Abstract
Respiratory syncytial virus (RSV) infection causes bronchiolitis and pneumonia in infants. RSV has a linear single-stranded RNA genome encoding 11 proteins, 2 of which are nonstructural (NS1 and NS2). RSV specifically downregulates STAT2 protein expression, thus enabling the virus to evade the host type I interferon response. Degradation of STAT2 requires proteasomal activity and is dependent on the expression of RSV NS1 and NS2 (NS1/2). Here we investigate whether RSV NS proteins can assemble ubiquitin ligase (E3) enzymes to target STAT2 to the proteasome. We demonstrate that NS1 contains elongin C and cullin 2 binding consensus sequences and can interact with elongin C and cullin 2 in vitro; therefore, NS1 has the potential to act as an E3 ligase. By knocking down expression of specific endogenous E3 ligase components using small interfering RNA, NS1/2, or RSV-induced STAT2, degradation is prevented. These results indicate that E3 ligase activity is crucial for the ability of RSV to degrade STAT2. These data may provide the basis for therapeutic intervention against RSV and/or logically designed live attenuated RSV vaccines.
Human respiratory syncytial virus (RSV) is the leading cause of severe lower respiratory tract infections in infants and young children (28, 31). RSV belongs to the genus Pneumovirus in the subfamily Pneumovirinae of the family Paramyxoviridae. It is an enveloped, nonsegmented negative-strand RNA virus encoding 11 proteins, including nucleocapsid proteins (N, P, and L), surface proteins (F and G), and a matrix protein (M). In addition, the genome encodes two nonstructural proteins (NS1 and NS2), the functions of which are less clearly defined. RSV primarily infects epithelial cells of the respiratory tract and replicates exclusively in the cytoplasm. Progeny RSV particles exit the host cell by budding through the apical surfaces of polarized cells (35).
In order to combat such infections, the immune system has evolved a potent antiviral response. Mediators, known as the type I interferons (alpha interferon [IFN-α] and IFN-β), stimulate the production of a range of antiviral gene products that limit virus replication and spread (4, 22). The type I IFN receptor consists of two subunits, IFNAR1 and IFNAR2, which are associated with the Janus kinases JAK1 and TYK2, respectively (23). Activation of these receptor tyrosine kinases results in tyrosine phosphorylation of signal transducer and activator of transcription 2 (STAT2) and STAT1. Activated STAT2 and STAT1 associate with interferon regulatory factor 9 (IRF-9) to form the transcriptional activator complex interferon-stimulated gene factor 3 (ISGF-3). These complexes translocate to the nucleus and bind IFN-stimulated response elements (ISRE) to initiate gene transcription and therefore antiviral immunity (8).
Wild-type RSV induces a weak type I IFN response following infection (27), suggesting that it has the capacity to evade this host defense mechanism in order to establish a successful infection. RSV is thought to block IFN-α and -β signaling by decreasing STAT2 protein levels (19). Recently, it has been reported that loss of STAT2 expression occurs via the 26S proteasome, since the addition of MG132, a proteasome inhibitor, can rescue STAT2 protein following RSV infection (19, 29). Recombinant RSV lacking the NS1 and NS2 genes induced high levels of IFN-α and -β in human pulmonary epithelial cells (A549) as well as in macrophages derived from primary human peripheral blood monocytes, highlighting the importance of these proteins in suppressing an antiviral response (29). Results with NS1 and NS2 single- and double-gene deletion viruses indicated that the two proteins function independently as well as coordinately to achieve the full inhibitory effect, with NS1 having a greater independent role (11, 29, 30). In contrast, a recent report by Ramaswamy et al. suggests that proteasomal degradation of STAT2 is dependent on NS2 protein only and that expression of NS2 in airway epithelial cells is sufficient to block type I IFN-dependent gene expression (25). However, these studies cannot formally exclude the involvement of other viral proteins in STAT2 degradation. Therefore, the relative importance of NS1 and NS2 in subverting the antiviral response and the precise mechanism by which proteasome-mediated degradation of STAT2 occurs remain unknown.
Proteasome-mediated degradation of proteins usually occurs after chains of four or more ubiquitin moieties have been linked to the target via lysine 48 of ubiquitin. Substrate ubiquitination requires a cascade of enzymatic reactions involving an E1 activating enzyme, an E2 conjugating enzyme, and finally an E3 ligating enzyme which covalently attaches ubiquitin to lysine side chains of the substrate protein (18). The ECS (elongin C-cullin-SOCS box)-type E3 ubiquitin ligases include the suppressor of cytokine signaling (SOCS) family of proteins and the Von Hippel-Lindau (VHL) tumor suppressor protein (9, 12, 17). These proteins assemble a multisubunit complex consisting of elongin BC, a cullin (Cul) family member, and the RING finger-containing protein Rbx (also referred to as Roc or Hrt), the latter of which stabilizes the interaction of cullin with the ubiquitin-conjugating enzyme (E2). Both SOCS proteins and VHL interact with elongin C via the BC box, a VxxLxxxCxxx(A/I/L/V) degenerate sequence. Elongin B binds to a short N-terminal region of elongin C and does not appear to interact directly with the BC box (10, 14, 17). Immediately downstream of the elongin C binding site, VHL interacts with Cul2, while some SOCS proteins are believed to interact with Cul5 (15).
Since RSV NS proteins lead to the proteasome-mediated degradation of STAT2, we investigated whether NS proteins possess the capacity to form a functional E3 ligase like VHL and SOCS proteins. Bioinformatic analysis identified a putative BC box and Cul binding site within NS1. Here we investigate whether NS proteins can interact with elongin C and Cul family members to form an active E3 ligase. We hypothesized that if NS proteins could form a competent E3 protein ligase, this may be the mechanism by which RSV downregulates STAT activity and the type I IFN response, thereby overcoming the host's antiviral defense mechanism.
MATERIALS AND METHODS
Constructs, antibodies, and reagents.
