Abstract
Human papillomavirus (HPV) E6 oncoprotein targets certain tumor suppressors such as MAGI-1 and SAP97/hDlg for degradation. A short peptide at the C terminus of E6 interacts specifically with the PDZ domains of these tumor suppressors, which is a property unique to high-risk HPVs that are associated with cervical cancer. The detailed recognition mechanisms between HPV E6 and PDZ proteins are unclear. To understand the specific binding of cellular PDZ substrates by HPV E6, we have solved the crystal structures of the complexes containing a peptide from HPV18 E6 bound to three PDZ domains from MAGI-1 and SAP97/Dlg. The complex crystal structures reveal novel features of PDZ peptide recognition that explain why high-risk HPV E6 can specifically target these cellular tumor suppressors for destruction. Moreover, a new peptide-binding loop on these PDZs is identified as interacting with the E6 peptide. Furthermore, we have identified an arginine residue, unique to high-risk HPV E6 but outside the canonical core PDZ recognition motif, that plays an important role in the binding of the PDZs of both MAGI-I and SAP97/Dlg, the mutation of which abolishes E6's ability to degrade the two proteins. Finally, we have identified a dimer form of MAGI-1 PDZ domain 1 in the cocrystal structure with E6 peptide, which may have functional relevance for MAGI-1 activity. In addition to its novel insights into the biochemistry of PDZ interactions, this study is important for understanding HPV-induced oncogenesis; this could provide a basis for developing antiviral and anticancer compounds.
Human papillomaviruses (HPVs) are small double-stranded DNA tumor viruses that induce hyperproliferative lesions in epithelial tissues (28). A subset of HPV types that act as the etiological agent of cervical cancers are called “high-risk” types; these include HPV16, HPV18, HPV31, and HPV45 among others (26, 27, 29, 45). The oncogenic potential of the high-risk types is dependent on the cooperative action of the two early viral gene products, E6 and E7, which bind and alter the activity of cell cycle regulatory proteins.
The targeting of p53 for degradation by E6 has extensively been studied, and this is an important feature for the oncogenic activity of the high-risk papillomaviruses that cause cancer (19, 36, 37). However, a large amount of evidence suggests the existence of p53-independent functions of E6 that are also necessary and important for transformation. Support for this idea comes from the observation that the transformation of cells or the induction of epithelial hyperproliferation in transgenic animals by E6 does not always correlate with its ability to degrade p53 (22, 33).
Numerous cellular proteins have been identified as targets of HPV E6 in cell transformation. These proteins are involved in a variety of cellular processes, such as calcium signaling (4), cell adhesion (41), transcriptional control (5), DNA synthesis (25), apoptosis (8), cell cycle control (10), DNA repair (20), and small G-protein signaling (11). One group of these targets of E6 includes the tumor suppressor proteins that harbor multiple copies of PDZ (for “PSD95/Discs Large/ZO-1”) domains specialized for protein-protein interaction, which include the membrane-associated guanylate kinase (MAGUK) homology proteins. Among the members of MAGUK proteins that are targeted by E6 are SAP97/Dlg, which is a human homolog of Drosophila melanogaster discs large (24), MAGI-1, -2 and -3 (14, 40a), and the non-MAGUKs MUPP1 (26a) and hScrib, which is a human homolog of the Drosophila scribble (40b).
In epithelial cells of vertebrates, SAP97/Dlg is associated with the adherens junction (AJ) (35), while MAGI-1, MUPP1, and ZO-2 are associated with the tight junction (TJ) (16). The AJ is responsible for cell-cell adhesion (17), and the TJ acts as an impermeable barrier that divides epithelial cells into functionally distinct apical and basolateral membrane domains (44). The disruption of AJs decreases the phosphorylation of E-cadherin by protein kinase CK2, and this process of downregulation is treated as a common event in carcinogenesis (38). TJ disruption and apobasical activity directly contribute to carcinogenesis by deregulating normal proliferation and differentiation programs in epithelial cells (32). High-risk papillomavirus E6 interacts with the PDZ domains of this class of tumor suppressor proteins and targets them for proteosome-mediated degradation (1, 7, 30).
The detail of the interactions between E6 and PDZ proteins is unclear. It is known that PDZ domains generally recognize a four-amino-acid peptide, X-T/S-X-V, that is usually found at the extreme carboxyl terminus of a protein (6). Previous studies have established the key role of the last valine residue (V-1 for V at the -1 position) and the T/S at the -3 position (or T/S-3) of the peptide for recognizing PDZs, whereas X-2 and X-4 can be any residue (39). The E6 proteins of high-risk HPVs have a conserved six-residue peptide at the C terminus (R-R/T/N/Q-E-T-Q/E-V/L), with T at the -3 position and V/L at the -1 position that is characteristic for PDZ-binding peptides (9). The E6 proteins from different HPV types appear to have various specificities and affinities for different PDZ domains. For example, HPV16 E6 binds to SAP97/Dlg mainly by PDZ domain 2 (PDZ2) (24), whereas HPV18 E6 binds to all three PDZ domains (13). This difference in binding PDZs by different HPV E6 proteins may be associated with the pathogenicity and prevalence of a particular virus type (2).
To understand the specific binding to different PDZs by HPV E6 in targeting the MAGUK PDZ-containing tumor suppressors for degradation, we have solved three crystal structures of the complexes containing MAGI-1 PDZ1, SAP97/Dlg PDZ2, and PDZ3 domains bound to a seven-residue polypeptide, R-R-R-E-T-Q-V, from the C terminus of HPV18 E6. The complex crystal structures reveal new insights into the general rules of the specificity in PDZ recognition and provide detailed molecular mechanisms of targeting MAGI-1 and SAP97/Dlg (referred to as SAP97 hereafter) for degradation. Surprisingly, all six residues of the E6 peptide participate in binding PDZ. A point mutation of R-5 of HPV18 E6 abolished the E6-mediated degradation of both SAP97 and MAGI-1, demonstrating the important role of residues outside the canonical PDZ-recognizing sequence that contains four amino acids. Moreover, for the PDZ1 domain of MAGI-1, which represents the first crystal structure for a MAGI family protein, we have identified a dimer form of PDZ1. These results illustrate the molecular mechanisms by which a viral oncoprotein, E6, specifically targets cellular tumor suppressors for inactivation in order to promote cell growth and transformation.
