Abstract
The processivity subunit of the herpes simplex virus DNA polymerase, UL42, is essential for viral replication and possesses both Pol- and DNA-binding activities. Previous studies demonstrated that the substitution of alanine for each of four arginine residues, which reside on the positively charged surface of UL42, resulted in decreased DNA binding affinity and a decreased ability to synthesize long-chain DNA by the polymerase. In this study, the effects of each substitution on the production of viral progeny, viral DNA replication, and DNA replication fidelity were examined. Each substitution mutant was able to complement the replication of a UL42 null mutant in transient complementation assays and to support the replication of plasmid DNA containing herpes simplex virus type 1 (HSV-1) origin sequences in transient DNA replication assays. Mutant viruses containing each substitution and a lacZ insertion in a nonessential region of the genome were constructed and characterized. In single-cycle growth assays, the mutants produced significantly less progeny virus than the control virus containing wild-type UL42. Real-time PCR assays revealed that these UL42 mutants synthesized less viral DNA during the early phase of infection. Interestingly, during the late phase of infection, the mutant viruses synthesized larger amounts of viral DNA than the control virus. The frequencies of mutations of the virus-borne lacZ gene increased significantly in the substitution mutants compared to those observed for the control virus. These results demonstrate that the reduced DNA binding of UL42 is associated with significant effects on virus yields, viral DNA replication, and replication fidelity. Thus, a processivity factor can influence replication fidelity in mammalian cells.
DNA polymerases frequently rely upon processivity factors. The best understood processivity factors, the “sliding clamps,” tether their corresponding catalytic subunits (Pols) to the DNA template to promote efficient DNA synthesis. Examples of sliding clamps include proliferating cell nuclear antigen (PCNA) (18), the processivity factor for several eukaryotic DNA polymerases, and gp45 (22), the clamp subunit of the bacteriophage T4 replisome. These sliding clamps are similar in both structure and activity, in which a trimer or dimer of the sliding clamp protein forms a ring to encircle the DNA. ATP-dependent clamp loader proteins are required for the association of sliding clamps with DNA to facilitate processive DNA synthesis (reviewed in reference 11).
HSV DNA polymerase consists of a heterodimer with a catalytic subunit Pol and an accessory subunit, UL42. UL42 functions as a processivity factor for the synthesis of long-chain DNA in vitro and, like Pol, is essential for viral replication (10, 20, 27, 28). Distinct from other sliding clamps, UL42 interacts with DNA directly, with high affinity as a monomer (29). UL42 increases the binding affinity of the polymerase to the primer/template 10- to 20-fold (9, 37), due primarily to a decrease in the dissociation rate (9). Insertion of four amino acids at residue 203 or 206 of UL42 abolished DNA binding without affecting binding to Pol, and each insertion mutant lost the ability to stimulate long-chain DNA synthesis (6). Furthermore, these insertion mutations failed to support the replication of a UL42 null mutant in Vero cells in a transient complementation assay. The results suggested that the DNA binding activity of UL42 is essential for the processive DNA synthesis by the Pol and that the failure of the UL42 mutants to bind to DNA is lethal for viral replication.
Processivity factors have also been shown to influence the fidelity of DNA replication. Mutations affecting PCNA can lead to elevated mutation rates in Saccharomyces cerevisiae (5, 7). In vitro studies have demonstrated that the addition of PCNA can increase misincorporation errors by mammalian Pol δ on defined primer/templates (24) and reduce the rate of deletion mutations during the synthesis of repeated sequences by yeast Pol δ (8). T4 gp45 also can influence replication fidelity in vitro (2, 3). UL42 can increase DNA replication fidelity in vitro, most likely by decreasing the dissociation of polymerase from the primer/template (4). However, we are unaware of any study demonstrating the influence of a processivity factor on replication fidelity in mammalian cells.
Recently, we showed that substitutions of alanine for each of four conserved arginine residues (113, 182, 279, and 280), which reside on the positively charged surface of UL42, led to reduced DNA binding and a corresponding decreased ability to stimulate long-chain DNA synthesis without affecting binding to Pol (31). This correlation further supports the model in which the binding of UL42 to DNA is important for the processivity of HSV DNA polymerase. Combining the four substitutions reduced, but did not abolish, the ability to complement the replication of a UL42 null mutant in Vero cells (31).
In order to address the effect of the binding of UL42 to DNA on viral replication and replication fidelity, we tested the effects of each of the four arginine-to-alanine substitutions for complementation of the replication of a UL42 null mutant. We also constructed and characterized recombinant viruses harboring each of the substitutions. Although none of the mutations abolished viral replication or DNA synthesis, they did affect these processes. Moreover, they exerted substantial effects on the fidelity of DNA replication.
MATERIALS AND METHODS
Cells and viruses.
Vero cells (obtained from the American Type Culture Collection) and their derivative strain, V42.3, which was constructed as described below, were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% newborn calf serum. HSV-1 strains 17 syn+ (a generous gift from D. Parris) and KOS and recombinant viruses containing UL42 point mutations (see below) were propagated on Vero cells. The UL42 null mutant virus, CgalΔ42 (a generous gift from P. Johnson and D. Parris) (17), which contains an inserted lacZ gene in a nonessential region of the Us region and a partial deletion of sequences from the UL42 gene, was propagated on V42.3 cells.
