Abstract
During viral entry, the paramyxovirus fusion (F) protein fuses the viral envelope to a cellular membrane. Similar to other class I viral fusion glycoproteins, the F protein has two heptad repeat regions (HRA and HRB) that are important in membrane fusion and can be targeted by antiviral inhibitors. Upon activation of the F protein, HRA refolds from a spring-loaded, crumpled structure into a coiled coil that inserts a hydrophobic fusion peptide into the target membrane and binds to the HRB helices to form a fusogenic hairpin. To investigate how F protein conformational changes are regulated, we mutated in the Sendai virus F protein a highly conserved 10-residue sequence in HRA that undergoes major structural changes during protein refolding. Nine of the 15 mutations studied caused significant defects in F protein expression, processing, and fusogenicity. Conversely, the remaining six mutations enhanced the fusogenicity of the F protein, most likely by helping spring the HRA coil. Two of the residues that were neither located at “a” or “d” positions in the heptad repeat nor conserved among the paramyxoviruses were key regulators of the folding and fusion activity of the F protein, showing that residues not expected to be important in coiled-coil formation may play important roles in regulating membrane fusion. Overall, the data support the hypothesis that regions in the F protein that undergo dramatic changes in secondary and tertiary structure between the prefusion and hairpin conformations regulate F protein expression and activation.
Paramyxoviruses have evolved two surface glycoproteins that cause membrane fusion during viral entry: a receptor-binding (HN, H, or G) protein and a fusion (F) protein (31). During viral entry, the receptor-binding protein attaches to its cellular receptor and then transduces a signal to the F protein to initiate membrane fusion (12, 22, 40, 46, 57). As a class I viral fusion protein, the paramyxovirus F protein is synthesized as a single precursor protein (F0), folded as a homotrimer, glycosylated, and cleaved into an active form consisting of a small amino-terminal subunit (F2) and a larger carboxy-terminal subunit (F1) (31). The ectodomain of the F1 subunit contains a hydrophobic fusion peptide at its amino terminus and two 4-3 heptad repeat regions, HRA and HRB (Fig. 1A). Upon activation, the F protein is thought to insert its hydrophobic fusion peptide into the target membrane and form a coiled-coil hairpin structure with its HRA and HRB regions (32, 48) in order to actively drive membrane fusion (35, 46).
FIG. 1.
The paramyxovirus F protein. (A) Domain structure of the F protein. HRA and HRB are shown in red and blue, respectively. The signal peptide (SP), fusion peptide (FP), transmembrane (TM), and cytoplasmic tail (CT) regions are also labeled. Structural domains DI, DII, and DIII are represented by solid lines. (B) Sequence alignment of HRA regions of SeV, Nipah virus, hPIV3, PIV5, and NDV. Identical residues are highlighted in red, and similar residues are highlighted in yellow. The black box identifies the sequence of the 10 conserved residues that were mutated in this study. The secondary structures of the region in the prefusion (native) and hairpin (final) conformations of the F protein are shown, with bars representing α-helices and arrows representing β-strands. Heptad repeat a and d residues are shown underneath the sequence alignment. The underlines correspond to a stutter in the heptad repeat (1). (C) Structure of the PIV5 F protein ectodomain in its uncleaved, prefusion form (64). (D) Structure of the hPIV3 F protein ectodomain in its hairpin form (63). In both panels C and D, HRA is shown in red and HRB in blue. The insets show the structures formed by one monomer of HRA, and the boxes identify the residues investigated in this study. Panels C and D were rendered in MOLMOL (30).
Peptides derived from the HRA and HRB regions of the paramyxovirus F protein inhibit membrane fusion and virus replication (3, 33, 39, 46, 62) by mechanisms similar to those of HR-derived peptides of human immunodeficiency virus (HIV) type 1 gp41 (6, 21, 25). HRA-derived peptides are thought to bind HRB in an early intermediate of the F protein, and HRB-derived peptides bind to HRA in a prehairpin intermediate (46, 47). In both cases, the binding of an HR-derived peptide to its complementary HR region in the F protein prevents formation of the hairpin that is needed to drive membrane fusion (35, 46). HRB-derived peptides are shorter, more soluble, and approximately 1,000 times as potent as HRA-derived peptides (19). Therefore, antiviral strategies often use HRB-derived peptides and small molecules to target the HRA coiled coil in the prehairpin intermediate (14).
HRB-derived peptides usually inhibit membrane fusion mediated only by the F protein from which they are derived (31). However, inhibition of closely related F proteins has been demonstrated (3), and an HRB-derived peptide from human parainfluenza virus type 3 (hPIV3) has been shown to inhibit Hendra virus fusion (39). The mechanism of cross-species inhibition is unresolved but may involve the topography of the nonpolar side chains on the hydrophobic face of the HRB-derived hPIV3 peptide allowing favorable packing interactions with the HRA coiled coil in the Hendra virus F protein. An implication of this finding is that the F protein hairpin may tolerate mutations, as long as the periodicity of nonpolar and polar residues is conserved and packing interactions are favorable. Conservative mutations to HR residues in the F protein of PIV5 (formerly known as simian virus 5) slightly reduce hairpin stability (17, 47) but more dramatically affect the stability and activation of the prefusion conformation of the F protein (47, 60). Thus, the tolerance of multiple conformations of the F protein to conservative mutations in the HR regions remains an unresolved issue, despite these residues being targeted in drug design efforts.
X-ray crystal structures have been determined for two conformations of the F protein ectodomain: an uncleaved prefusion form and a hairpin form (Fig. 1C and D). The prefusion form of the PIV5 F protein has a mushroom-like shape formed by a large globular head attached to a rod-like stalk (64). The stalk consists of a triple-stranded coiled coil formed by HRB, and the head is formed by three domains: DI, DII, and DIII. At the base of the head, DI forms a highly twisted β-barrel-like assembly, and DII forms an immunoglobulin-like β-sandwich domain. The fusion peptide is located on the side of the head and is held in place by interactions with other residues in DIII and with residues in DII. At the top of the head, DIII contains HRA in a crumpled, spring-loaded structure consisting of α-helices, β-strands, and turns (Fig. 1C, inset). In the hairpin structures of the F proteins of Newcastle disease virus (NDV) (7) and hPIV3 (63), HRA forms a triple-stranded coiled coil that is buttressed by antiparallel HRB helices. DI and DII remain intact in both forms of the F protein, undergoing changes in relative orientation to each other.
Many questions remain about how the paramyxovirus F protein regulates its irreversible structural change from a prefusion to a hairpin structure to cause membrane fusion at the right time and place. Mutations to isolated residues in the fusion peptide (27, 45), in HRA (28, 60), between DI and DIII (52), and in HRB (37, 47) of the F proteins of PIV5, NDV, and hPIV3 have been found to increase membrane fusion activity, presumably by destabilizing the prefusion conformation of the F protein. A retrospective analysis of the mutational studies, in light of the recently determined high-resolution structures of the F protein (7, 63, 64), showed that these residues are located in regions of the F protein that undergo significant conformational changes during the transition from the prefusion to the hairpin structure.
On the basis of these observations, we hypothesized that F protein residues that undergo dramatic changes in secondary and tertiary structure between the prefusion and hairpin conformations are important in regulating the membrane fusion activity of the F protein. To test this hypothesis, we performed a mutational analysis on residues in a highly conserved portion of the HRA domain of the Sendai virus (SeV) F protein (Fig. 1). The 10 residues in this sequence form part of a β-strand/turn/α-helix structure in the prefusion conformation and part of the triple-stranded coiled coil in the hairpin conformation (Fig. 1C and D). Consistent with this portion of the HRA region playing an important role in the folding and stability of the prefusion F conformation, nine of the mutations significantly disrupted F protein expression, processing, and fusogenicity. Conversely, the other six mutations caused the SeV F protein to have hyperactive membrane fusion activity, most likely by destabilizing the spring-loaded conformation of HRA. Overall, these HRA residues that undergo significant structural changes during protein refolding were observed to be key regulators of F protein folding and fusogenicity.
MATERIALS AND METHODS
Plasmids.
The pCAGGS SeV F (Enders strain) wild-type plasmid has been described previously (56). The pGEM3X SeV F wild-type plasmid was generated by using the EcoRI and XhoI restriction sites to subclone into the pGEM3X vector, a derivative of the pGEM3 vector that contains an XhoI site (37). Mutants were generated in the background of pGEM3X plasmids by using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions and subcloning those mutations into pCAGGS plasmids by using EcoRI and XhoI restriction sites. The mutant F protein designated Fcl, which is cleaved intracellularly by furin or furin-like proteases, contains the mutations N104Q, V112R, P113R, Q114R, and S115K.