MATERIALS AND METHODS
Cloning, protein expression, and purification.
The DNA encoding SAP97 PDZ2 (residues 318 to 401), PDZ3 (residues 459 to 543) and MAGI1b PDZ1(residues 451 to 534) domains was amplified by PCR and cloned into pGEX-6P-1 vector as a glutathione S-transferase (GST) fusion. All clones were verified by DNA sequencing. The expression of the proteins was induced by 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) in Escherichia coli overnight at room temperature. The cell pellet was sonicated in lysis buffer (50 mM Tris-Cl, pH 8.0, 250 mM NaCl). The cell lysate was purified by affinity chromatography using glutathione-Sepharose 4B (Amersham Pharmacia Biotech). After on-column cleavage with precision protease at 4°C, the proteins were purified through Resource Q, followed by gel filtration on a Superdex G75 column (Amersham Pharmacia Biotech). Purified PDZ domain proteins were pooled and concentrated in a buffer containing 20 mM Tris-HCl (pH 8.0) and 50 mM NaCl.
Crystallization and data collection.
Purified PDZ domains of SAP97 and MAGI-1 were concentrated to 10 to 16 mg/ml in a buffer containing 20 mM Tris (pH 8.0), 50 mM NaCl, and 1 mM dithiothreitol (DTT) and mixed with an HPV18 E6 C-terminal seven-residue peptide (R-R-R-E-T-Q-V, over 98% purity; Celtek Bioscience, Nashville, TN) in a molar ratio of 1:1.5 prior to crystallization. SAP97 PDZ3 crystallized in 22.5% polyethylene glycol 4000 and 0.1 M MES (morpholineethanesulfonic acid), pH 6.5. MAGI-1 PDZ1 crystallized in 21% polyethylene glycol 2000 monomethyl ether, 0.1 M sodium acetate (pH 4.6), and 0.2 M ammonium sulfate. SAP97 PDZ2 crystallized in 2.2 M ammonium sulfate and 0.1 M sodium citrate. Diffraction data were processed using HKL2000, and data statistics are shown in Table 1.
TABLE 1.
Crystallographic statisticsa
| Parameter | MAGI-1 PDZ1 (with E6 peptide) | SAP97 PDZ2 (with E6 peptide) | SAP97 PDZ3 (with E6 peptide) |
|---|---|---|---|
| Crystal cell parameters | |||
| Space group | P21 | C2 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 48.9, 27.6, 59.5 | 90.9, 44.2, 52.5 | 94.2, 61.9, 57.1 |
| β (°) | 105.4 | 121.8 | 123.3 |
| Data collection statistics | |||
| Resolution range (Å) | 50.0-2.1 | 50-2.3 | 50-2.8 |
| No. of observations | 35,814 | 24,166 | 21,871 |
| No. of unique reflections | 9,135 | 6,986 | 6,843 |
| % Completeness | 98.0 (84.3) | 89.0 (93.5) | 93.7 (82.0) |
| Rsym of last bin (%) | 3.9 (5.6) | 9.1 (33.1) | 5.3 (17.1) |
| I/σ of last bin | 35.8 (21.6) | 12.5 (2.2) | 21.4 (6.6) |
| Refinement statistics | |||
| Rcryst (%) | 21.4 | 23.7 | 21.7 |
| Rfree (%) | 26.7 | 29.8 | 28.4 |
| Root mean square deviation | |||
| Bond length (Å) | 0.010 | 0.0089 | 0.0066 |
| Bond angle (°) | 1.8 | 1.7 | 1.4 |
| Average B factor (Å2) | 22.56 | 28.75 | 48.56 |
Rsym = ∑ij|Ii(j) − < I(j) >|/ ∑ij Ii(j), where Ii(j) is the ith measurement of reflection j and < I(j) > is the overall weighted mean of i measurements. Rcryst = Σhkl ∥ Fo<1/2>|−<1/2>|Fc ∥ / Σhkl |<1/2>Fo<1/2>|. Seven percent of the reflections were excluded for the Rfree calculation.
Structure determination and refinement.
The structures of SAP97 PDZ2, PDZ3, and MAGI-1 PDZ1 were solved by molecular replacement using MOLREP of the CCP4 suite. The second PDZ domain of discs large homologue 2 (Protein Data Bank [PDB] code, 2BYG) worked as a search model for solving the structure of SAP97 PDZ2 in space group (spg) C2. SAP97 PDZ3 crystal was also in the C2 spg with three molecules per asymmetric unit, and the structure was solved using the coordinates of the human discs large protein PDZ3 (PDB code, 1PDR). For MAGI PDZ1, a polyalanine model from the nuclear magnetic resonance structure of human atrophin-1 PDZ1 (PDB code, 1UEQ) was used as a search model and yielded two molecules per asymmetric unit in spg P21. The structures were refined by alternating cycles of model rebuilding and refinement using O and CNS, respectively. Progressive inclusions of side chains, water molecules, and the bound E6 peptides coupled with a few cycles of refinement yielded good refinement statistics (Table 1).
HPV18 E6-mediated degradation assays.
The plasmids expressing SAP97, MAGI-1, p53, and HPV18 E6 have been described previously (40). The R154G mutation of HPV18 E6 was introduced using the Invitrogen GeneTailor kit and verified by DNA sequencing. The proteins were expressed in vitro using the Promega TNT kit and were radiolabeled with [35S]cysteine (Amersham). The target proteins were incubated in the presence or absence of wild-type (wt) or mutant E6 for 30 min (MAGI-1), 1 h (p53), or 2 h (SAP97) and then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by autoradiography.
GST pull-down assays.
