Abstract
Hepatitis C virus (HCV) enters cells via a pH- and clathrin-dependent endocytic pathway. Scavenger receptor BI (SR-BI) and CD81 are important entry factors for HCV internalization into target cells. The SR-BI gene gives rise to at least two mRNA splice variants, SR-BI and SR-BII, which differ in their C termini. SR-BI internalization remains poorly understood, but SR-BII is reported to endocytose via a clathrin-dependent pathway, making it an attractive target for HCV internalization. We demonstrate that HCV soluble E2 can interact with human SR-BI and SR-BII. Increased expression of SR-BI and SR-BII in the Huh-7.5 hepatoma cell line enhanced HCV strain J6/JFH and JFH infectivity, suggesting that endogenous levels of these receptors limit infection. Elevated expression of SR-BI, but not SR-BII, increased the rate of J6/JFH infection, which may reflect altered intracellular trafficking of the splice variants. In human plasma, HCV particles have been reported to be complexed with lipoproteins, suggesting an indirect interaction of the virus with SR-BI and other lipoprotein receptors. Plasma from J6/JFH-infected uPA-SCID mice transplanted with human hepatocytes demonstrates an increased infectivity for SR-BI/II-overexpressing Huh-7.5 cells. Plasma-derived J6/JFH infectivity was inhibited by an anti-E2 monoclonal antibody, suggesting that plasma virus interaction with SR-BI was glycoprotein dependent. Finally, anti-SR-BI antibodies inhibited the infectivity of cell culture- and plasma-derived J6/JFH, suggesting a critical role for SR-BI/II in HCV infection.
Hepatitis C virus (HCV) is an enveloped positive-strand RNA virus and the sole member of the genus Hepacivirus, within the Flaviviridae. Approximately 170 million individuals are infected with HCV worldwide, and the majority are at risk of developing serious progressive liver disease. The principal reservoir for viral replication is believed to be hepatocytes within the liver, and until recently, minimal information was available on the mechanism(s) of HCV entry. However, the last 3 years have seen several advances that contribute to our ability to study HCV hepatotropism. First, the development of the retrovirus pseudoparticle system, in which cell entry is dependent upon the expression of HCV glycoproteins (HCVpp) (4, 20), and secondly, the ability of the JFH strain of HCV to release infectious particles in cell culture (HCVcc) (25, 51, 55).
Early studies with a truncated soluble version(s) of HCV E2 (sE2) allowed the identification of a number of interacting cellular proteins, including the tetraspanin CD81 (16, 37), scavenger receptor class B type I (SR-BI) (43), and DC-specific ICAM-3-grabbing nonintegrin (DC-SIGN) and the related molecule DC-SIGN(R), or L-SIGN (15, 18, 27, 40). The availability of HCVpp and infectious HCVcc has provided tools for validating these receptor candidates.
CD81 is a nonglycosylated member of the tetraspanin family of proteins. Both HCVpp and HCVcc infectivities are inhibited by soluble forms of CD81 and by anti-CD81 monoclonal antibodies (MAbs), suggesting that CD81 is required for HCV infection (6, 20, 25). Definitive experiments showing that expression of CD81 in a CD81-negative human liver cell line, HepG2, confers infectivity support a critical role of CD81 in HCV cell entry (24, 25, 54, 55).
SR-BI is expressed within the liver, steroidogenic tissue, and macrophages and is considered to be the major receptor for high-density lipoprotein (HDL) (23). SR-BI mediates the traffic of cholesterol to and from lipoproteins by selective cholesterol uptake, cholesterol efflux, and receptor-mediated endocytosis (1, 34, 42, 44). The SR-BI gene gives rise to at least two mRNA splice variants. The SR-BII isoform differs from SR-BI at the C terminus, which is reported to confer intracellular localization on SR-BII (14, 33, 52).
Experiments to validate the role of SR-BI in HCV infection have proven difficult, since all cell types studied to date express SR-BI, and small interfering RNA silencing has a modest effect on HCVpp infectivity (6, 24, 48). The native lipoprotein ligands have differential effects on HCV infectivity: HDL enhances infectivity, low-density (LDL) and very low-density lipoproteins (VLDL) have no effect (5, 48), and oxidized LDL abrogates infectivity (50), suggesting a complex interplay between SR-BI, lipoproteins, and HCV. Treatment of target cells with inhibitors of SR-BI-dependent selective cholesterol uptake, BLT-2 and BLT-4, abrogates HDL-enhanced viral infectivity (5, 12), suggesting a role for this selective process in HCV entry. A recent study demonstrated that anti-SR-BI and anti-CD81 antibodies inhibit JFH infectivity in a synergistic manner, suggesting cooperativity between the receptors in mediating viral infection (21).
In human plasma, HCV particles have been reported to be complexed with lipoproteins, suggesting an indirect interaction of the virus with lipoprotein receptors (2, 28, 35, 45). However, the significance of the virus-lipoprotein interaction for the virus life cycle is unknown. Several laboratories have purified HCV from plasma to study virus-cell interactions; however, these experiments are difficult to interpret, since they are unable to measure viral infectivity. The recent observation that HCVcc is infectious for uPA-SCID mice with transplanted human hepatocytes provides a source of plasma that is infectious for cultured cells and allows in vitro experimentation (26, 31, 32).
