Abstract
Hepatitis E virus (HEV) is a positive-strand RNA virus that is prevalent in much of the developing world. ORF2 is the major capsid protein of HEV. Although ORF2 is an N-linked glycoprotein, it is abundantly located in the cytoplasm in addition to having membrane and surface localization. The mechanism by which ORF2 protein obtains access to the cytoplasm is unknown. In this report, we prove that initially all ORF2 protein is present in the endoplasmic reticulum and a fraction of it becomes retrotranslocated to the cytoplasm. The ability of ORF2 to be retrotranslocated is dependent on its glycosylation status and follows the canonical dislocation pathway. However, in contrast to general substrates of the dislocation pathway, retrotranslocated ORF2 protein is not a substrate of the 26S proteasome complex and is readily detectable in the cytoplasm in the absence of any protease inhibitor, suggesting that the retrotranslocated protein is stable in the cytoplasm. This study thus defines the pathway by which ORF2 obtains access to the cytoplasm.
Cells are equipped with various checkpoints and quality control mechanisms to maintain proper homeostasis, which is essential for their survival and proliferation. One of the most critical quality control mechanisms operates in the endoplasmic reticulum (ER); this mechanism ensures that nascent proteins are properly folded and only proteins with a biologically active conformation are allowed to move beyond the ER (2). Improperly folded or unassembled proteins are retrotranslocated from the ER and degraded by a process called ER-associated degradation (ERAD) (20). This mechanism also helps the cell alleviate ER overload in case of viral infection, whereby expression of viral proteins creates an extra burden over the cell organelle (6). Pathogens too have evolved strategies to exploit the retrotranslocation pathway to degrade host molecules in order to create a favorable environment for them. For example, the US11 gene product of human cytomegalovirus virus induces ERAD of major histocompatibility complex class I molecules to evade the host immune response (19). The retrotranslocation pathway has also been used by toxic proteins of several extracellular pathogens to gain access to the cytosol. Notable examples of these are cholera toxin, ricin, and AB toxins (3, 11, 16). Once in the cytosol, these proteins play strategic roles to further interfere with the cellular machinery.
Hepatitis E virus (HEV) is the causative agent of viral hepatitis, which is a major concern for much of the developing world. The virus carries a positive-strand RNA as its genome, which carries three known open reading frames (ORFs). While ORF1 codes for nonstructural proteins essential for viral replication, ORF2 codes for the major capsid protein which is predicted to encapsulate the genome, and ORF3 codes for a small phosphoprotein whose in vivo function is yet to be defined. However, in cell culture-based assays, it has been shown to perturb various cellular signaling intermediates, such as inhibiting mitogen-activated protein kinase (MAPK) activity (5, 7), and it enhances the secretion of an immunosuppressant called α1-microglobulin (14, 18).
ORF2 is an N-linked glycoprotein which is cotranslationally translocated into the ER (23). It is glycosylated at three amino acid residues, as judged by mutational analysis. A fraction of it is also observed on the cell surface. Despite being cotranslationally translocated into the ER, ORF2 has been found to exist as both glycosylated and nonglycosylated forms in the total cell lysate, and a significant fraction of it is also observed in the cytoplasm (15). The mechanism behind this phenomenon remains unknown.
In this article, we describe the pathway by which ORF2 protein localizes to the cytoplasm. Heterologous expression of ORF2 protein results in its accumulation in the ER initially. Gradually, a fraction of this protein is translocated back to the cytoplasm. This retrotranslocation could be blocked by treatment of cells with various biochemical and genetic inhibitors of the ER-associated degradation pathway, thus suggesting that it may be a canonical ERAD substrate. However, ORF2 protein is not a substrate of the 26S proteasome. Rather, retrotranslocated ORF2 protein is stable in the cytoplasm, indicating that it mimics an ERAD substrate to get access to the cytoplasm. In addition, the protein levels of several ER stress-regulated chaperones were found to be upregulated, suggesting that ER stress was induced in ORF2-expressing cells.
MATERIALS AND METHODS
Plasmids and reagents.
