Abstract
Jaagsiekte sheep retrovirus (JSRV) uses hyaluronidase 2 (Hyal2) as a cell entry receptor. By making inactivating mutations to the catalytic residues of human Hyal2, we found that hyaluronidase activity was dispensable for its receptor function. The affinities of the JSRV envelope glycoprotein for Hyal2 and the Hyal2 mutant were similar, and hyaluronan did not block either high-affinity interaction or virus infection. While generating the Hyal2 mutant, we discovered that our previous analysis of the hyaluronidase activity of Hyal2 was affected by a contaminating hyaluronan lyase, which we have identified as the occlusion-derived baculovirus E66 protein of the recombinant baculovirus used to produce Hyal2. We now report that purified human Hyal2 is a weak acid-active hyaluronidase.
Hyaluronidase 2 (Hyal2) is a glycosylphosphatidylinositol-anchored cell surface protein that serves as the cell entry receptor for jaagsiekte sheep retrovirus (JSRV) (10). Hyal2 belongs to the hyaluronidase family of proteins (EC 3.2.1.35) and can digest hyaluronan (3, 7, 13), a major polysaccharide component of the extracellular matrix. It is unknown whether the interaction of hyaluronan with Hyal2 can inhibit virus entry and, if so, whether virus entry is facilitated by the hyaluronidase activity of Hyal2. Most studies characterizing the receptor activity of Hyal2 have been carried out using tissue culture fibroblasts that produce little hyaluronan. In contrast, the natural target of JSRV is the lung epithelium, where hyaluronan is known to be present (5). A previous study of the receptor functions of a series of Hyal2 mutants suggests that the virus-binding site on Hyal2 does not overlap with the hyaluronan-binding groove (4). However, it is still possible that long hyaluronan chains might affect binding of JSRV to surfaces of Hyal2 located at some distance from the surfaces responsible for hyaluronan binding.
Here, we have examined the role of the hyaluronidase activity of Hyal2 in its function as the JSRV receptor. Residues important for hyaluronidase catalysis have been described for sperm hyaluronidase (2) and bee venom hyaluronidase (8) and correspond to amino acids D133 and E135 of human Hyal2. We mutated these residues to generate a hyaluronidase-dead version of Hyal2, Hyal2-HD, and a soluble carboxy-terminal-deleted form of the protein, sHyal2-HD. Initial attempts to purify sHyal2-HD revealed a contaminating hyaluronidase present in preparations of sHyal2-HD and in previously generated (13) preparations of sHyal2. Elimination of this contaminant allowed us to confirm that sHyal2-HD has little if any hyaluronidase activity and showed that sHyal2 is actually an acid-active hyaluronidase, in contrast to our previous results indicating that sHyal2 was active over a broad pH range. Hyal2 and Hyal2-HD functioned equally well as receptors for JSRV, and soluble versions of both proteins could inhibit JSRV vector transduction with similar efficiencies. The presence of hyaluronan in cell culture medium did not exert a significant specific effect on JSRV entry into cells expressing either wild-type Hyal2 or the Hyal2-HD mutant. Furthermore, using surface plasmon resonance (SPR) spectroscopy, we found that the interaction kinetics between the JSRV Env surface (SU) domain and sHyal2-HD are virtually identical to those between JSRV Env SU and sHyal2 and that the presence of short hyaluronan oligomers does not block either interaction.
MATERIALS AND METHODS
Cell culture.
Mammalian cell lines were maintained in Dulbecco's modified Eagle medium with high glucose (4.5 g per liter) and 10% fetal bovine serum at 37°C in a 10% CO2-air atmosphere unless otherwise noted. Insect cell lines were maintained at 27°C in air. Sf9 cells were grown in SF-900 II serum-free medium, and Drosophila Schneider 2 (S2) and High5 cells were grown in Express Five serum-free medium (Invitrogen).
Expression and purification of HyalX.