NS1, NS2, SOCS2, and Zap70 His-tagged fusion proteins, glutathione S-transferase (GST)-SOCS3, and cDNA encoding NS1 and NS2 were produced by Fusion Antibodies (Belfast, Northern Ireland). The NS1 and NS2 cDNA clones were derived from recent clinical isolates of RSV, kindly provided by Peter Coyle (Regional Virus Laboratory, RVH, Belfast, Northern Ireland). Both strands of the clones were sequenced using BigDye Terminator (version 3.1) cycle sequencing kits (Applied Biosciences, Cheshire, United Kingdom) according to the manufacturers' instructions. Global-Ref sequence alignments (Align plus 5, version 5.11, Clone Manager Professional Suite; Sci Ed Central, Cary, NC) between NS1 and NS2 clones used in this study and sequences obtained from GenBank indicated that the current clones were derived from RSV subgroup A. Plasmids for the expression of MYC-elongin C and FLAG-elongin B were obtained from Joan Conaway, while hemagglutinin (HA)-cullin 2, HA-cullin 5, and Rbx1 cDNA were kind gifts from Karen Murphy (The Walter and Eliza Institute of Medical Research, Melbourne, Australia). FLAG-STAT1 and STAT2 cDNA was obtained from Curt Horvath (Northwestern University, Evanston, IL), and HA-ubiquitin was a gift from Y. Yarden (Weizmann Institute, Israel). Small interfering RNA (siRNA) constructs directed against Cul2 (Cul2-1 and Cul2-2), Cul5 (Cul5-1 and Cul5-2), Rbx1 (Rbx1-1 and Rbx1-2), Rbx2 (Rbx2-1 and Rbx2-2), and a control siRNA empty vector were generously donated by Keiichi I. Nakayama (Kyushu University, Fukuoka, Japan) (15). FLAG M2, anti-MYC, anti-His, anti-STAT1, and anti-phospho-STAT1 monoclonal antibodies were purchased from Sigma Aldrich (Poole, United Kingdom). Monoclonal anti-hemagglutinin (HA) was purchased from Roche (Indianapolis, IN). Anti-STAT2 and anti-phospho-STAT2 were obtained from BD Biosciences (San Jose, CA) and Upstate, respectively. MG132 and LLnL (Calbiochem) were both used at a final concentration of 0.5 μM and were added to cells 3 h before lysing to block proteasomal activity. IFN-β was obtained from Sigma Aldrich (Poole, United Kingdom) and used to treat cells as stated in figure legends.
Protein alignments.
Protein alignments between RSV NS proteins, elongin C-interacting proteins (SOCS1 to -3 and VHL), and Cul2-interacting proteins (VHL, LRR1, and FEM1B) were performed using CLUSTALW sequence alignment software (http://www.ebi.ac.uk/clustalw/).
Cells and transfections.
293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco-BRL) supplemented with 10% fetal calf serum (FCS), 2 mM glutamine, 100 U/ml penicillin, and 100 U/ml streptomycin and were transfected using the calcium phosphate method described previously (2). HEp-2 cells (human laryngeal carcinoma derived; kindly provided by Ralph Tripp, University of Georgia) were grown in high-glucose DMEM with GlutaMax and pyruvate and supplemented with 5% FCS and gentamicin. HEp-2 cells were transfected using Lipofectamine 2000 (Invitrogen, Paisley, United Kingdom) according to the manufacturer's instructions.
Virus stocks.
Stocks of RSV subgroup A (strain A2) (kindly provided by Geraldine Taylor, Institute of Animal Health, Compton, United Kingdom) were cultured on HEp-2 cells in 175-cm2 tissue culture flasks by infecting with multiplicities of infection (MOI) of 0.1 to 0.5 for 1.5 to 2 h, rinsing twice with 5 ml phosphate-buffered saline (PBS), and feeding with DMEM supplemented with 1% FCS. At 24 h postinfection, the medium was removed, and the cells were rinsed twice with 5 ml PBS and then refed with DMEM containing 1% FCS. When an extensive cytopathic effect was observed, attached cells were scraped into the medium, and the suspension was pooled in 50-ml centrifuge tubes, vortexed thoroughly, sonicated for 10 min, and centrifuged at 400 × g for 10 min at 4°C. Supernatants were recovered, vortexed thoroughly, aliquoted into cryovials, and snap-frozen in liquid nitrogen. Virus titers were determined by a 50% tissue culture infectious dose assay, as previously described (24).
HEp-2 cell infection and IFN-β stimulation.
HEp-2 cells were seeded into 10-cm tissue culture dishes at 2 × 106 cells/dish. The following day, the monolayers were rinsed twice with 3 ml PBS and infected with RSV A2 at an MOI of 15. Controls were mock infected with maintenance medium. At 24 h postinfection, the cells were rinsed twice with 3 ml PBS and then 3 ml serum-free DMEM was added, with or without IFN-β, per plate. The cells were then incubated at 37°C for 30 min and lysed as described below.
Immunoprecipitation and immunoblotting.
293T and HEp-2 cells were washed in PBS supplemented with Na3VO4 (0.2 mM) and lysed by adding a buffer composed of 0.875% (vol:vol) Brij 97, 50 mM Tris, pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride (PMSF). For analysis of NS2 protein expression, 293T cells were lysed in 8 M urea supplemented with protease inhibitors. After 15 min, cell lysates were clarified by centrifugation (13,400 × g). A sample of the whole-cell lysate (WCL) was resuspended in Laemmli sample buffer containing 5% β-mercaptoethanol and boiled under reducing conditions for 5 min. For immunoprecipitation analysis, the remainder of the lysates were incubated for 1.5 h with beads that had been precoupled to antibody. The beads were subsequently collected by centrifugation and washed three times in lysis buffer before being resuspended in Laemmli sample buffer as described above. For immunoblot analysis, proteins were subjected to electrophoresis on 7.5% or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to Polyscreen polyvinylidene difluoride transfer membranes. The membranes were blocked in PBS supplemented with 0.2% Tween 20 and either 3% bovine serum albumin or 5% Marvel Milk and then incubated for 1 h (or overnight at 4°C) with a primary antibody. The following dilutions were used: 1:500 for anti-HA; 1:1,000 for anti-MYC, anti-STAT2, and anti-pSTAT2; 1:2,000 for anti-FLAG; 1:3,000 for anti-HIS, anti-STAT1, and anti-pSTAT1. Thereafter, the membranes were washed three times for 15 min in PBS supplemented with 0.2% Tween and were incubated for 1 h with peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin Gs (1:10,000) in PBS supplemented with 0.2% Tween 20 and 3% bovine serum albumin or 5% Marvel milk. The blots were extensively washed, and antibody binding was visualized by enhanced chemiluminescence (ECL; Amersham Biosciences, Chalfont, St, Giles, United Kingdom).
Fusion protein pulldown experiments.