HCVpp and HCVcc enter cells via a pH- and clathrin-dependent endocytic pathway (7, 10, 20, 30, 47). SR-BI internalization remains poorly understood, but SR-BII is reported to endocytose via a clathrin-dependent pathway, making it an attractive target for the study of HCV attachment and entry (13). In this study, we demonstrate that HCV sE2 can interact with human SR-BI and SR-BII expressed in CHO cells. Increased expression of SR-BI and SR-BII in Huh-7.5 cells enhances the infectivity of cell culture- and plasma-derived J6/JFH, suggesting that endogenous levels of these receptors limit HCV infection. Anti-SR-BI antibodies inhibit the infectivity of cell culture- and plasma-derived J6/JFH, supporting a critical role for SR-BI/II in HCV infection.
MATERIALS AND METHODS
Cells, antibodies, and plasmids.
293T and Chinese hamster ovary (CHO) cells were obtained from the American Type Culture Collection and propagated according to their recommendations. Huh-7.5 cells (provided by Charles Rice, The Rockefeller University, New York) (8) were propagated in Dulbecco's modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% nonessential amino acids. All cells were grown at 37°C in 5% CO2. CHO cells stably expressing SR-BI were generated and propagated as previously described (28). Anti-SR-BI sera were generated by immunizing rabbits (Mirus Corporation, Madison, WI) with a pcDNA expression vector encoding full-length SR-BI (28). Antisera specific for the SR-BI and SR-BII C-terminal peptides were generated by immunizing rabbits with peptides corresponding to amino acids 496 to 509 of murine SR-BI (46) and amino acids 493 to 514 of human SR-BII (33). The cloning of pCM5 expressing murine SR-BI tagged at the N terminus with enhanced green fluorescent proteins (eGFP) (provided by Deneys van der Westhuyzen, University of Kentucky) was previously reported (13, 14).
Generation of lentiviral vectors expressing SR-BI and SR-BII.
Fully sequenced human SR-BI and SR-BII genes were transfer cloned into pTRIP lentiviral packaging plasmids capable of directing their packaging into TRIP lentiviral gene delivery vectors (53).
HDL-SR-BI interactions.
Labeling of HDL with the fluorescence probe 1,1′-dioctadecyl-3,3,3-,3-tetramethylindocarbocyanine perchlorate (DiI) was carried out as previously described (38). Briefly, 5 × 105 CHO or CHO-SR-BI cells in suspension were preincubated with an irrelevant rabbit serum or with the anti-SR-BI serum for 1 h at 4°C, washed, and incubated with DiI-labeled HDL (50 μg protein/ml) for 1 h at 4°C. The cells were fixed with 5% formalin neutral buffered solution (Sigma), and DiI fluorescence was measured by flow cytometry (Epics XL; Beckman Coulter).
Expression of SR-BI and SR-BII.
TRIP lentiviruses expressing SR-BI, SR-BII, or CD9 were generated by cotransfecting 293T cells with plasmids encoding vesicular stomatitis virus G protein, human immunodeficiency virus Gag-Pol, and the pTRIP construct (1:3:3 ratio). CHO and Huh-7.5 cells were seeded at 8 × 105 cells per well in a six-well plate and infected 24 h later with the packaged lentivirus diluted in DMEM supplemented with 3% FBS. After 12 h, the cells were washed, trypsinized, and seeded into appropriate plates for HCV infection or flow cytometry. Murine eGFP-SR-BI was expressed in Huh-7.5 cells by transient transfection with Lipofectamine (Invitrogen, California)-based delivery of plasmid into cells.
Flow cytometry.
Cell surface expression of SR-BI/II was monitored by live-cell staining with antisera specific for the SR-BI extracellular region and preimmune or irrelevant-species isotype-matched antibodies, as previously described (17). To detect the intracellular C-terminal regions of SR-BI and SR-BII, cells were fixed with 1% paraformaldehyde and permeabilized with 0.05% saponin prior to being stained with specific antisera as previously described (8, 20). sE2 binding to cells was assayed as previously described (50). Analyses were performed using a FACScalibur flow cytometer (BD Biosciences) and FlowJo software (Tree Star, San Carlos, CA).
HCVcc genesis and infection assays.
J6/JFHcc and JFHcc particles were generated as previously described (25). Briefly, RNA was transcribed in vitro from full-length genomes using the Megascript T7 kit (Ambion, Austin, TX) and electroporated into Huh-7.5 cells. Seventy-two and 96 h postelectroporation, the supernatants were collected, pooled, and stored at −80°C. Three uPA+/+ SCID mice were transplanted with human hepatocytes and infected with cell culture-derived J6/JFH by intraperitoneal injection, as described previously (26, 32). COBAS Ampliprep TaqMan analysis (Roche Diagnostics, Mannheim, Germany) of mouse plasma demonstrated high-level infection, with a mean viremia of 4.72 × 106 IU/ml within 2 weeks. An acute-phase plasma-derived virus stock was generated by subsequent twice-weekly blood sampling for 4 weeks, pooled, aliquoted, and stored at −80°C. Huh-7.5 cells were seeded at 1.5 × 104 cells per well in 48-well plates and the following day were infected with virus diluted in 3% FBS-DMEM for 1 hour. At 72 h postinoculation, viral infection was detected by methanol fixation and staining for NS5A antigen using the anti-NS5A MAb 9E10 and Alexa 488-conjugated anti-mouse immunoglobulin G (IgG) (Invitrogen, California) (25). Huh-7.5 cells expressing murine eGFP-SR-BI (48 h posttransfection) were infected with JFHcc as described above, and after 72 h, the infected cells were visualized with anti-NS5A MAb and Alexa 633-conjugated anti-mouse IgG (Invitrogen, California).
RESULTS
SR-BI and SR-BII can interact with HCV sE2.