Recombinant baculovirus stocks encoding proteins unrelated to hyaluronidases were generated as previously described for sHyal2-encoding viruses (13). Supernatants from High5 cultures infected with these viruses were harvested by centrifugation to remove cells at 4 days postinfection, supplemented with 0.02% sodium azide and 1 mM EDTA to prevent microbial contamination and inhibit metalloprotease activity, and dialyzed against 20 mM ethanolamine, pH 9.5. The resulting protein solution was applied to a 1-ml HiTrapQ column (Pharmacia) and eluted using a 0 to 500 mM NaCl linear gradient. Fractions containing hyaluronidase activity (eluting at ∼60 mM NaCl) were pooled, concentrated, and size fractionated on a Superdex 200 HR 10/30 sizing column (Amersham Biosciences AB) using PNEA buffer {20 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 150 mM NaCl, 1 mM EDTA, and 0.02% sodium azide}. Fractions containing hyaluronidase activity were pooled and concentrated.
Expression and purification of baculoviral ChiA.
The ChiA coding region was amplified from genomic DNA of recombinant baculovirus (derived from Autographa californica multicapsid nucleopolyhedrovirus) using forward (5′-AGA ATT CAT GTT GTA CAA ATT GTT AAA CG-3′) and reverse (5′-AGG ATC CTT AAT GGT GAT GGT GAT GGT GAT GCA GTT CAT CTT TAG GTT T-3′) primers, which added 5′ EcoRI and 3′ BamHI restriction sites, as well as a sequence encoding a His6 tag, to the 3′ end. The PCR product was cloned into the Cu-inducible pRmHa3 insect cell expression vector using the EcoRI and BamHI sites to make the pRmHa3-ChiA vector. One million S2 cells were cotransfected with 10 μg of pRmHa3-ChiA and 0.5 μg of a blasticidin resistance gene vector, using FuGENE 6 (Roche) according to the manufacturer's instructions. Following selection in 25 μg/ml blasticidin (Invitrogen), the cells were induced with 1 mM CuSO4 for 4 to 7 days, and the culture supernatant was collected and treated as described previously for production of sHyal2 (13).
Hyaluronidase assay.
Hyaluronidase activity in protein samples was assessed by the agarose electrophoretic mobility assay as previously described (13). Briefly, protein samples were incubated with human umbilical chord hyaluronan (Sigma) at 37°C in the presence of 0.1 M buffer at the indicated pH. The gels were stained in 0.005% Stains-All (Sigma) in 50% ethanol overnight and then photographed on a fluorescent-light transilluminator.
Hyaluronidase zymography.
Protein samples were separated in 7.5% polyacrylamide gels using an ammonia-3-cyclohexylamino-1-propanesulfonic acid (CAPS) continuous buffer system, as previously described (9), with the following modifications. The gels were polymerized in the presence of 33 μg/ml human umbilical chord hyaluronan and the buffer contained 20 mM CAPS and 270 mM ammonia. Electrophoresis was carried out at 100 V for 2 h at 4°C to avoid heating of the proteins. After electrophoresis, the gels were extensively washed with 0.1 M MES (morpholineethanesulfonic acid), pH 5.0, incubated at 37°C overnight, stained in 0.005% Stains-All (Sigma) in 50% ethanol, and photographed on a fluorescent-light transilluminator. Next, the gels were photobleached under fluorescent light to degrade the Stains-All and were then counterstained using Coomassie blue to detect protein.
Protein interaction analysis by surface plasmon resonance.
Preparation of JSRV Env SU-human immunoglobulin G constant region (JSU-IgG) fusion protein, SPR measurements, and subsequent data analysis were carried out as previously described (13). Briefly, purified JSU-IgG was immobilized on a CM5 research grade sensor chip (Biacore AB) by amine coupling. This surface was exposed for 200 s to various concentrations of either sHyal2 or sHyal2-HD alone or in a mixture with purified 10-mer or 6-mer hyaluronan oligosaccharides (kind gifts from Francis C. Szoka, Jr., University of California, San Francisco), followed by a 1-h dissociation phase during which protein-free buffer was allowed to flow across the chip surface. Data analysis and curve fitting were performed using BIAevaluation software (version 3.2) and reported as fit value ± standard error of fit.
RESULTS
Generation of a hyaluronidase-dead mutant of sHyal2.
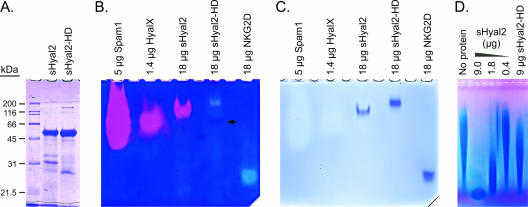
D133N and E135Q mutations were made in putative critical active-site residues of human sHyal2 to make sHyal2-HD (hyaluronidase dead). The mutant protein was produced by using a baculovirus expression system as previously described for sHyal2 (13). Like sHyal2, purified sHyal2-HD was found to be a 50-kDa monomer by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 1A) and size exclusion chromatography (data not shown). Initial sHyal2-HD preparations had significant hyaluronidase activity, which we found was due to a contaminating hyaluronidase (HyalX [see below]) that could be removed by increased washing of the affinity resin during sHyal2-HD purification.
