the signature motif P-X-[DE]-X-X-[KR]. Computational sequence analysis of the mimivirus genome showed that the protein encoded by the L276 gene contained three copies of the mitochondrial carrier family motif (PF00153), as defined by Pfam (14). It had been annotated as a putative oxoglutarate/malate carrier protein on the basis of sequence similarity (25). With 237 residues, the viral mitochondrial carrier VMC1 encoded by the L276 gene is significantly shorter than eukaryotic mitochondrial carriers. An alignment of VMC1 with the human ADP/ATP carrier AAC1 (3), the bovine oxoglutarate carrier (28), and the human uncoupling protein UCP1 (2) shows that it possesses all three repeats, all six transmembrane α-helices and the signature motifs of the family but that it lacks the matrix α-helix in the third repeat (Fig. 1). The mitochondrial S-adenosylmethionine transporter lacks the first matrix α-helix, showing that this type of loss is not unique (20).
FIG. 1.
Multiple-sequence alignment of the three repeats (R1, R2, and R3) of the bovine AAC1, the human oxoglutarate carrier (OGC), the human uncoupling protein 1 (UCP1), and the mimivirus VMC1. The residue numbering and secondary structure annotation are taken from the first repeat of AAC1, and the cyan boxes above the alignment indicate residue positions that face the aqueous cavity in the structure of AAC1 (22). Regions of high sequence similarity are outlined with a black box, and the contact points of the proposed binding site are outlined with a red box.
The L276 gene of mimivirus was cloned and expressed in L. lactis using the NICE system (21). This expression system has been applied successfully in the functional expression of 11 mitochondrial carriers from Saccharomyces cerevisiae (21) and the mitosomal adenine nucleotide transporter from Entamoeba histolytica (7), which also belongs to the mitochondrial carrier family.
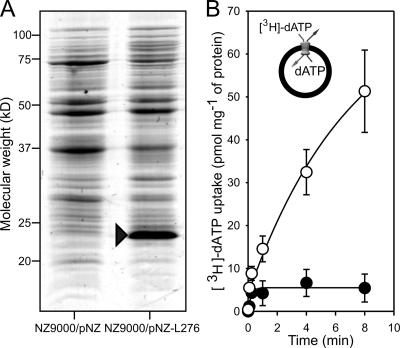
The viral carrier was expressed in lactococcal membranes to 5% of total membrane protein (Fig. 2A). The identity of the band in Fig. 2A was investigated with peptide mass fingerprinting and tandem mass spectrometry. Of the 45 peptide masses comprising the fingerprint, 17 matched the protein sequence of VMC1, representing 50% sequence coverage with a score of 110/77, and 9 peptide masses matched the ribosomal protein S4 from L. lactis (gi 62464342) representing 49% sequence coverage with a score of 96/77. Eight peptides matching the mimivirus sequence were chosen and fragmented, producing spectra whose fragment ions supported this assignment with a combined score of 390/54 and representing a sequence coverage of 38%, showing highly convincing evidence for its presence.
FIG. 2.
(A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of lactococcal membranes stained with Coomassie brilliant blue showing expression of the viral mitochondrial carrier from the recombinant L. lactis (NZ9000/pNZ-L276) and a control strain with an empty plasmid (NZ9000/pNZ). (B) Uptake rate of [3H]dATP substrate into lactococcal membrane-liposome vesicles containing the viral mitochondrial carrier (open circles) and controls without it (filled circles), adjusted to that of the total amount of lactococcal membrane protein in micrograms. Error bars are standard errors of the means from three independent experiments. The insert in panel B illustrates the main principle of the transport assays with the vesicles, incorporated substrate (dATP), and external substrate ([3H]dATP).
Transport assays.
Three sets of transport assays were carried out with the expressed VMC1 protein in lactococcal membranes. Mitochondrial transporters most often carry out transport steps in which one substrate is exported in exchange for another substrate that is imported. Thus, in the first set of experiments, unlabeled substrate was incorporated into lactococcal membrane vesicles, and the exchange with the external radiolabeled substrate of the same type was monitored (as depicted for [3H]dATP/dATP exchange in the insert of Fig. 2B). Significant uptake into vesicles was observed for dATP (Fig. 2B), whereas uptake of malate, malonate, 2-oxoglutarate, lactate, pyruvate, glutamine, oxoisocaproate, citrate, aminolevulinic acid, ATP, ADP, GTP, GDP, cAMP, CTP, cysteine, glutamate, histidine, lysine, alanine, glycine, ornithine, leucine, methionine, tyrosine, tryptophan, proline, biotin, glutathione, pyridoxine, pantothenate, choline, pyridoxal phosphate, putrecine, riboflavin, nicotinate, NAD+, nicotinamide, S-adenosylmethionine, and folate was not detectable. Transport of dATP was unaffected by the classical inhibitors of the ADP/ATP carriers, carboxyatractyloside (cATR) and bongkrekic acid (Fig. 3A). The radiolabeled dATP was not transported into vesicles that did not contain dATP, indicating that VMC1 operates as a strict homo-exchanger of dATP rather than a uniporter.