The HRA mutants of SeV Fcl were generated in the background of pGEM3X plasmids and subcloned into the pCAGGS plasmid as described above. The nucleotide sequences of all the plasmid constructs were verified by DNA sequencing, which was performed in the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital.
Cell culture.
Monolayer cultures of Vero cells (ATCC CCL-81), BHK-21 cells (ATCC CCL-10), and BSR-T7/5 cells (4) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 1% glutamine, 1% penicillin, and 1% streptomycin. BSR-T7/5 cells were also grown in the presence of Geneticin (1 mg/ml, final concentration), which was added to the DMEM every other passage. BHK-21 cells were also supplemented with 10% tryptose phosphate broth. CV-1 cells (ATCC CCL-70) were grown in DMEM supplemented with 10% Nu-serum, 1% glutamine, 1% penicillin, and 1% streptomycin.
Transient expression of viral envelope glycoproteins.
Monolayers of Vero cells in six-well dishes (85% to 95% confluence) were transiently transfected with 1 μg of pCAGGS F protein DNA, 1 μg pCAGGS HN protein DNA, or both by using the Lipofectamine Plus expression system (Invitrogen) according to the manufacturer's instructions. Transfected Vero cells were incubated for 4 h at 37°C. DMEM (containing 10% fetal bovine serum and 1% glutamine) was then added to the cells, which were subsequently incubated for 16 h at 37°C. Cells were then treated as indicated for each experiment.
Flow cytometry.
At 16 h posttransfection, the Vero cells were washed five times with phosphate-buffered saline containing calcium and magnesium at 0.1 g/liter(PBS+) solution, overlaid with PBS+ solution containing primary antibody, and incubated at 4°C for 30 min. For quantification of the cell surface expression of the SeV F protein, the primary monoclonal antibody M16 (dilution, 1:200) (41) was used. The transfected cells were subsequently washed five times with PBS+, overlaid with PBS+ solution containing fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody (dilution, 1:100), incubated at 4°C for 30 min, and washed five times with PBS+. Cells were removed from six-well dishes with PBS deficient in calcium and magnesium (PBS−) and containing 50 mM EDTA and were fixed in suspension by adding methanol-free formaldehyde (final concentration, 0.5%). The cell surface fluorescence of 10,000 cells was measured using a FACSCalibur flow cytometer (Becton Dickinson). Mean fluorescence intensities (MFIs) were normalized to the MFI of the SeV F wild-type protein.
Generation of a monoclonal antibody against the cytoplasmic tail of the SeV F protein.
B6 mice were immunized with 25 μg keyhole limpet hemocyanin (KLH)-conjugated peptide from the cytoplasmic tail of the SeV F protein (sequence, NPDDRIPRDTYTLEPKIR). One month later, mice received a 25-μg KLH-peptide boost with complete Freund's adjuvant. Three days prior to cell fusion, mice received a final 25-μg boost of KLH-peptide in PBS. Hybridomas were produced by fusion of spleen cells to P3x63-Ag8.653 mouse myeloma cells (ATCC CRL-158). Monoclonal antibodies were selected from tissue culture supernatant plated on 96-well plates coated with nonconjugated SeV F cytoplasmic tail peptide. Hybridoma SeV F tail 31705-1C12 was produced in larger quantities by Harlan Bioproducts for Science (Madison, WI).
Radioimmunoprecipitation.
Vero cells expressing F proteins (wild type or mutant) were maintained in culture for 30 min in methionine- and cysteine-free medium and then labeled for 15 min with 50 μCi [35S]Promix (Amersham Pharmacia Biotech) in 0.5 ml of DMEM lacking methionine and cysteine and containing 20 mM HEPES buffer (pH 7.3). The cells were then washed once with PBS+ and chased with 3 ml of DMEM containing 2 mM methionine, 2 mM cysteine, and 20 mM HEPES buffer (pH 7.3) for the reported time. Samples were lysed with ice-cold radioimmunoprecipitation assay (RIPA) buffer containing 0.15 M NaCl, 9.25 mg/ml iodoacetamide, 1.7 mg/ml aprotinin, and 10 mM phenylmethylsulfonyl fluoride (36). The lysate was spun at 67,000 × g in an Optima TLX ultracentrifuge (Beckman Coulter).
The supernatant was incubated overnight (18 to 22 h) at 4°C with 5 μl mouse anti-F tail peptide monoclonal antibody (dilution, 1:200). Immune complexes were adsorbed to protein G-Sepharose 4 Fast Flow (GE Healthcare) for 1 h at 4°C. Samples were washed three times with RIPA buffer containing 0.3 M NaCl, three times with RIPA buffer containing 0.15 M NaCl, and once with 50 mM Tris buffer (0.25 mM EDTA, 0.15 M NaCl, pH 7.4). The samples were resuspended in 50 μl of sample dye buffer containing 200 mM Tris, 8% sodium dodecyl sulfate (SDS), 0.2% bromophenol blue, 40% glycerol, and 12% β-mercaptoethanol. The samples were then boiled for 5 min, centrifuged at high speed for 1 min, and fractionated on 12% NuPAGE bis-Tris polyacrylamide-SDS gels (Invitrogen). Protein bands were visualized using a Typhoon 9200 phosphorimager (GE Healthcare) and quantified using ImageQuant 5.2 software (Molecular Dynamics).
Total F0 protein expression and cleavage.
Five hours after transfection, the cells were washed twice with PBS+ solution. Cells were then chased for 0 or 180 min and radioimmunoprecipitated as described above. F0 bands were visualized as described above. For analysis of F protein cleavage, at 5 h posttransfection cells were pulsed for 15 min and chased for 3 h as described above. During the last hour of the chase, TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-treated trypsin (5 μg/μl; Sigma) was added to each sample in 0.5 ml chase medium (DMEM, 1% penicillin, 1% streptomycin, 2 mM methionine, 2 mM cysteine, and 20 mM HEPES). The samples were then radioimmunoprecipitated as described above. F0 and F1 bands were visualized and quantified as described above.
Biotinylation of surface-expressed F protein.
At 16 h posttransfection, the Vero cells were washed twice with PBS+ solution. Cells were then radiolabeled and chased for 180 min. The samples were subsequently biotinylated twice for 15 min at 4°C with 2 mg of EZ-Link Sulfo-NHS-SS-biotin (Pierce) in 1 ml of PBS at pH 8. Excess biotin was washed off with PBS containing 50 mM glycine. Samples were then radioimmunoprecipitated as described above. Following the wash with 50 mM Tris buffer (0.25 mM EDTA, 0.15 M NaCl, pH 7.4), the samples were resuspended in 100 μl of 50 mM Tris buffer (0.5% SDS, pH 7.4), boiled for 5 min, and centrifuged at high speed for 1 min. The supernatants were split into two 50-μl fractions. One fraction was saved for direct loading onto the SDS-polyacrylamide gel for polyacrylamide gel electrophoresis (PAGE). The other fraction was diluted to 1 ml with streptavidin buffer (20 mM Tris, 0.15 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.2% bovine serum albumin, pH 8) and incubated with streptavidin-agarose overnight at 4°C. The samples were washed as described above for radioimmunoprecipitation, resuspended in SDS-PAGE sample buffer containing 12% β-mercaptoethanol, and fractionated on SDS-12% polyacrylamide gels. The F0 bands were visualized and quantified as described above.
Endo H digestion.
To test for the conversion of N-linked carbohydrate chains from the high-mannose form to the complex form, we radioimmunoprecipitated the F protein from transfected cells at chase times of 0, 45, 90, 135, and 180 min. The immune complexes were dissociated by boiling in 50 mM Tris-HCl (pH 7.4) and 0.5% (wt/vol) SDS. To each sample, a solution of 0.1 M sodium citrate (pH 5.3) containing 1 mM phenylmethylsulfonyl fluoride and 2 mU of endoglycosidase H (endo H) (Roche Diagnostics) was added, and digestion was carried out for 18 to 24 h at 37°C. The reaction was terminated by the addition of sample dye buffer. Samples were fractionated in a 12% polyacrylamide-SDS gel under nonreducing conditions. Results were visualized as described above.
Luciferase reporter gene assay for cell-cell membrane fusion.