FIG. 1.
Characterization of sHyal2, sHyal2-HD, and HyalX. (A) SDS-PAGE analysis of sHyal2 and sHyal2-HD. (B) Native-PAGE zymography of HyalX, sHyal2, and sHyal2-HD. Pink clearings indicate hyaluronidase activity, and light-blue clearings are caused by protein. A faint pink band in the sHyal2-HD lane (arrow) likely represents residual HyalX activity. (C) Restaining of the gel shown in panel B with Coomassie blue reveals the positions of the protein species. Note that the amounts of Spam1 and HyalX listed above the gel refer to total protein of these relatively impure protein preparations, and the actual amounts of Spam1 and HyalX were below the limit of detection using Coomassie blue stain. (D) Hyaluronidase assay. Fifty-microgram samples of hyaluronan were incubated at 37°C at pH 5.6 in the presence of the indicated proteins for 14 h. The samples were then separated in a 0.5% agarose gel by electrophoresis and visualized with Stains-All.
The activities of sHyal2 and sHyal2-HD were analyzed by native-PAGE zymography in nondenaturing gels containing hyaluronan (Fig. 1B). This technique separates protein species according to charge and molecular weight while preserving enzyme activity. Following electrophoresis, the gels were incubated in an appropriate buffer to allow digestion of the hyaluronan in the gels by the migrated proteins. Pink clearings in the native-PAGE zymograms indicated areas where the substrate was digested to fragments capable of diffusing out of the gel, as seen with the well-characterized hyaluronidase Spam1 (Fig. 1B). Stains-All, which was used in these experiments to visualize hyaluronan (blue stain), also interacts with protein to produce a light-blue clearing (Fig. 1B, sHyal2-HD and NKG2D lanes) (NKG2D [11] is a nonhyaluronidase protein control). After zymographic analysis, the Stains-All was bleached and the gels were stained with Coomassie blue to reveal the positions of the protein bands (Fig. 1C). Unlike wild-type sHyal2, sHyal2-HD did not produce a pink clearing (Fig. 1B) near its position on the gel (Fig. 1C). Note that the migration of sHyal2-HD by native-PAGE was slower than that of sHyal2 due to the mutations present in the former, which make it less negatively charged. A faint pink band was present below the sHyal2-HD band (Fig. 1B) with mobility corresponding to that of the HyalX hyaluronidase, showing that this sHyal2-HD preparation was mostly but not completely, free of HyalX.
We also examined the hyaluronidase activity of sHyal2-HD by incubation with hyaluronan and separation of the reaction products by agarose gel electrophoresis. In this assay, partially digested hyaluronan migrates as a low-molecular-weight band at the bottom of the gel, whereas the undigested hyaluronan appears as a long smear at higher molecular weights. We saw little hyaluronidase activity in sHyal2-HD samples compared with that observed for sHyal2 (Fig. 1D). Trace activity of Hyal2-HD in this highly sensitive assay is likely accounted for by the presence of residual HyalX.
Baculovirus infection of High5 cells leads to production of a secreted hyaluronidase.
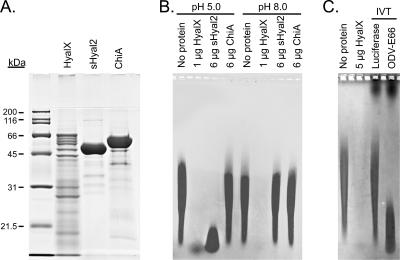
Initial troubleshooting of the sHyal2-HD purification led us to check for hyaluronidase activity in media from High5 cultures infected by several recombinant baculoviruses encoding proteins unrelated to hyaluronidases. Unlike media from uninfected High5 cultures, which showed no hyaluronidase activity, media from all infected cultures contained a readily detectible hyaluronidase (data not shown). We partially purified this hyaluronidase (HyalX), using its enzymatic activity as a marker. SDS-PAGE analysis of the HyalX preparation revealed multiple protein bands in the range of 14 to 70 kDa (Fig. 2A). HyalX is active over a broad pH range with an optimum of about pH 8 (Fig. 3A). This pH activity profile is identical to that previously reported by us for sHyal2 (13).