Plasmids.
To construct UL42-expressing cell lines, the plasmid pBE-5.1 (Fig. 1) was constructed to contain the 5.1-kbp BamHI-EcoRI fragment of HSV-1 (isolated from DNA prepared from KOS-infected Vero cells), which includes the entire UL42 and UL43 genes and partial sequences of UL41 and UL44, and was inserted into the cloning vector pGEM7Zf(+). The following plasmids were constructed for transient complementation and oriS-dependent DNA replication assays and for construction of recombinant viruses. Plasmid pHC700 (Fig. 1) was constructed by deleting a 1.1-kbp SacII fragment, which contains partial sequences of UL43 and UL44 genes, from pBE-5.1. This deletion also removed a PstI site. Plasmids pHC-R113A, pHC-R182A, pHC-R279A, and pHC-R280A were constructed by replacing the 737-bp PstI fragment of pHC700 with the corresponding fragment derived from pMal-PP-UL42Δ340 containing mutations R113A, R182A, R279A, and R280A, respectively (41). Each plasmid was sequenced to confirm the expected mutation.
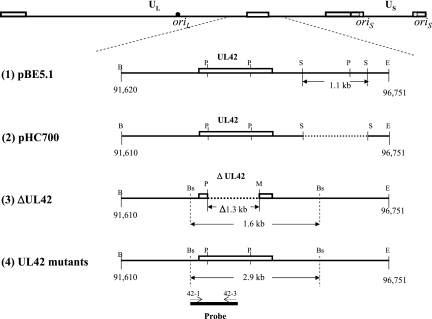
FIG. 1.
Map of UL42 plasmids and viruses. The top line shows the genomic structure of HSV-1. The relative locations of UL42 and its flanking sequences are enlarged and the region of sequences cloned and modified in plasmids (1 and 2) and recombinant viruses (3 and 4) are shown (1). pEB5.1 contains a 5.1-kbp BamHI-EcoRI DNA fragment of HSV-1 strain KOS, which includes UL42 and UL43 and partial UL41 and UL44 sequences, inserted into pGEM7zf(+) (2). pHC700 was constructed from pEB5.1 by deleting a 1.1-kbp SacII fragment (3). Recombinant virus CgalΔ42, derived from HSV-1 strain syn+, contains an inserted lacZ gene between Us9 and Us10 and a deletion of 1,258 bp from within UL42. (4). UL42 recombinants were constructed from CgalΔ42 using pHC to introduce the corresponding UL42 mutation. The probe used for Southern blotting is shown at the bottom of the figure. The open boxes above the line represent the open reading frame of UL42. Dotted lines represent deleted regions. Oligonucleotides 42-1 and 42-3 were used to amplify and label the DNA fragment. B, BamHI; Bs, BstEII; E, EcoRI; M, MluI; P, PstI; S, SacII.
For transient oriS-dependent DNA replication assays, plasmid pHOS9.2 was constructed by substituting the kanamycin resistance gene for the ampicillin resistance gene of pHOS1, which also contains the oriS sequences of HSV-1, the ColE1 sequences for propagation in Escherichia coli, and the supF gene (12). pHOS9.2 also contains the EGFP gene, inserted under the control of the simian virus 40 promoter/enhancer.
Construction of cell lines and recombinant viruses.
Approximately 5 × 105 Vero cells were preseeded into a 60-mm2 dish, incubated overnight, and cotransfected with 2 μg of pBE-5.1 and 0.2 μg of pSV2-neo using LipofectAmine 2000 transfection reagent according to the manufacturer's protocol. Forty-eight hours after transfection, cells were placed under selection conditions by adding 400 μg/ml of G418. More than 200 foci formed after 2 weeks of G418 selection, and 12 foci were randomly isolated and expanded. Ten of twelve G418-resistant lines were able to support the growth of the UL42 null mutant, CgalΔ42, and one in particular, V42.3, was used for the study. A Southern blotting hybridization experiment demonstrated the presence of integrated UL42 sequences in this cell line (data not shown).
To construct recombinant viruses harboring either the wild-type or the mutant UL42 gene, approximately 5 × 105 V42.3 cells were plated on a 60-mm2 dish, incubated overnight, and transfected with 2 μg of the relevant pHC plasmid DNA using LipofectAmine 2000 reagent. Twenty-four hours later, the transfected cells were infected with 1,000 PFU of CgalΔ42 and incubated for an additional 3 to 4 days or until 75% of cells showed cytopathic effect. Progeny viruses harvested from the transfection/superinfection procedure were plated on Vero cells to isolate individual plaques, which presumably had recombined with the UL42 sequences, either from the transfected clone or from the endogenous copy within the cells. Two independent transfection experiments were performed for the wild-type UL42 plasmid, pHC700, and each of the four mutants (pHC-R113A, etc.) to obtain independent recombinants, designated A and B. Individual plaques were amplified on Vero cells. Total DNA was isolated from the infected cells and subjected to PCR to amplify the UL42 sequences. Oligonucleotide primers UL42-1 and UL42-4, which correspond to sequences immediately upstream and downstream, respectively, of the UL42 open reading frame, were used. PCR products then were subjected to restriction enzyme digestion to identify the gain or loss of the appropriate restriction site corresponding to the altered nucleotide(s) for each arginine-to-alanine substitution. PCR products were sequenced to confirm that the expected mutation(s) was the sole change occurring in each recombinant. Southern blot hybridization using a NEBlot Phototope-labeled probe and a chemiluminescent detection kit (New England Biolabs) was performed to verify restoration of the full-length UL42 gene and flanking regions and the homogeneity of the recombinant.