Cloning of pSGI ORF2 and pSGI 35 ORF2 has been described earlier (4, 23). pSGI KDEL ORF2 was prepared by modifying the native C-terminal sequence of ORF2, KTREL, to KDEL by site-directed mutagenesis. The clone was verified by sequencing. Wild-type and ATPase-dead p97 expression constructs (pCDNA 3.1 vector) (21) were gifts from T. A. Rapoport. All the DNA constructs used for mammalian cell transfection were purified by cesium chloride gradient centrifugation (10). All antibodies except those for ORF2 and ORF3 were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). ORF2 and ORF3 antibodies were raised in our laboratory and have been described earlier (4). MG132 and kifunensine were purchased from Calbiochem Chemicals (San Diego, CA). Tunicamycin, cycloheximide, NH4Cl, and monensin were from Sigma-Aldrich (St. Louis, MO).
Cell culture and transfection.
Huh7 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with penicillin, streptomycin, and 10% fetal bovine serum. Cells were transfected with Lipofectin/Lipofectamine 2000 reagent (Invitrogen Corp., Carlsbad, CA) as per the manufacturer's instructions. For mock transfection, cells were transfected with the respective empty vectors.
Metabolic labeling, immunoprecipitation, and immunoblotting.
Radiolabeling of cells with [35S]Cys/Met promix, immunoprecipitation, and immunoblotting were done as described by Surjit et al. (13). The data reported are representative of at least three independent sets of experiments conducted. All the inhibitors were added during the starvation period and maintained throughout the labeling period. The effective concentration of the different inhibitors are as follows: MG132, 50 μM; cycloheximide, 100 μg/ml; tunicamycin, 10 μg/ml; monensin, 5 μM; NH4Cl, 30 mM; and kifunensine, 0.2 mM.
Pulse-chase assay.
At 44 h posttransfection, cells transfected in 60-mm culture dishes were pulse-labeled for 20 min with 250 μCi of [35S]Cys/Met labeling mix mixed with 1 ml Cys/Met-deficient DMEM and chased in complete medium (DMEM containing Cys/Met and serum) for the indicated time periods, followed by immunoprecipitation, as described by Surjit et al. (12). The approximate cell density at the time of labeling was 85 to 90%.
Membrane fractionation.
Cytoplasmic proteins were separated from the membrane fraction by digitonin permeabilization of the plasma membrane as described by Afshar et al. (1). Membrane fractionation by ultracentrifugation was done as described by Surjit et al. (12).
RESULTS
A fraction of the ORF2 protein is retrotranslocated from the ER to the cytoplasm.
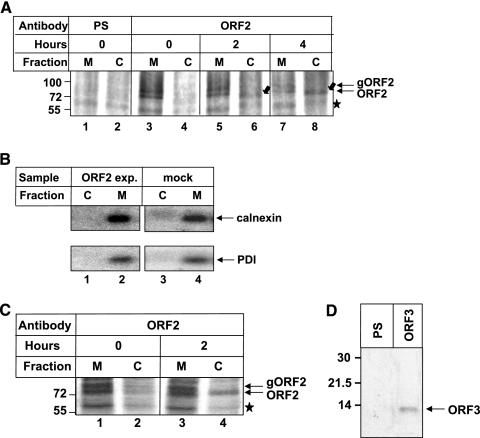
In an attempt to understand the mechanism of the intracellular distribution of the ORF2 protein, we systematically checked its appearance in the cytoplasmic fraction after entry into the ER. There are two possible ways for an N-linked glycoprotein to gain access to the cytoplasm: (i) by direct retrotranslocation from the ER to the cytoplasm or (ii) through retrograde transport from the cell surface. To test the first possibility, we blocked protein transport to the cell surface by treating cells with monensin, pulse-labeled the ORF2 protein with [35S]Cys/Met promix, separated cytoplasmic and membrane fractions from the cell lysate, and examined each fraction for the presence of the ORF2 protein at different time points. Cytoplasmic proteins were released from the cells by permeabilizing the plasma membrane with digitonin, which has earlier been proved to maintain the integrity of the ER membrane (1). As seen in Fig. 1A, at time zero of chase, the membrane fraction contained two bands specific to the ORF2 protein, which correspond to glycosylated and nonglycosylated types of ORF2 protein. No ORF2 protein was detected in the cytoplasm at that time point. However, ORF2 presence in the cytoplasm gradually increased with time (compare 2 h to 4 h; Fig. 1A, lanes 6 to 8). These cells were also treated with monensin to block anterograde transport beyond the Golgi. This experiment indicated that initially all ORF2 protein is translocated to the ER compartment and a fraction of it is retrotranslocated back to the cytoplasm. Cross contamination between ER and cytoplasm fractions was ruled out by immunoprecipitating aliquots of cell lysate with antibodies to calnexin and protein disulfide isomerase (PDI), which are known to be membrane associated and soluble ER resident proteins, respectively (Fig. 1B). Next, we checked whether ORF2 is able to retrotranslocate in the presence of other viral proteins. For this, we coexpressed ORF2 along with ORF3 protein, which is a small phosphoprotein produced by hepatitis E virus. These cells were pulse-labeled for 20 min, followed by a 2-h chase in complete medium and immunoprecipitation of ORF2 from the membrane and cytoplasmic fractions. As expected, a fraction of ORF2 protein was detected in the cytoplasmic fraction after the 2-h chase period (Fig. 1C). Expression of ORF3 protein in these samples was verified by immunoprecipitating an aliquot of the sample with anti-ORF3 antibody (Fig. 1D).