293T cells were transfected as described previously (2). Cells were lysed in buffer containing 50 mM Tris-HCl, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM Na3VO4, 10 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mM PMSF. Lysates were centrifuged at 13,400 × g for 10 min at 4°C, and supernatants were incubated with 5 μg of His-tagged NS1, NS2, or the control His-tagged fusion protein SOCS2 or Zap70 (Fusion Antibodies, Belfast, Northern Ireland) which had been precoupled to 50 μl of 20% nickel-nitrilotriacetic acid (NTA) beads (Promega, Madison, WI). To study the association of NS1 and NS2 with elongin C, reaction mixtures were incubated for 2 to 4 h at 4°C in protein interaction buffer containing 50 mM NaH2PO4, 300 mM NaCl, and 10 mM imidazole. A more stringent protein interaction buffer containing 50 mM NaH2PO4, 400 mM NaCl, and 20 mM imidazole was used to examine the association of NS1 and NS2 with Cul2. After the resulting precipitates were washed in protein interaction buffer, the beads were resuspended in Laemmli sample buffer containing 5% β-mercaptoethanol and analyzed by SDS-PAGE.
RNA isolation and RT-PCR analysis.
RNA samples were extracted from 10 × 106 293T or HEp-2 cells using Stat60 reagent (Tel-Test Inc., Friendswood, TX). Reverse transcription-PCR (RT-PCR) was carried out using the One Step RT-PCR system (QIAGEN, Crawley, United Kingdom). The forward (F) and reverse (R) primers used to amplify cDNA are as follows: Cul5 (F), GATGATAAAGGCCCAGCAAA; Cul5 (R), GCTGCCCTGTTTACCCATTA; Rbx1 (F), TGCAGGAACCACATTATGGA; Rbx1 (R), TGGACACACCTGTCGTG TTT; Rbx2 (F), CTCCCTCAAGAAGTGGAACG; Rbx2 (R), ACATGCAGCAGTTGTGGAAG; Cul2 (F), CTTACTCCGTGCTGTGTCCA; Cul2 (R), GCCTTATCCAACGCACTCAT; β-actin (F), GGACTTCGAGCAAGAGATGG; β-actin (R), AGCACTGTGTTGGCGTACAG. RT-PCR products were analyzed by agaraose gel electrophoresis followed by ethidium bromide staining.
RESULTS
NS1 contains putative elongin C and Cul2 binding sites.
By directing proteasome-mediated degradation of STAT2, RSV induces a poor type I IFN response in host cells, thus allowing the virus to establish successful infections (19, 25). Although expression of RSV-encoded nonstructural proteins NS1 and NS2 is required for regulating STAT2 levels, the relative contribution of each NS protein and the precise mechanism by which STAT2 degradation occurs remain unknown. Since protein degradation is usually preceded by polyubiquitination of target substrates by E3 ubiquitin ligases, we investigated whether RSV NS proteins have the potential to form an E3 protein ligase through association with elongin C and a Cul family member.
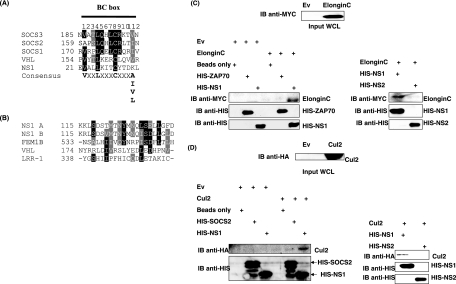
To establish whether NS proteins contain putative elongin C and Cul binding sites, CLUSTALW protein alignments between NS proteins and known elongin C/Cul-interacting proteins were performed (Fig. 1). Bioinformatic analysis revealed a putative elongin C-interacting site within NS1. Archetypal E3 ligases such as SOCS1 to -3 and VHL associate with elongin C through VxxLxxxCxxx(A/I/L/V) motifs (16, 17). NS1 also contains a VxxLxxxC sequence but possesses a lysine residue at position 12 which does not match the consensus of A/I/L/V (Fig. 1A). Despite this, NS1 may still have the ability to interact with elongin C. Interestingly, the putative NS1 elongin C binding motif is conserved among several recent clinical isolates and all NS1 genes currently deposited in GenBank (data not shown).
FIG. 1.
NS1 interacts with elongin C and Cul2. (A and B) CLUSTALW protein alignments of RSV NS1 protein and elongin C-interacting proteins (VHL and SOCS1 to -3) (A) and (B) Cul2-interacting proteins (VHL, FEM1B, LLR1). Identical (black) and similar (gray) amino acids are highlighted. (C) MYC-elongin C expressed in WCLs from 293T cells (top panel) were incubated with HIS-NS1 or HIS-Zap70 (control) precoupled to nickel-NTA beads. Membranes were immunoblotted (IB) with anti-MYC (left side, upper panel) to detect elongin C or with anti-HIS to detect HIS-Zap70 or HIS-NS1 protein (left side, lower panels). Association between MYC-elongin C and HIS-NS1 or HIS-NS2 was analyzed as above (right panels). (D) HA-Cul2 expressed in WCLs from 293T cells (top panel) was incubated with HIS-NS1 or HIS-SOCS2 (control) precoupled to nickel-NTA beads. Membranes were IB with anti-HA (left side, upper panel) to detect Cul2 or with anti-HIS to detect HIS-SOCS2 or HIS-NS1 (left side, lower panels). Association between HA-Cul2 and HIS-NS1 or HIS-NS2 was analyzed as above (right panels).
ECS ubiquitin ligases are classified into two groups depending on which Cul family member they interact with. SOCS proteins possess a Cul5 binding site (LPxP motif), while VHL is thought to interact with Cul2 (15). Since NS1 lacks an LPxP motif, we investigated whether homology existed between NS1 and the Cul2-interacting proteins VHL, FEM1B, and LRR-1, which had previously been identified by Kamura et al. The Cul2 binding site has been mapped only for VHL, and mutation of the LDIV residues prevents the VHL-Cul2 interaction (15). However, FEM1B and LRR-1 can still associate with Cul2 even though these proteins lack this LDIV motif, questioning the stringency of the Cul2-interacting site. Despite this, CLUSTALW protein alignments suggested some homology between these Cul2-interacting proteins and NS1 (Fig. 1B). Furthermore, the putative NS1 Cul2-interacting domain is conserved among recent clinical isolates and RSV strain A NS1 genes in GenBank (data not shown). RSV strain B NS1 sequence also shows homology to Cul2 binding proteins, most notably FEM1B (Fig. 1B). Therefore, as well as elongin C, NS1 may have the potential to associate with Cul2. We failed to find any homology between NS2 and elongin C/Cul-interacting proteins (data not shown), suggesting that NS2 cannot form an ECS-type E3 ligase. The possible function of NS2 will be discussed later.
NS1 associates with elongin C and Cul2 in vitro.