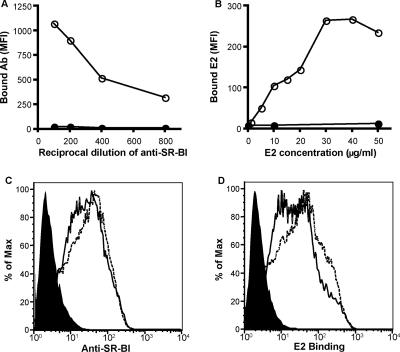
SR-BI expression in CHO cells confers sE2 binding (43). To address whether SR-BII can interact with sE2, CHO cells were transduced with lentiviral vectors expressing human SR-BI or SR-BII and transgene expression was assessed by anti-SR-BI serum reactivity. Antisera were raised by genetic immunization of rabbits with a plasmid encoding full-length SR-BI and were shown to bind CHO cells stably expressing SR-BI (Fig. 1A). HCV sE2 strain H77-bound CHO cells expressed SR-BI, showing saturation at 30 μg/ml (Fig. 1B). Anti-SR-BI serum bound to CHO cells transduced with TRIP-SR-BI or TRIP-SR-BII, confirming expression of the receptors at the cell surface (Fig. 1C). SR-BI and SR-BII bound comparable levels of sE2 at 30 μg/ml (Fig. 1D), suggesting that both molecules have the capacity to bind and mediate HCV infection.
FIG. 1.
Expression of SR-BII in CHO cells confers sE2 binding. (A) Anti-SR-BI serum reactivity for CHO (closed symbols) and CHO-SR-BI (open symbols). (B) sE2 reactivity for CHO (closed symbols) and CHO-SR-BI (open symbols). Bound E2 antigen was detected with rat anti-E2 monoclonal antibody 9/75 and Alexa Fluor 488 anti-rat Ig. The data are expressed as the mean fluorescence intensity (MFI). CHO cells were transduced with TRIP lentiviral vectors expressing human SR-BI or SR-BII and assessed for their reactivities with anti-SR-BI (1:400 dilution) (C) and sE2 (30 μg/ml) (D). Each panel displays parental CHO (filled), CHO-SR-BI (solid line), and CHO-SR-BII (dashed line).
Scavenger receptor BI and BII expression levels modulate hepatitis C virus infectivity.
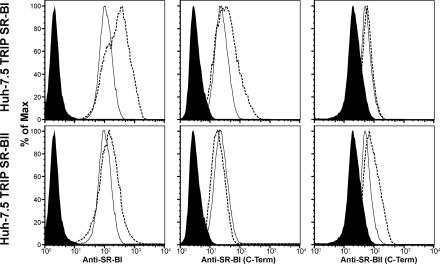
To study the effect of SR-BI/II overexpression on viral infectivity, Huh-7.5 cells were transduced with the TRIP viruses characterized in Fig. 1. Live-cell staining with the anti-SR-BI antiserum demonstrated ∼3-fold and ∼1.8-fold increases in cell surface-expressed SR-BI and SR-BII, respectively (Fig. 2). C-terminus-specific antibodies capable of differentiating between SR-BI and SR-BII demonstrated increased reactivity with permeabilized cells, confirming expression of the differentially spliced receptors (Fig. 2). SR-BI/II overexpression did not affect cell proliferation (data not shown). As controls, Huh-7.5 cells were transduced to express CD9, a tetraspanin with no role in HCV infection, showing an ∼13-fold increase in cell surface expression and eGFP-tagged murine SR-BI (data not shown). CD81 cell surface expression levels were comparable in all of the transduced cells (data not shown).
FIG. 2.
Overexpression of SR-BI and SR-BII in Huh-7.5 cells. Huh-7.5 cells were transduced with lentiviruses expressing human SR-BI or SR-BII. SR-BI/II expression was determined by flow cytometry using extracellular-domain-specific anti-SR-BI serum (on live cells) and C-terminus-specific antisera capable of discriminating between SR-BI and SR-BII (on fixed and permeabilized cells). Each panel displays the isotype control (filled), parental (solid line), and transduced (dashed line) Huh-7.5 cells.
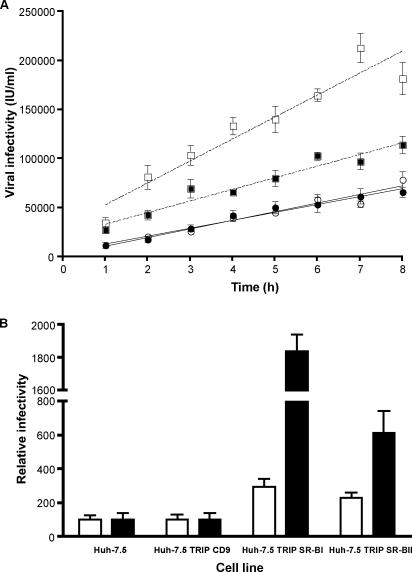
Parental and transduced cells were studied for the ability to support J6/JFHcc infection. We previously noted that the time allowed for virus adsorption can affect HCVcc infectivity, suggesting that viral attachment and/or internalization is slow in Huh-7.5 cells (30). We compared the abilities of the parental and transduced cells to support J6/JFH infection with increasing times of viral adsorption from 1 to 8 h. Infectivity for parental cells increased linearly over time, with a rate of 8,800 infections/h/ml (Fig. 3A). Overexpression of SR-BI increased J6/JFH infectivity approximately threefold and significantly increased the rate of infection, 22,360 infections/h/ml. In contrast, overexpression of SR-BII increased J6/JFH infectivity 2.5-fold with no significant change in the rate of infection, 11,280 infections/h/ml. Huh-7.5 cells overexpressing CD9 and eGFP murine SR-BI showed no change in susceptibility to HCVcc infection (data not shown). To determine if the same result was observed with a different strain of HCV, we assessed the infectivity of JFH-1 for parental and transduced Huh-7.5 cells following a 1-h period of adsorption. JFH infectivity for SR-BI- and SR-BII-overexpressing cells increased 18- and 6-fold, respectively, compared to parental cells (Fig. 3B). Several independent JFH and J6/JFH viral stocks showed the same boosting of infectivity, with JFH being more sensitive to SR-BI/II expression levels than J6/JFH (data not shown). The increased infectivity of J6/JFH and JFH for cells overexpressing SR-BI and SR-BII demonstrates that the level(s) of both receptors limits HCV infection of Huh-7.5 cells.