FIG. 2.
Determination of the identity of HyalX. (A) SDS-PAGE analysis of HyalX purified from medium conditioned by recombinant baculovirus-infected High5 cells, as well as sHyal2 and ChiA proteins purified from Drosophila S2 stable cell lines transfected with respective constructs. Each lane contains 10 μg of protein. (B) The protein samples shown in panel A were incubated for 14 h with 50-μg samples of hyaluronan at the indicated pH. Hyaluronan was then separated in a 0.5% agarose gel by electrophoresis, and the digestion products were visualized using Stains-All. (C) Fifty-microgram samples of hyaluronan were diluted in phosphate-buffered saline (pH 7.4), mixed with the indicated protein samples, and analyzed as described for panel B; 20 μl of coupled in vitro transcription/translation reaction mixtures were used per lane (lanes labeled IVT).
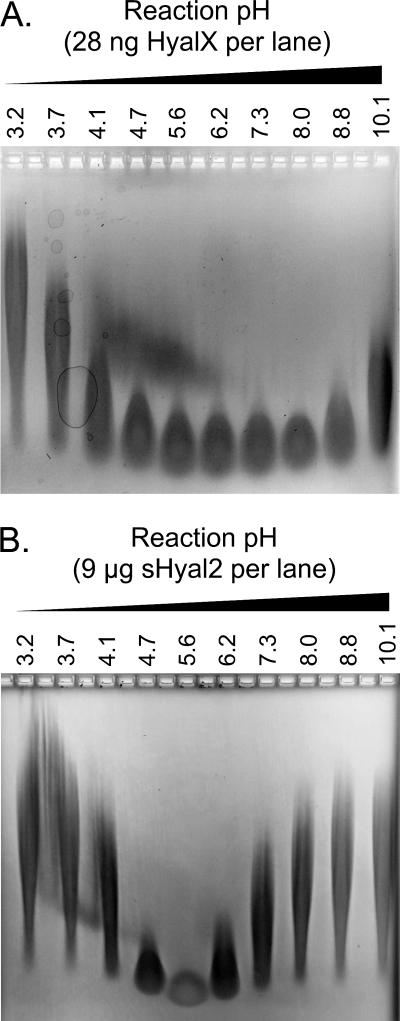
FIG. 3.
sHyal2 is a weak hyaluronidase with a pH optimum of ∼5.6. Fifty-microgram samples of hyaluronan were incubated at 37°C at various pH levels with either 28 ng HyalX (A) or 9 μg sHyal2 (B). The samples were then separated in a 0.5% agarose gel by electrophoresis and visualized with Stains-All.
We used the native-PAGE migration pattern of HyalX (Fig. 1B) to assist in identifying this hyaluronidase. The protein band corresponding to active HyalX was excised from the gel and was sequenced by matrix-assisted laser desorption ionization-time of flight mass spectrometry, revealing baculoviral chitinase A (ChiA) and occlusion-derived baculovirus E66 (ODV-E66) protein as the major species present. We purified ChiA from S2 cells transfected with a ChiA expression vector and found its chitinase activity, using 4-methylumbelliferyl β-d-N,N′,N"-triacetylchitotrioside as a substrate, to be similar to those of other enzymes of its class: kcat = 22 ± 1 s−1, Vmax = 23 ± 1 μmol/min/mg ChiA, and Km = 0.88 ± 0.17 μM. However, we found no evidence of hyaluronidase activity (Fig. 2B) when 6 μg of ChiA was incubated with 50 μg of hyaluronan. In contrast, 28 ng of HyalX was sufficient for detection of its hyaluronidase activity (Fig. 3A) and 1 μg of HyalX almost completely digested the hyaluronan (Fig. 2B).
To examine the hyaluronidase activity of ODV-E66, we used a coupled in vitro transcription/translation rabbit reticulocyte kit (Promega), together with an ODV-E66 expression construct (a kind gift from M. D. Summers and S. C. Braunagel, Texas A&M University). As seen in Fig. 2C, the in vitro-translated product of the ODV-E66 construct had significant hyaluronidase activity, while the control construct encoding firefly luciferase did not. These results indicate that the hyaluronidase produced by baculovirus-infected insect cells is due to production of the baculoviral protein ODV-E66.
sHyal2 is a weak hyaluronidase with a moderately acidic pH optimum.