Following the discovery of putative elongin C and Cul2 binding sites within NS1, we next investigated whether NS1 could associate with elongin C and Cul2 in vitro by using a full-length His-tagged NS1 fusion protein. The HIS-ZAP70, HIS-SOCS2, or HIS-NS2 fusion protein was used as a control. Lysates from 293T cells expressing an empty vector (Ev), MYC-elongin C, or HA-Cul2 were incubated with 5 μg His-tagged fusion protein (4°C for 2 h) that had been precoupled to nickel-NTA beads. The beads were washed and the resulting interacting proteins visualized by SDS-PAGE. Elongin C expressed in lysates from 293T cells specifically associated with the HIS-NS1 fusion protein. Control reactions confirmed that elongin C did not bind to the nickel-NTA beads or the His tag of the fusion protein as demonstrated by HIS-ZAP70 (Fig. 1C, upper panel). Observations from CLUSTALW protein alignments suggesting that NS2 lacked an elongin C-interacting site were also reinforced in vitro using protein pulldown assays. Again, while HIS-NS1 bound elongin C, HIS-NS2 failed to interact with MYC-elongin C in lysates from 293T cells (Fig. 1C, upper panel). Equal quantities of HIS-ZAP70, HIS-NS1, and HIS-NS2 were present in all cases (Fig. 1C, lower panels).
Protein pulldown assays also confirmed that NS1 can interact with Cul2 in vitro (Fig. 1D, upper panel). Again, controls demonstrated that HA-Cul2 expressed in lysates from 293T cells did not bind nonspecifically to the nickel-NTA beads or to the His tag of the fusion protein. A HIS-SOCS2 fusion protein was used as a negative control in this instance, because SOCS2 does not contain a Cul2-interacting motif. We next investigated whether Cul2 could bind to a HIS-NS2 fusion protein. Again, Cul2 interacted with HIS-NS1 but failed to interact with HIS-NS2 (Fig. 1D, upper right panel). Equal quantities of HIS-SOCS2, HIS-NS1, and HIS-NS2 fusion proteins were present (Fig. 1D, lower panels) in each lane. These results suggest that NS1 has the potential to form part of an E3 ubiquitin ligase complex through its interaction with elongin C and Cul2. We hypothesized that this may be a potential mechanism by which RSV can earmark STAT2 for degradation.
As a further control, we investigated whether NS1 could bind to Cul5 in vitro. Using a GST-SOCS3 fusion protein as a positive control, we demonstrated that while SOCS3 could associate with both elongin C and Cul5 in vitro, NS1 was unable to associate with Cul5 in lysates from 293T cells (data not shown), suggesting that NS1 specifically interacts with Cul2 but not Cul5.
Effects of NS1 and NS2 on STAT1 and STAT2 in 293T cells.
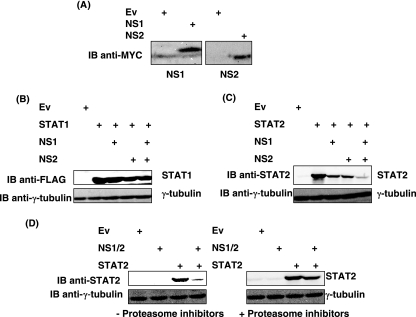
Previously, it has been reported that RSV infection results in a proteasome-dependent loss of STAT2 protein. However, the specific roles of NS1 and NS2 in decreasing STAT2 levels during RSV infection are controversial. While Ramaswamy et al. proposed that NS2 alone was sufficient to cause loss of STAT2 protein, Lo et al. suggested that both NS1 and NS2 were involved in STAT2 degradation (19, 25). To determine the relative contributions of RSV NS1 and NS2 to STAT2 degradation in coexpression studies, MYC-tagged cDNAs expressing NS1 or NS2 were constructed. These constructs were expressed transiently in 293T cells, but, in agreement with observations made by other groups (19), NS1 and NS2 proteins proved difficult to detect using standard cell lysis conditions and Western blotting techniques. However, expression of NS1 and NS2 proteins was observed when cells were treated with proteasome inhibitors or when cells were lysed using 8 M urea, respectively (Fig. 2A), suggesting that NS1 and NS2 proteins are incredibly unstable or insoluble. NS1 and NS2 cDNAs were expressed individually or in combination in 293T cells transfected with cDNA encoding FLAG-STAT1 (control) or STAT2. As shown in Fig. 2B (upper panel), expression of NS1, NS2, or both NS proteins did not cause a marked reduction in STAT1 protein levels. In comparison, while NS1 or NS2 alone did decrease STAT2 levels, the addition of both NS1 and NS2 together caused the greatest decrease in STAT2 protein levels, with STAT2 being almost undetectable (Fig. 2C, upper panel). In both experiments, immunoblots were reprobed with γ- tubulin to show equal loading (Fig. 2B and C, lower panels). Therefore, while NS1 and NS2 alone are sufficient to cause a decrease in STAT2 levels, loss of STAT2 is enhanced by the presence of both NS proteins. The possible roles of NS2 in STAT2 degradation are discussed later.
FIG. 2.
Effects of NS1 and NS2 on STAT1 and STAT2 in 293T cells. (A) Ev, or NS1 or NS2 cDNA, was transfected into 293T cells. Cells were treated with proteasome inhibitors (MG132 and LLnL) 4 h prior to lysis to confirm NS1 expression (left panel) or lysed in 8 M urea to analyze NS2 expression (right panel). (B and C) Cells were transfected with cDNA encoding FLAG-STAT1 (B) or STAT2 (C) in the presence of NS1, NS2, or both NS1 and NS2. Membranes were probed with anti-FLAG to detect STAT1 or anti-STAT2 (upper panels) and reprobed with anti-γ-tubulin to confirm equal protein loading in all lanes (lower panels). (D) Loss of STAT2 protein by NS1 and NS2 can be prevented by blocking proteasomal activity. 293T cells were transfected with vectors expressing STAT2 in the absence and presence of NS1/2. Cells were treated with and without proteasome inhibitors (MG132 and LLnL) 3 h before lysing. STAT2 levels were analyzed by immunoblotting membranes with anti-STAT2 (upper panels). Membranes were reprobed with anti-γ-tubulin (lower panels).
Since robust loss of STAT2 in 293T cells was observed when both NS1 and NS2 proteins were present, we next investigated whether loss of STAT2 protein by NS1/2 in 293T cells was proteasome mediated. To do this, 293T cells were transfected with cDNA encoding STAT2 in the presence or absence of NS1 and NS2 (NS1/2). Cells were then treated with and without proteasome inhibitors (MG132 and LLnL) for 3 h to block proteasome activity. As before, STAT2 levels were markedly reduced in the presence of NS1/2 alone. However, STAT2 protein levels were rescued by the addition of proteasome inhibitors (Fig. 2D, upper panels). Again, immunoblots were reprobed with anti-γ-tubulin to demonstrate equal loading in all lanes (Fig. 2D, lower panels). This confirms previous results suggesting that NS1/2-dependent loss of STAT2 occurs via proteasomal degradation.
