Abstract
Genetic and biochemical studies have suggested the existence of a bacteriophage-like, DNA-packaging/ejecting portal complex in herpesviruses capsids, but its arrangement remained unknown. Here, we report the first visualization of a unique vertex in the Kaposi's sarcoma-associated herpesvirus (KSHV) capsid by cryoelectron tomography, thus providing direct structural evidence for the existence of a portal complex in a gammaherpesvirus. This putative KSHV portal is an internally localized, umbilicated structure and lacks all of the external machineries characteristic of portals in DNA bacteriophages.
Kaposi's sarcoma-associated herpesvirus (KSHV) is a DNA tumor virus and the most recently discovered human pathogen in the Herpesviridae family (4). It is the causative agent of all forms of Kaposi's sarcoma and is closely linked to AIDS-associated lymphomas, including primary effusion lymphoma and multicentric Castleman's disease (6). KSHV is a member of the gammaherpesvirus subfamily, which is a group of tumorigenic herpesviruses. Biochemical characterization and structural analyses of these viruses and their capsid assembly have been limited due to technical difficulties in viral particle isolation. Similar to the widely studied herpes simplex virus type 1 (HSV-1), an alphaherpesvirus and the prototypical herpesvirus, KSHV has a multilayered architecture, with a pleomorphic envelope, a middle or tegument layer, and an inner icosahedral capsid. Infection with KSHV, as with all herpesviruses, depends on the successful insertion of its linear genome from the intact icosahedral capsid through the nuclear pore into the host cell nucleus. During productive infection, newly synthesized viral DNA enters preformed capsids. Genetic and biochemical studies have suggested the existence of a bacteriophage-like, DNA-packaging/ejection portal complex, presumably located at only 1 of the 12 vertices of the capsid of herpesviruses (13, 19). Using cryoelectron microscopy (cryoEM), Trus and colleagues have recently resolved an isolated portal-like structure that can self-assemble in vitro from purified UL6 protein of HSV-1 (19). Based on sequence comparison and mass spectrometry studies of purified virions, gammaherpesviruses also contain a portal protein (PORT), which is encoded by ORF43 in KSHV or its rhesus homolog (14) and by BBRF1 in Epstein-Barr virus (9). However, despite rapid progress on herpesvirus capsid structure determination by icosahedral cryoEM reconstruction toward higher resolution (e.g., reference 25), conventional single-particle cryoEM studies have failed to detect this putative portal complex following image reconstructions of any of the herpesvirus capsids. To date, attempts to visualize the putative herpesvirus portal complex in its native capsid environment have failed, and, thus, our understanding of its orientation in and connection with the capsid has remained speculative.
Earlier studies suggest that the HSV-1 portal exists at a single vertex of the capsid (13), a finding that is analogous to the portal complex within DNA bacteriophages (16, 21). In bacteriophages, the portal connects to a large tail outside the capsid (21), which has allowed its visualization by asymmetric single-particle reconstruction (3, 8, 11). However, the capsids of herpesviruses do not have such protrusions, making it difficult to locate this unique vertex by conventional cryoEM methods. In gammaherpesviruses, the existence of the portal complex was never established due to difficulties in isolating adequate amounts of purified capsid materials for biochemical analyses and the lack of antibodies against PORT of any gammaherpesviruses. Recent advances in cryoelectron tomography (cryoET) have made it possible to reconstruct a three-dimensional (3D) structure from a single particle and capture relatively subtle features of asymmetrically arranged structures at near molecular resolution. In this study, we took advantage of this technology to identify a unique vertex that is different from the other 11 pentons in the KSHV capsid, thus providing the first evidence of the existence of a portal complex in gammaherpesvirus capsids.