FIG. 1.
A fraction of the ORF2 protein is retrotranslocated from the ER to the cytoplasm. (A) Full-length ORF2-expressing cells were treated with monensin, pulse-labeled for 20 min with [35S]cysteine/methionine labeling mix, and harvested (lanes 1 to 4) or chased for 2 or 4 h in complete medium (lane 5 to 8). Cytoplasmic (C) and membrane (M) fractions were separated by digitonin permeabilization and immunoprecipitated with preimmune serum (lanes 1 and 2) or anti-ORF2 antibody (lane 3 to 8), and radioactive bands were visualized by fluorography. The star indicates a nonspecific band present in all samples. Block arrows indicate the ORF2 band in the cytoplasm fraction, and line arrows indicate ORF2 bands (glycosylated [gORF2] and nonglycosylated [ORF2]). (B) ORF2-expressing (lanes 1 and 2) or mock (lanes 3 and 4) cell lysates processed simultaneously and immunoprecipitated with anticalnexin (upper panel) or anti-PDI (lower panel) antibody. (C) Huh7 cells cotransfected with full-length ORF2 and ORF3 expression plasmids were treated with monensin, pulse-labeled for 20 min with [35S]cysteine/methionine labeling mix, and harvested (lanes 1 and 2) or chased for 2 h in complete medium (lanes 3 and 4). Cytoplasmic and membrane fractions were separated by digitonin permeabilization and immunoprecipitated with anti-ORF2 antibody, and bands were detected as described above. (D) Huh7 cells cotransfected with full-length ORF2 and ORF3 expression plasmids were treated with monensin, pulse-labeled for 20 min with [35S]cysteine/methionine labeling mix, harvested, and immunoprecipitated with preimmune serum (PS) (lane 1) or anti-ORF3 antibody (lane 2). Samples were resolved in 15% sodium dodecyl sulfate-polyacrylamide gels and bands detected by fluorography.
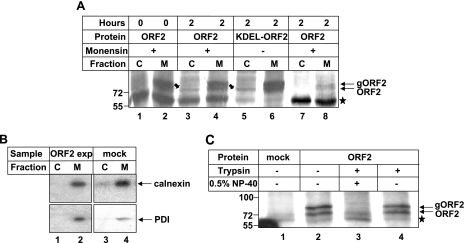
The ability of the ORF2 protein to retrotranslocate from the ER was further verified by using a different approach to separate cytoplasmic and membrane fractions, i.e., separating both fractions by ultracentrifugation. In addition to monensin-treated wild-type ORF2-expressing cells, we also simultaneously processed KDEL-ORF2-expressing cells to further ensure that anterograde trafficking of ORF2 is blocked. KDEL signature residues are frequently present at the C termini of many ER resident proteins, which are responsible for their ER retention. We modified the KTREL sequence present at the C terminus of ORF2 protein to KDEL to ensure that this protein does not move beyond the ER by the anterograde transport pathway (data not shown). Thus, by using this mutant, we would be able to further confirm that any ORF2 protein detectable in the cytoplasm comes directly from the ER compartment and not through any other pathway. This experiment revealed identical results (Fig. 2A), whereby both wild-type and KDEL-ORF2 proteins appeared in the cytoplasmic fraction after the 2-h chase period. Corresponding controls were used to rule out the possibility of cross contamination (Fig. 2B). Further, to prove that the ORF2 protein present in the membrane fraction actually represented the ORF2 protein inside the ER compartment, we conducted a trypsin protection assay. Aliquots of the membrane fraction of ORF2-expressing cells were incubated with 100 μg/ml trypsin for 30 min in either the presence or absence of 0.5% NP-40. In the absence of detergent, the ORF2 band could be detected, whereas in samples treated with detergent, no band or a very faint band specific to ORF2 was detected (Fig. 2C). This proved that the ORF2 protein present in the membrane fraction represents the protein inside the ER compartment. Thus, it was clear that a fraction of the ORF2 protein is directly translocated from the ER to the cytoplasm.
