Abstract
Expression of Ebola virus nucleoprotein (NP) in mammalian cells leads to the formation of helical structures, which serve as a scaffold for the nucleocapsid. We recently found that NP binding with the matrix protein VP40 is important for nucleocapsid incorporation into virions (T. Noda, H. Ebihara, Y. Muramoto, K. Fujii, A. Takada, H. Sagara, J. H. Kim, H. Kida, H. Feldmann, and Y. Kawaoka, PLoS Pathog. 2:e99, 2006). To identify the region(s) on the NP molecule required for VP40 binding, we examined the interaction of a series of NP deletion mutants with VP40 biochemically and ultrastructurally. We found that both termini of NP (amino acids 2 to 150 and 601 to 739) are essential for its interaction with VP40 and for its incorporation into virus-like particles (VLPs). We also found that the C terminus of NP is important for nucleocapsid incorporation into virions. Of interest is that the formation of NP helices, which involves the N-terminal 450 amino acids of NP, is dispensable for NP incorporation into VLPs. These findings enhance our understanding of Ebola virus assembly and in so doing move us closer to the identification of targets for the development of antiviral compounds to combat Ebola virus infection.
Ebola viruses, together with Marburg viruses, comprise the family Filoviridae in the order Mononegavirales. The single-stranded, nonsegmented, negative-sense RNA genome of Ebola virus encodes seven structural proteins that form filamentous virions of approximately 80 nm in diameter but of various lengths (37). The glycoprotein GP sits on the surface of the lipid-enveloped virions, where it is responsible for virion binding to cellular receptors and for the fusion of the viral and cellular lipid bilayers (40). The matrix protein VP40 is the major structural component of the virions and is critical for virion formation (37). A minor membrane-associated protein, VP24, is involved in nucleocapsid formation (12) but also functions as an interferon antagonist (35). The nucleocapsid, a viral genomic RNA-protein complex, is composed of the nucleoprotein NP, polymerase protein L, and VP30 and VP35, which are regulators of replication and transcription (25, 45), and resides in the long axis of the filamentous virion (37).
The matrix protein of enveloped and negative-sense RNA viruses generally associates with lipid bilayers and oligomerizes to form macromolecular complexes. These properties enable matrix proteins to facilitate their own release, when expressed in mammalian cells, in the form of virus-like particles (VLPs) enclosed in lipid bilayers (2, 5, 8, 10, 14, 20, 31, 36, 41, 42). In addition, matrix proteins are thought to mediate virion formation through interactions with viral genome-protein complexes (5, 6, 22, 24, 27, 33, 46). Such interactions are thought to assist in the budding process and also to promote the efficiency of virion formation (39). Thus, matrix proteins, including Ebola virus VP40, play a key role in viral assembly, budding, and virion formation.
Recently, we demonstrated that Ebola virus NP forms oligomers and self-assembles in cells expressing NP into helical tubular structures approximately 20 to 25 nm in diameter (30, 43). These structures are morphologically different from Ebola virus nucleocapsids, which are approximately 50 nm in diameter. Because NP helices serve as the core of the nucleocapsid and are incorporated into VP40-induced VLPs (30), the association of NP helices with VP40 is likely important for the incorporation of NP helices into VLPs and for nucleocapsid incorporation into virions. Licata et al. (21) biochemically showed that the C-terminal 50 amino acids of Ebola virus NP are necessary for its incorporation into VLPs; however, the importance or significance of the other regions of NP was not assessed. To better understand Ebola virus assembly, we mapped the regions in the NP molecule that are responsible for its binding to VP40 both biochemically and ultrastructurally.
MATERIALS AND METHODS
Cells.
Human embryonic kidney 293T cells and 293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, l-glutamine, and penicillin-streptomycin-amphotericin B. The cells were grown in an incubator at 37°C under 5% CO2.
Plasmids and transfection.
The open reading frame encoding Zaire ebolavirus NP, VP40, VP24, and VP35 was cloned into the expression vector pCAGGS/MCS (18, 29) as described previously (44). A series of NP deletion constructs (see Fig. 2) were generated as described previously (43). FLAG-tagged VP40 at the N terminus and VP40 mutants (F125A, R134A, and F125A/R134A) were generated by PCR and standard cloning techniques. 293T cells (approximately 106 cells/well) grown in six-well plates were transfected with plasmids by using Trans IT 293 reagent (Mirus Corporation, Madison, WI) according to the manufacturer's instructions.