KSHV capsids were purified from the media of lytically induced KSHV-infected BCBL-1 cells as previously described (12, 24). An aliquot of 3 μl of the purified capsids was placed onto a carbon-coated grid with holes in it and quickly frozen to liquid nitrogen temperature so that the capsid particles were embedded in a thin layer of vitreous ice. Tilt series were recorded with an electron dose of ∼1 electron/Å2/micrograph at a magnification of ×38,200 and 8-μm underfocus in an FEI 300-kV G2 Polara cryoelectron microscope (FEI Co., Hillsboro, OR), equipped with a 16-megapixel charge-coupled device camera and the EMMENU data acquisition software (TVIPS GmbH, Gauting, Germany). A total of 18 sets of tilt series (average tilt angle range of −63° to +63°; see, e.g., Fig. 1a) were processed by using a marker-free alignment approach and back-projection 3D reconstruction method (23). The final 3D tomograms were filtered to a 50-Å resolution using a Gaussian low-pass filter. For averaging of cryoET tomograms, 3D alignments of individual capsid structures were first adjusted manually and subsequently refined computationally by using the Foldhunter program (7). Surface and volume rendering were performed using the Chimera visualization software package (17).
FIG. 1.
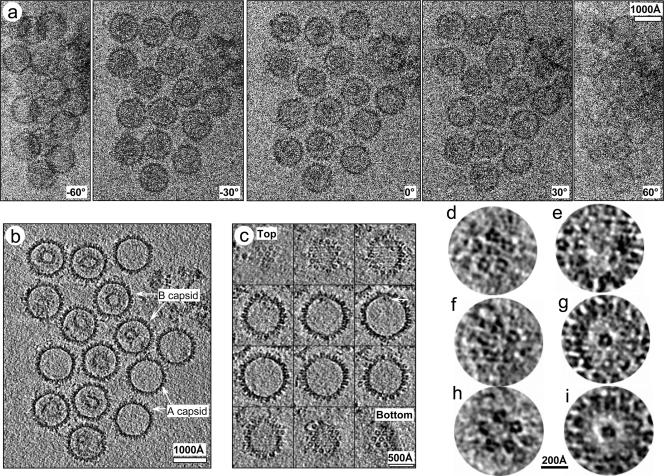
CryoET of KSHV capsids. (a) Representative original images (at tilt angles of 0°, ±30°, and ± 60°) from a −72° to 66° cryoET tilt series recorded at 8-μm underfocus and 300 kV. (b) Representative density slice from the 3D tomographic reconstruction showing A and B capsids. (c) Serial slices 7.9 Å thick parallel to the unique vertex axis of an A capsid. The arrow points to a density attached only to one of the 12 vertices. (d to g) Slices 7.9 Å thick perpendicular to the vertex axis of either a penton (d and e) or the unique vertex (f and g). (h and i) Same as panels f and g, except that the slices are shown after averaging three A capsids at different orientations. The radial distance between the two slices (d and e, f and g, or h and i) is 100 Å.
We focused our in-depth 3D structural analysis on the empty (or A) capsids to minimize ambiguity and complexity in structure identification that might arise from internal densities, such as the scaffolding or viral DNA inside B and C capsids, respectively (Fig. 1b and c and see Movie S1 in the supplemental material). Our approach led to cryoET reconstructions that permitted the unambiguous assignments of pentons and hexons based on their locations on the well-established icosahedral lattice (20, 24). In particular, we identified 12 vertices for each capsid by careful 3D interactive examination of the cryoET tomogram. Among these, one vertex was unique, lacking an outward-protruding penton density and, instead, having an umbilicated density pointing inward and a corresponding concavity on the capsid surface (Fig. 1c to i). This absence of penton density in the unique vertex was further confirmed by comparing corresponding vertical slices of the unique vertex with other vertices at the same radial height in individual cryoET reconstructions (c.f., Fig. 1d versus f). Below the capsid floor, there is no internal density under the pentons, as expected (Fig. 1e). Under the unique vertex, however, there is a discernible cylindrical structure with a diameter of ∼140 Å and height of ∼150 Å attached to the capsid floor (Fig. 1c and g). This umbilicated density is slightly smaller than the penton (∼140 Å in diameter and ∼160 Å in height) and has a central axial channel that measures approximately 50 Å in diameter and 70 Å in depth (Fig. 1g to i). To eliminate structural artifacts and misinterpretation that might arise from the limited tilt range inherent in cryoET analysis, we averaged cryoET reconstructions of three A capsids with different orientations relative to the specimen stage (Fig. 1h and i). The averaged 3D tomogram (Fig. 2 and see Movie S2 in the supplemental material) showed the penton and hexon arrangement characteristic of the T = 16 icosahedral lattice evident in cryoEM icosahedral reconstructions (20, 24), demonstrating the validity of our cryoET analysis. Most notably, it clearly confirmed the absence of a penton and the presence of an internal density with a central channel at the unique vertex (Fig. 1h and i and Fig. 2; also see the movies in the supplemental material). Thus, in the absence of biochemical data, our structural data firmly established that the KSHV capsid contains one unique vertex and 150 hexons and 11 pentons arranged on a T = 16 icosahedral lattice. We proposed that, like HSV-1 capsid, KSHV capsid also contains a bacteriophage-like portal complex and that the unique vertex identified in our cryoET structure is the KSHV portal.