The expression levels of GST alone, GST-Dlg, and GST-M1P1 (MAGI-1 PDZ domain 1) have been reported previously (40). The expressed proteins were immobilized on glutathione-agarose (Sigma), and after extensive washing, they were incubated with wild-type and mutant E6 proteins, translated in vitro as described above. After 1 h at room temperature, the glutathione-agarose was washed extensively with phosphate-buffered saline containing 2% Triton X-100 and 0.5% Nonidet P-40. The remaining proteins were separated by SDS-PAGE and subjected to analysis by radiography and phosphorimaging.
RESULTS AND DISCUSSION
Overall organization of PDZ-E6 peptide complexes.
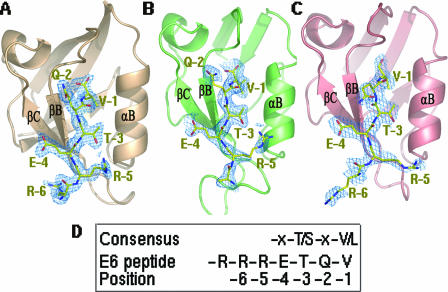
HPV18 E6 proteins target a number of MAGUK proteins for degradation by binding through their PDZ domains. We have solved the crystal structures of MAGI-1 PDZ1, SAP97 PDZ2, and PDZ3 domains, each in complex with a seven-residue peptide (R-R-R-E-T-Q-V) from the C terminus of HPV18 E6, at resolutions of 2.1 Å, 2.3 Å, and 2.8 Å, respectively (Fig. 1; Table 1). In the following discussion, the C-terminal Val residue of the E6 peptide will be referred to as V-1 and the remaining E6 peptide residues will be numbered in ascending order as Q-2, T-3, E-4, R-5, etc.
FIG. 1.
Structures of three PDZ domains bound to the C-terminal peptide of HPV18 E6. (A) MAGI-1 PDZ1-E6 peptide complex (PDB identification [ID] no. 2I04). (B) SAP97/Dlg PDZ2-E6 peptide complex (PDB ID no. 2I0L). (C) SAP97/Dlg PDZ3-E6 peptide complex (PDB ID no. 2I0I). The PDZ structure is drawn as ribbons. The electron density corresponding to the E6 peptide was calculated before the peptide was built in (at a contour level of 1σ). A stick model of the bound peptide (shown as yellow sticks) fits nicely into the density in each complex. Clear electron density maps are seen for the side chains of six peptide residues in MAGI-1 PDZ1 and SAP97/Dlg PDZ3 complexes, while side chains of five residues are identified in the SAP97/Dlg PDZ2-peptide complex. (D) Consensus of the PDZ-binding ligand and the E6 peptide sequence used in this work. The position assignment (positions -1, -2, -3, etc.) is labeled below each amino acid.
For all three PDZ complexes, the E6 peptide ligand is positioned in the “substrate” groove of PDZs between the strand βB and the helix αB (Fig. 1). This substrate groove is also used for binding other peptides by different PDZ domains (reviewed in reference 39). However, in contrast to the previously characterized peptide-PDZ complexes where only four residues from the peptide were found to interact with PDZ, we saw six of the seven HPV E6 peptide residues bound in all three PDZ-E6 complexes (Fig. 1A to C), even though the contribution of the side chain of R-6 is less defined in the SAP97 PDZ2-peptide complex (Fig. 1B). These additional interactions with the PDZs by the two extra Rs, together with other new interaction features to be discussed later, provide the viral E6 protein with an upper hand in competing with cellular ligands for binding these PDZ domains and may therefore allow the virus to deregulate the activity of these PDZ-containing tumor suppressors.
Detailed interactions of the E6 peptide with PDZ.
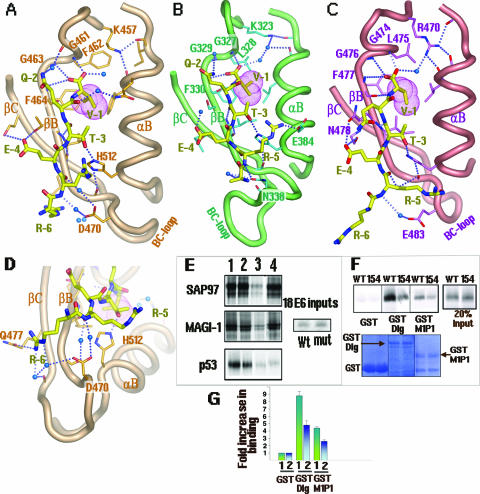
It is well understood how the short four-residue peptide sequence X-T/S-X-V/L can interact with the PDZ domains of a few proteins (6, 15, 42). Our complex crystals not only show the basic features revealed by the common four-residue peptide sequence X-T/S-X-V/L but also uncover several novel features about the specific interactions of the seven-residue HPV18 E6 peptide with the three PDZ domains from MAGI-1 and SAP97. The HPV18 E6 peptide bound to the groove of all three PDZ domains forms an additional β-strand that is antiparallel with βB in a so-called β-augmentation process. The main chain carboxyl group of the E6 peptide is well anchored with the main chain nitrogens of the GLGF motif of PDZ (Fig. 2A to C), which is GFGF (residues 461 to 464) in the case of the MAGI-1 PDZ domains. Other common features include the interactions of two methyl groups of V-1 with a hydrophobic pocket within all three PDZ domains (Fig. 2A to C).
FIG. 2.