FIG. 3.
Overexpression of SR-BI and SR-BII in Huh-7.5 cells enhances HCVcc infection. (A) Parental (open circles) and transduced (filled circles) Huh-7.5 cells overexpressing human CD9, SR-BI (open squares), or SR-BII (filled squares) were incubated with J6/JFHcc for various times between 1 and 8 h; unbound virus was removed by washing, and the infection was allowed to proceed for 72 h. Both SR-BI- and SR-BII-transduced cells showed significantly elevated levels of infection (P < 0.05), with SR-BI cells showing a significantly higher rate of infection (22,360 infected cells/h/ml for SR-BI versus 8,800 to 11,000 for the other cell lines; P < 0.05; F test). Virus infectivity is expressed as the number of NS5A-positive cells or infected units (IU)/ml. (B) Parental and transduced cells were incubated with J6/JFHcc (white bars) or JFHcc (black bars) for 1 h, followed by a 72-h infection. Infected cells were visualized by staining them for intracellular NS5A, and virus infectivity for transduced cells was expressed relative to the parental cells. J6/JFH and JFH infectivities for parental Huh-7.5 cells were 11,000 ± 3,000 IU/ml and 2,400 ± 900 IU/ml, respectively. The error bars indicate the standard deviation above the mean of five individual infections.
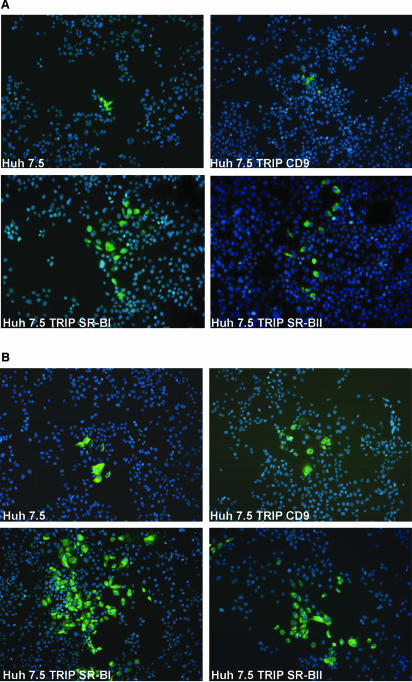
Overexpression of SR-BI/II increased the size of J6/JFH- and JFH-infected cell foci; this was particularly striking for JFH infection (Fig. 4). At 72 h, a focus of infected cells (a group of four or more infected cells) represents “local” virus spread from an initial primary infection event. We interpret the focus count to represent the number of primary infection events and the infected-cell count to reflect primary and secondary infection events. We observed a threefold increase in the number of J6/JFH-infected Huh-7.5 TRIP SR-BI cells compared to parental cells, with a twofold increase in infected foci, whereas JFH infection of Huh-7.5 TRIP SR-BI led to an 18-fold increase in the number of infected cells, with a 5-fold increase in the number of foci. These data suggest that increased SR-BI/II levels enhance HCV infectivity by promoting secondary/local virus spread, in addition to increasing the number of primary infection events.
FIG. 4.
Overexpression of SR-BI and SR-BII in Huh-7.5 cells increases infected-focus size. Parental and Huh-7.5 cells overexpressing human CD9, SR-BI, or SR-BII were infected with (A) J6/JFHcc or (B) JFHcc for 1 h, and the infection was allowed to proceed for 72 h. Infected cells were visualized by staining them for NS5A (green), and the nuclei were counterstained with DAPI. The images were taken at ×100 magnification.
Scavenger receptor BI and BII expression levels modulate plasma-derived J6/JFH infectivity.
The observation that J6/JFH is infectious for uPA-SCID mice with transplanted human hepatocytes provides us with a source of infectious plasma for infectivity studies (31). Transplanted hepatocytes within the chimeric mice secrete human lipoproteins (32), providing virus that closely mimics the virus-lipoprotein complexes circulating in HCV-infected patients. We tested the infectivity of plasma- and cell culture-derived J6/JFH for parental and transduced Huh-7.5 cells. Overexpression of SR-BI and SR-BII increased the infectivity of plasma-derived J6/JFH by 4-fold and 2.5-fold, respectively (Fig. 5A), consistent with the scavenger receptor-mediated enhancement of J6/JFH. Maillard and colleagues (28) suggested that plasma-derived HCV bound SR-BI independently of the viral glycoproteins. To test this model, we investigated the infectivity of plasma- and cell culture-derived J6/JFH for Huh-7.5 cells in the presence of a neutralizing anti-E2 MAb, C1 (25). Infectivity of plasma- and cell culture-derived J6/JFH was neutralized by MAb C1, demonstrating that both sources of virus infect cells in an E2-dependent manner (Fig. 5B).
FIG. 5.