FIG. 2.
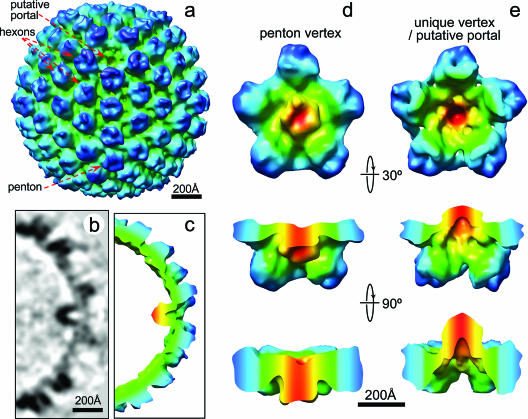
Structure of the KSHV portal. (a) Shaded surface representation of an averaged A capsid cryoET reconstruction, as viewed roughly along a two-fold axis. A cavity appears at one of the 12 vertices. The signal/noise ratio was improved by imposing a five-fold symmetry along the unique vertex axis identified in the above analysis. (b and c) A 7.9-Å-thick slice (b) and (c) cutaway shaded surface view of a 100-Å thick slab of the unique vertex. (d) One penton vertex with its five neighboring hexons. (e) The portal vertex and its five neighboring hexons. Maps in panels d and e are colored based on radial distance from the vertex axis. The densities closest to the vertex axis are shown in red and then change to green and blue at the farthest distance from the axes. The second and third rows in panels d and e are side views obtained by rotating forward from the first row with their upper portions of the structure removed for clarity.
Similar to cryoEM reconstructions of bacteriophages recently determined by single-particle cryoEM averaging (8, 11), our cryoET reconstructions of KSHV A capsids demonstrate that the putative portal of this human tumorigenic herpesvirus also contains a defined density protruding inward from the inner capsid floor (Fig. 2b and c). Although both the KSHV portal and bacteriophage connector have a similar central channel, the putative KSHV portal is markedly longer than the analogous connector structure in bacteriophages, which has a diameter of 140 Å but a height of only 70 Å (18). Fitting of the density map derived from the bacteriophage φ29 connector (18) (Fig. 3a) into our KSHV portal vertex in our cryoET capsid reconstruction (Fig. 3b) confirmed the match of their diameters, while demonstrating that the bacteriophage connector is significantly shorter.
FIG. 3.
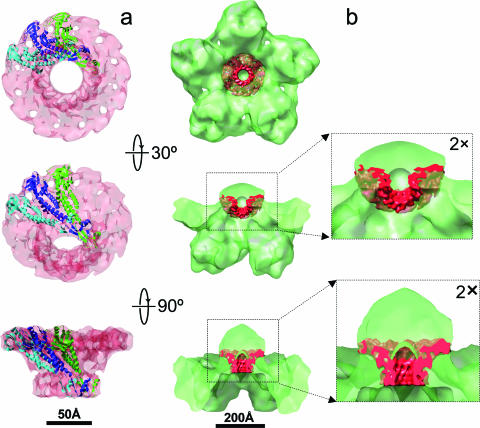
Fitting of the bacteriophage φ29 portal connector complex (18) into the portal of KSHV capsid. (a) φ29 portal determined by X-ray crystallography (Protein Data Bank no. 1FOU) shown as density map filtered to 10-Å resolution (semitransparent pink) superimposed with three subunits in ribbon representation. (b) φ29 portal (red) fitted into the KSHV portal. The orientations of the side views are the same as those in Fig. 2d and e.