Complementation and transient DNA replication assays.
Complementation assays were performed as described previously (6, 31). Briefly, each pHC plasmid was transfected into Vero cells, which subsequently were infected with the CgalΔ42 virus at a multiplicity of infection (MOI) of 1. The titer for progeny virus was determined using V42.3 cells as well as Vero cells to check for recombinants. The titer for Vero cells was at least 10,000-fold less than the titer obtained from V42.3 cells. The percentage of complementation of mutant plasmid was calculated as [(titer for V42.3 − titer for Vero)mutant/(titer for V42.3 − titer for Vero)wild type] × 100%.
Transient oriS-dependent DNA replication assays were performed using Vero cells. Briefly, cells were cotransfected with 1 μg of each UL42 plasmid together with 1 μg of pHOS9.2, which contains oriS sequences that direct DNA replication. Twenty-four hours after transfection, cells were infected with CgalΔ42 at an MOI of 1 and incubated overnight. Total infected cell DNA was isolated and purified. One-third of the DNA was restriction digested with EcoRI, which linearizes pHOS9.2 DNA; another one-third of DNA was treated with EcoRI plus DpnI, which digests DNA originating from E. coli into small fragments to distinguish the input DNA from that replicated in mammalian cells. Enzyme-treated DNA was then fractionated on a 0.8% agarose gel, transferred onto a nylon membrane, and probed with a 32P-labeled DNA fragment corresponding to E. coli ColE1 sequences. The result was analyzed using a PhosphorImager (Molecular Dynamics).
Measurements of single-cycle replication kinetics, burst size, and plaque size and quantification of viral DNA.
To measure single-cycle replication kinetics and burst sizes, Vero cells (1 × 105) were infected with each virus at an MOI of 3. In three different experiments, virus yields at 48 and 72 h after infection were less than those obtained at 24 h after infection (data not shown). Therefore, it was assumed that virus yields peaked between 24 and 48 h. The infected cells were harvested at 2, 5, 8, 12, 24, and 36 h postinfection, and the yield of progeny was titrated with Vero cells. The burst size was determined as the ratio of the peak titer of progeny/number of cells. Plaque sizes were measured by digital imaging using a Spot digital camera and the accompanying software (Nikon).
To prepare DNA for real-time PCR quantification, infected cells and the medium containing cell-free virus were freeze-thawed three times, followed by incubation with 0.6% sodium dodecyl sulfate and 10 μg/ml protease K at 56°C for 2 h and purified by phenol-chloroform extractions. The amount of viral DNA was determined via real-time PCR using a QuantiTect SYBR Green PCR kit (QIAGEN). Oligonucleotide primers used for PCRs were DBP-3261 and DBP-3315, corresponding to nucleotides 61,522 to 61,541 and 61,595 to 61,576, respectively, of HSV-1 (21). PCR amplification included initial denaturation at 95°C for 15 min, followed by 36 cycles of 94°C for 15 seconds, 58°C for 20 seconds, and 72°C for 20 seconds. Each PCR assay contained a negative control and a series of pUL29-OriL DNA dilutions (1 × 101 to 1 × 106 copies per μl) (14), which can be amplified efficiently, to generate the standard curve. Valid PCRs had an r2 value of 0.993 or 0.992 and a slope of −3.386 or −3.387, respectively. The PCR efficiency was 97%.
lacZ mutagenesis.
Each virus stock was prepared by inoculating 100 PFU onto 2 × 105 Vero cells. Progeny virus was harvested at 4 days postinfection and, after the titer was determined, was assayed for mutagenesis of the lacZ gene, as described previously (13, 14). Briefly, Vero cells plated on 10-cm2 culture dishes were infected with virus at a density of ∼1,200 plaques/plate. One hour after inoculation, the cell monolayer was washed with DMEM and overlaid with 0.75% methylcellulose containing DMEM/2% newborn calf serum. Forty-eight hours after infection, cells were washed, fixed, and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), using the β-galactosidase reporter gene staining kit (Sigma) according to the manufacturer's protocol. Clear or light-blue plaques were identified by using an inverted microscope, and the mutation frequency was determined as the ratio of the number of clear plaques plus the number of light-blue plaques over the sum of the total number of plaques. A chi-square test was used to calculate the statistical significance of the differences in mutation frequencies.