Next, we compared the hyaluronidase activities of sHyal2 and HyalX. sHyal2 had a sharp pH optimum of 5.6, in contrast to the broad activity of HyalX, with an optimum at pH 8.0 (Fig. 3). The activity of 9 μg of sHyal2 at ph 5.6 was comparable to that of 28 ng of HyalX at pH 8.0 (Fig. 3), indicating that sHyal2 is ∼300-fold less active than HyalX. No hyaluronidase activity was found at any pH using 28 ng of sHyal2 (data not shown). Titration of the amount of sHyal2 present in reaction allowed us to conclude that the activity of 1.8 μg sHyal2 (Fig. 1D) is approximately equivalent to that of 4.4 ng Spam1 (13), showing that sHyal2 is ∼400-fold less active than Spam1.
Hyal2-HD serves as a JSRV receptor.
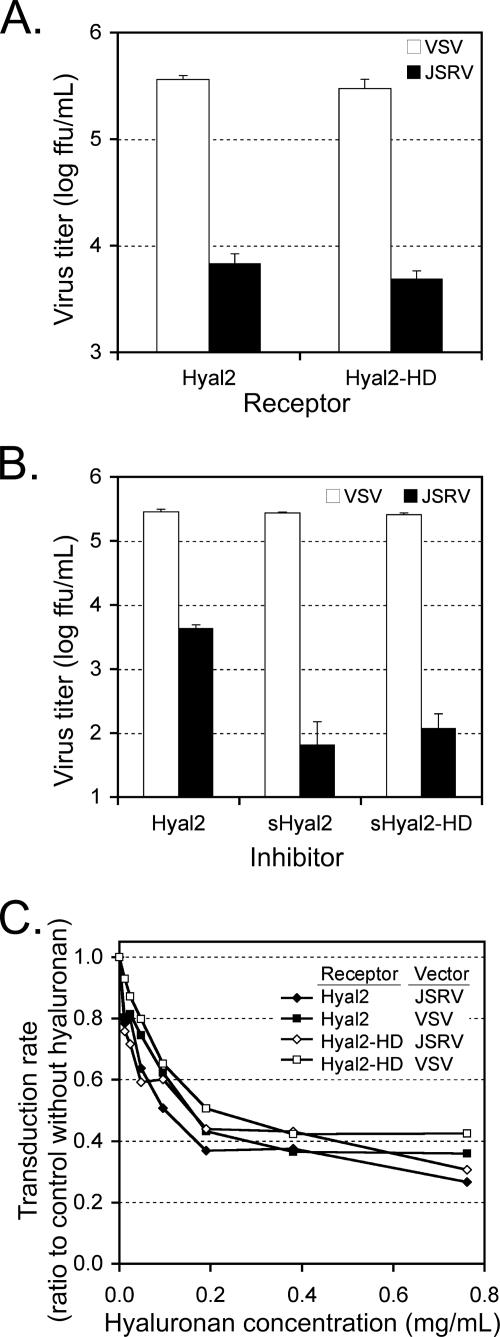
Mouse cells cannot be transduced by JSRV vectors due to lack of a functional JSRV receptor (10). The receptor function of Hyal2-HD was compared to that of Hyal2 by using mouse cell lines expressing either protein in a transduction assay. Briefly, JSRV pseudotype and vesicular stomatitis virus G protein (VSV-G) pseudotype retroviral vectors encoding human placental alkaline phosphatase (AP) were used to transduce NIH 3T3 cells expressing Hyal2 (NIH 3T3/LH2SN) or Hyal2-HD (NIH 3T3/LH2HDSN). Both cell lines were transduced equally well by the JSRV vector, showing that Hyal2 and Hyal2-HD are equally functional as JSRV receptors (Fig. 4A). The VSV-G pseudotype vector does not use Hyal2 as a receptor and, as expected, transduced both cell lines equally well (Fig. 4A).
FIG. 4.