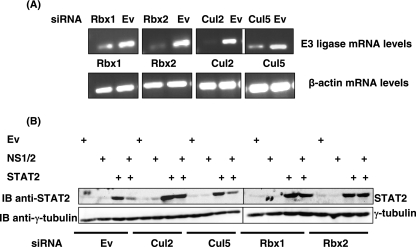
NS1/2-dependent loss of STAT2 is inhibited by a siRNA directed against E3 ligase components.
Since NS1 has the capacity to form part of an E3 ligase complex through its association with elongin C and Cul2 (Fig. 1), we proposed that this may be the mechanism by which NS1/2 can target STAT2 for proteasome-mediated degradation. Our hypothesis predicts that by knocking down the expression of E3 ligase components (which are ubiquitously expressed in 293T cells [data not shown]) using siRNA, STAT2 levels will be rescued in the presence of NS1/2. Initially, siRNA constructs directed against Cul2 (Cul2-1 and Cul2-2), Cul5 (Cul5-1 and Cul5-2), Rbx1 (Rbx1-1 and Rbx1-2), Rbx2 (Rbx2-1 and Rbx2-2), or a siRNA empty vector as a control were transfected into 293T cells. RNA was isolated 48 h later, and RT-PCR was performed using primers specific to cDNA encoding each protein or β-actin (control) to determine the effects of individual siRNAs on transcript levels. mRNA levels were analyzed, since commercially available antibodies to detect protein levels of endogenous E3 ligase components were unsatisfactory. As shown in Fig. 3A, siRNA constructs effectively and specifically reduced the levels of mRNA for each E3 ligase component (top panel), while β-actin mRNA levels were unaltered (lower panel).
FIG. 3.
NS1/2-dependent loss of STAT2 is inhibited by a siRNA directed against E3 ligase components. (A) Effect of siRNA on E3 ligase mRNA levels in 293T cells. 293T cells were transfected with a siRNA empty vector (Ev) control or with siRNA directed against Cul2 (Cul2-1 and Cul2-2), Cul5 (Cul5-1 and Cul5-2), Rbx1 (Rbx1-1 and Rbx1-2), or Rbx2 (Rbx2-1 and Rbx2-2). At 48 h after transfection, RNA was isolated and RT-PCR performed using primer pairs specific to each E3 ligase component (upper panels). Control reactions with β-actin primers confirmed equal quantities of RNA in each reaction (lower panels). (B) Loss of STAT2 by NS1/2 can be inhibited by downregulating Cul and Rbx expression. 293T cells were transfected with a siRNA empty vector (Ev) control or with a siRNA directed against Cul2 (Cul2-1 and Cul2-2), Cul5 (Cul5-1 and Cul5-2), Rbx1 (Rbx1-1 and Rbx1-2) or Rbx2 (Rbx2-1 and Rbx2-2) in the presence of STAT2 and/or NS1/2. Lysates were analyzed for STAT2 expression by immunoblotting with a STAT2 antibody (upper panel) and were reprobed with an antibody against γ-tubulin (lower panel).
Since these constructs successfully downregulated mRNA transcript levels for each Cul and Rbx protein, we investigated their effects on STAT2 protein in the presence and absence of NS1/2. siRNA constructs directed against Cul2 (Cul2-1 and Cul2-2), Cul5 (Cul5-1 and Cul5-2), Rbx1 (Rbx1-1 and Rbx1-2), Rbx2 (Rbx2-1 and Rbx2-2), or a siRNA empty vector as a control were transfected into 293T cells in the presence of STAT2 and/or NS1/2 (Fig. 3B). STAT2 protein was significantly reduced in the presence of NS1/2 alone, while siRNA directed against Cul2, Rbx1, and Rbx2 restored STAT2 protein levels. The presence of Cul5 siRNA did not hamper the ability of NS1/2 to degrade STAT2 (Fig. 3B). These observations suggest that loss of STAT2 in association with NS1/2 expression is dependent on Cul2 and either Rbx1 or Rbx2. In addition, these data are consistent with our findings that NS1 interacts with Cul2 (Fig. 1D). Essentially, upon downregulating expression of Cul2, NS1/2 is unable to direct STAT2 for degradation.
Loss of STAT2 protein in RSV-infected HEp-2 cells occurs via the proteasome.
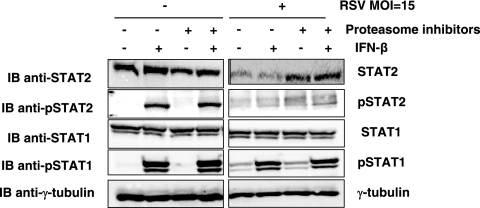
An important extension of this work was to determine whether observations made in coexpression studies could be reproduced during RSV infection. To explore this, HEp-2 cells were mock infected or infected with RSV (strain A2) at an MOI of 15. Cells were subsequently treated in the presence or absence of proteasome inhibitors (0.5 μM MG132 and LLnL for 3 h) and with and without IFN-β (1,000 U/ml for 30 min). Cells were lysed, and the levels of STAT1, STAT2, phosphorylated STAT1 (pSTAT1), and phosphorylated STAT2 (pSTAT2) were determined by SDS-PAGE followed by immunoblotting. RSV infection at an MOI of 15 caused a decrease in detectable STAT2 protein, while the levels of STAT1 remained unaltered (Fig. 4). The reduction of STAT2 protein levels following RSV infection occurred independently of IFN-β treatment but required proteasomal activity, as the addition of proteasome inhibitors prevented STAT2 degradation (Fig. 4, top panel). These observations are consistent with RSV specifically causing a proteasome-dependent loss of STAT2.
FIG. 4.
Loss of STAT2 expression in HEp-2 cells infected with RSV. HEp-2 cells were either mock infected or infected with RSV at an MOI of 15. After 24 h, cells were treated with and without IFN-β (1,000 U/ml for 30 min) both in the presence and in the absence of proteasome inhibitors (MG132 and LLnL), which were added 3 h prior to lysis. Whole-cell lysates were analyzed for STAT1, STAT2, phosphorylated STAT1 (pSTAT1), phosphorylated STAT2 (pSTAT2), or γ-tubulin.
RSV-dependent loss of STAT2 in HEp-2 cells is rescued by siRNA directed against Rbx1 and Cul2.