RESULTS
Previous studies suggested that UL42 tethers Pol to DNA and mediates the processivity of Pol during DNA synthesis (6, 10, 28, 30, 41). This mechanism is further supported by a recent in vitro study showing that the substitution of arginine residues on the positively charged surface of UL42 with alanine leads to decreased DNA binding and that these reductions correlated with a reduced ability to synthesize long-chain DNA (31). We wished to assess the effects of these substitutions on viral yield and DNA replication in virus-infected cells and to investigate whether alterations in UL42 would affect the fidelity of DNA replication. As we had previously found that a quadruple substitution of arginine residues in UL42 would still permit some complementation of a UL42 null mutant (31), we anticipated that single substitutions would cause less impairment and not be lethal when incorporated into virus. In the present study, we assessed the effects of single substitutions in transient DNA replication assays and for complementation of a UL42 null mutant with Vero cells. Recombinant viruses were then constructed to characterize the effects of the mutations on viral replication and fidelity.
The mutants permitted DNA replication and complementation in transient assays.
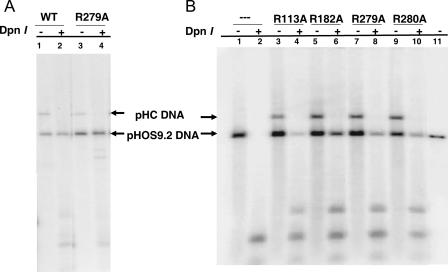
As an initial test of the effects of each arginine-to-alanine substitution, we performed both transient DNA replication and complementation assays. For these assays, we constructed pHC700, which contains UL42 and its flanking sequences, including regions of the UL41 and UL43 genes. Each substitution mutation was then transferred from pMal-ppΔ340-based E. coli expression plasmids (31, 41) into pHC700 (Fig. 1) to obtain pHC-R113A, pHC-R182A, pHC-R279A, and pHC-R280A, which contain the arginine-to-alanine substitution at residues 113, 182, 279, and 280, respectively. Transient DNA replication assays were performed by cotransfecting each pHC plasmid with pHOS9.2, which contains HSV-1 oriS sequences, and by infecting the transfected cells with the UL42 null mutant virus, CgalΔ42. Like the control plasmid pHC700, which encodes wild-type UL42, each plasmid containing the UL42 mutation permitted DNA replication of the pHOS9.2 plasmid in Vero cells induced by CgalΔ42, while CgalΔ42 alone failed to induce the replication of pHOS9.2 (Table 1). Examples of the Southern blots are shown in Fig. 2. These results demonstrated that these UL42 substitutions permitted replication of pHOS9.2 by the UL42 null mutant. Thus, these UL42 substitutions were functionally able to support the replication of a plasmid containing oriS in transfection-infected Vero cells.
TABLE 1.
Complementation of CgalΔ42 and transient oriS-dependent DNA replication on Vero cells by UL42 in trans
| Plasmid | Mutation | Complementationa | oris-dependent DNA replicationb |
|---|---|---|---|
| pHC700 | Wild type | + | + |
| pHC-R113A | R113A | + | + |
| pHC-R182A | R182A | + | + |
| pHC-R279A | R279A | + | + |
| pHC-R280A | R280A | + | + |
| No DNA | − | − |
Three independent experiments were performed for the complementation assays as described in Materials and Methods.
The oris-dependent DNA replication assay was performed as described in Materials and Methods. At least two independent experiments were performed for each mutant plasmid, using different batches of DNA for each plasmid. Examples of Southern blots are shown in Fig. 2.
FIG. 2.
Southern blots of transient DNA replication supported by UL42 mutant. The transient DNA replication of the oriS-containing plasmid (pHOS9.2) supported by the UL42 mutants was performed as described in Materials and Methods. Aliquots of purified DNA samples were digested with either EcoRII alone or with EcoRII plus DpnII, fractionated on a 0.8% agarose gel, and hybridized to a probe corresponding to the ColE1 sequence. (A) Results shown from the wild-type UL42 control (lanes 1 and 2) and the mutation R279A (lanes 3 and 4). (B) Results are shown of no UL42 DNA (lanes 1 and 2) and those of each mutant as well as the linearized pHOS1 (lane 11). The positions of linearized pHOS9.2 (bottom arrows) and pHC plasmids (top arrows) are indicated.
The effects of the plasmid-borne substitution mutations on virus replication were examined using a transient complementation assay. In this assay, V42.3 cells were used to measure the yield of CgalΔ42 progeny propagated with Vero cells upon the transfection of the UL42 mutant DNA and subsequent infection with CgalΔ42. At least two independent experiments were analyzed for each UL42 mutant in parallel with wild-type UL42. These mutants were able to support the replication of CgalΔ42 virus in Vero cells (Table 1). These results are in agreement with those of the oriS-dependent transient DNA replication and further demonstrate that these UL42 substitutions are competent in supporting transient virus replication assay.
Construction of viable recombinant viruses with UL42 substitutions.