To quantify membrane fusion, we performed a luciferase reporter gene assay as described previously (47). Briefly, six-well dishes containing Vero cells (70% to 80% confluence) were transfected with 1.0 μg luciferase control DNA (Promega), 1.0 μg pCAGGS F DNA, and 1.0 μg pCAGGS HN DNA. At 16 h posttransfection, Vero cells were treated with TPCK-treated trypsin (5 μg/ml) for 1 h at 37°C. Soybean trypsin inhibitor (20 μg/ml; Sigma) was then added, and the samples were incubated at 37°C for 30 min. BSR-T7/5 target cells (expressing T7 RNA polymerase) were overlaid onto the Vero cells expressing the SeV F and HN proteins. After an 8-h incubation at 37°C, the monolayers were washed, lysed in reporter lysis buffer (Promega), and clarified by centrifugation at 15,000 × g in a tabletop centrifuge (5417C; Eppendorf) at room temperature. From each clarified lysate, a 150-μl sample was transferred into a 96-well plate (Lumitrac 200; Promega). The luciferase activity resulting from fusion of the two cell populations was quantified with a Veritas luminometer (Promega); 100 μl luciferase assay substrate (Promega) was injected into each sample.
Dye transfer fusion assay for lipid mixing and content mixing.
To measure whether any of the SeV F protein mutants were deficient in either lipid mixing or content mixing, we performed a standard dye transfer fusion assay as described previously (45), with a few modifications. Monolayers of CV-1 cells were grown in six-well plates and transfected with pGEM3X SeV F and HN plasmids by using the vaccinia virus-bacteriophage T7 RNA polymerase (vTF7.3) expression system (20). Before infection of the cells, the vaccinia virus was inactivated with psoralen (Trioxsalen, 4′-aminomethyl-, hydrochloride; 1 μl per well) (Calbiochem) and irradiated with long-wavelength UV light for 2 min. At 16 to 20 h posttransfection, CV-1 cells were incubated in the presence of TPCK-treated trypsin (5 μg/ml) for 30 min at 37°C. The Fcl mutants were tested for lipid mixing and content mixing in the absence of trypsin treatment. Human red blood cells (RBCs) were double labeled with octadecyl rhodamine B chloride (R18) and 6-carboxyfluorescein (6CF) (both from Molecular Probes) as described previously (45). Membrane fusion was quantified by counting 6CF or R18 dye transfer events from labeled RBCs to CV-1 cells. To measure the temperature dependence of cell-cell fusion by the different SeV F protein mutants, we labeled CV-1 effector cells with the red fluorescent nucleic acid stain SYTO 17 (Molecular Probes). RBCs were single labeled with 6CF. Fusion was promoted by incubation for 15 min at 25, 29, 33, or 37°C followed by washing with PBS+ at 4°C and a second incubation on ice. Dye transfer was visualized using a Nikon eclipse TE300 fluorescence microscope, and random fields were captured using the IPLab 3.7 software (BD Biosciences).
Syncytium assay for cell-cell fusion by Fcl mutants.
Monolayers of BHK-21 cells grown in six-well plates were transfected with 1 μg pCAGGS SeV Fcl and 1 μg pCAGGS HN DNA as described above. At 24 h posttransfection, cells were fixed and stained with Hema 3 solution (Fisher) per the manufacturer's instructions. Representative fields were captured with a Nikon D70 digital camera attached to a Nikon Eclipse TS100 inverted microscope.
RESULTS
Many of the conservative mutations to HRA residues reduce expression of the SeV F protein.
Mutation of nonpolar “a” and “d” residues in HRA of the NDV F protein to lysine residues decreases cell surface expression and eliminates membrane fusion activity (51, 53). In contrast, mutation of a and d residues in HRA of the PIV5 F protein to methionine residues does not, in many cases, disrupt the α-helix formation, protein expression and processing, or membrane fusion activity (60). Therefore, we investigated the functional importance of the 10-residue HRA sequence of the SeV F protein by mutating each residue individually to the residues of Nipah virus, hPIV3, PIV5, and NDV (Fig. 1; Table 1). The 15 conservative mutations maintained the periodicity of hydrophobic and polar residues in HRA and thus were not expected to disrupt significantly the HRA coiled coil in the prehairpin intermediate or the hairpin structure.
TABLE 1.
Phenotypes of wild-type and mutant Sendai virus F proteinsa
| Se F mutation | HRA 10 | Total F0b | Cleaved F1/(F1 + F0)c | Expression
|
Endo H resistanceg | Cell-cell membrane fusionh | ||
|---|---|---|---|---|---|---|---|---|
| % Positive cellsd | MFIe | Biotinylationf | ||||||
| None (wild type) | ALKTLQDFVN | 100 | 0.9 | 64 | 100 ± 4 | 100 | 4.6 | 100 ± 10 |
| A178V | V......... | 62 | 0.9 | 87 | 101 ± 2 | 73 | 3.2 | 155 ± 23 |
| L179I | .I........ | 60 | 0.9 | 80 | 95 ± 9 | 277 | 4.8 | 244 ± 13 |
| L179V | .V........ | 125 | 0.9 | 81 | 120 ± 11 | 178 | 6.9 | 403 ± 36 |
| K180G | ..G....... | 67 | 0.9 | 61 | 44 ± 2 | 22 | 1.2 | 50 ± 20 |
| K180Q | ..Q....... | 60 | 0.9 | 68 | 81 ± 5 | 49 | 1.2 | 65 ± 12 |
| K180T | ..T....... | 149 | 0.9 | 84 | 82 ± 2 | 201 | 5.6 | 271 ± 47 |
| T181A | ...A...... | 73 | 0.8 | 67 | 51 ± 3 | 13 | 0.9 | 41 ± 4 |
| T181K | ...K...... | 21 | 0.0 | 2 | 0 ± 0 | 1 | 1.4 | 3 ± 4 |
| T181S | ...S...... | 90 | 0.9 | 71 | 113 ± 3 | 146 | 3.7 | 236 ± 15 |
| L182M | ....M..... | 93 | 0.2 | 15 | 3 ± 0 | 3 | 0.2 | 9 ± 13 |
| L182V | ....V..... | 25 | 0.1 | 1 | 0 ± 0 | 11 | 0.2 | 8 ± 5 |
| D184Q | ......Q... | 43 | 0.4 | 7 | 1 ± 0 | 4 | 0.3 | 9 ± 1 |
| F185H | .......H.. | 33 | 0.0 | 1 | 0 ± 0 | 3 | 0.3 | 8 ± 7 |
| F185Y | .......Y.. | 87 | 0.9 | 55 | 101 ± 8 | 81 | 4.4 | 79 ± 15 |
| V186I | ........I. | 109 | 0.9 | 53 | 42 ± 2 | 23 | 0.4 | 38 ± 17 |
Sendai virus F proteins expressed from pCAGGS DNA in Vero cells.
Total F0 expression after 15 min of [35S]methionine pulse-labeling and a 3-h chase. Data are normalized to wild-type SeV F.
SeV F protein cleavage efficiency after 15 min of [35S]methionine pulse-labeling and a 3-h chase with exogenous trypsin (5 μg/μl) added in the last hour of chase.
Percentage of cells expressing the SeV F protein as determined by flow cytometry using monoclonal antibody M16.
Cell surface expression determined by flow cytometry using monoclonal antibody M16. Data are normalized to wild-type SeV F. The reported error indicates two standard deviations from triplicate experiments.
Cell surface expression determined by biotinylation. Data are normalized to wild-type SeV F.
Ratio of F protein resistant to endoglycosidase H digestion to F protein sensitive to digestion at 180 min.
Cell-cell fusion as determined by a gene reporter assay. Data are normalized to wild-type SeV F. The reported error indicates two standard deviations from triplicate experiments.
To study the effects of the mutations on F protein expression, we measured via radioimmunoprecipitation the total F0 protein expression after 180 min. All of the mutant F0 proteins were expressed, though many at different levels from that of the wild-type F protein (Fig. 2A). The levels of expression of F protein mutants L179V and K180T were significantly higher than that of the wild-type F protein (Table 1). Conversely, the levels of expression of the F protein mutants T181K, L182V, D184Q, and F185H were less than 50% of that of the wild-type F protein (Table 1).
FIG. 2.