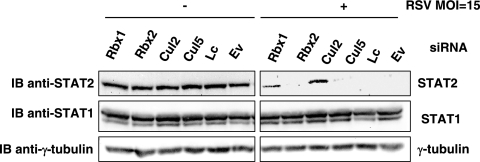
To determine if loss of STAT2 in HEp-2 cells following RSV infection is dependent on E3 ligase activity, siRNA directed against Cul2 (Cul2-1 and Cul2-2), Cul5 (Cul5-1 and Cul5-2), Rbx1 (Rbx1-1 and Rbx1-2), Rbx2 (Rbx2-1 and Rbx2-2), or a siRNA empty vector as a control were transfected into HEp-2 cells and cells were mock infected or infected with RSV at an MOI of 15. Cells were subsequently lysed and STAT2, STAT1, and γ-tubulin levels analyzed by SDS-PAGE. When siRNA directed against Rbx1 or Cul2 was introduced in combination with RSV infection, STAT2 protein was visibly rescued (Fig. 5). In comparison, a siRNA against Cul5 or Rbx2 did not prevent STAT2 degradation following RSV infection. Immunoblots were reprobed with anti-γ-tubulin to confirm equal protein in all lanes (lower panel). These observations suggest that RSV-dependent loss of STAT2 requires Cul2 and Rbx1 but not Cul5 or Rbx2.
FIG. 5.
Loss of STAT2 protein in RSV-infected HEp-2 cells is rescued with a siRNA against Rbx1 or Cul2. HEp-2 cells were transfected with a siRNA empty vector (Ev) or a siRNA directed against Cul2 (Cul2-1 and Cul2-2), Cul5 (Cul5-1 and Cul5-2), Rbx1 (Rbx1-1 and Rbx1-2), or Rbx2 (Rbx2-1 and Rbx2-2). A Lipofectamine transfection control was also included (Lc). Cells were then either mock infected or infected with RSV at an MOI of 15. Whole-cell lysates were analyzed for STAT2 (upper panels), STAT1 (middle panels), or γ- tubulin (lower panels) expression.
STAT2 is modified by ubiquitin.
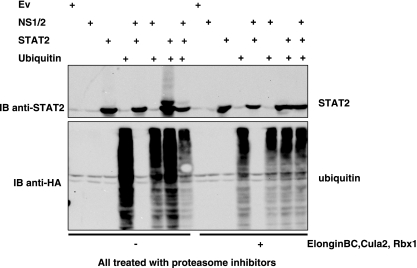
We next investigated whether STAT2 could be modified by ubiquitin in 293T cells. Cells were transfected with combinations of STAT2, NS1/2, and HA-ubiquitin both in the presence and in the absence of coexpressed E3 ligase components elongin BC, Cul2, and Rbx1. In all cases, cells were treated with proteasome inhibitors (MG132 and LLnL) to prevent NS1/2-induced proteasome-mediated STAT2 degradation. Lysates were analyzed for STAT2 and HA-ubiquitin expression (Fig. 6). When STAT2 and ubiquitin were coexpressed in the absence of exogenous E3 ligase components, a monoubiquitinated form of STAT2 was apparent and HA-ubiquitin remained in lower-molecular-weight forms, like HA-ubiquitin expressed alone. Interestingly, in the presence of NS1/2, monoubiquitinated STAT2 levels were significantly reduced and the majority of ubiquitin resided in higher-molecular-weight complexes, suggesting that NS1/2 can drive polyubiquitination. We next investigated the ubiquitin status of STAT2 when the E3 ligase components elongin BC, Cul2, and Rbx1 were coexpressed with combinations of NS1/2, STAT2, and HA-ubiquitin. When E3 ligase activity was enhanced by the introduction of exogenous E3 ligase components, monoubiquitinated STAT2 was not detected, and again HA-ubiquitin was incorporated into higher-molecular-weight complexes. This result was indistinguishable from that observed when NS1/2 was coexpressed with STAT2 and HA-ubiquitin alone (Fig. 6). These observations suggest that expression of NS1/2 or expression of exogenous elongin BC, Cul2, and Rbx1 both decrease STAT2 monoubiquitination and force HA-ubiquitin into higher complexes. They also imply that E3 ligase activity (as measured by the HA-ubiquitin banding pattern) can be driven by expressing either NS1/2 or the E3 ligase component elongin BC, Cul2, or Rbx1.
FIG. 6.
STAT2 is modified by ubiquitin. 293T cells were transfected with vectors expressing NS1/2, STAT2, and HA-ubiquitin both in the presence and in the absence of exogenous E3 ligase components elongin BC, Cul2, and Rbx1. Cells were treated with proteasome inhibitors (MG132 and LLnL), which were added 3 h prior to lysis. Whole-cell lysates were analyzed for STAT2 (upper panel) or ubiquitin (lower panel).
DISCUSSION
Due to the potent antiviral effects of the host type I interferon response, it is not surprising that viruses have evolved several mechanisms to downregulate this signaling pathway in order to enable successful viral replication. STAT degradation is a common mechanism by which paramyxoviruses overcome the antiviral effects of interferons (6, 7). Both mumps virus and simian virus 5 (SV5) degrade STAT1, while human parainfluenza virus type 2 (hPIV2) degrades STAT2, thereby hampering IFN-α/β signal transduction (34). The mechanisms by which these viruses induce STAT degradation are well established and are dependent on the formation of multisubunit E3 ligase complexes assembled around the virus-encoded V protein (21). Active E3 ligase activity also requires host-encoded factors including DDB1 (a UV-damaged DNA binding protein) (1) and members of the cullin family. These active virus degradation complexes can then polyubiquinate target STAT proteins, resulting in their effective degradation via the proteasome (32, 7).
Like hPIV2, RSV also subverts the host type I interferon response by mediating STAT2 degradation, and it has been shown, in this and in previous studies, that degradation is dependent on proteasomal activity (19, 29). However, since RSV lacks a V protein, the mechanism by which RSV directs STAT2 to the proteasome has remained elusive. Although the expression of RSV nonstructural proteins NS1 and/or NS2 have been implicated in mediating STAT2 degradation, the relative contribution of each protein and the mechanism by which they target STAT2 has not been explored. Using a bioinformatics approach, we have identified putative elongin C and cullin 2 binding sites within NS1 and confirmed that NS1 can specifically interact with these proteins in vitro. This result suggests that NS1 has the capacity to act as an ECS-like E3 ubiquitin ligase, a capacity similar to that reported for other viral proteins. For example, Vif (viral infectivity factor) encoded by HIV-1 interacts with elongin C and cullin 5 to direct the ubiquitination and proteasome-dependent degradation of APOBEC3G, a host factor that causes hypermutation in newly synthesized viral DNA (26). Like the putative elongin C binding site within NS1 (VxxLxxxCxxxK), the elongin C binding site in Vif (VxxLxxxAxxxL) does not conform precisely to the consensus interacting sequence [VxxLxxxCxxx(A/I/L/V), with NS1 and Vif each differing by a single amino acid. Despite this, Vif can still associate with elongin C in coimmunoprecipitation experiments (20), and this supports our findings that NS1 can also interact with elongin C in vitro. In combination, these data therefore raise questions concerning how stringent the elongin C binding site must be for an interaction to occur.