The transient oriS-dependent DNA replication and complementation assays described above demonstrated that plasmid constructs with these substitutions were competent in supporting virus replication. Therefore, we constructed recombinant viruses for further characterization of the effects of these mutations on viral and DNA replication in infected cells. Two recombinant viruses of each mutation were constructed in two independent transfection experiments. After two to three rounds of plaque purification, each recombinant virus was amplified with Vero cells to prepare viral stocks. DNA was prepared and purified from infected cells and used for Southern blotting analysis. Southern blotting results demonstrated the relative size of the BstEII restriction fragment containing UL42 and the flanking sequences (data not shown). Southern blotting also demonstrated the homogeneity of each recombinant virus (not shown). In addition, the UL42 gene was PCR amplified using oligonucleotide primers 42-1 and 42-4, corresponding to nucleotides 93,076 to 93,094 and 94,622 to 94,601, respectively, of the viral genome (21). Sequencing analyses confirmed that the only mutated bases within this segment in each recombinant corresponded to the engineered arginine-to-alanine substitution. Obtaining these recombinant viruses further demonstrated that these UL42 base mutations, associated with reduced DNA binding and a decreased ability to synthesize long-chain DNA, were not lethal. Moreover, the UL42 mutants formed similar sized plaques, as did the recombinants containing wild-type UL42 (data not shown).
Effects of the mutations on virus yield.
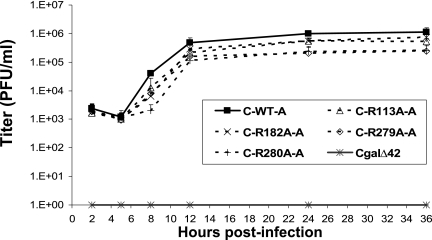
Each recombinant virus was examined for single-cycle replication kinetics. The UL42 null mutant CgalΔ42 did not produce any detectable progeny virus in Vero cells. Results from four different experiments demonstrated that the mutant viruses exhibited significant decreases in virus yields (Student's t test, P, <0.05) compared with the that of the control virus containing wild-type UL42 (Fig. 3 and Table 2). The C-R113A and C-R182A recombinants produced twofold lower yields, and the C-R279A and C-R280A recombinants produced fivefold lower yields. Therefore, these UL42 mutants exhibited modest defects in virus replication.
FIG. 3.
Single-cycle growth assay of UL42 recombinants. Vero cells (1 × 105) were inoculated with UL42 recombinants at an MOI of 3. Infected cells were harvested at 2, 5, 8, 12, 24, and 36 h postinfection and titrated with Vero cells to determine the yield of progeny viruses. Since recombinant A and B of each mutant exhibited similar growth kinetics, only results of recombinant A are shown to clearly demonstrate the difference of virus yields.
TABLE 2.
Burst size and genomic DNA copy/PFU of UL42 recombinants
| Virus | Burst size (PFU/cell)a | DNA copies/PFUb |
|---|---|---|
| C700-A | 12.5 ± 3.0 | 200 ± 100 |
| C700-B | 11.5 ± 4.2 | 300 ± 80 |
| C-R113A-A | 6.0 ± 2.0 | 600 ± 100 |
| C-R113A-B | 5.3 ± 1.5 | 700 ± 200 |
| C-R182A-A | 8.2 ± 2.1 | 1000 ± 40 |
| C-R182A-B | 7.4 ± 1.8 | 1,100 ± 400 |
| C-R279A-A | 2.6 ± 1.9 | 4,600 ± 40 |
| C-R279A-B | 2.4 ± 1.6 | 2,700 ± 100 |
| C-R280A-A | 2.5 ± 0.7 | 3,100 ± 400 |
| C-R280A-B | 2.6 ± 1.1 | 2,900 ± 500 |
| CgalΔ42 | NDc | ND |
Burst size was calculated by the ratio of peak titer (36 h postinfection) over the number of cells. Data are averages and standard deviations of four experiments.
DNA copies/PFU and standard deviations were calculated by the ratio of numbers of DNA copies present in cell-free medium over viral titers at 36 h postinfection.
ND, not determined.
The mutants exhibit altered kinetics of viral DNA synthesis.
Since UL42 is a processivity factor critical for viral DNA synthesis, we wished to determine whether these mutations could affect the levels of DNA synthesized at various times during viral infection. Vero cells were infected with recombinant virus at an MOI of 3, and an aliquot of DNA prepared from total infected cells including cell-free virus was purified. Isolated DNA was then subjected to real-time PCR quantification using 200 bp of ICP8 sequences as the target DNA. Each experiment also included reactions to quantify control DNA to establish the standard curve for calculating the relative copy number of DNA present in each sample. The sensitivity of these assays was demonstrated to be between 1 and 10 copies of target DNA.