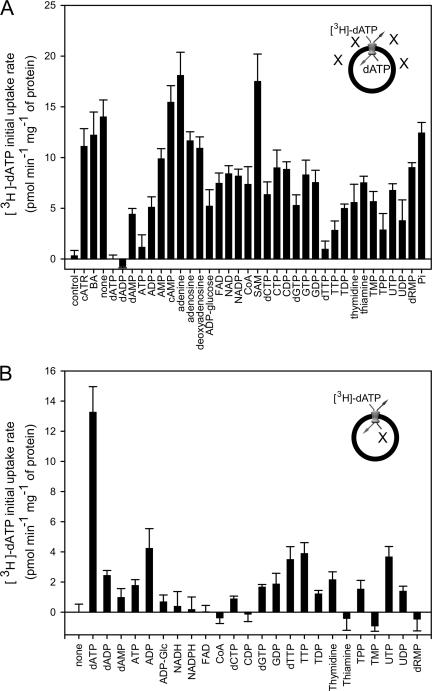
FIG. 3.
Transport assays with [3H]dATP. (A) Effect of excesses of different inhibitors and substrates on the initial uptake rate of [3H]dATP into lactococcal membrane-liposome vesicles. Error bars are standard errors of the means of triplicates from two independent experiments. (B) Initial uptake rates of external [3H]dATP into lactococcal membrane-liposome vesicles in exchange for different substrates. Error bars are standard errors of the means of triplicates from two independent experiments. Abbreviations: carboxyatractyloside, cATR; bongkrekic acid, BA; coenzyme A, CoA; S-adenosylmethionine, SAM; thiamine monophosphate, TMP; thiamine pyrophosphate, TPP; 2′-deoxyribose 5′-monophosphate, dRMP. The inserts in panels A and B illustrate the main principle of the transport assays with the alternative substrates, X.
In a second set of experiments, competitive transport assays were carried out in the presence of a 3,000-fold excess of a potential competitor to identify substances that affect the transport of dATP (Fig. 3A). If the unlabeled substrate competes with dATP for the same substrate binding site, uptake of [3H]dATP will be suppressed. The uptake of the dATP was reduced by several adenine, thymidine, and uridine nucleotides (Fig. 3A), suggesting that they are potential competitors for binding, but this does not mean that they are transported by the carrier.
A third set of experiments was performed to test active transport of the competing substrates by incorporating them into membrane vesicles and by determining the initial exchange rate with external radiolabeled dATP (Fig. 3B). The radiolabeled dATP is taken up into the vesicle only if the incorporated substrate is exported. These experiments showed that ADP, dTTP, TTP, and UTP are transported in exchange for dATP, although at a lesser rate than for the homo-exchange of dATP. Thus, the carrier does not facilitate ADP-ADP exchange, but it does catalyze dATP-ADP exchange.
DISCUSSION
The VMC1 protein has three copies of the signature motif of the eukaryotic mitochondrial carriers, but it is sufficiently diverged that there is no clear orthologue among the known eukaryotic mitochondrial carriers to indicate an ancestral sequence. In addition, the 73% A+T content of the L276 gene is consistent with the mimivirus genome as a whole and is significantly higher than the A+T content of most eukaryotes. Together, this suggests that the VMC1 gene is not a recent horizontal gene transfer from a eukaryotic host. The VMC1 protein is the smallest functional mitochondrial carrier yet identified. The NCLDV Paramecium bursari chlorella virus 1 has also been reported to possess enzymes that are either the smallest or among the smallest proteins of a family (11), including a functional K+ channel (23). These diminutive enzymes may represent either the ancestors of eukaryotic proteins or the effect of evolutionary pressures to reduce gene size following horizontal transfer (11). It is believed that the ancestral mitochondrial carrier was a single domain protein that assembled to form a carrier but that its gene later duplicated to form the three tandem repeats of the modern family members (29). The presence of a mitochondrial carrier in the mimivirus genome does not resolve the debate on the origin and evolution of either mimivirus or the mitochondrial carrier family.