Since bioinformatic analysis and in vitro protein pulldown experiments suggest that NS1 can act as a scaffold on which a multisubunit E3 ligase complex can be formed, we determined whether expression of these host-encoded E3 ligase components was essential for NS1/2-induced STAT2 degradation. Using a siRNA approach to knock down expression of cullin or Rbx family members, we confirmed that NS1/2-induced STAT2 degradation in coexpression studies and during RSV infection was dependent on Cul2 but not Cul5. This result, combined with our observation that NS1 interacts with Cul2 in vitro, suggests that NS1 forms an active E3 ligase complex containing Cul2 to direct STAT2 degradation. When a siRNA directed against Rbx1 or Rbx2 was introduced, NS1/2 failed to effectively degrade STAT2 in 293T cells, while during RSV infection, STAT2 degradation was dependent on Rbx1 only. Although Kamura et al. have attempted to classify ECS-type E3 ligases as requiring Cul2-Rbx1 or Cul5-Rbx2, Cul5 can utilize either Rbx member to form an active E3 (13, 15), and the same may be true for Cul2. The cellular environment or the availability of the E2 conjugating enzyme may dictate which Rbx family member is utilized by Cul2. In 293T cells, Cul2 proteins may be promiscuous and may associate with either Rbx family member to form active E3 ligases, but Cul2 may preferentially associate with Rbx1 to form an active E3 ligase in HEp-2 RSV-infected cells.
We also investigated the effect of NS1/2 on STAT2 ubiquitination. The presence of NS1/2 or coexpression of exogenous E3 ligase components (elongin BC, Cul2, and Rbx1) both resulted in a decrease in STAT2 monoubiquitination and accumulation of high-molecular-weight ubiquitin complexes. This suggests that E3 ligase activity in 293T cells may be enhanced by either expressing NS1/2 or coexpressing exogenous E3 ligase components, further reinforcing the ability of NS1/2 to function as an E3 ligase.
The involvement of NS1 and/or NS2 in antagonizing the host IFN response is unclear. In one report investigating the role of NS1 in RSV infection using a siRNA targeting the NS1 gene (siNS1), the expression of IFN-β and IFN-inducible genes in RSV-infected A549 cells was upregulated. In addition, mice treated intranasally with siNS1 nanoparticles before or after RSV infection showed substantially decreased virus titers in lungs and decreased airway inflammation (36, 3), suggesting that NS1 is a major RSV virulence factor. In contrast, airway epithelial cells infected with RSV lacking NS2 did not result in decreased STAT2 levels or loss of type 1 IFN signaling, indicating that NS2 is necessary for RSV effects on STAT2 (25). Similarly, deletion of the NS2 gene severely attenuates RSV by restricting viral replication both in RSV-seropositive and in RSV-seronegative humans (33). However, other reports using NS1 and NS2 single and double gene deletion viruses indicate that these proteins function independently as well as coordinately to downregulate IFN-α/β production in the human alveolar epithelial cell line A549, as well as in macrophages derived from primary human peripheral blood monocytes (29, 30). NS1 and NS2 may therefore form the basis of complementary as well as distinct functions whose combined effects ensure sufficient downregulation of the host antiviral response to facilitate replication.
We studied the relative contributions of NS1, NS2, or a combination of both in mediating STAT2 degradation in coexpression studies. In agreement with the results observed for A549 cells (29, 30), while NS1 and NS2 did induce loss of STAT2, a combination of NS1 and NS2 caused the greatest decrease in STAT2 levels. STAT1 levels remained unaffected by the presence of NS1 and/or NS2. This result implies that although NS1 may form an E3 ligase complex to target STAT2, effective STAT2 degradation requires both NS1 and NS2. It may be that NS2 is required to bring STAT2 into close proximity to the NS1 E3 ligase or to stabilize or regulate the multisubunit complex. Our attempts to demonstrate an interaction between NS proteins and STAT2 were unsuccessful and were hampered by the inherent instability of NS1 and NS2. Throughout this study, NS1/2 expression could be demonstrated only by the addition of proteasome inhibitors or by lysing cells in 8 M urea. Work is ongoing to define the conditions that mediate STAT2 binding to the E3 ubiquitin ligase. A complete NS1 E3 ligase may need to assemble before STAT2 associates, or other viral proteins may be necessary to mediate or stabilize STAT2 association with NS1. In support of the latter possibility, NS1 can associate with the matrix (M) protein in coimmunoprecipitation experiments (5). We also found that expression of NS2 alone caused a reduction in STAT2 protein levels, ssuggesting that NS2 provides RSV with an alternative mechanism to downregulate STAT2. Since NS2 cannot interact with elongin C and Cul2, we believe it cannot form an E3 ligase. Further work will identify NS2-interacting proteins, which may help us to define the mechanism by which NS2 expression results in STAT2 degradation.
Further understanding of the mechanisms that RSV employs to evade the host antiviral response will benefit the development of novel RSV therapies. Since NS1 forms a complex with E3 ligase components to degrade STAT2, interrupting the NS1-elongin C-Cul2 interaction may be an attractive therapeutic target to overcome RSV infection. In addition, polymorphisms identified within the NS1 elongin C and/or Cul2 binding sites may be useful RSV virulence determinants and could aid in the identification and development of novel vaccine strains.
Acknowledgments
This work has been supported by the Wellcome Trust (grant 070304/2/03/2 to J.A.J.) and Queens University Belfast (start-up package, to U.F.P.).
We thank G. Clarke and B. O'Loughlan for continuing support and excellent technical assistance.
Footnotes
Published ahead of print on 24 January 2007.
REFERENCES
- 1.Andrejeva, J., E. Poole, D. F. Young, S. Goodbourn, and R. E. Randall. 2002. The p127 subunit (DDB1) of the UV-DNA damage repair binding protein is essential for the targeted degradation of STAT1 by the V protein of the paramyxovirus simian virus 5. J. Virol. 76:11379-11386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Short protocols in molecular biology, 3rd ed. John Wiley and Sons, Inc., New York, NY.