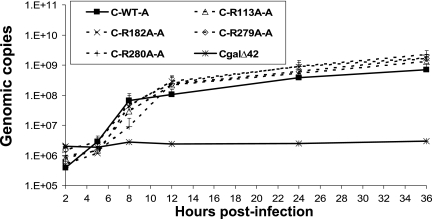
No increases of viral DNA were detected in Vero cells infected with the CgalΔ42 mutant (Fig. 4). In cells infected with recombinant viruses with wild-type UL42, viral DNA measurably increased between 2 and 5 h and continued to increase between 5 and 8 h after infection (Fig. 4). Interestingly, viral DNA increased only slightly, if at all, in cells infected with each of the four UL42 mutants between 2 and 5 h after infection, although increases of viral DNA became obvious between 5 and 8 h after infection. Furthermore, all four mutants synthesized less DNA compared to that of recombinants with wild-type UL42 at 8 h after infection (Fig. 4). Interestingly, the UL42 mutants replicated more viral DNA than the viruses with wild-type UL42 during the late phase of infection. Significant increases were found at 24 and 36 h postinfection (Student's t test, P < 0.05) (Fig. 4), despite the mutants' production of less infectious virus at those time points (Fig. 3). Figure 4 shows the average amounts of DNA detected during the infection from four different experiments. Thus, the relatively low yields of infectious virus of the UL42 mutants were accompanied by reduced DNA synthesis early in infection but increased DNA synthesis late in infection.
FIG. 4.
Viral DNA synthesized by UL42 recombinants. Vero cells (1 × 105) were inoculated with UL42 recombinants at an MOI of 3. Infected cells were harvested at different times postinfection. DNA was isolated from aliquots of infected cells, including both infected cells and medium containing cell-free virus. Purified DNA was serially diluted and quantified by real-time PCR as described in Materials and Methods to determine the relative copy number of viral DNA. Since recombinant A and B of each mutant replicated similar amounts of DNA, only results of recombinant A are shown to clearly demonstrate different amounts of DNA synthesized by these recombinants.
lacZ mutagenesis.
To test the hypothesis that UL42 influences replication fidelity, we took advantage of the inserted lacZ reporter gene in the CgalΔ42 recombinant and its derivatives. The frequency of mutations for lacZ was assayed using an established protocol (13, 14) in which viral stocks were prepared by inoculation of 100 PFU of the original virus stock of each recombinant virus onto Vero cells. This procedure effectively excludes preexisting mutants from the inocula. Table 3 shows the frequency of mutations that reduce lacZ activity following replication by each UL42 mutant recombinant. The control recombinant (both independent isolates, A and B) containing wild-type UL42 exhibited similar mutation frequencies (0.052% and 0.064%, respectively), which were only slightly higher than those observed in other studies, in which the lacZ reporter gene was inserted at different loci of the viral genome of a different HSV-1 strain (13, 14). Each UL42 mutant virus exhibited significantly higher mutation frequencies of lacZ, ranging from 0.174% to 0.774%, than that of the control virus. Interestingly, the R113A and R182A mutations, which caused milder defects in DNA binding by UL42 and in synthesizing long-chain DNA in vitro (31), exhibited higher mutation frequencies than those of the C-R280A mutant. It should be noted, however, that both C-R279A recombinants (C-R279A causes defects in DNA binding similar to those of C-R280A) contained a preexisting lacZ mutation(s); the recombinant C-R279A-A formed only light-blue plaques after X-Gal staining, and C-R279A-B formed clear plaques. Thus, these recombinants could not be assayed and may actually possess higher mutation frequencies. The lacZ mutagenesis studies demonstrated that UL42 mutations that decrease DNA binding also reduce the fidelity of replication of the lacZ gene.
TABLE 3.
Mutation frequency of the lacZ gene
| Virus | No. of total plaques | No. of clear and light-blue (LB) plaques | Mutation frequency (%) | P valuea |
|---|---|---|---|---|
| C700-A | 7,769 | 4 (0) | 0.05 | |
| C700-B | 9,305 | 6 (0) | 0.06 | |
| C-R113A-A | 8,770 | 32 (4) | 0.41 | <0.0001 |
| C-R113A-B | 10,636 | 57 (1) | 0.55 | <0.0001 |
| C-R182A-A | 7,756 | 56 (4) | 0.77 | <0.0001 |
| C-R182A-B | 6,770 | 17 (6) | 0.34 | <0.0001 |
| C-R279A-A | 8,592 | 10 (?) | ND | ND |
| C-R279A-B | NDb | ND | ND | ND |
| C-R280A-A | 10,334 | 6 (12) | 0.17 | <0.05 |
| C-R280A-B | 11,357 | 18 (15) | 0.29 | <0.001 |
The chi-square test was used to determine the P values, compared to the mutation frequency of C700-B. No significant differences were detected between the mutation frequencies induced by two independently isolated recombinants containing wild-type UL42.
ND, not determined due to the preexisting mutation.
DISCUSSION
Processivity factors of DNA polymerases play critical roles in replication by preventing the dissociation of polymerase from the primer/template. Earlier studies demonstrated that UL42 mutations that decrease DNA binding activity result in reduced synthesis of long-chain DNA in vitro and complementation of UL42 null mutants in cells (6, 31). However, the effect of UL42 mutations that affect DNA binding had not been characterized following their incorporation into virus. In the present study, recombinant viruses were constructed to contain single UL42 mutations that reduced DNA binding synthesis of long-chain DNA (31). This permitted us to examine the effects of these mutations of viral and DNA replication and to test the hypothesis that they would reduce replication fidelity.
Replication phenotype of UL42 mutants.