FIG. 2.
A fraction of the ORF2 protein is retrotranslocated from the ER to the cytoplasm. (A) Full-length ORF2-expressing cells were pulse-labeled for 20 min with [35S]cysteine/methionine labeling mix and harvested (lanes 1 and 2) or chased for 2 h in complete medium in the presence (lanes 1, 2, 3, 4, 7, and 8) or absence (lanes 5 and 6) of monensin. Cytoplasmic (C) and membrane (M) fractions were separated by ultracentrifugation and immunoprecipitated with anti-ORF2 antibody (lanes 1 to 6) or preimmune serum (lanes 7 and 8), and radioactive bands were visualized by fluorography. The star indicates a nonspecific band present in all samples. Block arrows and line arrows indicate cytoplasmic ORF2 and total ORF2 (both glycosylated [gORF2] and nonglycosylated) bands, respectively. (B) ORF2-expressing (lanes 1 and 2) or mock (lanes 3 and 4) cell lysates processed simultaneously and immunoprecipitated with anticalnexin (upper panel) or anti-PDI (lower panel) antibody. (C) Huh7 cells were transfected with empty vector (mock, lane 1) or with pSGI ORF2 expression plasmid (lanes 2 to 4). [35S]Cys/Met promix-labeled cells were harvested, and membrane fraction isolated and immunoprecipitated with anti ORF2 antibody. Equal aliquots of the sample were treated with trypsin (lane 4) or trypsin and NP-40 (lane 3) or were not treated (lane 2). Samples were resolved by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and bands detected by fluorography.
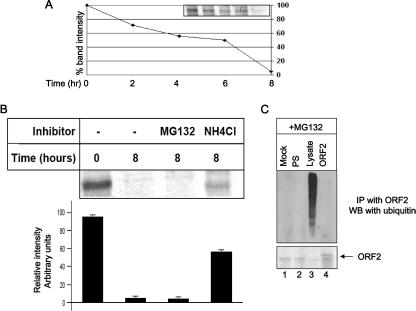
In the normal course of events, retrotranslocated substrates display a very short half-life in the cytoplasm since they are very efficiently degraded by the 26S proteasome (17). However, the ORF2 protein was stably present in the cytoplasm even after a 4-h chase period. Hence, we subsequently designed experiments to check whether ORF2 was a substrate of the 26S proteasome. A pulse-chase assay revealed that, at 8 h of chase, all the pulse-labeled ORF2 protein was completely degraded (Fig. 3A). Next, we checked the effect of lysosomal (NH4Cl) and proteasomal (MG132) inhibitors on the stability of the ORF2 protein. At 8 h of chase, MG132 had no effect; however, NH4Cl could stabilize approximately 60% of the total ORF2 protein (Fig. 3B). This suggested that ORF2 degradation was independent of proteasomal activity. This observation was further confirmed by checking for ubiquitination of the ORF2 protein. For this purpose, ORF2-expressing cells were treated with MG132 for 3 h, followed by immunoprecipitation using anti-ORF2 antibody and immunoblotting with antiubiquitin antibody. We were unable to detect any ubiquitinated species of the ORF2 protein (Fig. 3C). Thus, it was confirmed that the ORF2 protein is not a proteasome substrate and hence that the classical ERAD pathway is not responsible for its degradation.
FIG. 3.