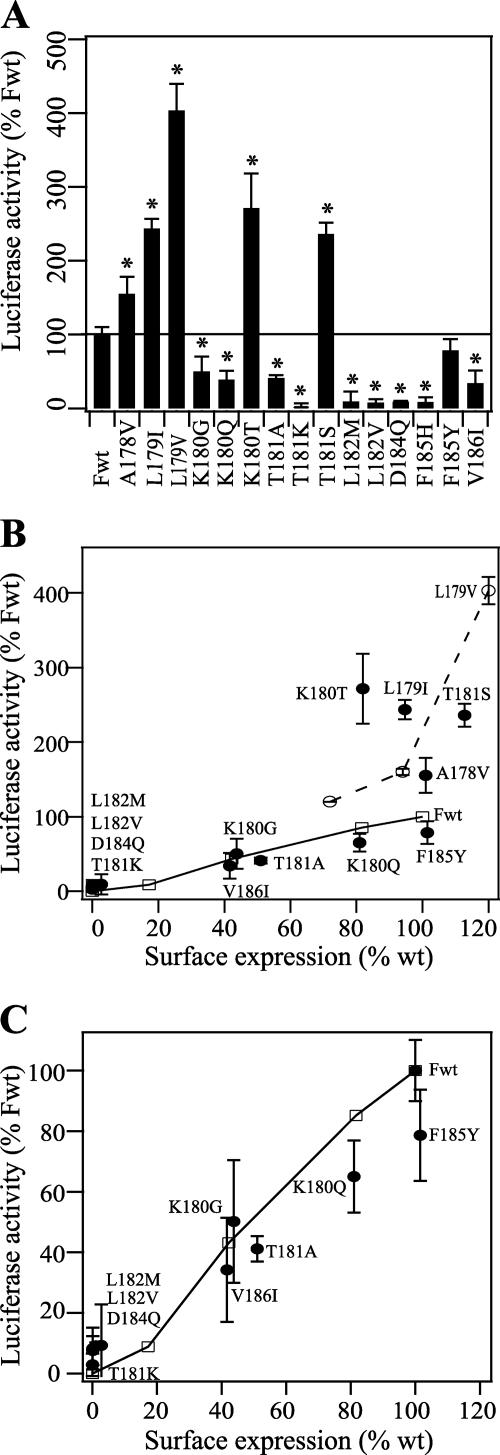
SeV F0 protein expression. (A) Total F0 protein expression. Vero cells expressing wild-type (Fwt) or mutant F proteins were labeled with [35S]Promix for 15 min and chased for 3 h. Polypeptides were immunoprecipitated with a monoclonal antibody against the cytoplasmic tail of the F protein and analyzed by SDS-PAGE under reducing conditions. (B) Surface expression of SeV F0 as analyzed by flow cytometry using M16, a conformation-specific monoclonal antibody to the SeV F protein ectodomain. Mean fluorescence intensities were normalized to 100% surface expression for Fwt. The solid line represents 100% Fwt expression. Error bars represent two standard deviations from triplicate experiments. Asterisks indicate a significant difference as measured by Student's t test (P < 0.05).
Flow cytometry and biotinylation studies were done to show how the mutations affected the cell surface expression of the F0 protein. For flow cytometry, we used the conformation-specific monoclonal antibody M16 (41). As shown in Fig. 2B, the cell surface expression levels of five mutants (A178V, L179I, L179V, T181S, and F185) were similar to or slightly higher than that of wild-type F protein. Five other mutants (K180G, K180Q, K180T, T181A, and V186I) had cell surface expression levels significantly lower than that of wild-type F protein, and five (T181K, L182M, L182V, D184Q, and F185H) did not show any binding to M16. For the biotinylation experiment, transiently expressed F proteins on the surface of transfected cells were conjugated with the biotinylation agent EZ-Link Sulfo-NHS-SS-biotin (Pierce) before lysis of cells, immunoprecipitation with the SeV F tail monoclonal antibody, pull-down with streptavidin agarose (Pierce), and analysis by SDS-PAGE. The results from densitometric analyses of the biotinylated F protein bands correlate to those obtained by flow cytometry, with the exception of K180T, which now shows cell surface expression levels slightly higher than that of wild-type F protein (Table 1). This discrepancy could perhaps be explained by a low binding affinity of the antibody M16 to the mutant protein. For both the biotinylation and flow cytometry experiments, 9 of the 15 mutations in this portion of the HRA domain significantly reduced cell surface expression, a finding that is consistent with these residues playing an important role in the folding and stability of the metastable prefusion form of the F protein.
Mutant F proteins that reach the cell surface are efficiently cleaved by trypsin.
To become active for membrane fusion, the precursor F0 protein must first be cleaved into F1 and F2 subunits (31). Many paramyxovirus F0 proteins contain an R-X-R/K-R consensus sequence for intracellular cleavage by furin or furin-like proteases. However, most isolates of SeV contain F proteins that lack a multibasic cleavage site and are cleaved by exogenous proteases while replicating in the lungs of mice (49). When expressed on the surface of tissue culture cells, the SeV F0 protein was efficiently cleaved by exogenous trypsin (Fig. 3A). A densitometric analysis of the relative abundance of the wild-type F0 and F1 bands showed that the wild-type F protein was cleaved by exogenous trypsin with an efficiency of 90% (Fig. 3B; Table 1). Because mutations to HRA residues could potentially interfere with proteolysis, we measured the extents to which the 15 mutant F proteins were cleaved by exogenous trypsin (Fig. 3). All of the mutant F0 proteins that reached the cell surface were cleaved to an extent comparable to that of wild-type F0 protein, whereas the mutant F0 proteins that did not reach the cell surface were not efficiently cleaved.
FIG. 3.
Cleavage of the mutant SeV F0 proteins. (A) Vero cells expressing wild-type (Fwt) or mutant F proteins were labeled with [35S]Promix (100 μCi/ml) for 15 min and chased for 3 h. During the last hour of the chase, TPCK-treated trypsin (tryp.) (5 μg/ml) was added. F proteins were immunoprecipitated with SeV F-tail-specific monoclonal antibody and analyzed by SDS-PAGE on a 12% gel under reducing conditions. (B) Ratios of cleaved F proteins. The intensities of the F0 and F1 bands from panel A were quantified, and the fraction of cleaved F1 was calculated by dividing F1 by total F (represented by F0 + F1 because the SeV F2 protein is not observed in the assay).
Mutant F proteins that do not reach the cell surface are not processed properly.
The SeV F protein contains three N-linked glycosylation sites that are utilized (N104, N245, and N449) (50). Endo H cleaves N-linked oligosaccharides of the high-mannose form after they are synthesized in the endoplasmic reticulum. However, endo H does not cleave complex-type oligosaccharides after maturation in the medial Golgi. To determine whether the HRA mutations affected the maturation of these carbohydrate chains, we measured the kinetics of endo H resistance of the mutant F proteins. For the wild-type F protein, a resistant form of the protein was detected within 90 min (Fig. 4A). For the five F protein mutants that do not reach the cell surface (e.g., L182V), the sensitive form continues to predominate, even after 180 min (Fig. 4B and C). These results are consistent with these five mutants not being processed properly in the medial Golgi and suggest that they may be misfolded and degraded intracellularly, thereby eliminating cell surface expression. The four mutants with decreased cell surface expression (i.e., K180G, K180Q, T181A, and V186I) had lower levels of endo H resistance than wild-type F protein (Table 1). As the F proteins containing K180G, K180Q, T181A, and V186I mutations reached the cell surface, albeit at lower levels than wild-type F protein, these mutants may undergo at least partial misfolding that reduces the extent of glycosylation maturation while trafficking to the cell surface. The six mutant F proteins that had cell surface expression levels similar to or greater than that of wild-type F protein (A178V, L179I, L179V, K180T, T181S, and F185Y) had kinetics of endo H resistance similar to that of wild-type F protein (Table 1).
FIG. 4.
Kinetics of endo H resistance. (A) Time course of endo H digestion of wild-type SeV F (Fwt) protein. Vero cells expressing Fwt protein were labeled with [35S]Promix for 15 min and chased for the times indicated. After immunoprecipitation with SeV F tail-specific monoclonal antibody, the protein was incubated with (+) or without (−) endo H and analyzed by SDS-PAGE on a 12% gel under nonreducing conditions. (B) Time course of endo H digestion of the L182V mutant SeV F protein. For panels A and B, the formation of complex carbohydrate chains resistant to endo H (FR) and the presence of high-mannose chains sensitive to endo H (FS) are indicated. (C) The kinetics of endo H resistance of Fwt and mutant F proteins were calculated as the ratio of FR bands to FS bands as a function of time.
The HRA mutations dramatically alter the fusogenicity of the SeV F protein.
To determine the effects of the HRA mutations on the fusogenicity of the SeV F protein, we performed a luciferase gene reporter assay (Fig. 5A). In this experiment, Vero effector cells that expressed the SeV HN and F proteins and contained a luciferase plasmid under control of a T7 promoter were overlaid with BSR-T7/5 cells that stably expressed T7 RNA polymerase (4). As would be expected, the five mutations that eliminated the proper expression and processing of the F protein (i.e., T181K, L182M, L182V, D184Q, and F185H) also eliminated membrane fusion activity. The four mutants that were expressed on the cell surface at levels lower than that of wild-type F protein (i.e., K180G, K180Q, T181A, and V186I) also promoted membrane fusion to a lesser extent than did wild-type F protein. In this assay, the F185Y mutant caused membrane fusion similar to that induced by wild-type F protein, and the other five mutant F proteins that had cell surface expression levels similar to or greater than that of wild-type F protein (i.e., A178V, L179I, L179V, K180T, and T181S) caused membrane fusion much more efficiently than wild-type F protein.