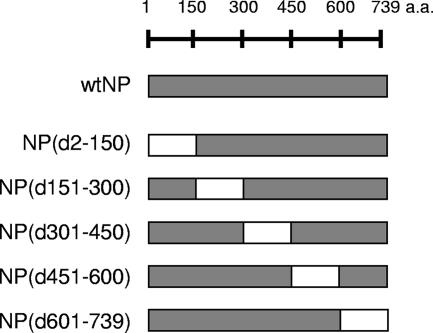
FIG. 2.
Schematic diagram of NP deletion mutants. A series of deletion mutants was constructed previously (43) by sequential deletion of 150 amino acids (a.a.). wt, wild type.
Immunoprecipitation.
293T cells cotransfected with plasmids expressing FLAG-tagged VP40 and wild-type NP or deletion mutant NP were collected at 24 h posttransfection and lysed in TNE buffer (10 mM Tris-HCl [pH 7.8], 1% NP-40, 0.15 M NaCl, 1 mM EDTA). After being clarified by centrifugation, the supernatants were incubated with mouse (M2) anti-FLAG antibody conjugated with Sepharose beads (Sigma-Aldrich, St. Louis, MO) overnight at 4°C. The Sepharose beads were then washed with TNE buffer and suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Samples were then subjected to SDS-PAGE, followed by Western blot analysis.
Protease protection assay.
Supernatants of 293T cells transfected with plasmids for the expression of VP40 and wild-type NP or deletion mutant NP were divided into three aliquots. One aliquot received no further treatment, and the others were treated with either trypsin at a final concentration of 0.1 mg/ml or both trypsin and Triton X-100 at a final concentration of 1%. Samples were then incubated at room temperature for 30 min and suspended in SDS-PAGE sample buffer.
Western blotting.
Plasmid-transfected cells and supernatants were lysed in SDS-PAGE sample buffer and separated on a 4% to 20% PAGE Tris-glycine gradient gel (Daiichi Pure Chemicals Co., Ltd., Tokyo). Blots were incubated with rabbit anti-NP and anti-VP40 serum; the binding of the primary antibodies was detected with the Vectastain ABC kit (Vector Laboratories, Inc., Burlingame, CA) by following the manufacturer's instructions. Proteins were then visualized by immunostaining (HRP-1000; Konica Minolta Holdings, Inc., Tokyo).
Immunofluorescence and confocal microscopy.
293 cells grown on glass coverslips were transfected with VP40 and wild-type NP or deletion mutant NP. The cells were fixed, at 24 h posttransfection, with 4% paraformaldehyde for 1 h, permeabilized with phosphate-buffered saline (PBS) containing 0.1% Triton X-100, and incubated with mouse anti-NP and rabbit anti-VP40 antibodies for 1 h at room temperature. After being washed with PBS, they were incubated with Alexa Fluor 488 goat anti-mouse immunoglobulin G (IgG) (Invitrogen/Molecular Probes, Eugene, OR) and Alexa Fluor 594 donkey anti-rabbit IgG (Invitrogen/Molecular Probes, Eugene, OR). Microscopic observation was performed using an LSM510 microscope (Carl Zeiss, Oberkochen).
Electron microscopy.
Ultra-thin-section electron microscopy and immunoelectron microscopy were done as described previously (31). Briefly, for ultra-thin-section electron microscopy, the plasmid-transfected cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer and postfixed with 2% osmium tetroxide in the same buffer. They were then dehydrated with a series of ethanol gradients followed by propylene oxide, before being embedded in Epon 812 Resin mixture (TAAB Laboratories Equipment Ltd., Berkshire). Thin sections were stained with 2% uranyl acetate and Raynold's lead and examined under a Hitachi H-7500 electron microscope at 80 kV. For immunoelectron microscopy, cells were fixed with 4% paraformaldehyde and 0.1% glutaraldehyde, dehydrated with a series of ethanol gradients, and then embedded in LR White resin (London Resin Company Ltd., Berkshire). The sections were absorbed to nickel grids, treated with a mouse anti-NP monoclonal antibody, and then incubated with a goat anti-mouse IgG conjugated to 5-nm gold beads (BBInternational, Cardiff).
RESULTS
Both ends of NP are important for its interaction with VP40.