The ORF2 protein is not a substrate of proteasome and is not ubiquitinated. (A) ORF2-expressing cells were pulse-labeled for 20 min with [35S]Cys/Met promix, chased for the indicated time period in complete medium, and immunoprecipitated with anti-ORF2 antibody, and radioactive bands were visualized by fluorography. Band intensities were quantified using the NIH Image program and plotted. The band intensity at time zero was treated as 100%. (B) ORF2-expressing cells were pulse-labeled for 20 min with [35S]Cys/Met promix, chased for 8 h in complete medium in the presence of the indicated inhibitor, and immunoprecipitated with anti-ORF2 antibody, and radioactive bands were visualized by fluorography. Band intensities were quantified using the NIH Image program and plotted. Data represent means ± standard errors of the means from three independent sets of experiments. (C) [35S]Cys/Met promix-labeled mock (lane 1) or ORF2 (lanes 2 and 4)-expressing cells were immunoprecipitated (IP) with anti-ORF2 antibody and immunoblotted (WB) with antiubiquitin antibody. Bands were detected by the ECL method. Lane 3 represents total cell lysate. All samples were maintained with MG132 for 3 h prior to lysis. PS, preimmune serum-immunoprecipitated sample. The lower panel shows ORF2 expression in the same gel.
ORF2 protein exploits the ERAD pathway to enter the cytoplasm.
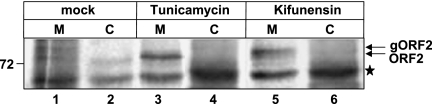
To gain further insight into the mechanism of retrotranslocation of the ORF2 protein, we utilized some known biochemical and dominant negative inhibitors of the ERAD pathway and monitored the appearance of ORF2 in the cytoplasmic fraction by pulse-chase analysis. We used two biochemical inhibitors, tunicamycin and kifunensine. Tunicamycin blocks N-linked glycosylation of proteins by inhibiting the transfer of N-acetylglucosamine 1-phosphate to dolichol monophosphate. Kifunensine is an inhibitor of ER mannosidase-1. In a 2-h chase of the ORF2 protein in tunicamycin-treated cells, no ORF2 protein was detected in the cytoplasmic fraction, indicating that ORF2 dislocation was dependent on the glycosylation status of the protein (Fig. 4, lanes 3 and 4). The fact that tunicamycin was active was reflected by the faster migration of ORF2 as a single band (lane 3). Similarly, treatment of cells with kifunensine blocked ORF2 dislocation (lane 6). Lanes 1 and 2 of Fig. 4 show control cells that were treated with vehicle only.
FIG. 4.
Biochemical inhibitors of the ERAD pathway block ORF2 retrotranslocation. Full-length ORF2-expressing cells were pulse-chased for 2 h in the presence of vehicle only (lanes 1 and 2) or the indicated inhibitor. Membrane (M) and cytoplasmic (C) fractions were isolated and immunoprecipitated with anti-ORF2 antibody, and radioactive bands were visualized by fluorography. The star denotes a nonspecific band pulled down by protein A-Sepharose.
Next, we tested whether ORF2 retrotranslocation is mediated in a p97-dependent manner, which has been shown to be crucial for retrotranslocation (9, 21). First, we looked for interaction of ORF2 with p97 in a coimmunoprecipitation assay. Both the full-length ORF2 and KDEL-ORF2 proteins were able to interact with p97 (Fig. 5A, upper panel, lanes 2 and 3). The interaction was further confirmed by immunoprecipitation from an aliquot of the same sample with anti-p97 antibody and immunoblotting with anti-ORF2 antibody (Fig. 5A, middle panel). The same blot was stripped and probed with anti-p97 to check for p97 levels (Fig. 5A, lower panel). Having shown that p97 can coprecipitate ORF2, we checked whether a dominant negative mutant of the p97 protein could block ORF2 retrotranslocation. Full-length ORF2 and the KDEL-ORF2 protein were expressed along with myc-tagged wild-type or dominant negative p97 (QQ-p97). The appearance of ORF2 in the cytoplasm was monitored by pulse-chase analysis as described above. Cells expressing full-length ORF2 were also treated with monensin to block anterograde transport. As expected, QQ-p97 could block dislocation of both full-length and KDEL-ORF2 (Fig. 5B, lanes 4 and 8). Aliquots of the total cell lysate were immunoblotted using anti-myc (9E10) antibody to check the expression of wild-type and dominant negative p97 (data not shown). Hence, it was confirmed that a fraction of the ORF2 protein was retrotranslocated from the ER to the cytoplasm in a p97-dependent manner.
FIG. 5.