FIG. 5.
Luciferase gene reporter assay of cell-cell fusion activities of the SeV F protein. (A) Membrane fusion activities of Vero cells that expressed wild-type (Fwt) or mutant F proteins and the HN protein and that were transfected with luciferase control DNA. At 16 h posttransfection, Vero cells were incubated with TPCK-treated trypsin (5 μg/ml) for 1 h at 37°C. To initiate fusion, Vero effector cells were overlaid with BSR-T7/5 target cells that express T7 RNA polymerase (4). The two cell populations were incubated for 8 h at 37°C. The reported luciferase activities were normalized to Fwt (∼2 × 106 relative light units). Error bars represent two standard deviations from triplicate experiments. The solid line represents 100% Fwt expression. Asterisks indicate a significant difference as measured by Student's t test (P < 0.05). (B) Cell-cell fusion versus cell surface expression. The open squares connected by a solid line represent a titration of increasing amounts of Fwt DNA. The open circles connected by a dotted line represent a titration of increasing amounts of F L179V DNA. The cell surface expression levels of the F proteins were determined by flow cytometry using the M16 monoclonal antibody. The data were normalized to 100% fusion and 100% expression, corresponding to the transfection of 1 μg of pCAGGS Fwt DNA. (C) Enlargement of panel B.
Previous work has shown that cell-cell fusion activity is proportional to cell surface density (16); therefore, we compared the fusogenicities of the mutant F proteins to that of the wild-type F protein when expressed at comparable levels (Fig. 5B and C). Smaller amounts of wild-type F DNA in the transfection mixtures yielded lower levels of wild-type F expression and membrane fusion. The efficiency of membrane fusion caused by K180G, K180Q, T181A, and V186I was either similar to or less than that caused by wild-type F protein at similar expression levels (Fig. 5C). These four mutants acquired lower levels of endo H resistance than wild-type F protein (Fig. 4C), perhaps because of partial folding differences. Unexpectedly, the presumed folding differences do not appear to significantly compromise the membrane fusion activities of these mutant F proteins beyond a reduction in overall cell surface expression levels. The membrane fusion activities of A178V, L179I, L179V, K180T, and T181S were much greater (150% to 400%) than that of wild-type F protein, even when we controlled for protein expression levels (Fig. 5B).
The dramatic degree to which the HRA mutations substantially increase, proportionally decrease, or completely eliminate membrane fusion activity confirms our original hypothesis that residues whose secondary and tertiary structures dramatically change during protein refolding are key regulators of the F protein's membrane fusion activity.
Wild-type and mutant F proteins with decreased fusogenicity do not have hemifusion phenotypes.
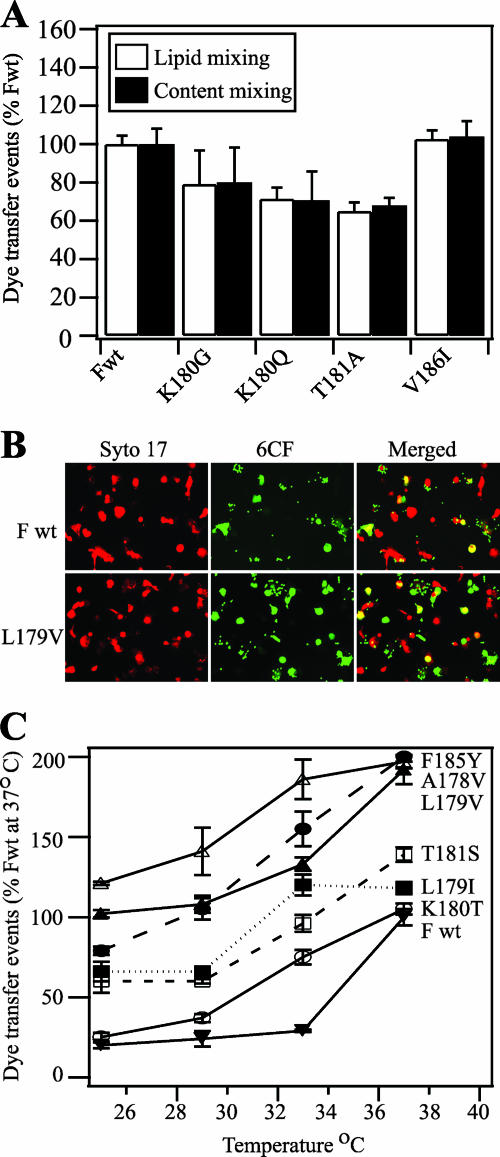
Next, we further examined the mechanism by which the above-mentioned mutations regulate the fusogenicity of the F protein. The lipid bilayers of enveloped viruses are thought to rearrange during membrane fusion in a series of discrete steps that include dimpling, lipid stalk formation, hemifusion, transient pore formation, and pore enlargement (9, 18). In some cases, mutations in the fusion peptide of the influenza virus hemagglutinin (HA) protein cause a hemifusion phenotype (lipid mixing without content mixing) (42, 54). Therefore, we investigated whether the wild-type or mutant F proteins arrest membrane fusion at the hemifusion stage. We performed a fluorescent dye transfer assay with target RBCs colabeled with the lipidic probe R18 and the aqueous probe 6CF. For wild-type and mutant F proteins that had decreased fusogenicities in the luciferase assay (i.e., K180G, K180Q, T181A, and V186I), lipid mixing and content mixing were coincident (Fig. 6A). The mutant F proteins with hyperfusogenic phenotypes also had equivalent levels of lipid mixing and content mixing in the dye transfer assay (data not shown). The data were consistent with wild-type and mutant F proteins not arresting membrane fusion during late stages when a fusion pore opens and enlarges.
FIG. 6.
Dye transfer assays of SeV F protein-mediated membrane fusion. (A) Lipid mixing (white bars) and content mixing (black bars) by SeV F proteins. Effector CV-1 cells were infected with vaccinia virus vTF7.3 and transfected with 1 μg each of SeV F DNA (wild-type [Fwt] or mutant) and SeV HN DNA. Target RBCs were colabeled with the lipidic dye R18 and the aqueous dye 6CF. At 16 h posttransfection, CV-1 cells were incubated with TPCK-treated trypsin (5 μg/ml) for 1 h at 37°C. Effector CV-1 cells were coincubated with target RBCs for 1 h at 4°C and incubated for 15 min at 37°C to promote dye transfer. The means and standard errors from three microscopic fields are reported. (B) Representative, cropped one-quarter-field images of dye transfer mediated by SeV Fwt or SeV F L179V. Effector CV-1 cells were infected and transfected as described above. At 16 h posttransfection, CV-1 cells were incubated with TPCK-treated trypsin (5 μg/ml) for 1 h at 37°C. Effector CV-1 cells were labeled with the nucleic acid dye SYTO 17 (red). RBC target cells were labeled with 6CF (green) only. Cell-cell fusion was monitored by the transfer of 6CF dye from the RBCs to the CV-1 effector cells (yellow). (C) Temperature dependence of SeV F-mediated dye transfer coexpressed with the SeV HN protein. CV-1 effector cells and RBC target cells were labeled and coincubated as described for panel B. At 16 h posttransfection, CV-1 cells were incubated with TPCK-treated trypsin (5 μg/ml) for 1 h at 37°C. After coincubation of effector and target cells at 4°C for 1 h, the cell complexes were incubated for 15 min at 25, 29, 33, or 37°C. Cell-cell fusion was monitored as described for panel B. Dye transfer events are normalized to SeV Fwt at 37°C. The means and standard errors from four microscopic fields are reported.
Hyperfusogenic mutations cause the F protein to be more readily activated only after HN binds to its receptor.
Mutations in the PIV5 and NDV F proteins that result in hyperfusogenic activity in the presence of HN coexpression and substantial fusogenic activity in the absence of HN coexpression have been discovered (37, 45, 47, 52). Thus, the hyperfusogenic mutations in HRA of the SeV F protein may also have short circuited the natural trigger for F protein activation, i.e., HN binding to its receptor and triggering a conformational change in the F protein (12, 22, 40, 46, 57). We repeated the luciferase and dye transfer assays with the SeV F proteins in the absence of HN coexpression by using uncleaved influenza virus HA at neutral pH as a surrogate receptor-binding protein. However, neither wild-type F protein nor any of the mutants caused membrane fusion without HN coexpression (data not shown).