Detailed interactions of the E6 peptide with the PDZ domains. (A) MAGI-1 PDZ1-peptide complex. The GFGF motif (G461 to F464) is labeled. Dashed lines denote hydrogen bonds. Blue spheres represent water molecules. (B) SAP97/Dlg PDZ2-peptide complex. The GLGF motif (G327 to F330) is labeled. (C) SAP97/Dlg PDZ3-E6 peptide complex. The GLGF motif (G474 to F477) is labeled. The E6 peptide is shown as a yellow stick in each complex. The involvement of the BC loop in E6 peptide binding is shown in each figure. (D) A close-up view of the interactions of R-6 with Q477 and the main chain carbonyl of MAGI-1 PDZ1. (E) Importance of the R-5 residue of E6 for the degradation of MAGI-1 and SAP97/Dlg. As shown above, R-5 contacts MAGI PDZ1 and hDlg/SAP97 PDZ2 through both the main chain and the side chain (also see Fig. 4D). In vitro-translated radiolabeled SAP97/Dlg, MAGI-1, and p53 proteins were incubated at 30°C alone (lane 2), with wild-type HPV18 E6 (lane 3), or with mutant (mut) HPV18 E6 that has R-5 mutated to G (R154G) (lane 4). The proteins in the assays were incubated for the times previously shown to give optimal degradation of the protein (2 h for SAP97/Dlg, 0.5 h for MAGI-1, and 1 h for p53). Note that the incubation time for MAGI-1 is four times shorter than for SAP97/Dlg, as the E6-mediated degradation for MAGI-1 is approximately four times faster. The input level of each target protein is shown in lane 1, and 20% of the input E6 protein is shown in the right-hand panel. (F) Reduced binding of the E6 R154G mutant to Dlg/SAP97 and MAGI-1 PDZ1. Equal amounts of in vitro-translated, radiolabeled wild-type or mutant E6 (R154G) protein were incubated with GST alone, GST-Dlg, or GST-M1P1 (MAGI-1 PDZ domain 1). The autoradiograph in the upper panel shows the amount of radiolabeled E6 protein retained, with 20% of the input in the right-hand panel. The lower panel shows the GST protein inputs, stained with Coomassie; the GST proteins are indicated with arrows. (G) A histogram showing the quantified results of the assays exemplified in panel F, determined by phosphorimaging analysis and expressed as increases (n-fold) of binding above binding to GST alone. Standard deviations are shown.
In all three PDZ-peptide complexes, the side chain of T-3 forms a hydrogen bond with a highly conserved PDZ His residue (Fig. 2A to C). Indeed, a study of Dlg degradation by HPV18 E6 showed that mutation of T-3 to an E residue reduced Dlg degradation, as this E6 mutant had undetectable Dlg binding (13). Interestingly, T-3 has previously been shown to be a critical functional residue in the case of the K+ ion channel peptide bound to PSD95 PDZ3, thus demonstrating a highly conserved interaction common to several PDZ domains of different proteins (6, 23). Protein kinase A-dependent phosphorylation at T-3 of E6 is found to be detrimental for PDZ binding, and this can now be explained by our structural observations. All of our complex structures (Fig. 2) show that the hydroxyl group of T-3 is only 3 Å away from the His belonging to αB of the PDZ, forming a hydrogen bond with its N-3 atom. Therefore, phosphorylation on T-3 is expected to result in a serious steric hindrance with the peptide binding groove of PDZ, which may eventually abrogate the β-augmentation of E6 within the PDZ domain.
The -4 position of HPV18 E6 is a Glu (E-4) (Fig. 1D). Even though this E-4 residue makes hydrogen bonds with all three PDZs, there are obvious differences in the details. For the PDZ1 of MAGI-1, the E6 peptide residue E-4 forms hydrogen bonds with two residues, T465 and S480, from strands βB and βC, respectively (Fig. 2A). Similar interactions are present for the same E-4 with N478 and S491 from strands βB and βC, respectively, for PDZ3 of SAP97 (Fig. 2C). However, in the SAP97 PDZ2-peptide complex, the residue E-4 makes hydrogen bonds with T350 from strand βC only. In the previously characterized PDZ-binding sequence of X-T/S-X-V/L, the -4 position was listed as X, which stands for any amino acid. However, the E-4 residue of the HPV E6 protein is conserved in all the high-risk HPV types (Fig. 3B). The fact that this E-4 residue makes hydrogen bonds with all three PDZs suggests that it contributes to the specific binding of HPV E6 to the PDZs of the two tumor suppressors. Conversely, all the PDZ domains with polar residues at the equivalent positions on βB and βC that can form hydrogen bonds with the E-4 residue of the HPV E6 protein are therefore potential binding targets of HPV E6.
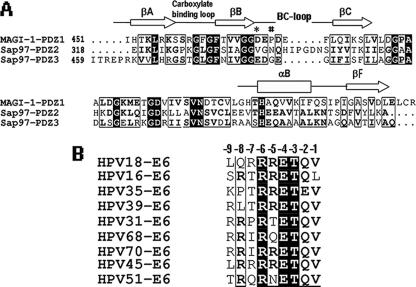
FIG. 3.
(A) Sequence alignment of MAGI-1 PDZ1 with SAP97/Dlg PDZ2 and PDZ3. Important secondary structural elements are highlighted; two key residues belonging to the BC loop, D470 of MAGI-1 PDZ1 and N338 of SAP97/Dlg PDZ2, are marked by * and #, respectively. (B) Sequence alignment of the C-terminal sequences from nine high-risk HPV types. The position number (-1, -2, -3, etc.) used for each residue is listed above the alignment.
E6 peptide residue Q-2 in PDZ binding.
In the previous crystal structure of PSD95 PDZ3 complexed with a four-residue peptide of CRIPT, the residue at the -2 position, S-2, did not make any interactions (6), which is consistent with the fact that the residue at position -2 is poorly conserved (labeled as X-2) among the C termini of membrane proteins and is expected to be functionally unimportant. However, the -2 position of the HPV E6 peptide is a Q (Q-2) that is highly conserved (Fig. 3B). Among the three PDZ-E6 peptide structures, the Q-2 of the E6 peptide makes hydrogen bond interactions with both MAGI PDZ1 (Fig. 2A) and SAP97 PDZ2 (Fig. 2B) but not with SAP97 PDZ3 (Fig. 2C). This differential interaction with PDZs at the -2 position of the E6 peptide may therefore contribute to the specificity of E6 for targeting a particular PDZ domain.