Overexpression of SR-BI and SR-BII in Huh-7.5 cells enhances the glycoprotein-dependent infectivity of cell culture- and plasma-derived J6/JFH. (A) Parental and Huh-7.5 cells overexpressing human CD9, SR-BI, or SR-BII were incubated with J6/JFH derived from cell culture (J6/JFHcc; white bars) or from the plasma of infected chimeric mice (J6/JFHplasma; gray bars) for 6 h, followed by a 72-h infection. Infected cells were visualized by staining them for intracellular NS5A, and infectivity for transduced cells was expressed relative to the parental cells. J6/JFHcc and J6/JFHplasma infectivities for parental Huh-7.5 cells were 35,000 (±1,300) cells/ml and 2,250 (±700) cells/ml, respectively. The error bars indicate the standard deviation above the mean of two individual infections. (B) Comparable levels of infectious virus from the two sources were incubated with anti-E2 MAb C1 and a control anti-dengue virus MAb at 10 μg/ml for 1 h prior to infecting Huh-7.5 cells. Infectivity was measured by quantifying NS5A-positive cells, and the percent neutralization of C1 was defined by comparing infectivity in the presence of the anti-dengue virus MAb.
Anti-SR-BI antibodies inhibit cell culture- and plasma-derived HCV infectivity.
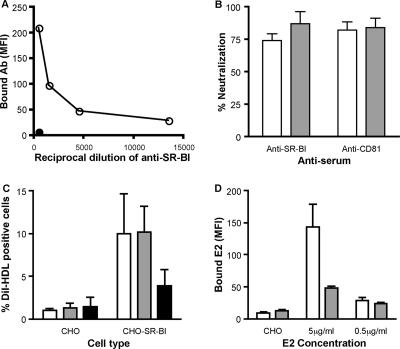
We assessed whether anti-SR-BI antibodies would modulate the ability of Huh-7.5 cells to support HCV replication. The polyclonal anti-SR-BI serum bound to Huh-7.5 cells and inhibited plasma- and cell culture-derived J6/JFH infectivity (Fig. 6A and B). Titration of the anti-SR-BI serum demonstrated a 50% neutralization endpoint of 1:1,550 for J6/JFHcc infection of Huh-7.5 cells (data not shown). Since we (17, 29, 54) and others (6, 24) have reported on the critical role of CD81 in HCV entry, we confirmed that infectivity of J6/JFH from both sources was inhibited with an anti-CD81 monoclonal antibody (MAb M38).
FIG. 6.
Anti-SR-BI serum inhibits cell culture- and plasma-derived J6/JFH infectivity. (A) Anti-SR-BI reactivity for Huh-7.5 cells (open circles). Control preimmune rabbit serum was tested at the highest dilution (filled circles). The data are expressed as the mean fluorescence intensity (MFI). (B) Huh-7.5 cells were incubated with anti-SR-BI and control preimmune sera at 1:500 or anti-CD81 MAb M38 and control isotype-matched Ig at 1 μg/ml for 1 h prior to infection with cell culture- (white bars) or plasma-derived (gray bars) J6/JFH. Infectivity was measured by quantifying NS5A-positive cells, and the percent neutralization of the receptor-specific antibodies was determined by comparing infectivity in the presence of the control antibodies. The error bars indicate the standard deviation of the mean of three replicate infections. (C) CHO or CHO-SR-BI cells were preincubated with no antibody (white bars), control rabbit serum (gray bars), or anti-SR-BI (black bars) at 1/400 prior to incubation with DiI-labeled HDL (50 μg protein/ml) for 1 h at 4°C. The data are shown as the percentage of DiI-HDL-positive cells and represent the mean plus standard deviation of four independent experiments. (D) CHO-SR-BI cells were preincubated with control rabbit serum (white bars) or anti-SR-BI (gray bars) at 1/400 prior to incubation with sE2 at 5 and 0.5 μg/ml. Cell-bound E2 was detected with rat anti-E2 MAb 9/75 and Alexa 488 anti-rat Ig. Data are shown for sE2 (5 μg/ml) binding to control CHO cells and are expressed as the mean fluorescence intensity (MFI) and standard deviation of duplicate experiments.
Several reports have demonstrated that HDL interaction with SR-BI can promote HCV infectivity (5, 12, 48, 49). Since hepatoma cells, including Huh-7, have been reported to assemble and secrete ApoA-containing high-density lipoprotein particles (9, 11, 19), HCV entry into Huh-7.5 may occur via lipoprotein-dependent and/or -independent pathways. In an effort to understand how the anti-SR-BI serum inhibits viral infectivity, we screened the serum for inhibition of HDL and sE2 binding to CHO-SR-BI. The antiserum inhibited HDL and sE2 interaction with SR-BI (Fig. 6C and D), suggesting that both activities may contribute to the neutralizing capacity of the polyclonal sera.
DISCUSSION
We have demonstrated that human SR-BI and SR-BII confer sE2 binding to CHO cells and, when overexpressed in Huh-7.5 cells, increase susceptibility to HCV infection. Antibodies specific for SR-BI could inhibit the infectivity of cell culture- and plasma-derived HCV, suggesting a critical role for the receptors in the HCV life cycle. Our findings were facilitated by the genesis of lentiviral vectors expressing SR-BI/II and the availability of antisera targeting both the extracellular regions and the C-terminus-specific regions of the two isoforms.