The D133N and E135Q mutations do not affect the ability of Hyal2 to interact with JSRV. (A) NIH 3T3 cell lines stably expressing either Hyal2 or Hyal2-HD were generated as previously described (10). The cell lines were exposed to vectors encoding human placental AP pseudotyped either with JSRV Env or with VSV-G glycoprotein. Two days postinfection, the cells were fixed and stained for AP, and AP+ foci were counted. (B) HEK 293 cells were transfected with 10 μg of plasmid DNA encoding sHyal2, sHyal2-HD, or Hyal2. The conditioned medium was harvested 3 days later, diluted 1:1 with fresh medium containing 8 μg Polybrene per ml, and added to dishes of subconfluent NIH 3T3/LH2SN cells. JSRV and VSV-G pseudotype vectors encoding AP were then added to the cultures, and vector titers were determined as described for panel A. (C) Cell lines and viral vectors shown in panel A were used to measure transduction efficiency in the presence of various concentrations of hyaluronan. All results are means of two experiments with triplicate determinations in each, and the errors bars indicate standard deviations.
sHyal2-HD inhibits entry of a JSRV vector into Hyal2-expressing host cells.
We previously showed that purified sHyal2 can inhibit JSRV pseudotype vector transduction when added to the medium of target cells (13). Here, we used transiently transfected HEK 293 cells to produce media containing either sHyal2-HD or sHyal2 to compare the ability of soluble receptors to block JSRV pseudotype vector transduction of human HTX cells expressing endogenous Hyal2. Medium containing either sHyal2 or sHyal2-HD significantly inhibited transduction by the JSRV vector compared to medium from HEK 293 cells expressing the natural membrane-anchored form of Hyal2 (Fig. 4B). In contrast, transduction by the VSV-G, pseudotype vector was unaffected by the presence of sHyal2 or sHyal2-HD (Fig. 4B).
Hyaluronan in the cell culture medium has little, if any, specific effect on JSRV vector entry.
We assessed the inhibitory potential of hyaluronan, as a natural ligand of Hyal2, on JSRV vector entry. As a control, we measured entry of an otherwise identical vector bearing the VSV-G protein that does not use Hyal2 as a receptor. Culture medium containing various concentrations of hyaluronan was added to NIH 3T3/LH2SN or NIH 3T3/LH2HDSN cells prior to infection with the JSRV or VSV pseudotype vectors. We saw similar decreases of infectivity for both vectors and for either target cell line as the concentration of hyaluronan was raised (Fig. 4C). The viscosity of the culture medium increased significantly as the concentration of hyaluronan was raised, and we assume that this nonspecific decrease in infectivity was due to inhibition of virus diffusion to the cell surface. These data indicate that the interaction of soluble hyaluronan with cell-associated Hyal2 or Hyal2-HD does not significantly alter the virus-receptor interaction.
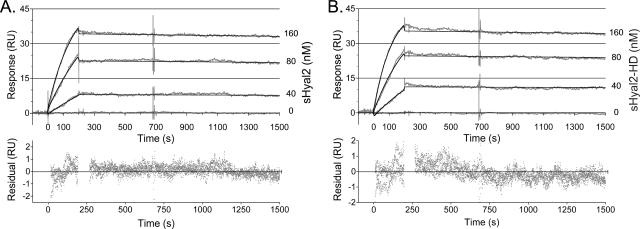
sHyal2-HD and sHyal2 bind to JSU-IgG with similar kinetics.
We used SPR to compare the interactions of sHyal2 and sHyal2-HD with the JSRV envelope SU domain, using previously described techniques (13). We found that the data sets obtained for sHyal2 and sHyal2-HD (Fig. 5) were virtually identical, and the interaction was characterized by a very low off rate for either protein. The data were used to build best-fit curves based on a 1:1 Langmuir interaction model yielding kinetic constants for the JSU-IgG-sHyal2 interaction (kon = [3.6 ± 0.1] × 104 M−1 s−1; koff = [2.46 ± 0.02] × 10−5 s−1) and for the JSU-IgG-sHyal2-HD interaction (kon = [5.1 ± 0.1] × 104 M−1 s−1; koff = [2.30 ± 0.02] × 10−5 s−1) with good fits (χ2 = 0.3 in each case) illustrated by small residual values. The calculated equilibrium dissociation constant (KD) for the JSU-IgG-sHyal2 interaction was 0.68 nM, and that for the JSU-IgG-sHyal2-HD interaction was 0.45 nM.
FIG. 5.