ORF2 retrotranslocation is dependent on p97 activity. (A) Full-length ORF2- or KDEL-ORF2-expressing cell lysate (lanes 1, 2 and 3) or control cell lysate (lane 4) was immunoprecipitated with preimmune serum (PS) (lane 1), anti-ORF2 antibody (lanes 2 and 3), or anti-p97 antibody (lane 4) and immunoblotted with anti-p97 antibody (upper panel). Mock (lane 1)-, ORF2 (lanes 2 and 4)-, or KDEL-ORF2 (lane 3)-transfected cells were immunoprecipitated with anti-p97 antibody (lanes 1, 2, and 3) or anti-ORF2 antibody (lane 4) and immunoblotted with anti-ORF2 antibody (middle panel). The same blot was stripped and probed with anti-p97 antibody (lower panel). (B) Huh7 cells were cotransfected with plasmids expressing full-length (fl) ORF2 or KDEL-ORF2 and wild-type p97 (WT P97) or mutant P97 (QQ P97) and pulse-chased for 2 h in the presence or absence of monensin as indicated. Membrane (M) and cytoplasmic (C) fractions were immunoprecipitated with anti-ORF2 antibody, and the radioactive bands were visualized by fluorography.
Heterologous expression of the ORF2 protein upregulates the expression of ER chaperones.
Most of the proteins that are misfolded, leading to induction of ER stress, are destined for degradation by the retrotranslocation pathway so as to maintain ER homeostasis. Since the ORF2 protein was observed to behave as a substrate of the retrotranslocation pathway, we checked whether it induces ER stress by monitoring the level of different ER stress-inducible chaperones. First, we conducted a chloramphenicol acetyltransferase (CAT) reporter assay to measure the promoter activity of an ER stress-inducible chaperone, GRP94, in ORF2-expressing cells. As expected, ORF2 expression resulted in upregulation of GRP94 promoter activity (Fig. 6A). Cycloheximide and tunicamycin were used as negative and positive controls for this experiment, respectively. Next, we tested the protein level of GRP94 by immunoblot analysis. There was an increase in the corresponding protein level in ORF2-expressing cells (Fig. 6B, top panel). We also tested the protein levels of other chaperones, i.e., PDI and calnexin. The PDI level was upregulated in ORF2-expressing cells (Fig. 6B). An aliquot of the cell lysate was immunoblotted with antiactin antibody as a loading control (Fig. 6B). The bottom panel of Fig. 6B shows the expression of ORF2 in the cell lysate. These experiments suggested that the ORF2 protein induced ER stress. Therefore, we conclude that the ORF2 protein induces ER stress and mimics a substrate of the retrotranslocation pathway to gain access to the cytoplasm.
FIG. 6.
Heterologous expression of ORF2 protein induces ER stress. (A) Huh7 cells were transfected in duplicate with the GRP 94 reporter construct alone or with full-length ORF2 or ORF3 expression constructs (pSGI ORF3 and pSGI ORF2, respectively), and CAT activity was assayed by measuring acetylation of [14C]chloramphenicol. The DNA amount was equalized for all samples with empty vector DNA. CAT activity was quantified and plotted, assuming the highest activity as being 100%. Data represent means ± standard errors of the means from three independent sets of experiments. Cycloheximide (CHX)- and tunicamycin (TUN)-treated samples were used as negative and positive controls, respectively. (B) Huh7 cells were transfected with empty vector (lane 1) or with pSGI ORF2 expression plasmid (lane 2). Cells were labeled with [35S]Cys/Met promix and harvested at 48 h posttransfection. Aliquots of the lysate were immunoblotted with GRP 94, PDI, calnexin, and actin antibodies. An aliquot of the sample was immunoprecipitated with ORF2 antibody to check the expression of ORF2 protein by fluorography.