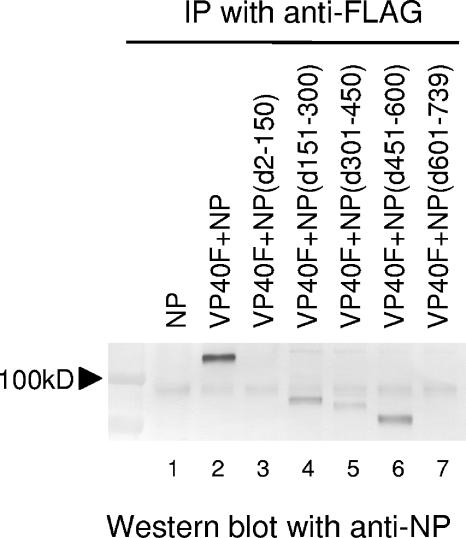
To identify the region(s) of NP responsible for the interaction with VP40, 293T cells expressing FLAG-tagged VP40 together with a wild-type NP or a deletion mutant NP were lysed at 24 h posttransfection and subjected to immunoprecipitation with an anti-FLAG antibody, followed by Western blotting with an anti-NP antibody. Under these conditions, wild-type NP was coimmunoprecipitated in cells that coexpressed FLAG-tagged VP40 but was not immunoprecipitated in cells expressing NP alone (Fig. 1, lanes 1 and 2), confirming previous reports (21, 30) and demonstrating that the FLAG tag at the N-terminal end of VP40 does not disturb the interaction between VP40 and NP. We then engineered a series of internal deletion mutant NPs (Fig. 2), all of which were expressed at a similar level as confirmed by Western blotting using an anti-NP antibody (data not shown) (43). NP(d151-300), NP(d301-450), and NP(d451-600) (Fig. 1, lanes 4 to 6), but not NP(d2-150) and NP(d601-739) (Fig. 1, lanes 3 and 7), coimmunoprecipitated with FLAG-tagged VP40. These results suggest that the region from amino acid 151 to 600 of NP is dispensable for VP40 binding and that both the N- and C-terminal ends of NP are important for the NP-VP40 interaction. The NP region from amino acid 301 to 450 could also contribute to the interaction of NP with VP40, since the signal for NP(d301-450) was weaker (Fig. 1, lane 5) than that for wild-type NP, NP(d151-300), or NP(d451-600) (Fig. 1, lanes 2, 4, and 6).
FIG. 1.
Interaction of VP40 with wild-type or mutant NPs. 293T cells expressing FLAG-tagged VP40 and wild-type NP or a deletion mutant were lysed and subjected to immunoprecipitation (IP) with an anti-FLAG antibody. The immunoprecipitated proteins were analyzed by Western blotting by use of an anti-NP antibody after SDS-PAGE.
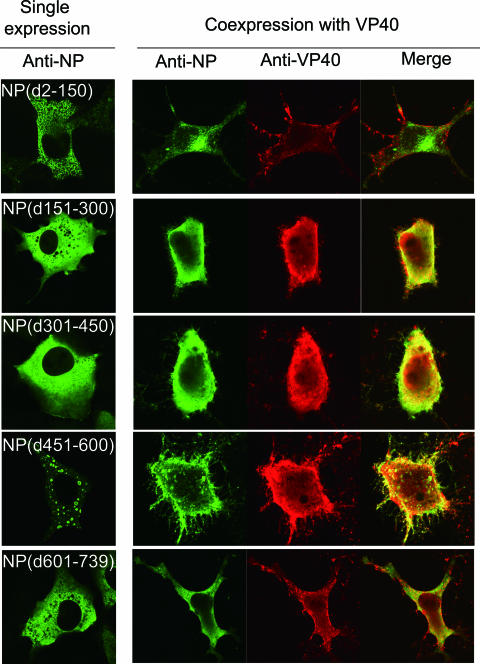
Intracellular localization of NP in the presence of VP40.
To assess whether the interaction of NP with VP40 affects the intracellular localization of these proteins, 293 cells coexpressing VP40 with wild-type NP or a deletion mutant NP were fixed at 24 h posttransfection and analyzed by indirect immunofluorescence microscopy. Expression of wild-type NP or NP(d451-600) alone resulted in the formation of large aggregates (Fig. 3) that resembled typical inclusion bodies in the perinuclear region (3, 19). NP(d151-300), NP(d301-450), and NP(d601-739) were diffusely distributed throughout the cytoplasm, while NP(d2-150) exhibited a punctuated pattern (Fig. 3). When expressed alone, VP40 was detected throughout the cytoplasm but was also strongly associated with the plasma membrane (Fig. 3). Coexpression with VP40 altered the localization of wild-type NP, causing it to colocalize with VP40 to the plasma membrane (Fig. 3). When NP(d151-300), NP(d301-450), or NP(d451-600), which bind to VP40, was coexpressed with VP40, they too colocalized with VP40 to the plasma membrane (Fig. 3). On the other hand, NP(d2-150) and NP(d601-739), which do not interact with VP40, were distributed in a diffuse pattern in the presence of VP40, although NP(d601-739) appeared to colocalize with VP40 in the cytoplasm (Fig. 3). These results indicated that NP interacts with VP40 intracellularly and that the NP-VP40 complex is present at the budding site.