The simplest mechanism by which the HRA mutations could increase fusogenicity is by destabilizing the spring-loaded conformation of HRA (Fig. 1C, inset) so that it has a lower activation energy barrier to form the coiled-coil structure (Fig. 1D, inset). To indirectly measure the activation energy of the F proteins containing hyperactive mutations (i.e., A178V, L179I, L179V, K180T, T181S, and F185Y), we compared their fusogenicities to that of the wild-type F protein as a function of incubation temperature (Fig. 6C). Wild-type F protein caused efficient dye transfer after incubation with target cells at 37°C but dropped to approximately 25% efficiency after incubation at 33°C. In contrast, the F proteins containing hyperfusogenic mutations caused dye transfer more efficiently at lower temperatures than did the wild-type F protein. In fact, the L179V and F185Y mutants caused dye transfer more efficiently at 25°C than the wild-type F protein did at 37°C.
In most cases, the results from the luciferase and dye transfer assays were roughly equivalent. However, the luciferase activity of the F185Y mutant was similar to that of wild-type F protein, whereas dye transfer caused by the F185Y mutant was almost twice as efficient. Target cells and effector cells were coincubated for 8 h in the luciferase assay and for 15 min in the dye transfer assay. This discrepancy between the phenotypes of the F185Y mutant may reflect fast activation kinetics that saturate over a longer period of time. However, further studies are needed to confirm this hypothesis.
Intracellular cleavage and delayed incubation with target cells do not inactivate F proteins carrying hyperfusogenic mutations.
Unlike the SeV F0 protein, the PIV5 F0 protein is cleaved intracellularly by resident proteases in the secretory pathway (31). Mutation of residue L477 or I449 in the HRB region of the PIV5 F protein to an aromatic residue results in hyperfusogenic fusion activity in assays where the F protein comes into contact immediately with target cells after reaching the cell surface (e.g., syncytium and luciferase assays) (47). However, these same mutations also cause the PIV F protein to become quickly inactivated following intracellular cleavage and delayed incubation with target cells (i.e., in the dye transfer assay). To investigate further the “do-or-die” phenotypes, the L447 and I449 mutations were introduced into the background of a PIV5 F protein that had a modified cleavage site. As a result, the mutant PIV5 F proteins were not cleaved intracellularly but instead were cleaved by exogenous trypsin immediately before incubation with target cells. Extracellular cleavage of the mutant PIV5 F proteins resulted in hyperactive membrane fusion activity in the dye transfer assay similar to that found in the luciferase assay. Thus, if the hyperfusogenic HRA mutations in the SeV F protein studied here have an effect similar to that of the hyperfusogenic HRB mutations in the PIV5 F protein (destabilizing the native F protein structure before HN binds to target cells), then intracellular cleavage of the SeV F protein might also result in do-or-die phenotypes for the SeV F protein mutants in the dye transfer assay.
To investigate whether the mutational effects of the HRA residues were influenced by extracellular cleavage, we generated an SeV F protein in which its cleavage site was mutated to R-Q-K-R (designated Fcl) (see Materials and Methods), which is similar to another SeV F protein construct that is intracellularly cleaved (26). The 10 HRA mutations that did not previously eliminate cell surface expression (Fig. 2B) were then subcloned into the background of the Fcl construct. The Fcl/HRA mutants were cleaved intracellularly similarly to wild-type Fcl, and they were also expressed at levels similar to those when they were in the trypsin-cleavable F background (data not shown). When coexpressed with SeV HN, the Fcl/HRA mutants promoted cell-cell fusion in syncytium, luciferase, and dye transfer assays after intracellular cleavage (Fig. 7), at levels comparable to those obtained after extracellular cleavage (Fig. 5 and 6). The L179V, K180T, T181S, and F185Y mutants in the Fcl background had hyperfusogenic phenotypes, and the A178V and L179I mutants caused fusion at levels similar to or slightly above that caused by the wild-type Fcl. Conversely, the K180G, K180Q, T181A, and V186I mutants in the Fcl background were generally less fusogenic than wild-type Fcl in the three assays. Interestingly, the F185Y mutation in the Fcl background caused a much larger increase in membrane fusion in the dye transfer assay than in the luciferase assay, similar to the effects of the F185Y mutation in the background of the F protein having a wild-type cleavage site. The results from the fusion assays of the Fcl mutants showed that the means by which the SeV F protein is cleaved does not significantly alter the effects of the HRA mutations. Moreover, the hyperfusogenic HRA mutations do not inactivate the SeV F protein before the binding of HN to target cells. Although not conclusive, these results are consistent with the hypothesis that hyperactive mutations enhance the activation of the F protein only after the HN protein triggers F protein refolding, most likely at the step when the HRA region springs into its coiled coil form.
FIG. 7.
Cell-cell fusion activity of SeV Fcl proteins that are cleaved intracellularly. (A) Representative photomicrographs of syncytia formed between BHK-21 cells coexpressing SeV Fcl proteins and SeV HN at 24 h posttransfection. Bar, 200 μm. (B) Cell-cell fusion activities of the SeV Fcl proteins as determined by the luciferase gene reporter assay. Vero cells that expressed wild-type (Fcl) or mutant F proteins and the HN protein and were transfected with luciferase control DNA. Vero cells were overlaid with BSR-T7/5 cells and incubated for 8 h at 37°C. After lysis, the luciferase activity was measured and normalized to 100% fusion corresponding to the luciferase activity of SeV HN and Fcl (∼7 × 105 relative light units [RLU]). Error bars represent two standard deviations from triplicate experiments. Asterisks indicate a significant difference as measured by Student's t test (P < 0.05). (C) Lipid mixing (white bars) and content mixing (black bars) by SeV Fcl proteins. Effector CV-1 cells were infected with vaccinia virus vTF7.3 and transfected with 1 μg each of SeV Fcl DNA (Fcl or mutant) and SeV HN DNA. Target RBCs were colabeled with the lipidic dye R18 and the aqueous dye 6CF. Effector CV-1 cells were coincubated with target RBCs for 1 h at 4°C and incubated for 15 min at 37°C to promote fusion. The means and standard errors from three or four microscopic fields are reported.
DISCUSSION
Upon activation, the paramyxovirus F protein undergoes a structural metamorphosis from a high-energy prefusion conformation to a low-energy hairpin structure, causing fusion of the viral and cellular membranes and allowing penetration of the viral genome into the host cell (32, 48). Central to this irreversible protein refolding process is the conversion of the HRA region from a crumpled, spring-loaded form to an extended, coiled-coil structure (Fig. 1C and D) (7, 63, 64). As has been well documented for the paramyxovirus F protein and other class I viral fusion proteins, nonpolar residues at the a and d positions in the HRA region are important in the formation of the coiled-coil hairpin (18, 19, 31, 55). Mutations to individual a and d residues in HRA also affect the folding and fusogenic activity of intact NDV and PIV5 F proteins (51, 53, 60), a finding that suggests that these residues are important in the formation and activation of the native conformation of the F protein. Many other HRA residues, in addition to a and d residues, are involved in structural interactions that maintain the prefusion conformation of the F protein (64).
To assess the importance of such residues in the functional mechanism of the F protein, we performed a mutagenesis analysis on a 10-residue sequence in HRA of the SeV F protein. This portion of HRA was selected because it is highly conserved, it undergoes a dramatic structural transformation during protein refolding, and it forms a prominent cavity that is both the nucleation site for HRB binding and a target in antiviral drug design. Nine of the 15 conservative mutations that were characterized either reduced or eliminated the expression and fusogenic activity of the SeV F protein. In contrast, the other six mutations induced hyperactive membrane fusion activity. Interestingly, various mutations made at residues K180 and T181, nonconserved residues that were not in a or d positions, either disrupted protein folding or led to hyperactive membrane fusion activity, depending on the substituted amino acid residue. This novel finding shows that residues not expected to be important in the formation of the coiled-coil structure may have crucial roles in regulating functional activity. Overall, these results demonstrate the importance of the entire 10-residue sequence in the folding, processing, and activation of the F protein. These results are also consistent with the hypothesis that residues undergoing significant structural changes during F protein refolding are important in regulating the functional activity of the F protein.