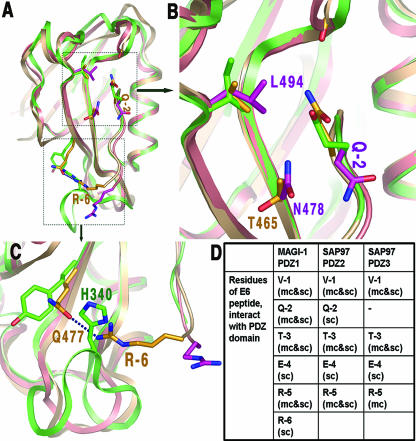
Superimposing the three PDZ structures from MAGI and SAP97 (Fig. 4A) reveals why the Q-2 residue of the E6 peptide can interact with MAGI1 PDZ1 and SAP97 PDZ2 but not with SAP97 PDZ3. SAP97 PDZ3 has two large residues, L494 and N478, which sterically keep the Q-2 side chain pointing away from PDZ, and no bond contacts can be made (Fig. 4B). However, these two large residues (L494 and N478) are replaced by smaller ones in MAGI-1 PDZ1 and SAP97 PDZ2 (Fig. 4B), which generates room for the Q-2 side chain to reorient, making hydrogen bonds with these two PDZs. This difference in the interactions of the Q-2 residue with PDZ2 and PDZ3 of SAP97 suggests that HPV18 E6 should have stronger binding to PDZ2 than to PDZ3. This observation is consistent with the fact that the PDZ2 domain of SAP97 seems to be the critical domain for the ability of E6 to degrade SAP97 (12).
FIG. 4.
Different interactions between E6 peptide residues Q-2 and R-6 with PDZs. (A) An overview of the superposition of MAGI-1 PDZ1 (silver brown) and SAP97/Dlg PDZ2 (green) on PDZ3 (salmon pink). (B) A close-up view of the marked box in panel A, showing the alternative orientations of Q-2 of the three E6 peptides bound to the three PDZs. Residues are in yellow for the MAGI-1 PDZ1-peptide complex, in green for the SAP97/Dlg PDZ2 complex, and in magenta for the SAP97/Dlg PDZ3 complex. (C) A close-up view of the area showing that the hydrogen bond between R-6 and Q477 in MAGI-1 PDZ1 with the E6 peptide complex would be obstructed in the SAP97/Dlg PDZ2-peptide complex by the presence of the longer BC loop and H340 of the PDZ2 domain. (D) Summary of the interactions of each residue of the HPV18 E6 peptide in the three PDZ complexes. mc, main chain interaction; sc, side chain interaction; mc&sc, interaction of both the main chain and the side chain.
E6 peptide residue R-6 in PDZ binding.
R-6, a conserved residue among high-risk HPVs (Fig. 3B), participates in PDZ binding in the MAGI-1 PDZ1-E6 complex. In this complex, the R-6 side chain interacts with the polar group of Q477 in MAGI PDZ1 (Fig. 4C). However, the side chain of R-6 is not making bonding contact with the PDZ2 and PDZ3 of SAP97, mainly because at the equivalent position of Q477 of MAGI PDZ1 is a hydrophobic residue (F) that cannot bond with R-6 (Fig. 4C). Additionally, the extended BC loop of SAP97 PDZ2 forces the side chain of R-6 to orient away from PDZ so that no bonding contact can be established (Fig. 4C).
A novel peptide-binding motif of PDZ: BC loop.
In addition to the structural elements previously identified in peptide binding, including the carboxylate GLGF loop, βB, and αB, our complex structures reveal a new peptide-binding structure element of PDZs which comprises the loop connecting the βB and βC strands (BC loop) (Fig. 2A to C). Residues on this BC loop interact with the E6 peptide directly (Fig. 2B and C) or via well-ordered water molecules (Fig. 2A, C, and D). In the case of MAGI-1 PDZ1, two waters with low temperature factors (14 and 24 Å2, respectively) mediate hydrogen bond interactions with the R-5 residue of the E6 peptide (Fig. 2D). Similar water-mediated hydrogen bond interactions are also observed in the SAP97 PDZ3-peptide complex (Fig. 2C).
Even though this BC loop of PDZs plays an important role in peptide binding, the loop region appears to be poorly conserved in length and sequence among the three PDZ domains (Fig. 3A). For example, D470 of MAGI PDZ1 and E384 of SAP97 PDZ3 located on the BC loop form hydrogen bonds with the R-5 residue of the E6 peptide (Fig. 2A and C). However, the position is not conserved in PDZ2 of SAP97, which is V336 in the alignment (Fig. 3A). Additionally, there is a five-residue insertion in the loop. Nonetheless, in the three-dimensional structure, the longer BC loop of PDZ2 is oriented in such a way that N338 of PDZ2, instead of V336, occupies the equivalent position of D470 of MAGI PDZ1 and bonds with the E6 peptide residue R-5 (Fig. 2B). Thus, the same type of peptide interaction is maintained in the SAP97 PDZ2-peptide complex by adopting a different BC-loop conformation to spatially align a similar residue to the equivalent position of D470 of MAGI PDZ1 in order to interact with the E6 peptide residue R-5.
Important role of E6 peptide residue R-5 in PDZ binding.
As discussed above, all three PDZ-peptide complexes show the BC loop as the new peptide-binding motif of PDZ to make contact with R-5. To test the functional relevance of the interactions of the BC loop with the R-5 residue of HPV E6, R-5 was mutated to G to generate an HPV18 E6 mutant, R154G, and the mutant was tested for its ability to mediate the degradation of MAGI-1 and SAP97. HPV18 E6 has been shown previously to induce the degradation of MAGI-1, SAP97, and p53 via the 26S proteasome pathway (13, 14, 37). We performed similar degradation assays using the wt and the R154G mutant of HPV18 E6 (Fig. 2E). The wt E6 protein was effective in inducing near-complete degradation of the three target proteins. However, the R154G mutant greatly reduced the ability to induce the degradation of MAGI-1 and SAP97 but was still as effective as wt E6 in degrading p53, which has no PDZs.
To confirm that the reduced ability of the E6 R154G mutant to induce the degradation of the PDZ target was the result of reduced binding by the E6 mutant, we performed GST pull-down assays. Figure 2F shows the autoradiograph and Coomassie-stained gel of a representative binding assay, while the graph in Fig. 2G shows the combined results for at least three assays, as determined by phosphorimaging. It is clear from these results that the R154G mutant has substantially lower binding to both full-length Dlg and MAGI-1 PDZ domain 1 than wild-type HPV18 E6. These results demonstrate that the interaction between R-5 of HPV E6 and the newly identified peptide-binding motif on the BC loop of the PDZs is functionally important in the degradation process.