FIG. 3.
Intracellular distribution of VP40 and NPs. 293T cells expressing NP alone, VP40 alone, or both VP40 and wild-type (wt) or mutant NP were fixed, permeabilized, and treated with a rabbit anti-VP40 and a mouse anti-NP antibody. Alexa Fluor 488-conjugated goat anti-mouse IgG (green) and Alexa Fluor 594-conjugated donkey anti-rabbit IgG (red) were used as secondary antibodies.
The NP-VP40 interaction is essential for nucleocapsid incorporation into VLPs.
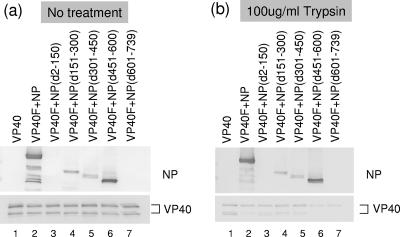
To determine whether mutant NPs are incorporated into VP40-induced VLPs, supernatants from cells coexpressing VP40 and a deletion mutant NP were subjected to a trypsin protection assay. As described previously (14), VP40 was detected in culture medium with either no treatment (Fig. 4a) or trypsin treatment alone (Fig. 4b) but not upon trypsin treatment in the presence of Triton X-100 (data not shown), indicating that VP40-induced VLPs, when released from cells, are enclosed in a lipid membrane. Wild-type NP, NP(d151-300), NP(d301-450), and NP(d451-600), all of which interact with VP40, were detected in supernatants (Fig. 4a, lanes 2 and 4 to 6) and were degraded by trypsin only in the presence of Triton X-100 (Fig. 4b, lanes 2 and 4 to 6, and data not shown), whereas NP(d2-150) and NP(d601-739), both of which do not coimmunoprecipitate with VP40 (Fig. 1), were not (Fig. 4a and b, lanes 3 and 7). These data demonstrate that the wild-type NP, NP(d151-300), NP(d301-450), and NP(d451-600) are enclosed in VLPs released into culture medium, and they suggest that both terminal regions of NP are necessary for the incorporation of NP into VLPs.
FIG. 4.
Protease protection assay of VLPs containing VP40 and wild-type or mutant NPs. VLPs were treated with (a) PBS or (b) trypsin. Samples were resolved by SDS-PAGE and detected by Western blotting with anti-NP and VP40 antibodies. NP(d301-450) exhibited two bands, possibly due to proteolysis.
Helix formation of NP is not essential for its incorporation into VLPs.
We recently demonstrated that wild-type NP, NP(d450-600), and NP(d601-739) form helical structures (43). Since NP helix formation appeared to be unrelated to the intracellular interaction of NP with VP40 (Fig. 2 and 3) or to the incorporation of NP into VLPs, as assayed biochemically (Fig. 4), we investigated the relationship between the NP helices and NP incorporation into VLPs ultrastructurally. As demonstrated previously (30), helical tubes of approximately 25 nm in diameter were formed in the cytoplasm upon expression of the wild-type NP alone and were incorporated into VP40-induced filamentous VLPs at the central axis when coexpressed with VP40 (Fig. 5a). When NP(d2-150) and VP40 were coexpressed, the distinctive structures of the NP tubes were not observed in the cytoplasm, and empty VLPs were found on the cell surface (Fig. 5b). When NP(d151-300) or NP(d301-450) was coexpressed with VP40, an electron-dense material, which reacted with an anti-NP antibody, was found along the central axis of the filamentous VLPs (Fig. 5c and d), even though these mutant NPs did not form helical tubes. In addition, in cells coexpressing NP(d301-450) and VP40, an electron-dense substance reacting with an anti-NP antibody was also found at the plasma membrane (Fig. 5d). When NP(d451-600) or NP(d601-739) was simultaneously expressed with VP40, helical tubes were found in the cytoplasm (Fig. 5e and f), as described previously (43). However, NP(d451-600), but not NP(d601-739), was incorporated into VLPs at the central axis of the filamentous structures (Fig. 5e). Taken together, our data demonstrate that both the N- and C-terminal 150 amino acids of NP are important for the interaction with VP40 and for NP incorporation into VLPs. In addition, the NP helices are not essential for NP incorporation into VLPs.
FIG. 5.