Although it is well recognized that herpesvirus capsids lack the tail machinery that exists in DNA bacteriophages, it is still quite surprising that the putative portal vertex of the KSHV capsid is completely devoid of all external components or structures (Fig. 2a and see Movie S2 in the supplemental material). The external tail machineries in bacteriophages (3, 8, 11) are essential for host recognition, penetration, and DNA ejection into bacterial cells. In herpesviruses such as KSHV, these functions are likely performed in a sequence of events involving many proteins in the outer compartments of the virus, including glycoproteins on the viral envelope for cell attachment and entry and tegument proteins for capsid translocation across the cytoplasm to the nuclear pore prior to DNA ejection.
The existence of an umbilicated density at a single vertex of the KSHV capsid seems to mirror the inner portion of the bacteriophage portal complex, suggesting at least some evolutionary conservation of function as well. In bacteriophages, in addition to the connector and the external tail complexes, there is an additional internal density that may facilitate the double-stranded DNA genome packaging and/or release (3, 8, 11). KSHV PORT is ∼8% smaller than HSV-1 PORT (UL6), with molecular masses of 74 kDa (15) and 68 kDa, respectively. Regardless, the mass of each of these herpesvirus proteins is nearly twice that of the bacteriophage φ29 connector protein (36 kDa) (18) and thus may account for the difference in length between the φ29 connector and the internal density of the unique vertex of KSHV capsid we observed in our map (Fig. 3b). Nevertheless, at the low resolution of the cryoET reconstruction, we cannot rule out the possibility that additional proteins may also contribute to the density we detected at the unique vertex (Fig. 3b). KSHV contributors could include the proteins encoded by ORF7 and ORF29, the homologs of HSV-1 UL28 and UL15, respectively. Indeed, HSV-1 PORT interacts with UL28 as well as UL15 (10, 22), and HCMV PORT (UL104) likewise interacts with pUL56 (KSHV ORF7 homolog) (5). In HSV-1, UL28 and UL15 are associated with PORT transiently during viral DNA packaging (reviewed in reference 1) and together form the terminase, a protein complex that possibly includes an additional protein, encoded by UL33 (2). Direct molecular evidence that the proteins encoded by KSHV ORF7 and ORF29 are also involved in DNA packaging is lacking, although by sequence homology to these proteins, it remains reasonable to hypothesize that these proteins may be components of the putative portal complex of KSHV capsid. Taken together with the conservation of the channel and the diameter of both bacteriophage connectors and herpesvirus portal complexes, our in situ identification of the unique vertex within the intact KSHV capsid provides compelling evidence for the existence of a bacteriophage-like portal in KSHV and similar mechanisms of DNA encapsidation and/or release across herpesviruses. Confirmation of these hypotheses resulting from our cryoET structural data, however, awaits future biochemical analyses, which are currently prohibited by technical limitations in KSHV capsid isolation.
Protein structure accession number.
The 3D structure reported in this paper has been deposited in the EBI database under accession number EMD-1320.
Supplementary Material
Acknowledgments
This research is supported in part by NIH grants CA094809 (Z.H.Z.) and CA088768 (D.H.K.). B.D. is supported by a training fellowship from the Keck Center Pharmacoinformatics Training Program of the Gulf Coast Consortia (NIH grant no. R90 DK071505).
We thank Ivo Atanasov for technical assistance with imaging.
Footnotes
Published ahead of print on 10 January 2007.
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Baines, J. D., and S. K. Weller. 2005. Cleavage and packaging of herpes simplex virus 1 DNA, p. 135-150. In C. Catalano (ed.), Viral genome packaging machines: genetics, structure, and mechanism. Landes Bioscience, Austin, TX.