Predicted difference in PDZ binding by HPV18 and HPV16 E6 proteins.
The organization of the hydrophobic residues around V-1 is critical in the PDZ1-E6 complexes reported here. Four hydrophobic residues, two from the GLGF motif, one from βB, and the other from βC, are closely packed around the V-1 side chain. A second layer of hydrophobic residues is found just beneath the hydrophobic pocket of V-1. This second layer restricts the conformational freedom of the residues involved in the hydrophobic pocket of V-1, and the packing patterns of the hydrophobic residues are very similar in all three complexes, despite their sequence differences, and look optimum for the Val side chain (Fig. 2A to C).
As HPV16 E6 has Leu at its carboxy terminus (Fig. 3B), we attempted to model Leu in place of V-1 in our complex structures. To fit the Leu side chain in the hydrophobic pocket, avoiding the short contacts, an outward shift of the peptide backbone has to be made in all three complexes, which eventually weakens the β-strand interactions between the peptide and βB of PDZ. The main chain interactions of βB of PDZ and the backbone of the peptide residues are responsible for stabilizing an extended peptide in the binding groove, thus increasing the affinity of interaction (6). Therefore, the weakening of the β-strand interactions that results from accommodating an E6 peptide with Leu at the -1 position could possibly be responsible for the lower affinity of HPV16 E6 towards these PDZ domains.
It is worth noting that in the case of a different PDZ domain-containing substrate of E6, HPV16 E6 is actually more efficient than HPV18 E6 in directing the degradation of the tumor suppressor scribble and the Leu/Val difference at the -1 position appears to be critical (40b). This implies that for other PDZ domain substrates of E6, Leu at -1 may in fact be optimal for binding. Therefore, the involvement of the other upstream C-terminal E6 residues in PDZ binding cannot be ruled out, and the structure determination of the HPV16 E6 peptide bound to the relevant PDZ domain should clarify this issue.
MAGI-1 PDZ1 dimer and peptide binding.
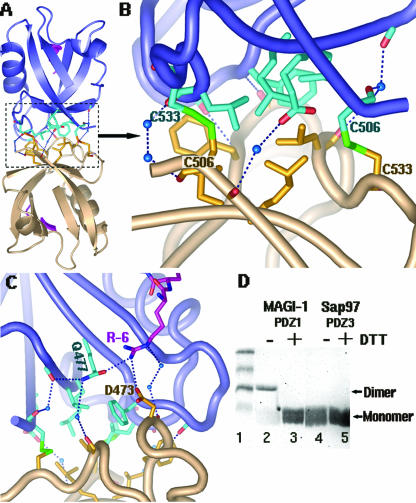
For the MAGI-1 PDZ1-E6 peptide complex, PDZ1 forms a dimer (Fig. 5A). There are extensive interactions at the dimeric interface, including hydrophobic packing and hydrogen bonds (Fig. 5B). Additionally, two intermolecular disulfide bonds form between C533 of one PDZ1 and C506 of another and effectively cross-link the two monomers (Fig. 5B).
FIG. 5.
Dimer formation of MAGI-I PDZ1. (A) An overview of the MAGI-1 PDZ1 homodimer, shown in a ribbon diagram. Each monomer is shown in a different color (salmon or blue). The residues involved in contacts at the interface are shown as sticks. (B) A close-up view of the interactions at the dimer interface: two disulfide bridges are shown as green sticks, hydrogen bonds as dashed lines, and water molecules as blue spheres. Hydrophobic residues were found to cluster at the core of the interface. (C) Direct interaction of R-6 with D473 of the bottom monomer (salmon). This R-6 reaches across the dimer interface from the E6 peptide bound to the substrate groove of the top monomer (in blue). (D) Dimer formation of MAGI-1 PDZ1 in solution. About 10 μl of purified MAGI-1 PDZ1 (3.0 μg/μl) was treated with 10 μl SDS loading buffer in the absence or presence of 10 mM DTT (lanes 2 and 3), followed by a 12.5% SDS-PAGE analysis. Purified SAP97/Dlg PDZ3 was used as a control (lanes 4 and 5). The dimer form was detected only for MAGI-1 PDZ1 under nonreducing conditions (without DTT) (lane 2). All the PDZ domains run as a relatively diffused band as monomers, possibly due to their small size (∼8 kDa).
To check whether MAGI-1 PDZ1 forms a disulfide-cross-linked dimer in solution, we analyzed the PDZ1 proteins through nonreducing SDS-PAGE (Fig. 5D). When the purified PDZ1 protein was treated with SDS loading buffer in the absence or presence of DTT, a protein band of ∼18 kDa or ∼8 kDa was detected, respectively (Fig. 5C, lanes 1 and 2), clearly demonstrating the presence of a dimer in the solution and suggesting the biochemical relevance of the dimer form. Because the PDZ domain proteins essentially have no UV absorption at an optical density at 280 nm, due to the lack of aromatic residues, no UV absorption profile from gel filtration chromatography can be obtained, even at high protein concentrations. However, SDS-PAGE analysis showed that there are proteins collected at the peak fractions expected for both the dimeric and the monomeric form, further confirming the dimer formation.
The role of oligomerization of the PDZ domain has already been implicated in the formation of intracellular signaling complexes (21). For example, INAD (inactivation no after potential D) multimerizes via PDZ3 and PDZ4 without interfering with PDZ ligand binding (43). In contrast, the PDZ-PDZ interaction-mediated nNOS association with syntrophin or PSD-95 is competitively inhibited by the peptide ligand (3). However, no direct evidence so far shows the dimer formation of MAGI-1 PDZs, and the biological significance of the dimer for MAGI-1 function requires further investigation. Nonetheless, the MAGI PDZ1-peptide complex shows that the dimer contains two E6 peptides, each one bound to one monomer, and each one of these E6 peptides interacts with both PDZ1 domains at the dimer interface. Each E6 peptide binds to the substrate groove of one PDZ1 through the interactions described above. In addition, it also uses its R-6 to interact directly with D473 of the other monomer as well as indirectly via water molecules (Fig. 5C). By interacting with both molecules of the PDZ dimer, the E6 peptide is expected to strengthen its association with MAGI PDZ1; this is consistent with the facts that the E6-mediated degradation of MAGI-1 is more efficient than that of SAP97 (Fig. 2E) (40) and that R-6 is a highly conserved residue for high-risk HPV E6 proteins (Fig. 3B), thus further emphasizing the relevance of this interaction.