Electron microscopy of cells expressing VP40 and wild-type or mutant NPs. (a) Cell expressing VP40 and wild-type (wt) NP. NP helices were observed in VLPs budding from the cell surface as well as in the cytoplasm (upper panel). NPs within VLPs were immunolabeled with an anti-NP antibody conjugated with 5-nm gold particles (lower panel). (b) Cells expressing VP40 and NP(d2-150). NP helices were not found in the cytoplasm. Empty VLPs were released from cells. (c) Cells expressing VP40 and NP(d151-300). NP helices were not found in the cytoplasm, but electron-dense material (arrow), which reacted with an anti-NP antibody conjugated with 5-nm gold particles (lower panel), could be seen in VLPs. (d) Cells expressing VP40 and NP(d301-450). Small electron-dense material, which reacted with an anti-NP antibody conjugated with 5-nm gold particles (lower panel), was found at the plasma membrane (upper left panel, arrowheads) and in VLPs (upper right panel, arrowhead). (e) Cells expressing VP40 and NP(d451-600). NP helices were formed in the cytoplasm and incorporated into VLPs. (f) Cells expressing VP40 and NP(d601-739). NP helices were found in the cytoplasm but not in VLPs. Bars, 200 nm.
The NP-VP40 interaction is essential for nucleocapsid incorporation into VLPs.
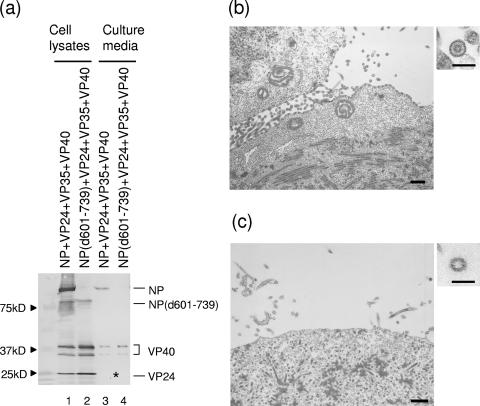
NP together with VP24 and VP35 forms nucleocapsid-like structures (12, 32). The C-terminal deletion mutant NP(d601-739) also forms nucleocapsid-like structures with VP24 and VP35 (43). We recently found that when VP40 is coexpressed with NP, VP24, and VP35, the nucleocapsid-like structures are transported to the plasma membrane and incorporated into VP40-induced VLPs (30). Here, we tested whether nucleocapsid-like structures composed of NP(d601-739), VP24, and VP35 are incorporated into VP40-induced VLPs. We found that in cells expressing wild-type NP, VP24, VP35, and VP40, the NP, VP24, and VP40 proteins were detected in the supernatant (Fig. 6a, lanes 1 and 3); however, only VP40 was detected in the supernatant when NP(d601-739) was expressed instead of the wild-type NP (Fig. 6a, lanes 2 and 4). By electron microscopy, we confirmed that nucleocapsids composed of wild-type NP were transported to the plasma membrane and incorporated into VLPs (Fig. 6b). On the other hand, the mutant nucleocapsids remained localized in the cytoplasm and were not found the beneath the plasma membrane, resulting in the release of empty VLPs (Fig. 6c). These results suggest that the NP-VP40 interaction at the C-terminal end of NP plays a critical role in nucleocapsid transport to plasma membrane and the incorporation into virions.
FIG. 6.
Impact of C-terminal deletion of NP on incorporation of nucleocapsid-like structures into VLPs. (a) Wild-type NP or NP(d601-739) was coexpressed with VP24, VP35, and VP40 in 293T cells. The cell lysates and VLPs from the supernatants were subjected to Western blotting with anti-NP, VP40, and VP24 antibodies. The asterisk indicates the position of VP24, which was detectable only when the immunostaining reaction was extended. (b) Electron micrograph of cells expressing wild-type NP, VP24, VP35, and VP40. Nucleocapsid-like structures were located at the plasma membrane and in VLPs. (c) Electron micrograph of cells expressing NP(d601-739), VP24, VP35, and VP40. Nucleocapsid-like structures remained in the cytoplasm and were not found in VLPs. Bars, 500 nm or 100 nm (insets).