- 2.Beard, P. M., N. S. Taus, and J. D. Baines. 2002. DNA cleavage and packaging proteins encoded by genes UL28, UL15, and UL33 of herpes simplex virus type 1 form a complex in infected cells. J. Virol. 76:4785-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, J., P. Weigele, J. King, W. Chiu, and W. Jiang. 2006. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure 14:1073-1082. [DOI] [PubMed] [Google Scholar]
- 4.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 5.Dittmer, A., J. C. Drach, L. B. Townsend, A. Fischer, and E. Bogner. 2005. Interaction of the putative human cytomegalovirus portal protein pUL104 with the large terminase subunit pUL56 and its inhibition by benzimidazole-d-ribonucleosides. J. Virol. 79:14660-14667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganem, D. 1998. Human herpesvirus 8 and its role in the genesis of Kaposi's sarcoma. Curr. Clin. Top. Infect. Dis. 18:237-251. [PubMed] [Google Scholar]
- 7.Jiang, W., M. L. Baker, S. J. Ludtke, and W. Chiu. 2001. Bridging the information gap: computational tools for intermediate resolution structure interpretation. J. Mol. Biol. 308:1033-1044. [DOI] [PubMed] [Google Scholar]
- 8.Jiang, W., J. Chang, J. Jakana, P. Weigele, J. King, and W. Chiu. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koslowski, K. M., P. R. Shaver, J. T. Casey II, T. Wilson, G. Yamanaka, A. K. Sheaffer, D. J. Tenney, and N. E. Pederson. 1999. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J. Virol. 73:1704-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lander, G. C., L. Tang, S. R. Casjens, E. B. Gilcrease, P. Prevelige, A. Poliakov, C. S. Potter, B. Carragher, and J. E. Johnson. 2006. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791-1795. [DOI] [PubMed] [Google Scholar]
- 12.Nealon, K., W. W. Newcomb, T. R. Pray, C. S. Craik, J. C. Brown, and D. H. Kedes. 2001. Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 75:2866-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Connor, C. M., and D. H. Kedes. 2006. Mass spectrometric analyses of purified rhesus monkey rhadinovirus reveal 33 virion-associated proteins. J. Virol. 80:1574-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Connor, C. M., and D. H. Kedes. 2006. Rhesus monkey rhadinovirus: a model for the study of KSHV. Curr. Top. Microbiol. Immunol. 312:43-69. [DOI] [PubMed] [Google Scholar]
- 16.Orlova, E. V., B. Gowen, A. Droge, A. Stiege, F. Weise, R. Lurz, M. van Heel, and P. Tavares. 2003. Structure of a viral DNA gatekeeper at 10 A resolution by cryo-electron microscopy. EMBO J 22:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 18.Simpson, A. A., Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trus, B. L., N. Cheng, W. W. Newcomb, F. L. Homa, J. C. Brown, and A. C. Steven. 2004. Structure and polymorphism of the UL6 portal protein of herpes simplex virus type 1. J. Virol. 78:12668-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trus, B. L., J. B. Heymann, K. Nealon, N. Cheng, W. W. Newcomb, J. C. Brown, D. H. Kedes, and A. C. Steven. 2001. Capsid structure of Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus. J. Virol. 75:2879-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valpuesta, J. M., and J. L. Carrascosa. 1994. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys. 27:107-155. [DOI] [PubMed] [Google Scholar]
- 22.White, C. A., N. D. Stow, A. H. Patel, M. Hughes, and V. G. Preston. 2003. Herpes simplex virus type 1 portal protein UL6 interacts with the putative terminase subunits UL15 and UL28. J. Virol. 77:6351-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler, H., and K. A. Taylor. 2006. Accurate marker-free alignment with simultaneous geometry determination and reconstruction of tilt series in electron tomography. Ultramicroscopy 106:240-254. [DOI] [PubMed] [Google Scholar]
- 24.Wu, L., P. Lo, X. Yu, J. K. Stoops, B. Forghani, and Z. H. Zhou. 2000. Three-dimensional structure of the human herpesvirus 8 capsid. J. Virol. 74:9646-9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, Z. H., M. Dougherty, J. Jakana, J. He, F. J. Rixon, and W. Chiu. 2000. Seeing the herpesvirus capsid at 8.5 Å. Science 288:877-880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.