Overexpression of SR-BI and -II in Huh-7.5 cells significantly increased their susceptibility to cell culture- and plasma-derived HCV, suggesting that SR-BI/II density plays an important role in HCV infection. Similar observations have been reported for CD4 and chemokine receptor expression levels influencing human immunodeficiency virus cell entry (3, 39). Enhanced expression of SR-BI increased the rate of J6/JFH infection, whereas increased SR-BII expression had no detectable effect(s) on the rate of virus infection. These differences may reflect the altered trafficking profiles reported for SR-BI and SR-BII and may offer alternative routes of entry into Huh-7.5 cells. It is noteworthy that Huh-7.5 cells naturally express both SR-BI and SR-BII, as detected by the C-terminus-specific antibodies (Fig. 2). Overexpression of SR-BI/II enhanced JFH infectivity to a much greater extent than J6/JFH; 18-fold and 3-fold, respectively (Fig. 3), suggesting strain-specific variation within genotypic clades. We reported a similar observation with HCVpp expressing diverse glycoproteins showing altered interaction(s) with rodent CD81 expressed in HepG2 cells, supporting a model of glycoprotein variants with different affinities for the coreceptors (17). Since J6/JFH and JFH differ in their Core-NS2 regions, it is possible that the observed differences may be due to an altered affinity of the envelope proteins for SR-BI/II.
Murine SR-BI and SR-BII have altered localization and trafficking profiles that are directed by their C-terminal domains (13, 42, 44). Our data are consistent with these reports, demonstrating that human SR-BII is predominantly expressed within the cell and shows reduced expression at the cell surface compared to SR-BI (Fig. 2 and data not shown). The reduced enhancement of infection by SR-BII compared to SR-BI most likely reflects its lower cell surface expression (Fig. 2). Our data show comparable levels of sE2 binding to CHO-expressed SR-BI and SR-BII, suggesting that the C-terminal region of SR-BI/II has no effect(s) on sE2 or ligand interactions with the extracellular region. However, at the present time, it is unclear whether sE2 is a reliable predictor of viral-SR-BI/II interactions. HCV particles are likely to interact with SR-BI/II and other components of the receptor complex in a cooperative manner that may depend on the extra- and intracellular regions of the coreceptors. Our data suggest that overexpression of either isoform does not affect expression of the other. However, we have been unable to study the localization and trafficking of heterologous SR-BI/II in the transduced cell lines, since N-terminal tagging of both receptors affects their localization (data not shown). Hence, we cannot rule out the possibility that overexpression of SR-BII may have consequences for SR-BI localization/trafficking and vice versa. SR-BI has been reported to form oligomers to facilitate selective cholesterol uptake (41). It will be interesting to ascertain if SR-BI/II can form hetero-oligomers and whether they act cooperatively to facilitate HCV infection.
Infection of the transduced cells resulted in increased size of infected cell foci that was particularly apparent in the JFH-infected cultures, suggesting improved local virus spread in the presence of excess SR-BI/II (Fig. 4). Current experiments suggest that HCV can be transmitted within a culture by extracellular virus infection of naive cells and through cell-cell contact(s) (J. M. Timpe and Z. Stamataki, unpublished observations), suggesting that SR-BI/II may promote the latter route of transmission. An increased proliferation rate of the Huh-7.5 SR-BI/II cells cannot explain the increased focal size, since the transduced cells showed no proliferation advantage (data not shown). JFH has a reduced capacity to assemble or release infectious particles compared to J6/JFH and other chimeric viruses (22, 36, 55). Zhong et al. (56) reported that long-term propagation of JFH in cell culture generated an adaptive variant that showed enhanced particle infectivity and transmission within a culture. It will be interesting to compare the infectivity and transmission of the wild type and the adaptive JFH variant in parental and SR-BI/II-transduced cells to ascertain if the reduced infectivity of JFH particles is compensated for by overexpressing SR-BI/II. Our data suggest that overexpression of SR-BI/II not only increases Huh-7.5 cell susceptibility to primary infection, but aids virus spread to adjacent cells, either at the point of the initial infection or after viral replication and de novo virion production.
The ability of the anti-SR-BI serum to inhibit the infectivity of plasma- and cell culture-derived J6/JFH suggests a critical role for SR-BI/II in virus entry. Several reports have demonstrated that HDL interaction with SR-BI can enhance HCV infectivity (5, 12, 48, 49). Since interactions between the viral glycoproteins, HDL, and SR-BI are likely to play a role in HCV entry, we were interested to know if the “neutralizing” anti-SR-BI antibodies inhibited either of these ligand-receptor interactions. As a model system to study SR-BI in isolation from the other receptor components expressed in human hepatoma cells, we studied sE2 and HDL interaction with SR-BI expressed in CHO cells. The anti-SR-BI serum inhibited sE2 and HDL interaction with SR-BI (Fig. 6). These data suggest that anti-SR-BI neutralization of viral infectivity may occur not only by inhibiting E2 binding, but through interference with SR-BI-HDL interactions. Further studies with defined anti-SR-BI MAbs are warranted to elucidate the mechanism(s) of virus neutralization.
There are many reports of HCV being complexed with VLDL/LDL in the plasma of infected patients, suggesting that viruses may interact with lipoprotein receptors indirectly through the associated lipoproteins and not via the virus-encoded glycoproteins (2, 28, 35, 45). Several recent observations demonstrating that (i) J6/JFH is infectious for uPA-SCID mice with transplanted human hepatocytes (26, 31), (ii) transplanted hepatocytes within the mice secrete human lipoproteins (32), and (iii) plasma-derived J6/JFH has an increased specific infectivity and altered buoyant density compared to J6/JFHcc (26) suggest that mouse-derived J6/JFH is the best mimic for the virus-lipoprotein complexes circulating in HCV-infected patients. Our data clearly demonstrate that the infectivities of plasma- and cell culture-derived J6/JFH for SR-BI/II-transduced cells were enhanced to comparable levels (Fig. 5). Furthermore, the infectivity of virus from both sources was inhibited by antibodies specific for E2 and SR-BI, suggesting a common entry pathway that is limited by SR-BI/II expression (Fig. 6).