DISCUSSION
Here, we have shown that both the N- and C-terminal regions of NP are important for NP interaction with VP40 and that the C-terminal region of NP is critical for the incorporation of nucleocapsids into the VLPs, an event that involves an interaction between NP and the matrix protein VP40. Vesicular stomatitis virus nucleoprotein interacts and colocalizes with its matrix protein at the plasma membrane (24, 33). Similarly, an interaction between Sendai virus NP and its matrix protein has been demonstrated in virus-infected cells by chemical cross-linking (22, 27, 46). Moreover, the C-terminal region of the NPs of human parainfluenza virus type 1 and Sendai virus (6) has been shown to be responsible for the interaction of NP with the matrix protein and for virion incorporation. Our finding is consistent with that of Licata et al. (21), who reported that the C-terminal 50 amino acids of Ebola virus NP are necessary for NP incorporation into VLPs, although in that study, other NP regions were not examined for their contributions to either NP or nucleocapsid incorporation into VLPs. Nevertheless, the accumulated data highlight the importance of the C-terminal region of NP for NP and nucleocapsid incorporation into virions via the interaction of NP with matrix proteins and suggest that this may be a common feature among the members of the Mononegavirales order. Because the C-terminal regions of the NPs of the members of the Mononegavirales order lack homology, unlike their N-terminal regions (37), the diversity of these C-terminal structures may be responsible for the specificity of the NP interactions with the individual matrix proteins.
As just stated, the N-terminal regions of NPs are conserved among the members of the Mononegavirales order. The N-terminal regions are thought to be important for the NP-NP interaction as well as for NP-RNA interactions (1, 4, 16, 17, 26, 28, 38). Our recent studies of Ebola virus NP (43) showed that the N-terminal 450 amino acids of NP are essential for the NP-NP interaction and that upon coexpression with VP24 and VP35, the mutant NP(d601-739), which does not bind to VP40, still forms transcription- and replication-competent nucleocapsids. Together with the current finding that the C-terminal region of NP plays an important role in NP incorporation into VLPs through the interaction of NP with VP40 (Fig. 1 and 3 to 5), these findings suggest that Ebola virus NP is composed of two domains, an NP-NP interaction domain at the N terminus and an NP-VP40 interaction domain at the C terminus. When NP forms its helical tube structures through the homologous interaction at the N terminus, the C-terminal domain may be exposed at the surface of NP helices to associate with VP40. Accordingly, these interacting domains may be targets for antiviral drug development.
Other viral proteins may also contribute to nucleocapsid incorporation into Ebola virions by interacting with VP40. Because VP35 is a component of nucleocapsids (12) and interacts with VP40 (15), it is a likely candidate for involvement in nucleocapsid incorporation into virions. However, when the NP-VP40 interaction was lost by the deletion of the C terminus of NP, nucleocapsid-like structures were not incorporated into VLPs even in the presence of VP35 (Fig. 6), suggesting that VP35 plays little or no role in nucleocapsid incorporation into virions. Another study reported that a single-stranded RNA with a specific sequence (5′-UGA-3′) binds to VP40 through phenylalanine at position 125 and arginine at position 134 of VP40 (9). Because the nucleocapsid-like structures would contain cellular RNA fragments, as do other viral nucleocapsid-like structures formed by NP expression (7, 13, 23, 34), it is possible that the interaction between the RNA fragment within the nucleocapsid-like structures and VP40 is involved in nucleocapsid incorporation into VLPs. To assess this hypothesis, we generated three VP40 mutants, one possessing a Phe-to-Ala mutation at position 125 (F125A), one possessing an Arg-to-Ala mutation at position 134 (R134A), and one with both mutations (F125A/R134A), none of which can interact with the RNA fragment (11), and coexpressed them with NP, VP24, and VP35. Biochemically and ultrastructurally, we found that nucleocapsid-like structures were incorporated into VLPs normally (data not shown), suggesting that the RNA-binding activity of VP40 is not involved in nucleocapsid incorporation. We therefore conclude that the NP-VP40 interaction plays a critical role in nucleocapsid incorporation into Ebola virus virions.
In conclusion, we have demonstrated that both terminal regions of NP are critical for the interaction of NP with VP40 and that the C-terminal region of NP is important for nucleocapsid incorporation into virions. The reciprocal interaction domain of VP40 (i.e., its NP-binding domain) has yet to be determined. Given the importance of the interaction between matrix proteins and nucleoproteins for nucleocapsid incorporation into virions, further characterization of this event is important for our understanding of the viral life cycle as well as for advancing strategies to combat Ebola virus infection.
Acknowledgments
We thank Susan Watson for editing the manuscript.