We have proposed previously a common substrate binding site for the mitochondrial carriers, which consists of three contact points that define selectivity between amino acid-, keto acid-, and adenine-containing substrates (27). The substrate binding site of the VMC1 protein has the characteristics of the keto acid carriers and nucleotide-binding uncoupling proteins. Uncoupling proteins have an aspartate or glutamate residue implicated in proton transport in the first transmembrane helix (12), and a glutamate residue is present in the corresponding position in the VMC1 protein (Fig. 1). This suggests that substrate binding to the VMC1 protein may be more similar to the nucleotide binding of uncoupling proteins than to that of the adenine nucleotide carriers (27). This suggestion is consistent with the lack of inhibition by cATR on dATP transport by VMC1 and with the residues of AAC1 that interact with CATR (22) not being conserved in the VMC1 protein.
Our results show that VMC1 can transport dATP, ADP, TTP, dTTP, and UTP in exchange for dATP. Thus, we speculate that mimivirus may exploit the mitochondrion of its host by harvesting its dATP and dTTP for the replication of its large A+T-rich genome, possibly in exchange for cytosolic ADP. Potential sources for the mitochondrial deoxynucleotides are its deoxynucleotide pool and the breakdown of mitochondrial DNA. Our speculation is in agreement with the proposal that the preferential affinity of the mimivirus nucleoside diphosphate kinase for deoxypyrimidine nucleotides is an evolutionary adaptation to accommodate the virus's dTTP requirement (17). Many viruses target the mitochondria of the host cell to control the cell death pathways, both through antiapoptotic pathways to ensure the host cell remains alive during their replication and proapoptotic proteins to induce cell lysis and allow the escape of viral particles once viral replication is completed (6). As well as VMC1, there are five other putative mitochondrial proteins encoded in the mimivirus genome: L359 (mitochondrial mismatch repair ATPase MutS), L572 and R776 (N-terminal half of the mitochondrial chaperone BCS1L), R596 (Evr1/Alr proteins of Fe/S cluster maturation), R740 (mitochondrial 18-kDa protein), and R824 [mitochondrial 5′(3′)-deoxyribonucleotidase]. In addition, there are four proteins, L81, R151, R900, and L908, with a potential mitochondrial targeting sequence but of unknown function. This suggests that mimivirus has evolved further means to exploit the mitochondrial physiology of its host for its biogenesis.
Acknowledgments
This work was supported by the United Kingdom Medical Research Council. M.M. was supported by an EC Marie Curie Fellowship (MCFI-2002-01203) and EMBO long-term fellowship (ALTF 43-2002).
We thank John Walker for many valuable suggestions and Didier Raoult for the kind donation of the mimivirus DNA.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aquila, H., T. A. Link, and M. Klingenberg. 1985. The uncoupling protein from brown fat mitochondria is related to the mitochondrial ADP/ATP carrier. Analysis of sequence homologies and of folding of the protein in the membrane. EMBO J. 4:2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aquila, H., D. Misra, M. Eulitz, and M. Klingenberg. 1982. Complete amino acid sequence of the ADP/ATP carrier from beef heart mitochondria. Hoppe-Seyler's Z. Physiol. Chem. 363:345-349. [PubMed] [Google Scholar]
- 4.Beitz, E. 2000. TEXshade: shading and labeling of multiple sequence alignments using LATEX2 epsilon. Bioinformatics 16:135-139. [DOI] [PubMed] [Google Scholar]
- 5.Berger, P., L. Papazian, M. Drancourt, B. La Scola, J. P. Auffray, and D. Raoult. 2006. Ameba-associated microorganisms and diagnosis of nosocomial pneumonia. Emerg. Infect. Dis. 12:248-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boya, P., A. L. Pauleau, D. Poncet, R. A. Gonzalez-Polo, N. Zamzami, and G. Kroemer. 2004. Viral proteins targeting mitochondria: controlling cell death. Biochim. Biophys. Acta 1659:178-189. [DOI] [PubMed] [Google Scholar]
- 7.Chan, K. W., D. J. Slotboom, S. Cox, T. M. Embley, O. Fabre, M. van der Giezen, M. Harding, D. S. Horner, E. R. Kunji, G. Leon-Avila, and J. Tovar. 2005. A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica. Curr. Biol. 15:737-742. [DOI] [PubMed] [Google Scholar]