Acknowledgments
We thank Takaji Wakita for the JFH clone, Brett Lindenbach and Charles Rice for J6/JFH and Huh-7.5 cells, Dennis Burton for the C1 MAb, Fedor Berditchevski for the anti-CD81 M38 MAb, and Deneys van der Westhuyzen for pCM5 expressing eGFP-murine SR-BI. We thank Ke Hu for expert technical assistance.
This work was supported by PHS grant AI50798, by the Wellcome Trust, and by Ghent University through Concerted Action Grant 1205023. Thomas Vanwolleghem is supported by a Ph.D. grant from the Research Foundation-Flanders.
Footnotes
Published ahead of print on 10 January 2007.
REFERENCES
- 1.Acton, S., A. Rigotti, K. T. Landschulz, S. Xu, H. H. Hobbs, and M. Krieger. 1996. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 271:518-520. [DOI] [PubMed] [Google Scholar]
- 2.Andre, P., F. Komurian-Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Brechot, G. Paranhos-Baccala, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannert, N., D. Schenten, S. Craig, and J. Sodroski. 2000. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J. Virol. 74:10984-10993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard, E., S. Belouzard, L. Goueslain, T. Wakita, J. Dubuisson, C. Wychowski, and Y. Rouille. 2006. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chisholm, J. W., E. R. Burleson, G. S. Shelness, and J. S. Parks. 2002. ApoA-I secretion from HepG2 cells: evidence for the secretion of both lipid-poor apoA-I and intracellularly assembled nascent HDL. J. Lipid Res. 43:36-44. [PubMed] [Google Scholar]
- 10.Codran, A., C. Royer, D. Jaeck, M. Bastien-Valle, T. F. Baumert, M. P. Kieny, C. A. Pereira, and J. P. Martin. 2006. Entry of hepatitis C virus pseudotypes into primary human hepatocytes by clathrin-dependent endocytosis. J. Gen. Virol. 87:2583-2593. [DOI] [PubMed] [Google Scholar]
- 11.Domitrovich, A. M., D. J. Felmlee, and A. Siddiqui. 2005. Hepatitis C virus nonstructural proteins inhibit apolipoprotein B100 secretion. J. Biol. Chem. 280:39802-39808. [DOI] [PubMed] [Google Scholar]
- 12.Dreux, M., T. Pietschmann, C. Granier, C. Voisset, S. Ricard-Blum, P. E. Mangeot, Z. Keck, S. Foung, N. Vu-Dac, J. Dubuisson, R. Bartenschlager, D. Lavillette, and F. L. Cosset. 2006. High density lipoprotein inhibits hepatitis C virus neutralising antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 281:18285-18295. [DOI] [PubMed] [Google Scholar]
- 13.Eckhardt, E. R., L. Cai, S. Shetty, Z. Zhao, A. Szanto, N. R. Webb, and D. R. Van der Westhuyzen. 2006. High density lipoprotein endocytosis by scavenger receptor SR-BII is clathrin-dependent and requires a carboxyl-terminal dileucine motif. J. Biol. Chem. 281:4348-4353. [DOI] [PubMed] [Google Scholar]
- 14.Eckhardt, E. R., L. Cai, B. Sun, N. R. Webb, and D. R. van der Westhuyzen. 2004. High density lipoprotein uptake by scavenger receptor SR-BII. J. Biol. Chem. 279:14372-14381. [DOI] [PubMed] [Google Scholar]
- 15.Falkowska, E., R. J. Durso, J. P. Gardner, E. G. Cormier, R. A. Arrigale, R. N. Ogawa, G. P. Donovan, P. J. Maddon, W. C. Olson, and T. Dragic. 2006. L-SIGN (CD209L) isoforms differently mediate trans-infection of hepatoma cells by hepatitis C virus pseudoparticles. J. Gen. Virol. 87:2571-2576. [DOI] [PubMed] [Google Scholar]
- 16.Flint, M., J. M. Thomas, C. M. Maidens, C. Shotton, S. Levy, W. S. Barclay, and J. A. McKeating. 1999. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J. Virol. 73:6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint, M., T. von Hahn, J. Zhang, M. Farquhar, C. T. Jones, P. Balfe, C. M. Rice, and J. A. McKeating. 2006. Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 80:11331-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higashi, Y., H. Itabe, H. Fukase, M. Mori, Y. Fujimoto, and T. Takano. 2003. Transmembrane lipid transfer is crucial for providing neutral lipids during very low density lipoprotein assembly in endoplasmic reticulum. J. Biol. Chem. 278:21450-21458. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapadia, S. B., H. Barth, T. Baumert, J. A. McKeating, and F. V. Chisari. 2006. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 81:374-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koutsoudakis, G., A. Kaul, E. Steinmann, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krieger, M. 2001. Scavenger receptor class B type I is a multiligand HDL receptor that influences diverse physiologic systems. J. Clin. Investig. 108:793-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41:265-274. [DOI] [PubMed] [Google Scholar]
- 25.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 26.Lindenbach, B. D., P. Meuleman, A. Ploss, T. Vanwolleghem, A. J. Syder, J. A. McKeating, R. E. Lanford, S. M. Feinstone, M. E. Major, G. Leroux-Roels, and C. M. Rice. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. USA 103:3805-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 28.Maillard, P., T. Huby, U. Andreo, M. Moreau, J. Chapman, and A. Budkowska. 2006. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/Cla1 is mediated by ApoB-containing lipoproteins. FASEB J. 20:735-737. [DOI] [PubMed] [Google Scholar]