DISCUSSION
The present study uncovers another interesting mode of a viral protein exploiting the cellular machinery to its benefit. The results define a possible mechanism by which ORF2 protein localizes to the cytoplasm despite being an N-linked glycoprotein. HEV is the causative agent of an acute, self-limiting, and icteric disease that is prevalent in much of the developing world. Although self-limiting infection occurs in adults with a mortality rate of ∼1 to 2%, a high (10 to 20%) mortality rate is observed during pregnancy (8). Due to the lack of a small animal model or an efficient replication-competent cell culture model for the propagation of HEV, the basic mechanism of pathogenesis mediated by the virus remains poorly understood. The ORF2 protein has been shown to be present initially as a glycosylated protein, which gradually becomes unglycosylated. The majority of this unglycosylated protein resides in the cytoplasm. Based on its distribution pattern, earlier studies had suggested that a fraction of the ORF2 protein is not translocated into the ER (15). However, that possibility is unlikely, since cotranslational translocation per se would send ORF2 protein into the ER compartment even before the complete protein is synthesized. Moreover, our membrane fractionation study has shown that initially all the ORF2 protein is localized in the membrane fraction only. Even if we presume that under our experimental setup, some undetectable amount of ORF2 does not enter the ER, there is a very clear increase in the band intensity of cytoplasmic ORF2 protein after 2-h and 4-h chase periods, and this increase coincides with a decrease in the band intensity of ORF2 present in the membrane fraction. Thus, clearly some fraction of ORF2 protein is being retrotranslocated from the ER. Hence, a more feasible hypothesis could be that the ORF2 protein accumulates in the ER and induces ER stress, which activates the cellular quality control mechanism to remove it from the ER by the retrotranslocation pathway. Accordingly, the levels of the ER chaperones were found to be upregulated in ORF2-expressing cells. Upregulation of the HSP70 (e.g., GRP78) and HSP90 (e.g., GRP94) family of chaperones is known to be associated with the response of unfolded protein, which binds to and retains the misfolded protein in the ER. This in turn releases their interaction with ATF6, which subsequently is cleaved to produce the active form of the transcription factor ATF6, which transmits the stress signal to the nucleus (22). This leads to the activation of ATF6-responsive promoters such as the GRP94 promoter. Thus, upregulation of GRP94-CAT activity in ORF2-expressing cells indicates that the ORF2 protein induces ER stress. Upregulation of PDI levels suggests that the ORF2 protein is misfolded in the ER. An increased PDI level may help ORF2 to attain the proper conformation by rearrangement of disulfide bonding. Alternatively, it may also be possible that ORF2 exploits PDI activity to become unfolded so as to be retrotranslocated, as has been demonstrated for cholera toxin (16).
ORF2 was found to follow the retrotranslocation pathway as judged by its inability to dislocate in the presence of a biochemical inhibitor of mannosidase I and a dominant negative mutant of p97. However, the property that was peculiar to ORF2 was its ability to avoid proteasomal degradation in the cytoplasm, since deglycosylated ORF2 could be readily detected in the cytosolic fraction. This may be due to the fact that the ORF2 protein was not ubiquitinated and hence was not a target of the proteasome (however, proteasomal degradation may also occur in the absence of ubiquitination). Alternatively, it may be possible that after being retrotranslocated, the ORF2 protein promptly refolds into such a conformation that the protease-sensitive or ubiquitination sites are masked. The protease-resistant property of the ORF2 protein has been described earlier, where in vitro-expressed ORF2 protein was found to be partially trypsin resistant despite having 50 trypsin cleavage sites (23). Our data also show that the ORF2 protein is a substrate of the lysosomal proteases. Further experiments need to be done to explain this process.
The ability of ORF2 to retrotranslocate was found to be dependent on its glycosylation status. The ORF2 protein bears three N-linked glycosylation sites at amino acid residues 137, 310, and 562 (23). Although these sites along with the signal sequence are conserved in all isolates of human as well as swine HEV, no functional significance of ORF2 glycosylation is yet defined. Based on our observation that glycosylation is an absolute requirement for cytoplasmic localization of wild-type ORF2, it is tempting to speculate that the observed phenomenon will have significance during the natural course of infection. However, further experiments using a model infection system need to be carried out to understand the actual phenomenon. Nevertheless, this study provides important insight into a unique property of the ORF2 protein.
Acknowledgments
We are thankful to Tom A. Rapoport for providing various p97 constructs, as mentioned in Materials and Methods.