- 8.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunigan, D. D., L. A. Fitzgerald, and J. L. Van Etten. 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res. 117:119-132. [DOI] [PubMed] [Google Scholar]
- 12.Echtay, K. S., E. Winkler, M. Bienengraeber, and M. Klingenberg. 2000. Site-directed mutagenesis identifies residues in uncoupling protein (UCP1) involved in three different functions. Biochemistry 39:3311-3317. [DOI] [PubMed] [Google Scholar]
- 13.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 14.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyer, L. M., S. Balaji, E. V. Koonin, and L. Aravind. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156-184. [DOI] [PubMed] [Google Scholar]
- 17.Jeudy, S., J. M. Claverie, and C. Abergel. 2006. The nucleoside diphosphate kinase from mimivirus: a peculiar affinity for deoxypyrimidine nucleotides. J. Bioenerg. Biomembr. 38:247-254. [DOI] [PubMed] [Google Scholar]
- 18.Kunji, E. R., D. J. Slotboom, and B. Poolman. 2003. Lactococcus lactis as host for overproduction of functional membrane proteins. Biochim. Biophys. Acta 1610:97-108. [DOI] [PubMed] [Google Scholar]
- 19.La Scola, B., S. Audic, C. Robert, L. Jungang, X. de Lamballerie, M. Drancourt, R. Birtles, J. M. Claverie, and D. Raoult. 2003. A giant virus in amoebae. Science 299:2033. [DOI] [PubMed] [Google Scholar]
- 20.Marobbio, C. M., G. Agrimi, F. M. Lasorsa, and F. Palmieri. 2003. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 22:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monné, M., K. W. Chan, D. J. Slotboom, and E. R. Kunji. 2005. Functional expression of eukaryotic membrane proteins in Lactococcus lactis. Protein Sci. 14:3048-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pebay-Peyroula, E., C. Dahout-Gonzalez, R. Kahn, V. Trezeguet, G. J. Lauquin, and G. Brandolin. 2003. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426:39-44. [DOI] [PubMed] [Google Scholar]
- 22a.Perkins, D. N., D. J. Pappin, D. M. Crensy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551-3567. [DOI] [PubMed] [Google Scholar]
- 23.Plugge, B., S. Gazzarrini, M. Nelson, R. Cerana, J. L. Van Etten, C. Derst, D. DiFrancesco, A. Moroni, and G. Thiel. 2000. A potassium channel protein encoded by chlorella virus PBCV-1. Science 287:1641-1644. [DOI] [PubMed] [Google Scholar]
- 24.Raoult, D. 2005. The journey from Rickettsia to mimivirus. ASM News 71:278-284. [Google Scholar]
- 25.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J. M. Claverie. 2004. The 1.2-megabase genome sequence of mimivirus. Science 306:1344-1350. [DOI] [PubMed] [Google Scholar]
- 26.Raoult, D., P. Renesto, and P. Brouqui. 2006. Laboratory infection of a technician by mimivirus. Ann. Intern. Med. 144:702-703. [DOI] [PubMed] [Google Scholar]
- 27.Robinson, A. J., and E. R. Kunji. 2006. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc. Natl. Acad. Sci. USA 103:2617-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Runswick, M. J., J. E. Walker, F. Bisaccia, V. Iacobazzi, and F. Palmieri. 1990. Sequence of the bovine 2-oxoglutarate/malate carrier protein: structural relationship to other mitochondrial transport proteins. Biochemistry 29:11033-11040. [DOI] [PubMed] [Google Scholar]
- 29.Saraste, M., and J. E. Walker. 1982. Internal sequence repeats and the path of polypeptide in mitochondrial ADP/ATP translocase. FEBS Lett. 144:250-254. [DOI] [PubMed] [Google Scholar]
- 30.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 31.Van Etten, J. L. 2003. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 37:153-195. [DOI] [PubMed] [Google Scholar]
- 32.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, L. Y. Geer, W. Helmberg, Y. Kapustin, D. L. Kenton, O. Khovayko, D. J. Lipman, T. L. Madden, D. R. Maglott, J. Ostell, K. D. Pruitt, G. D. Schuler, L. M. Schriml, E. Sequeira, S. T. Sherry, K. Sirotkin, A. Souvorov, G. Starchenko, T. O. Suzek, R. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2006. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 34:D173-D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, W. H., D. C. Schroeder, M. J. Allen, M. T. Holden, J. Parkhill, B. G. Barrell, C. Churcher, N. Hamlin, K. Mungall, H. Norbertczak, M. A. Quail, C. Price, E. Rabbinowitsch, D. Walker, M. Craigon, D. Roy, and P. Ghazal. 2005. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science 309:1090-1092. [DOI] [PubMed] [Google Scholar]