Kinetics of sHyal2 and sHyal2-HD binding to JSU-IgG. Reference-subtracted data (gray curves) illustrating the interaction between immobilized JSU-IgG and either sHyal2 (A) or sHyal2-HD (B) are shown, together with the corresponding fit (black curves) for several concentrations of analyte. Deviations of data from the proposed fit are shown below each panel in a residuals plot (gray dots). All data were analyzed and curve fits were generated using BIAevaluation software. RU, response units.
Hyaluronan fragments do not block the binding of sHyal2 or sHyal2-HD to JSU-IgG.
To examine the impact of hyaluronan on binding of Hyal2 and Hyal2-HD to JSRV Env glycoprotein, we used hyaluronan fragments of defined length in SPR experiments. From our estimates, sHyal2 and sHyal2-HD bind to 12-mer fragments of hyaluronan (biotinylated to allow coupling to the SPR chip) with a micromolar affinity (estimated KD, 1 to 10 μM [data not shown]). In order to ensure saturation of the substrate-binding site, we used the minimum detectible concentration of sHyal2 or sHyal2-HD (24 nM), together with up to 10,000-fold molar excess of 6-mer or 10-mer hyaluronan oligosaccharides, to examine the effect of these hyaluronan fragments on Hyal2 interaction with JSU-IgG. In all cases, we observed that the presence of hyaluronan fragments of either length was unable to block the binding of either sHyal2 or sHyal2-HD to JSU-IgG (data not shown).
DISCUSSION
Due to the presence of a contaminating hyaluronidase (HyalX) in our early preparations of sHyal2, we have had to reexamine our previously published characterization of the hyaluronidase activity of sHyal2 (13). Three lines of evidence support our current characterization. First, the pH profile of hyaluronidase activity observed for sHyal2 differs from that of the HyalX contaminant (Fig. 3) and agrees better with previously published data from others (3, 7). Second, the native-PAGE zymography migration pattern of sHyal2 is distinct from that of HyalX and colocalizes with the sHyal2 protein band (Fig. 1B and C). Finally, sHyal2 hyaluronidase activity can be greatly reduced, if not abolished, by the mutation of the predicted catalytic residues (Fig. 1B, C, and D). Given the acidic pH optimum and the very low activity of Hyal2 relative to other hyaluronidases, such as Spam1 and Hyal1, Hyal2 may not be a physiologically important hyaluronidase. However, additional factors may modulate Hyal2 activity or acidify the local environment to increase Hyal2 activity (3).
Mutations made to the catalytic residues of Hyal2 did not significantly affect the ability of JSRV to use it as a receptor in the presence or absence of soluble hyaluronan. We also found that the JSU-IgG-sHyal2 (wild type or HD) interaction could not be blocked, even in the presence of saturating concentrations of hyaluronan fragments. These findings suggest that either the Env-binding site lies outside the substrate-binding groove of Hyal2, as suggested by previous data (4), or the high-affinity Hyal2-JSRV Env interaction overwhelms weak binding of hyaluronan.
Here, we found a KD for the sHyal2-JSU-IgG interaction of 0.68 nM, different from our earlier reported value of 32 pM (13). This 20-fold difference is likely the result of several factors that are difficult to control, particularly variation in the state of the JSU-IgG protein immobilized on the Biacore chip and the difficulty in consistently measuring an extremely tight interaction that is near the limits of Biacore technology. Despite these variations, the data show an interaction of very high affinity. The comparison between sHyal2 and sHyal2-HD affinities for JSU-IgG reported here was made under conditions selected to minimize experimental variation, in particular, by using the same chip bearing high-quality JSU-IgG protein to measure the binding of both sHyal2 and sHyal-HD.
Hyaluronan is found as a component of the extracellular matrix in animals. Our results indicate that the presence of soluble hyaluronan has no specific effect on the ability of JSRV to infect cells in culture. Nevertheless, it is possible that epithelial monolayers covered by an extensive extracellular matrix may be protected from JSRV infection. Additionally, the ability of Hyal2 to locally degrade hyaluronan may be important for the ability of the virus to penetrate the extracellular matrix if the environment of Hyal2 is acidified by a mechanism such as that suggested by Bourguignon et al. (3). Further studies with animals or with cell lines that produce an extracellular matrix containing hyaluronan are required to address these possibilities.