Concluding remarks.
The three PDZ-peptide complex crystal structures reported here have provided molecular details and new insights about the specificity of HPV E6 oncoproteins in binding to PDZs from MAGI-1 and SAP97/Dlg PDZ2. In contrast to the normal four-residue peptide X-T/S-X-V/L for PDZ binding, these crystal structures reveal that six out of the seven residues of the E6 peptide (R-R-R-E-T-Q-V) are involved in PDZ recognition. In addition to the new interactions of the longer peptide, the residues at positions -2 (Q-2) and -4 (E-4) of the peptide, which are X in the canonical PDZ binding peptide but are highly conserved in high-risk HPV E6 proteins, make strong contacts with PDZs in two complexes. A new peptide-binding motif on PDZ is revealed in all of these complex crystal structures and comprises the BC loop that specifically interacts with the arginine residue at the -5 position (R-5). The functional relevance of these newly identified R-5 and BC-loop interactions is supported by our mutational study, in which an R-5-to-G mutation abolished the HPV18 E6-mediated degradation of MAGI-1 and SAP97. Moreover, we have identified a dimer form of MAGI-1 PDZ1 in the crystal structure and in solution. The E6 peptide in the complex structure interacts with both PDZ molecules, which may contribute to a stronger PDZ recognition by HPV E6 and provide a plausible explanation for a more efficient E6-mediated degradation of MAGI-1 than of SAP97. This study therefore highlights the importance of additional residues outside the canonical PDZ recognition motif in determining substrate specificity in this class of protein-protein interactions. These results also illustrate the detailed molecular mechanisms by which HPV E6 can specifically recognize and compete for the PDZs of cellular tumor suppressors in the process of cell transformation and oncogenesis.
Acknowledgments
We thank members of the X. S. Chen lab for their help and input and the staff at synchrotron beamlines 8.2.1 and 8.2.2 in ALS at Berkeley for assistance in data collection.
This work is supported by NIH R01 (AI048747) to X.S.C. and funding from the Associazione Italiana per la la Ricerca sul Cancro to L.B.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Reference deleted.
- 2.Berger, T., E. Stockfleth, T. Meyer, F. Kiesewetter, and J. O. Funk. 2005. Multiple disseminated large-cell acanthomas of the skin associated with human papillomavirus type 6. J. Am. Acad. Dermatol. 53:335-337. [DOI] [PubMed] [Google Scholar]
- 3.Brenman, J. E., D. S. Chao, S. H. Gee, A. W. McGee, S. E. Craven, D. R. Santillano, Z. Wu, F. Huang, H. Xia, M. F. Peters, S. C. Froehner, and D. S. Bredt. 1996. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell 84:757-767. [DOI] [PubMed] [Google Scholar]
- 4.Das, K., J. Bohl, and S. B. Vande Pol. 2000. Identification of a second transforming function in bovine papillomavirus type 1 E6 and the role of E6 interactions with paxillin, E6BP, and E6AP. J. Virol. 74:812-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Degenhardt, Y. Y., and S. J. Silverstein. 2001. Gps2, a protein partner for human papillomavirus E6 proteins. J. Virol. 75:151-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doyle, D. A., A. Lee, J. Lewis, E. Kim, M. Sheng, and R. MacKinnon. 1996. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85:1067-1076. [DOI] [PubMed] [Google Scholar]
- 7.Reference deleted.
- 8.Filippova, M., H. Song, J. L. Connolly, T. S. Dermody, and P. J. Duerksen-Hughes. 2002. The human papillomavirus 16 E6 protein binds to tumor necrosis factor (TNF) R1 and protects cells from TNF-induced apoptosis. J. Biol. Chem. 277:21730-21739. [DOI] [PubMed] [Google Scholar]
- 9.Flores, R., M. Papenfuss, W. T. Klimecki, and A. R. Giuliano. 2006. Cross-sectional analysis of oncogenic HPV viral load and cervical intraepithelial neoplasia. Int. J. Cancer 118:1187-1193. [DOI] [PubMed] [Google Scholar]
- 10.Gao, Q., A. Kumar, S. Srinivasan, L. Singh, H. Mukai, Y. Ono, D. E. Wazer, and V. Band. 2000. PKN binds and phosphorylates human papillomavirus E6 oncoprotein. J. Biol. Chem. 275:14824-14830. [DOI] [PubMed] [Google Scholar]
- 11.Gao, Q., S. Srinivasan, S. N. Boyer, D. E. Wazer, and V. Band. 1999. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol. Cell. Biol. 19:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardiol, D., S. Galizzi, and L. Banks. 2002. Mutational analysis of the discs large tumour suppressor identifies domains responsible for human papillomavirus type 18 E6-mediated degradation. J. Gen. Virol. 83:283-289. [DOI] [PubMed] [Google Scholar]
- 13.Gardiol, D., C. Kuhne, B. Glaunsinger, S. S. Lee, R. Javier, and L. Banks. 1999. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene 18:5487-5496. [DOI] [PubMed] [Google Scholar]
- 14.Glaunsinger, B. A., S. S. Lee, M. Thomas, L. Banks, and R. Javier. 2000. Interactions of the PDZ-protein MAGI-1 with adenovirus E4-ORF1 and high-risk papillomavirus E6 oncoproteins. Oncogene 19:5270-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grembecka, J., T. Cierpicki, Y. Devedjiev, U. Derewenda, B. S. Kang, J. H. Bushweller, and Z. S. Derewenda. 2006. The binding of the PDZ tandem of syntenin to target proteins. Biochemistry 45:3674-3683. [DOI] [PubMed] [Google Scholar]