- 3.Bitko, V., A. Musiyenko, O. Shulyayeva, and S. Barik. 2005. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 11:50-55. [DOI] [PubMed] [Google Scholar]
- 4.Decker, T., M. Muller, and S. Stockinger. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 5:675-687. [DOI] [PubMed] [Google Scholar]
- 5.Evans, J. E., P. A. Cane, and C. R. Pringle. 1996. Expression and characterisation of the NS1 and NS2 proteins of respiratory syncytial virus. Virus Res. 43:155-161. [DOI] [PubMed] [Google Scholar]
- 6.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2002. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 12:337-357. [DOI] [PubMed] [Google Scholar]
- 7.Horvath, C. M. 2004. Silencing STATs: lessons from paramyxovirus interferon evasion. Cytokine Growth Factor Rev. 15:117-127. [DOI] [PubMed] [Google Scholar]
- 8.Horvath, C. M., G. R. Stark, I. M. Kerr, and J. E. Darnell, Jr. 1996. Interactions between STAT and non-STAT proteins in the interferon-stimulated gene factor 3 transcription complex. Mol. Cell. Biol. 16:6957-6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ivan, M., and W. G. Kaelin, Jr. 2001. The von Hippel-Lindau tumor suppressor protein. Curr. Opin. Genet. Dev. 11:27-34. [DOI] [PubMed] [Google Scholar]
- 10.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. USA 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin, H., H. Zhou, X. Cheng, R. Tang, M. Munoz, and N. Nguyen. 2000. Recombinant respiratory syncytial viruses with deletions in the NS1, NS2, SH, and M2-2 genes are attenuated in vitro and in vivo. Virology 273:210-218. [DOI] [PubMed] [Google Scholar]
- 12.Johnston, J. A. 2004. Are SOCS suppressors, regulators, and degraders? J. Leukoc. Biol. 75:743-748. [DOI] [PubMed] [Google Scholar]
- 13.Kamura, T., D. Burian, Q. Yan, A. L. Schmidt, W. S. Lane, E. Querido, P. E. Branton, A. Shilatifard, R. C. Conaway, and J. W. Conaway. 2001. Muf1, a novel elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276:29748-29753. [DOI] [PubMed] [Google Scholar]
- 14.Kamura, T., M. N. Conrad, Q. Yan, R. C. Conaway, and J. W. Conaway. 1999. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13:2928-2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamura, T., K. Maenaka, S. Kotoshiba, M. Matsumoto, D. Kohda, R. C. Conaway, J. W. Conaway, and K. I. Nakayama. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, Jr., R. C. Conaway, and J. W. Conaway. 1998. The elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kile, B. T., B. A. Schulman, W. S. Alexander, N. A. Nicola, H. M. Martin, and D. J. Hilton. 2002. The SOCS box: a tale of destruction and degradation. Trends Biochem. Sci. 27:235-241. [DOI] [PubMed] [Google Scholar]
- 18.Liu, Y. C., J. Penninger, and M. Karin. 2005. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5:941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo, M. S., R. M. Brazas, and M. J. Holtzman. 2005. Respiratory syncytial virus nonstructural proteins NS1 and NS2 mediate inhibition of Stat2 expression and alpha/beta interferon responsiveness. J. Virol. 79:9315-9319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 21.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 22.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 23.Platanias, L. C., S. Uddin, and O. R. Colamonici. 1994. Tyrosine phosphorylation of the alpha and beta subunits of the type I interferon receptor. Interferon-beta selectively induces tyrosine phosphorylation of an alpha subunit-associated protein. J. Biol. Chem. 269:17761-17764. [PubMed] [Google Scholar]
- 24.Power, U. F., H. Plotnicky-Gilquin, T. Huss, A. Robert, M. Trudel, S. Stahl, M. Uhlen, T. N. Nguyen, and H. Binz. 1997. Induction of protective immunity in rodents by vaccination with a prokaryotically expressed recombinant fusion protein containing a respiratory syncytial virus G protein fragment. Virology 230:155-166. [DOI] [PubMed] [Google Scholar]
- 25.Ramaswamy, M., L. Shi, S. M. Varga, S. Barik, M. A. Behlke, and D. C. Look. 2006. Respiratory syncytial virus nonstructural protein 2 specifically inhibits type I interferon signal transduction. Virology 344:328-339. [DOI] [PubMed] [Google Scholar]
- 26.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2004. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10:291-297. [DOI] [PubMed] [Google Scholar]
- 27.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sigurs, N. 2001. Epidemiologic and clinical evidence of a respiratory syncytial virus-reactive airway disease link. Am. J. Respir. Crit. Care Med. 163:S2-S6. [DOI] [PubMed] [Google Scholar]
- 29.Spann, K. M., K. C. Tran, B. Chi, R. L. Rabin, and P. L. Collins. 2004. Suppression of the induction of alpha, beta, and lambda interferons by the NS1 and NS2 proteins of human respiratory syncytial virus in human epithelial cells and macrophages. J. Virol. 78:4363-4369. (Erratum, 78:6705.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spann, K. M., K. C. Tran, and P. L. Collins. 2005. Effects of nonstructural proteins NS1 and NS2 of human respiratory syncytial virus on interferon regulatory factor 3, NF-κB, and proinflammatory cytokines. J. Virol. 79:5353-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tripp, R. A., C. Oshansky, and R. Alvarez. 2005. Cytokines and respiratory syncytial virus infection. Proc. Am. Thorac. Soc. 2:147-149. [DOI] [PubMed] [Google Scholar]
- 32.Ulane, C. M., and C. M. Horvath. 2002. Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304:160-166. [DOI] [PubMed] [Google Scholar]
- 33.Wright, P. F., R. A. Karron, S. A. Madhi, J. J. Treanor, J. C. King, A. O'Shea, M. R. Ikizler, Y. Zhu, P. L. Collins, C. Cutland, V. B. Randolph, A. M. Deatly, J. G. Hackell, W. C. Gruber, and B. R. Murphy. 2006. The interferon antagonist NS2 protein of respiratory syncytial virus is an important virulence determinant for humans. J. Infect. Dis. 193:573-581. [DOI] [PubMed] [Google Scholar]
- 34.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, L., M. E. Peeples, R. C. Boucher, P. L. Collins, and R. J. Pickles. 2002. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J. Virol. 76:5654-5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, W., H. Yang, X. Kong, S. Mohapatra, H. San Juan-Vergara, G. Hellermann, S. Behera, R. Singam, R. F. Lockey, and S. S. Mohapatra. 2005. Inhibition of respiratory syncytial virus infection with intranasal siRNA nanoparticles targeting the viral NS1 gene. Nat. Med. 11:56-62. [DOI] [PubMed] [Google Scholar]