This work was supported by CREST (Japan Science and Technology Agency) and by grants-in-aid from the Ministries of Education, Culture, Sports, Science, Japan; by National Institute of Allergy and Infectious Diseases Public Health Service research grants; and by the NIH/NIAID Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (RCE) Program.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Bankamp, B., S. M. Horikami, P. D. Thompson, M. Huber, M. Billeter, and S. A. Moyer. 1996. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272-277. [DOI] [PubMed] [Google Scholar]
- 2.Bavari, S., C. M. Bosio, E. Wiegand, G. Ruthel, A. B. Will, T. W. Geisbert, M. Hevey, C. Schmaljohn, A. Schmaljohn, and M. J. Aman. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J. Exp. Med. 195:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, S., C. Rinne, U. Hofsass, H. D. Klenk, and E. Muhlberger. 1998. Interactions of Marburg virus nucleocapsid proteins. Virology 249:406-417. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, C. J., D. Spehner, R. Drillien, W. J. Neubert, and H. E. Homann. 1993. The conserved N-terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J. Virol. 67:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coronel, E. C., K. G. Murti, T. Takimoto, and A. Portner. 1999. Human parainfluenza virus type 1 matrix and nucleoprotein genes transiently expressed in mammalian cells induce the release of virus-like particles containing nucleocapsid-like structures. J. Virol. 73:7035-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronel, E. C., T. Takimoto, K. G. Murti, N. Varich, and A. Portner. 2001. Nucleocapsid incorporation into parainfluenza virus is regulated by specific interaction with matrix protein. J. Virol. 75:1117-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fooks, A. R., J. R. Stephenson, A. Warnes, A. B. Dowsett, B. K. Rima, and G. W. Wilkinson. 1993. Measles virus nucleocapsid protein expressed in insect cells assembles into nucleocapsid-like structures. J. Gen. Virol. 74:1439-1444. [DOI] [PubMed] [Google Scholar]
- 8.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomis-Ruth, F. X., A. Dessen, J. Timmins, A. Bracher, L. Kolesnikowa, S. Becker, H. D. Klenk, and W. Weissenhorn. 2003. The matrix protein VP40 from Ebola virus octamerizes into pore-like structures with specific RNA binding properties. Structure 11:423-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harty, R. N., M. E. Brown, G. Wang, J. Huibregtse, and F. P. Hayes. 2000. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc. Natl. Acad. Sci. USA 97:13871-13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoenen, T., V. Volchkov, L. Kolesnikova, E. Mittler, J. Timmins, M. Ottmann, O. Reynard, S. Becker, and W. Weissenhorn. 2005. VP40 octamers are essential for Ebola virus replication. J. Virol. 79:1898-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307-316. [DOI] [PubMed] [Google Scholar]
- 13.Iseni, F., A. Barge, F. Baudin, D. Blondel, and R. W. Ruigrok. 1998. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J. Gen. Virol. 79:2909-2919. [DOI] [PubMed] [Google Scholar]
- 14.Jasenosky, L. D., G. Neumann, I. Lukashevich, and Y. Kawaoka. 2001. Ebola virus VP40-induced particle formation and association with the lipid bilayer. J. Virol. 75:5205-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, R. F., S. E. McCarthy, P. J. Godlewski, and R. N. Harty. 2006. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J. Virol. 80:5135-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kho, C. L., W. S. Tan, B. T. Tey, and K. Yusoff. 2003. Newcastle disease virus nucleocapsid protein: self-assembly and length-determination domains. J. Gen. Virol. 84:2163-2168. [DOI] [PubMed] [Google Scholar]
- 17.Kingston, R. L., W. A. Baase, and L. S. Gay. 2004. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78:8630-8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobasa, D., M. E. Rodgers, K. Wells, and Y. Kawaoka. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J. Virol. 71:6706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolesnikova, L., E. Muhlberger, E. Ryabchikova, and S. Becker. 2000. Ultrastructural organization of recombinant Marburg virus nucleoprotein: comparison with Marburg virus inclusions. J. Virol. 74:3899-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y., L. Luo, M. Schubert, R. R. Wagner, and C. Y. Kang. 1993. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J. Virol. 67:4415-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Licata, J. M., R. F. Johnson, Z. Han, and R. N. Harty. 2004. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 78:7344-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markwell, M. A., and C. F. Fox. 1980. Protein-protein interactions within paramyxoviruses identified by native disulfide bonding or reversible chemical cross-linking. J. Virol. 33:152-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mavrakis, M., L. Kolesnikova, G. Schoehn, S. Becker, and R. W. Ruigrok. 2002. Morphology of Marburg virus NP-RNA. Virology 296:300-307. [DOI] [PubMed] [Google Scholar]