- 16.Gumbiner, B., T. Lowenkopf, and D. Apatira. 1991. Identification of a 160-kDa polypeptide that binds to the tight junction protein ZO-1. Proc. Natl. Acad. Sci. USA 88:3460-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumbiner, B. M. 1996. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell 84:345-357. [DOI] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Huibregtse, J. M., M. Scheffner, and P. M. Howley. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 10:4129-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iftner, T., M. Elbel, B. Schopp, T. Hiller, J. I. Loizou, K. W. Caldecott, and F. Stubenrauch. 2002. Interference of papillomavirus E6 protein with single-strand break repair by interaction with XRCC1. EMBO J. 21:4741-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im, Y. J., J. H. Lee, S. H. Park, S. J. Park, S. H. Rho, G. B. Kang, E. Kim, and S. H. Eom. 2003. Crystal structure of the Shank PDZ-ligand complex reveals a class I PDZ interaction and a novel PDZ-PDZ dimerization. J. Biol. Chem. 278:48099-48104. [DOI] [PubMed] [Google Scholar]
- 22.James, M. A., J. H. Lee, and A. J. Klingelhutz. 2006. Human papillomavirus type 16 E6 activates NF-κB, induces cIAP-2 expression, and protects against apoptosis in a PDZ binding motif-dependent manner. J. Virol. 80:5301-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, E., M. Niethammer, A. Rothschild, Y. N. Jan, and M. Sheng. 1995. Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 378:85-88. [DOI] [PubMed] [Google Scholar]
- 24.Kiyono, T., A. Hiraiwa, M. Fujita, Y. Hayashi, T. Akiyama, and M. Ishibashi. 1997. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl. Acad. Sci. USA 94:11612-11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kühne, C., and L. Banks. 1998. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J. Biol. Chem. 273:34302-34309. [DOI] [PubMed] [Google Scholar]
- 26.Laimins, L. A. 1993. The biology of human papillomaviruses: from warts to cancer. Infect. Agents Dis. 2:74-86. [PubMed] [Google Scholar]
- 26a.Lee, S. S., B. Glaunsinger, F. Mantovani, L. Banks, and R. T. Javier. 2000. Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74:9680-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longuet, M., P. Cassonnet, and G. Orth. 1996. A novel genital human papillomavirus (HPV), HPV type 74, found in immunosuppressed patients. J. Clin. Microbiol. 34:1859-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowy, D. R., and P. M. Howley. 2001. Fields virology, vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 29.Lowy, D. R., R. Kirnbauer, and J. T. Schiller. 1994. Genital human papillomavirus infection. Proc. Natl. Acad. Sci. USA 91:2436-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Massimi, P., N. Gammoh, M. Thomas, and L. Banks. 2004. HPV E6 specifically targets different cellular pools of its PDZ domain-containing tumour suppressor substrates for proteasome-mediated degradation. Oncogene 23:8033-8039. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto, K., H. Yoshikawa, S. Nakagawa, X. Tang, T. Yasugi, K. Kawana, S. Sekiya, Y. Hirai, I. Kukimoto, T. Kanda, and Y. Taketani. 2000. Enhanced oncogenicity of human papillomavirus type 16 (HPV16) variants in Japanese population. Cancer Lett. 156:159-165. [DOI] [PubMed] [Google Scholar]
- 32.Matter, K., and M. S. Balda. 2003. Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4:225-236. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen, M. L., M. M. Nguyen, D. Lee, A. E. Griep, and P. F. Lambert. 2003. The PDZ ligand domain of the human papillomavirus type 16 E6 protein is required for E6's induction of epithelial hyperplasia in vivo. J. Virol. 77:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pim, D., M. Thomas, and L. Banks. 2002. Chimaeric HPV E6 proteins allow dissection of the proteolytic pathways regulating different E6 cellular target proteins. Oncogene 21:8140-8148. [DOI] [PubMed] [Google Scholar]
- 35.Reuver, S. M., and C. C. Garner. 1998. E-cadherin mediated cell adhesion recruits SAP97 into the cortical cytoskeleton. J. Cell Sci. 111:1071-1080. [DOI] [PubMed] [Google Scholar]
- 36.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75:495-505. [DOI] [PubMed] [Google Scholar]
- 37.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 38.Serres, M., O. Filhol, H. Lickert, C. Grangeasse, E. M. Chambaz, J. Stappert, C. Vincent, and D. Schmitt. 2000. The disruption of adherens junctions is associated with a decrease of E-cadherin phosphorylation by protein kinase CK2. Exp. Cell Res. 257:255-264. [DOI] [PubMed] [Google Scholar]
- 39.Sheng, M., and C. Sala. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24:1-29. [DOI] [PubMed] [Google Scholar]
- 40.Thomas, M., B. Glaunsinger, D. Pim, R. Javier, and L. Banks. 2001. HPV E6 and MAGUK protein interactions: determination of the molecular basis for specific protein recognition and degradation. Oncogene 20:5431-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.Thomas, M., R. Laura, K. Hepner, E. Guccione, C. Sawyers, L. Lasky, and L. Banks. 2002. Oncogenic human papillomavirus E6 proteins target the MAGI-2 and MAGI-3 proteins for degradation. Oncogene 21:5088-5096. [DOI] [PubMed] [Google Scholar]
- 40b.Thomas, M., P. Massimi, C. Navarro, J. P. Borg, and L. Banks. 2005. The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24:6222-6230. [DOI] [PubMed] [Google Scholar]
- 41.Tong, X., and P. M. Howley. 1997. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc. Natl. Acad. Sci. USA 94:4412-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilken, C., K. Kitzing, R. Kurzbauer, M. Ehrmann, and T. Clausen. 2004. Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell 117:483-494. [DOI] [PubMed] [Google Scholar]
- 43.Xu, X. Z., A. Choudhury, X. Li, and C. Montell. 1998. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J. Cell Biol. 142:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeaman, C., K. K. Grindstaff, and W. J. Nelson. 1999. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol. Rev. 79:73-98. [DOI] [PubMed] [Google Scholar]
- 45.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]