The available structural and biochemical data are consistent with the following sequence of events occurring after the F protein is triggered by the receptor-binding protein (48, 64): first, the HRB coiled coil melts, pulling the transmembrane domains apart; second, the crumpled HRA structure extends into a coiled coil, propelling the fusion peptide into the target membrane; and finally, the HRB segments swing around the base of the head formed by DI and DII, thereby binding to the HRA coiled coil and forming the fusogenic hairpin structure. The hyperfusogenic SeV F mutations studied here could, therefore, affect any or all of these structural transitions. The direct effects of the HRA mutations on F protein conformational changes have not yet been investigated. However, the location of the mutations and the low-temperature activation are consistent with their predominant effects occurring during the second step, the springing of the HRA coil.
After cleavage, the first step in F protein refolding is thought to involve a conformational change in HRB (46). Moreover, a peptide derived from HRB interacts physically with a peptide derived from the stalk region of the HN receptor-binding protein (22). Mutations to residues 447 and 449 in HRB cause the PIV5 F protein to become activated at lower temperatures, to be more easily triggered in the absence of the HN protein, and to become inactivated after delayed incubation with target cells in a dye transfer assay (47). In the prefusion structure of the PIV5 F protein, residues 447 and 449 interact with the F protein head at the nucleation site of the HRB coiled coil (64). Therefore, the mutations in HRB appear to affect the first step in F protein refolding, i.e., the melting of the HRB coil. Although the HRA mutations studied here also cause the SeV F protein to become activated at lower temperatures, they do not result in membrane fusion activity in the absence of HN coexpression, and they do not lead to F protein inactivation after intracellular cleavage and delayed incubation with target cells in the dye transfer assay. Thus, the hyperfusogenic HRA mutations in the SeV F protein do not appear to enhance F protein activation until after the HN protein is bound to target cells. The data are also inconsistent with hyperfusogenic HRA mutations alleviating a potential inefficiency by the wild-type SeV F protein during late steps in membrane fusion. Neither wild-type F protein nor any of the mutants studied here have a hemifusion phenotype. The hyperfusogenic HRA mutations could perhaps stabilize the hairpin structure, thereby coupling more energy to membrane fusion. Future biophysical studies directly measuring the effects of the HRA mutations on the thermostability of the coiled-coil hairpin could rule out this possibility. However, previous studies on HR-derived peptides from the PIV5 F protein have shown that mutations to HRA and HRB residues generally do not increase coiled-coil stability (17, 47).
An L161M mutation in HRA of the PIV5 F protein causes normal expression and fusogenicity at 37°C but results in enhanced fusogenicity at 30°C (60). Residue L161 is located three amino acid residues upstream of the 10-residue sequence in the SeV F protein studied here. In the prefusion structure of the PIV5 F protein (64), the L161 residue is located in a β-strand that may also include SeV F protein residues A178 and L179, if analogous residues in the F proteins of PIV5 and SeV both form similar β-strands in their prefusion forms. These results suggest that destabilizing the β-strands in the prefusion structure of HRA decreases the activation energy barrier of the F protein, thereby resulting in increased membrane fusion activity at lower temperatures. Conversely, stabilizing the β-strands and turns in HRA may perhaps favor the prefusion structure and consequently increase the activation energy barrier. Just as residues in the HRA region of the paramyxovirus F protein play an important role in regulating protein folding and membrane fusion, residues in and around the helix-loop-helix structure in the HRA region of the influenza virus HA protein also regulate its activation (13). Moreover, proline mutations predicted to stabilize the spring-loaded conformation of the HA protein prevent its conformational rearrangement and functional activity (24, 43).
Membrane fusion inhibitors that block virus replication can be clinically effective. For example, an HRB-derived peptide from HIV gp41 (T-20, Enfuvirtide) has been successfully used in drug combination therapies against HIV infection (23). HRB-derived peptides and small-molecule mimetics are also potent inhibitors of F protein-mediated membrane fusion and virus replication (3, 11, 33, 39, 46, 62). HRB-derived peptides block membrane fusion by binding to the HRA coiled coil in a prehairpin intermediate of the F protein, consequently preventing formation of the fusogenic hairpin (47). A small molecule designed to bind a prominent cavity in the HRA coiled coil inhibits replication of respiratory syncytial virus (RSV) (11). A similar potential drug target exists in the HRA regions of the parainfluenza viruses, including hPIV1, whose residues are identical to those in SeV that were mutated in this study. Some conservative mutations to HRA drug target residues might allow for efficient virus replication, just as they allowed efficient membrane fusion after expression from plasmid DNA in the present study. If such is the case, then resistance mutations could potentially arise in the HRA region of the F protein, analogous to those in the HRA region of HIV gp41 resulting from T-20 treatment (2, 34, 44). Previous studies on a small-molecule inhibitor that binds to the HRA region of the RSV F protein have shown that a K394R mutation in DII of the F protein can also lead to resistance (10). Although the mechanism of resistance has not yet been determined, we believe that the mutation most likely alters the kinetics of accessibility of the HRA coiled coil in the prehairpin intermediate form of the F protein. Future studies characterizing the effects of F protein mutations on virus replication by Sendai virus, RSV, and other paramyxoviruses will be needed to explore the importance of HRA residues and other F protein residues in virus pathogenesis and susceptibility to fusion inhibitors.
Prefusion structures have been determined for two class I viral fusion proteins, the influenza virus HA protein (61) and the paramyxovirus F protein (64). In the HA protein, HRA forms a helix-loop-helix structure. In the F protein, HRA forms an 11-segment structure consisting of α-helices, β-strands, and turns. In both structures, the HRA region is in a spring-loaded conformation, which helps position the fusion peptide on the side of the molecule until the fusion protein becomes activated. Just as the HRA residues in the SeV F protein studied here play an important role in regulating expression and fusogenicity, residues in and around the helix-loop-helix structure in the HA protein also regulate its activation (13). Based on similarities between paramyxovirus F and influenza virus HA, class I viral fusion proteins may, in general, be expected to regulate protein expression and activation with HRA residues that are spring loaded in the prefusion form. Mutations to conserved HRA residues in HIV gp41, analogous to those in SeV F studied here, are accommodated in the hairpin structure of gp41 (29). However, these same mutations eliminate membrane fusion activity by gp41 (5), a finding that suggests that the HRA mutations may compromise other conformations of the protein. The importance of HRA residues in the membrane fusion activities of HIV gp160 (5, 8, 15, 59), Ebola virus GP (58), and severe acute respiratory syndrome coronavirus S (38) may perhaps suggest that the HRA regions of class I viral fusion proteins may, in general, undergo significant conformational changes during protein refolding. However, determinations of the prefusion structures of these proteins are needed to confirm this hypothesis and reveal whether their HRA regions also adopt spring-loaded conformations analogous to those found in the prefusion structures of paramyxovirus F and influenza virus HA. Ultimately, further structural, biochemical, and virological studies will be needed to understand better how regulated conformational changes by class I viral fusion proteins contribute to virus replication and pathogenesis.
Acknowledgments
We thank Nancy Hutson and Jean-Baptiste Penigault for help with molecular cloning. We thank Robert Sealy and Julia Hurwitz for the generation of the Sendai virus F tail monoclonal antibody. We thank Allen Portner for the Sendai virus plasmids and the M16 monoclonal antibody. We thank Robert Lamb for the pGEM3X vector and the pCAGGS influenza virus Udorn HA plasmid. We thank Karl-Klaus Conzelmann for the BSR-T7/5 cells and Bernard Moss for the vTF7.3 vaccinia virus. We thank Angela McArthur for editing the manuscript.
This work was supported by a Cancer Center support grant (CA 21765), the American Lebanese Syrian Associated Charities (ALSAC), and the Children's Infection Defense Center (CIDC) at St. Jude Children's Research Hospital.