- 24.McCreedy, B. J. Jr., and D. S. Lyles. 1989. Distribution of M protein and nucleocapsid protein of vesicular stomatitis virus in infected cell plasma membranes. Virus Res. 14:189-205. [DOI] [PubMed] [Google Scholar]
- 25.Muhlberger, E., M. Weik, E. V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication system. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy, L. B., C. Loney, J. Murray, D. Bhella, P. Ashton, and R. P. Yeo. 2003. Investigations into the amino-terminal domain of the respiratory syncytial virus nucleocapsid protein reveal elements important for nucleocapsid formation and interaction with the phosphoprotein. Virology 307:143-153. [DOI] [PubMed] [Google Scholar]
- 27.Nagai, Y., T. Yoshida, M. Hamaguchi, M. Iinuma, K. Maeno, and T. Matsumoto. 1978. Cross-linking of Newcastle disease virus (NDV) proteins. Arch. Virol. 58:15-28. [DOI] [PubMed] [Google Scholar]
- 28.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, S. Kusagawa, H. Komada, and Y. Ito. 1999. Mapping of domains on the human parainfluenza virus type 2 nucleocapsid protein (NP) required for NP-phosphoprotein or NP-NP interaction. J. Gen. Virol. 80:2017-2022. [DOI] [PubMed] [Google Scholar]
- 29.Niwa, H., M. K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 30.Noda, T., H. Ebihara, Y. Muramoto, K. Fujii, A. Takada, H. Sagara, J. H. Kim, H. Kida, H. Feldmann, and Y. Kawaoka. 2006. Assembly and budding of Ebolavirus. PLoS Pathog. 2:e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noda, T., K. Aoyama, H. Sagara, H. Kida, and Y. Kawaoka. 2005. Nucleocapsid-like structures of Ebola virus reconstructed using electron tomography. J. Vet. Med. Sci. 67:325-328. [DOI] [PubMed] [Google Scholar]
- 33.Odenwald, W. F., H. Arnheiter, M. Dubois-Dalcq, and R. A. Lazzarini. 1986. Stereo images of vesicular stomatitis virus assembly. J. Virol. 57:922-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prehaud, C., R. D. Harris, V. Fulop, C. L. Koh, J. Wong, A. Flamand, and D. H. Bishop. 1990. Expression, characterization, and purification of a phosphorylated rabies nucleoprotein synthesized in insect cells by baculovirus vectors. Virology 178:486-497. [DOI] [PubMed] [Google Scholar]
- 35.Reid, S. P., L. W. Leung, A. L. Hartman, O. Martinez, M. L. Shaw, C. Carbonnelle, V. E. Volchkov, S. T. Nichol, and C. F. Basler. 2006. Ebola virus VP24 binds karyopherin α1 and blocks STAT1 nuclear accumulation. J. Virol. 80:5156-5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakaguchi, T., T. Uchiyama, Y. Fujii, K. Kiyotani, A. Kato, Y. Nagai, A. Kawai, and T. Yoshida. 1999. Double-layered membrane vesicles released from mammalian cells infected with Sendai virus expressing the matrix protein of vesicular stomatitis virus. Virology 263:230-243. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez, A., A. S. Khan, S. R. Zaki, G. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae: Marburg and Ebola viruses, p. 1279-1304. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, PA.
- 38.Sanchez, A., M. P. Kiley, H. D. Klenk, and H. Feldmann. 1992. Sequence analysis of the Marburg virus nucleoprotein gene: comparison to Ebola virus and other non-segmented negative-strand RNA viruses. J. Gen. Virol. 73:347-357. [DOI] [PubMed] [Google Scholar]
- 39.Schmitt, A. P., and R. A. Lamb. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145-196. [DOI] [PubMed] [Google Scholar]
- 40.Takada, A., C. Robimson, H. Goto, A Sanchez, K. G. Murti, M. A Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 94:14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takimoto, T., K. G. Murti, T. Bousse, R. A. Scroggs, and A. Portner. 2001. Role of matrix and fusion proteins in budding of Sendai virus. J. Virol. 75:11384-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Timmins, J., S. Scianimanico, G. Schoehn, and W. Weissenhorn. 2001. Vesicular release of Ebola virus matrix protein VP40. Virology 283:1-6. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe, S., T. Noda, and Y. Kawaoka. 2006. Functional mapping of the nucleoprotein of Ebola virus. J. Virol. 80:3743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watanabe, S., T. Watanabe, T. Noda, A. Takada, H. Feldmann, L. D. Jasenosky, and Y. Kawaoka. 2004. Production of novel Ebola virus-like particles from cDNAs: an alternative to Ebola virus generation by reverse genetics. J. Virol. 78:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weik, M., J. Modrof, H. D. Klenk, S. Becker, and E. Muhlberger. 2002. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J. Virol. 76:8532-8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida, T., Y. Nagai, S. Yoshii, K. Maeno, and T. Matsumoto. 1976. Membrane (M) protein of HVJ (Sendai virus): its role in virus assembly. Virology 71:143-161. [DOI] [PubMed] [Google Scholar]