- 29.McKeating, J. A., L. Q. Zhang, C. Logvinoff, M. Flint, J. Zhang, J. Yu, D. Butera, D. D. Ho, L. B. Dustin, C. M. Rice, and P. Balfe. 2004. Diverse hepatitis C virus glycoproteins mediate viral infection in a CD81-dependent manner. J. Virol. 78:8496-8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meertens, L., C. Bertaux, and T. Dragic. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J. Virol. 80:11571-11578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mercer, D. F., D. E. Schiller, J. F. Elliott, D. N. Douglas, C. Hao, A. Rinfret, W. R. Addison, K. P. Fischer, T. A. Churchill, J. R. Lakey, D. L. Tyrrell, and N. M. Kneteman. 2001. Hepatitis C virus replication in mice with chimeric human livers. Nat. Med. 7:927-933. [DOI] [PubMed] [Google Scholar]
- 32.Meuleman, P., L. Libbrecht, R. De Vos, B. de Hemptinne, K. Gevaert, J. Vandekerckhove, T. Roskams, and G. Leroux-Roels. 2005. Morphological and biochemical characterization of a human liver in a uPA-SCID mouse chimera. Hepatology 41:847-856. [DOI] [PubMed] [Google Scholar]
- 33.Mulcahy, J. V., D. R. Riddell, and J. S. Owen. 2004. Human scavenger receptor class B type II (SR-BII) and cellular cholesterol efflux. Biochem. J. 377:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieland, T. J., M. Ehrlich, M. Krieger, and T. Kirchhausen. 2005. Endocytosis is not required for the selective lipid uptake mediated by murine SR-BI. Biochim. Biophys. Acta 1734:44-51. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen, S. U., M. F. Bassendine, A. D. Burt, C. Martin, W. Pumeechockchai, and G. L. Toms. 2006. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 80:2418-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 38.Pitas, R. E., T. L. Innerarity, J. N. Weinstein, and R. W. Mahley. 1981. Acetoacetylated lipoproteins used to distinguish fibroblasts from macrophages in vitro by fluorescence microscopy. Arteriosclerosis 1:177-185. [DOI] [PubMed] [Google Scholar]
- 39.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reaven, E., Y. Cortez, S. Leers-Sucheta, A. Nomoto, and S. Azhar. 2004. Dimerization of the scavenger receptor class B type I: formation, function, and localization in diverse cells and tissues. J. Lipid Res. 45:513-528. [DOI] [PubMed] [Google Scholar]
- 42.Rhainds, D., and L. Brissette. 2004. The role of scavenger receptor class B type I (SR-BI) in lipid trafficking. Defining the rules for lipid traders. Int. J. Biochem. Cell Biol. 36:39-77. [DOI] [PubMed] [Google Scholar]
- 43.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silver, D. L. 2004. SR-BI and protein-protein interactions in hepatic high density lipoprotein metabolism. Rev. Endocrine Metabolic Disorders 5:327-333. [DOI] [PubMed] [Google Scholar]
- 45.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. 182:329-334. [DOI] [PubMed] [Google Scholar]
- 46.Treguier, M., C. Doucet, M. Moreau, C. Dachet, J. Thillet, M. J. Chapman, and T. Huby. 2004. Transcription factor sterol regulatory element binding protein 2 regulates scavenger receptor Cla-1 gene expression. Arterioscler. Thromb. Vasc. Biol. 24:2358-2364. [DOI] [PubMed] [Google Scholar]
- 47.Tscherne, D. M., C. T. Jones, M. J. Evans, B. D. Lindenbach, J. A. McKeating, and C. M. Rice. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voisset, C., N. Callens, E. Blanchard, A. Op De Beeck, J. Dubuisson, and N. Vu-Dac. 2005. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 280:7793-7799. [DOI] [PubMed] [Google Scholar]
- 49.Voisset, C., A. O. de Beeck, P. Horellou, M. Dreux, T. Gustot, G. Duverlie, F. L. Cosset, N. Vu-Dac, and J. Dubuisson. 2006. High-density lipoproteins reduce the neutralizing effect of hepatitis C virus (HCV)-infected patient antibodies by promoting HCV entry. J. Gen. Virol. 87:2577-2581. [DOI] [PubMed] [Google Scholar]
- 50.von Hahn, T., B. D. Lindenbach, A. Boullier, O. Quehenberger, M. Paulson, C. M. Rice, and J. A. McKeating. 2006. Oxidized low-density lipoprotein inhibits hepatitis C virus cell entry in human hepatoma cells. Hepatology 43:932-942. [DOI] [PubMed] [Google Scholar]
- 51.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Webb, N. R., P. M. Connell, G. A. Graf, E. J. Smart, W. J. de Villiers, F. C. de Beer, and D. R. van der Westhuyzen. 1998. SR-BII, an isoform of the scavenger receptor BI containing an alternate cytoplasmic tail, mediates lipid transfer between high density lipoprotein and cells. J. Biol. Chem. 273:15241-15248. [DOI] [PubMed] [Google Scholar]
- 53.Zennou, V., C. Serguera, C. Sarkis, P. Colin, E. Perret, J. Mallet, and P. Charneau. 2001. The HIV-1 DNA flap stimulates HIV vector-mediated cell transduction in the brain. Nat. Biotechnol. 19:446-450. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, J., G. Randall, A. Higginbottom, P. Monk, C. M. Rice, and J. A. McKeating. 2004. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 78:1448-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]