M.S. is supported by a research fellowship from the Council of Scientific and Industrial Research (CSIR), India. S.J. is an international senior research fellow of The Wellcome Trust. This work was supported by internal funds from ICGEB.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Afshar, N., B. E. Black, and B. M. Paschal. 2005. Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell. Biol. 25:8844-8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond, C., and A. Helenius. 1995. Quality control in the secretory pathway. Curr. Opin. Cell Biol. 7:523-529. [DOI] [PubMed] [Google Scholar]
- 3.Hazes, B., and R. J. Read. 1997. Accumulating evidence suggests that several AB-toxins subvert the endoplasmic reticulum-associated protein degradation pathway to enter target cells. Biochemistry 36:11051-11054. [DOI] [PubMed] [Google Scholar]
- 4.Jameel, S., M. Zafrullah, M. H. Ozdener, and S. K. Panda. 1996. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J. Virol. 70:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kar-Roy, A., H. Korkaya, R. Oberoi, S. K. Lal, and S. Jameel. 2004. The hepatitis E virus open reading frame 3 protein activates ERK through binding and inhibition of the MAPK phosphatase. J. Biol. Chem. 279:28345-28357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopito, R. R. 1997. ER quality control: the cytoplasmic connection. Cell 88:427-430. [DOI] [PubMed] [Google Scholar]
- 7.Korkaya, H., S. Jameel, D. Gupta, S. Tyagi, R. Kumar, M. Zafrullah, M. Mazumdar, S. K. Lal, L. Xiaofang, D. Sehgal, S. R. Das, and D. Sahal. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 276:42389-42400. [DOI] [PubMed] [Google Scholar]
- 8.Panda, S. K., and S. Jameel. 1997. Hepatitis E virus: from epidemiology to molecular biology. Viral Hepatitis Rev. 3:227-251. [Google Scholar]
- 9.Rabinovich, E., A. Kerem, K. U. Frohlich, N. Diamant, and S. Bar-Nun. 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22:626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 11.Simpson, J. C., L. M. Roberts, K. Römisch, J. Davey, D. H. Wolf, and J. M. Lord. 1999. Ricin A chain utilises the endoplasmic reticulum-associated protein degradation pathway to enter the cytosol of yeast. FEBS Lett. 459:80-84. [DOI] [PubMed] [Google Scholar]
- 12.Surjit, M., R. Kumar, R. N. Mishra, M. K. Reddy, V. T. Chow, and S. K. Lal. 2005. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 79:11476-11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Surjit, M., B. Liu, S. Jameel, V. T. Chow, and S. K. Lal. 2004. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 383:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surjit, M., R. Oberoi, R. Kumar, and S. K. Lal. 2006. Enhanced alpha1 microglobulin secretion from hepatitis E virus ORF3-expressing human hepatoma cells is mediated by the tumor susceptibility gene 101. J. Biol. Chem. 281:8135-8142. [DOI] [PubMed] [Google Scholar]
- 15.Torresi, J., F. Li, S. A. Locamini, and D. A. Anderson. 1999. Only the non-glycosylated fraction of hepatitis E virus capsid (open reading frame 2) protein is stable in mammalian cells. J. Gen. Virol. 80:1185-1188. [DOI] [PubMed] [Google Scholar]
- 16.Tsai, B., C. Rodighiero, W. I. Lencer, and T. A. Rapoport. 2001. Protein disulfide isomerase acts as a redox-dependent chaperone to unfold cholera toxin. Cell 104:937-948. [DOI] [PubMed] [Google Scholar]
- 17.Tsai, B., Y. Ye, and T. A. Rapoport. 2002. Retrotranslocation of proteins from the endoplasmic reticulum into the cytosol. Nat Rev. Mol. Cell. Biol. 3:246-255. [DOI] [PubMed] [Google Scholar]
- 18.Tyagi, S., M. Surjit, A. K. Roy, S. Jameel, and S. K. Lal. 2004. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor alpha1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J. Biol. Chem. 279:29308-29319. [DOI] [PubMed] [Google Scholar]
- 19.Wiertz, E. J. H., T. R. Jones, L. Sun, M. Boygo, H. J. Geuze, and H. L. Ploegh. 1996. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84:769-779. [DOI] [PubMed] [Google Scholar]
- 20.Wiertz, E. J. H., D. Tortorella, M. Boygo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]
- 21.Ye, Y., H. H. Meyer, and T. A. Rapoport. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652-656. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida, H., T. Okada, K. Haze, H. Yanagi, T. Yura, M. Negishi, and K. Mori. 2000. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 20:6755-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zafrullah, M., M. H. Ozdener, R. Kumar, S. K. Panda, and S. Jameel. 1999. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J. Virol. 73:4074-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]