A previous study showed that each single arginine-to-alanine substitution does not quantitatively affect the binding of UL42 to the C terminus of Pol (31) and that a UL42 mutant containing all four arginine-to-alanine mutations retains the ability to support replication in transient complementation assays (31). Thus, it was not surprising that each UL42 mutant with a single arginine-to-alanine substitution was able to support virus replication, albeit with reduced virus yield. Importantly, although the mutations decreased virus yield only modestly, the mutations that had the larger effects on DNA binding (R279A and R280A) had the larger effects on virus yield. As might have been expected, all of the mutants exhibited less DNA synthesis than the wild-type UL42 control virus during the early phase of DNA replication. This result could be due to the lower affinities of the mutant proteins for DNA (31), which may result in the dissociation of the Pol holoenzyme and, thus, result in reduced amounts of DNA synthesized. Alternatively, these mutant viruses could have a delayed onset of DNA replication due to defects in the initiation of DNA synthesis. In this regard, interactions have been reported between UL42 and the original binding protein, UL9 (23, 35). Further studies will be necessary to distinguish among these possibilities.
More surprising was that the UL42 mutants synthesized more DNA than the wild-type UL42 control virus during the late phase of infection, with an inverse relationship between the total amounts of viral DNA synthesized and the yields of progeny virus. One possibility is that dissociation of polymerase holoenzyme results in partially single-stranded regions that promote recombination. There is evidence that HSV utilizes recombination-dependent mechanisms to synthesize viral DNA during the late phase of DNA replication (reviewed in reference 38 and references therein). Therefore, it is reasonable to hypothesize that the UL42 mutants may induce more recombination-dependent DNA replication and this may explain the greater amount of DNA synthesis during the late stage of infection. Additionally, UL42 mutants with a lower affinity for DNA are mutagenic (Table 3), which may result in the higher DNA copy/PFU ratio observed, perhaps due to particles containing lethal mutations.
Mutagenic effects of UL42 mutants.
The UL42 mutants replicated the virus-borne lacZ gene with lower fidelity than the wild-type UL42 control virus. These observations demonstrate that UL42 plays an important role in DNA replication fidelity. Although PCNA has been linked to replication fidelity in yeast, PCNA and gp45 have been linked to DNA replication fidelity in vitro (2, 3, 5, 7, 8, 24), and UL42 can increase fidelity in vitro (4), to our knowledge, this is the first study to demonstrate an effect of a processivity factor in the replication fidelity of a virus in mammalian cells.
Previous in vitro studies demonstrated that PCNA can affect replication fidelity of polymerase δ (8, 24). Furthermore, biochemical studies support a role for PCNA in promoting DNA synthesis past template lesions by mammalian polymerase δ (24-26). Studies of T4 gp45 also demonstrate that gp45 can affect site- and type-specific fidelity in vitro (3). It will be interesting to examine the spectrum of mutations produced by UL42 mutants and to test biochemically how UL42 substitutions result in decreased fidelity.
Several possible mechanisms may account for the mutagenic effect of UL42 mutants. UL42 may promote switching from the polymerase site to the exonuclease active site of Pol (32). If mutant UL42s with impaired DNA binding are defective in this process, more substitutions may occur due to less efficient proofreading. Alternatively, UL42 mutants may induce more displaced strands or flaps during lagging-strand synthesis, based on a report that UL42 reduces the formation of such flaps (40). This could prevent the maturation of Okazaki fragments and lead to genome instability (1, 15, 16) and the formation of complex changes, such as duplications (16). It is also possible that UL42 mutants that decrease DNA binding may increase deletion mutations in the viral genome due to the formation of frayed primers. Such a model has been recently proposed to explain the formation of large deletions during the synthesis of repeat sequences and proposes that PCNA may play a role in suppressing frayed primer formation and deletion errors (8). Another possibility is that UL42 mutants with less processive DNA synthesis may result in the formation of single-stranded gaps, which can be the substrates of error-prone, recombination-dependent repair (39).
It is also possible that UL42 mutants may mediate less efficient mismatch repair (MMR). A direct link between PCNA and MMR has been found: alterations of PCNA can result in a mutator phenotype in yeast (5, 19, 36). Interestingly, direct or indirect interactions between UL42, the HSV-1 single-stranded DNA binding protein, ICP8, and two MMR proteins (MSH3 and MSH6) have been found (34). Furthermore, MSH6 expression has been reported to be up-regulated in HSV-1-infected cells (33). Therefore, it is possible that UL42 may be involved in MMR, which would predict an increased frequency of point mutations in UL42 mutants. Further studies will be necessary to examine the mutation spectra produced by UL42 mutants, which may help to define the possible mechanisms of how UL42 mutations result in decreased fidelity.
Acknowledgments
We thank J. W. Drake for useful discussions. We are grateful for the technical assistance from Johnny Wang and Tashieka Hucey.