Finally, we identified the contaminating hyaluronidase in our sHyal2 and sHyal2-HD preparations as ODV-E66, which is an envelope protein of occlusion-derived particles of the baculovirus Autographa californica multicapsid nucleopolyhedrovirus (6). The function of ODV-E66 was not previously known; however, PSI-BLAST analysis (1) of its sequence reveals signatures of bacterial hyaluronan lyase catalytic domains. Hyaluronan digestion by HyalX, but not by any mammalian hyaluronidase we tested, resulted in reaction products absorbing at 235 nm (data not shown), characteristic of lyase-type enzymes (EC 4.2.2.1) (12). These data indicate that baculoviral ODV-E66 is a hyaluronan lyase. The fact that ODV is the form of the virus that is released from the dead insect and is subsequently eaten by new hosts suggests that this hyaluronan lyase activity may be important for penetration of extracellular barriers during virus release or in the course of new infection in the insect gut epithelium.
Acknowledgments
We thank Francis C. Szoka, Jr. (Biopharmaceutical Sciences and Pharmaceutical Chemistry, UCSF), for his kind gift of hyaluronan fragments, Max Summers and Sharon Braunagel (Texas A&M University) for providing us with the ODV-E66 constructs, and Phil Gafken (Proteomics, Fred Hutchinson Cancer Research Center) for performing the matrix-assisted laser desorption ionization-time of flight analysis.
This work was supported by National Cancer Institute training grant NIH T32 CA80416 (V.V.), grant DK47754 (A.D.M.), and grant AI48675 (R.K.S.) from the NIH.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arming, S., B. Strobl, C. Wechselberger, and G. Kreil. 1997. In vitro mutagenesis of PH-20 hyaluronidase from human sperm. Eur. J. Biochem. 247:810-814. [DOI] [PubMed] [Google Scholar]
- 3.Bourguignon, L. Y., P. A. Singleton, F. Diedrich, R. Stern, and E. Gilad. 2004. CD44 interaction with Na+-H+ exchanger (NHE1) creates acidic microenvironments leading to hyaluronidase-2 and cathepsin B activation and breast tumor cell invasion. J. Biol. Chem. 279:26991-27007. [DOI] [PubMed] [Google Scholar]
- 4.Duh, F. M., C. Dirks, M. I. Lerman, and A. D. Miller. 2005. Amino acid residues that are important for Hyal2 function as a receptor for jaagsiekte sheep retrovirus. Retrovirology 2:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forteza, R., T. Lieb, T. Aoki, R. C. Savani, G. E. Conner, and M. Salathe. 2001. Hyaluronan serves a novel role in airway mucosal host defense. FASEB J. 15:2179-2186. [DOI] [PubMed] [Google Scholar]
- 6.Hong, T., S. C. Braunagel, and M. D. Summers. 1994. Transcription, translation, and cellular localization of PDV-E66: a structural protein of the PDV envelope of Autographa californica nuclear polyhedrosis virus. Virology 204:210-222. [DOI] [PubMed] [Google Scholar]
- 7.Lepperdinger, G., B. Strobl, and G. Kreil. 1998. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J. Biol. Chem. 273:22466-22470. [DOI] [PubMed] [Google Scholar]
- 8.Markovic-Housley, Z., G. Miglierini, L. Soldatova, P. J. Rizkallah, U. Muller, and T. Schirmer. 2000. Crystal structure of hyaluronidase, a major allergen of bee venom. Struct. Fold Des. 8:1025-1035. [DOI] [PubMed] [Google Scholar]
- 9.McLellan, T. 1982. Electrophoresis buffers for polyacrylamide gels at various pH. Anal. Biochem. 126:94-99. [DOI] [PubMed] [Google Scholar]
- 10.Rai, S. K., F.-M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, and A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 98:4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steinle, A., P. Li, D. L. Morris, V. Groh, L. L. Lanier, R. K. Strong, and T. Spies. 2001. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics 53:279-287. [DOI] [PubMed] [Google Scholar]
- 12.Sutherland, I. W. 1995. Polysaccharide lyases. FEMS Microbiol. Rev. 16:323-347. [DOI] [PubMed] [Google Scholar]
- 13.Vigdorovich, V., R. K. Strong, and A. D. Miller. 2005. Expression and characterization of a soluble, active form of the jaagsiekte sheep retrovirus receptor, Hyal2. J. Virol. 79:79-86. [DOI] [PMC free article] [PubMed] [Google Scholar]