Footnotes
Published ahead of print on 24 January 2007.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, C. E., R. W. Sanders, Y. Deng, S. Jurriaans, J. M. Lange, M. Lu, and B. Berkhout. 2004. Emergence of a drug-dependent human immunodeficiency virus type 1 variant during therapy with the T20 fusion inhibitor. J. Virol. 78:12428-12437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossart, K. N., B. A. Mungall, G. Crameri, L.-F. Wang, B. T. Eaton, and C. C. Broder. 2005. Inhibition of Henipavirus fusion and infection by heptad-derived peptides of the Nipah virus fusion glycoprotein. Virol. J. 2:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao, J., L. Bergeron, E. Helseth, M. Thali, H. Repke, and J. Sodroski. 1993. Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J. Virol. 67:2747-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Cambridge) 9:255-266. [DOI] [PubMed] [Google Scholar]
- 8.Chen, S. S., C. N. Lee, W. R. Lee, K. McIntosh, and T. H. Lee. 1993. Mutational analysis of the leucine zipper-like motif of the human immunodeficiency virus type 1 envelope transmembrane glycoprotein. J. Virol. 67:3615-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chernomordik, L. V., and M. M. Kozlov. 2003. Protein-lipid interplay in fusion and fission of biological membranes. Annu. Rev. Biochem. 72:175-207. [DOI] [PubMed] [Google Scholar]
- 10.Cianci, C., N. Meanwell, and M. Krystal. 2005. Antiviral activity and molecular mechanism of an orally active respiratory syncytial virus fusion inhibitor. J. Antimicrob. Chemother. 55:289-292. [DOI] [PubMed] [Google Scholar]
- 11.Cianci, C., K. L. Yu, K. Combrink, N. Sin, B. Pearce, A. Wang, R. Civiello, S. Voss, G. Luo, K. Kadow, E. V. Genovesi, B. Venables, H. Gulgeze, A. Trehan, J. James, L. Lamb, I. Medina, J. Roach, Z. Yang, L. Zadjura, R. Colonno, J. Clark, N. Meanwell, and M. Krystal. 2004. Orally active fusion inhibitor of respiratory syncytial virus. Antimicrob. Agents Chemother. 48:413-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corey, E. A., A. M. Mirza, E. Levandowsky, and R. M. Iorio. 2003. Fusion deficiency induced by mutations at the dimer interface in the Newcastle disease virus hemagglutinin-neuraminidase is due to a temperature-dependent defect in receptor binding. J. Virol. 77:6913-6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daniels, R. S., J. C. Downie, A. J. Hay, M. Knossow, J. J. Skehel, M. L. Wang, and D. C. Wiley. 1985. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell 40:431-439. [DOI] [PubMed] [Google Scholar]
- 14.Debnath, A. K. 2006. Prospects and strategies for the discovery and development of small-molecule inhibitors of six-helix bundle formation in class 1 viral fusion proteins. Curr. Opin. Investig. Drugs 7:118-127. [PubMed] [Google Scholar]
- 15.Dubay, J. W., S. J. Roberts, B. Brody, and E. Hunter. 1992. Mutations in the leucine zipper of the human immunodeficiency virus type 1 transmembrane glycoprotein affect fusion and infectivity. J. Virol. 66:4748-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dutch, R. E., G. P. Leser, and R. A. Lamb. 1999. Paramyxovirus fusion protein: characterization of the core trimer, a rod-shaped complex with helices in anti-parallel orientation. Virology 254:147-159. [DOI] [PubMed] [Google Scholar]
- 18.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 20.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 22.Gravel, K. A., and T. G. Morrison. 2003. Interacting domains of the HN and F proteins of Newcastle disease virus. J. Virol. 77:11040-11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenberg, M., N. Cammack, M. Salgo, and L. Smiley. 2004. HIV fusion and its inhibition in antiretroviral therapy. Rev. Med. Virol. 14:321-337. [DOI] [PubMed] [Google Scholar]
- 24.Gruenke, J. A., R. T. Armstrong, W. W. Newcomb, J. C. Brown, and J. M. White. 2002. New insights into the spring-loaded conformational change of influenza virus hemagglutinin. J. Virol. 76:4456-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, Y., R. Vassell, M. Zaitseva, N. Nguyen, Z. Yang, Y. Weng, and C. D. Weiss. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 77:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heminaway, B. R., Y. Yang, Y. Tanaka, M. Panin, Y. T. Huang, and M. S. Galinski. 1995. Role of basic residues in the proteolytic activation of Sendai virus fusion glycoprotein. Virus Res. 36:15-35. [DOI] [PubMed] [Google Scholar]
- 27.Horvath, C. M., and R. A. Lamb. 1992. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J. Virol. 66:2443-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ito, M., M. Nishio, H. Komada, Y. Ito, and M. Tsurudome. 2000. An amino acid in the heptad repeat 1 domain is important for the haemagglutinin-neuraminidase-independent fusing activity of simian virus 5 fusion protein. J. Gen. Virol. 81:719-727. [DOI] [PubMed] [Google Scholar]
- 29.Ji, H., W. Shu, F. T. Burling, S. Jiang, and M. Lu. 1999. Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion. J. Virol. 73:8578-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph 14:51-55, 29-32. [DOI] [PubMed] [Google Scholar]
- 31.Lamb, R. A., and D. Kolakofsky (ed.). 2001. Paramyxoviridae: the viruses and their replication, 4th ed., vol. 1. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 32.Lamb, R. A., R. G. Paterson, and T. S. Jardetzky. 2006. Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. R. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 78:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paterson, R. G., and R. A. Lamb. 1993. The molecular biology of influenza viruses and paramyxoviruses, p. 35-73. In A. Davidson and R. M. Elliott (ed.), Molecular virology: a practical approach. IRL Oxford University Press, Oxford, United Kingdom.
- 37.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 38.Petit, C. M., J. M. Melancon, V. N. Chouljenko, R. Colgrove, M. Farzan, D. M. Knipe, and K. G. Kousoulas. 2005. Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion. Virology 341:215-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porotto, M., L. Doctor, P. Carta, M. Fornabaio, O. Greengard, G. E. Kellogg, and A. Moscona. 2006. Inhibition of Hendra virus fusion. J. Virol. 80:9837-9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porotto, M., M. Murrell, O. Greengard, L. Doctor, and A. Moscona. 2005. Influence of the human parainfluenza virus 3 attachment protein's neuraminidase activity on its capacity to activate the fusion protein. J. Virol. 79:2383-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Portner, A., R. A. Scroggs, and C. W. Naeve. 1987. The fusion glycoprotein of Sendai virus: sequence analysis of an epitope involved in fusion and virus neutralization. Virology 157:556-559. [DOI] [PubMed] [Google Scholar]
- 42.Qiao, H., R. T. Armstrong, G. B. Melikyan, F. S. Cohen, and J. M. White. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell 10:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiao, H., S. L. Pelletier, L. Hoffman, J. Hacker, R. T. Armstrong, and J. M. White. 1998. Specific single or double proline substitutions in the “spring-loaded” coiled-coil region of the influenza hemagglutinin impair or abolish membrane fusion activity. J. Cell Biol. 141:1335-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reeves, J. D., F. H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79:4991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2004. Conserved glycine residues in the fusion peptide of the paramyxovirus fusion protein regulate activation of the native state. J. Virol. 78:13727-13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell, C. J., K. L. Kantor, T. S. Jardetzky, and R. A. Lamb. 2003. A dual-functional paramyxovirus F protein regulatory switch segment: activation and membrane fusion. J. Cell Biol. 163:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell, C. J., and L. E. Luque. 2006. The structural basis of paramyxovirus invasion. Trends Microbiol. 14:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheid, A., and P. W. Choppin. 1974. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology 57:475-490. [DOI] [PubMed] [Google Scholar]
- 50.Segawa, H., T. Yamashita, M. Kawakita, and H. Taira. 2000. Functional analysis of the individual oligosaccharide chains of Sendai virus fusion protein. J. Biochem. (Tokyo) 128:65-72. [DOI] [PubMed] [Google Scholar]
- 51.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2001. Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J. Virol. 75:7934-7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sergel, T. A., L. W. McGinnes, and T. G. Morrison. 2000. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J. Virol. 74:5101-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sergel-Germano, T., C. McQuain, and T. Morrison. 1994. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J. Virol. 68:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skehel, J. J., K. Cross, D. Steinhauer, and D. C. Wiley. 2001. Influenza fusion peptides. Biochem. Soc. Trans. 29:623-626. [DOI] [PubMed] [Google Scholar]
- 55.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 56.Takimoto, T., K. G. Murti, T. Bousse, R. A. Scroggs, and A. Portner. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J. Virol. 75:11384-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takimoto, T., G. L. Taylor, H. C. Connaris, S. J. Crennell, and A. Portner. 2002. Role of the hemagglutinin-neuraminidase protein in the mechanism of paramyxovirus-cell membrane fusion. J. Virol. 76:13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Watanabe, S., A. Takada, T. Watanabe, H. Ito, H. Kida, and Y. Kawaoka. 2000. Functional importance of the coiled-coil of the Ebola virus glycoprotein. J. Virol. 74:10194-10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.West, D. S., M. S. Sheehan, P. K. Segeleon, and R. E. Dutch. 2005. Role of the simian virus 5 fusion protein N-terminal coiled-coil domain in folding and promotion of membrane fusion. J. Virol. 79:1543-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 62.Yao, Q., and R. W. Compans. 1996. Peptides corresponding to the heptad repeat sequence of human parainfluenza virus fusion protein are potent inhibitors of virus infection. Virology 223:103-112. [DOI] [PubMed] [Google Scholar]
- 63.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]