This study was supported by NIH grants AI056359 (to C.B.C.H.) and AI19838 (to D.M.C.). J.C.W.R. was a predoctoral fellow of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Ayyagari, R., X. V. Gomes, D. A. Gordenin, and P. M. J. Burgers. 2003. Okazaki fragment maturation in yeast. I. Distribution of functions between FEN1 and DNA2. J. Biol. Chem. 278:1618-1625. [DOI] [PubMed] [Google Scholar]
- 2.Bebenek, A., G. T. Carver, H. K. Dressman, F. A. Kadyrov, J. K. Haseman, V. Petrov, W. H. Konigsberg, J. D. Karam, and J. W. Drake. 2002. Dissecting the fidelity of bacteriophage RB69 DNA polymerase: site-specific modulation of fidelity by polymerase accessory proteins. Genetics 162:1003-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bebenek, A., G. T. Carver, F. A. Kadyrov, G. E. Kissking, and J. W. Drake. 2005. Processivity clamp gp45 and ssDNA-binding-protein gp32 modulate the fidelity of bacteriophage RB69 polymerase in a sequence-specific manner, sometime enhancing and sometimes compromising accuracy. Genetics 169:1815-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudhuri, M., L. Song, and D. S. Parris. 2003. The herpes simplex virus type 1 DNA polymerase processivity factor increases fidelity without altering pre-steady-state rate constants for polymerization or excision. J. Biol. Chem. 278:8996-9004. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C., B. J. Merrill, P. J. Lau, C. Holm, and R. D. Kolodner. 1999. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol. Cell. Biol. 19:7801-7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chow, C. S., and D. M. Coen. 1995. Mutations that specifically impair the DNA binding activity of the herpes simplex virus protein UL42. J. Virol. 69:6965-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eissenberg, J. C., R. Ayyagari, X. V. Gomes, and P. M. Burgers. 1997. Mutations in yeast proliferating cell nuclear antigen define distinct sites for interaction with DNA polymerase δ and DNA polymerase ɛ. Mol. Cell. Biol. 17:6367-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortune, J. M., C. M. Stith, G. E. Kissling, P. M. J. Burgers, and T. A. Kunkel. 2006. RPA and PCNA suppress formation of large deletion errors by yeast DNA polymerase δ. Nucleic Acids Res. 34:4335-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb, J., and M. D. Challberg. 1994. Interaction of herpes simplex virus type 1 DNA polymerase and the UL42 accessory protein with a model primer template. J. Virol. 68:4937-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gottlieb, J., A. I. Marcy, D. M. Coen, and M. D. Challberg. 1990. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J. Virol. 64:5976-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hingorani, M. M., and M. Donnell. 2000. Sliding clamps: a (tail)ored fit. Curr. Biol. 10:R25-R29. [DOI] [PubMed] [Google Scholar]
- 12.Hwang, Y. T., B.-Y. Liu, C.-Y. Hong, E. J. Shillitoe, and C. B. C. Hwang. 1999. Effects of exonuclease activity and nucleotide selectivity of the herpes simplex virus DNA polymerase on the fidelity of DNA replication in vivo. J. Virol. 73:5326-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang, Y. T., Y. A. Wang, Q. Lu, and C. B. Hwang. 2003. Thymidine kinase of herpes simplex virus type 1 strain KOS lacks mutator activity. Virology 305:388-396. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, C., Y. T. Hwang, and C. B.-C. Hwang. 2006. Herpes simplex virus type 1 recombinants without the oriL sequence replicate DNA with increased fidelity. Virology 347:277-285. [DOI] [PubMed] [Google Scholar]
- 15.Jin, Y. H., R. Ayyagari, M. A. Resnick, D. A. Gordenin, and P. M. J. Burgers. 2003. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 278:1626-1633. [DOI] [PubMed] [Google Scholar]
- 16.Jin, Y. H., R. Obert, P. M. J. Burgers, T. A. Kunkel, M. A. Resnick, and D. A. Gordenin. 2001. The 3′right-arrow5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl. Acad. Sci. USA 98:5122-5127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, P. A., M. G. Best, T. Friedmann, and D. S. Parris. 1991. Isolation of a herpes simplex virus type 1 mutant deleted for the essential UL42 gene and characterization of its null phenotype. J. Virol. 65:700-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishna, T. S. R., X.-P. Kong, S. Gary, P. M. Burgers, and J. Kuriyan. 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79:1233-1243. [DOI] [PubMed] [Google Scholar]
- 19.Lau, P. J., H. Flores-Rozas, and R. D. Kolodner. 2002. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol. Cell. Biol. 22:6669-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchetti, M. E., C. A. Smith, and P. A. Schaffer. 1988. A temperature-sensitive mutation in a herpes simplex virus type 1 gene required for viral DNA synthesis maps to coordinates 0.609 through 0.614 in UL. J. Virol. 62:715-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGeoch, D. J., M. A. Dalrymple, A. J. Dasison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 22.Moarefi, I., D. Jeruzalmi, J. Turner, M. Donnell, and J. Kuriyan. 2000. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 296:1215-1223. [DOI] [PubMed] [Google Scholar]
- 23.Monahan, S. J., L. A. Grinstead, W. Olivieri, and D. S. Parris. 1998. Interaction between the herpes simplex virus type 1 origin-binding and DNA polymerase accessory proteins. Virology 241:122-130. [DOI] [PubMed] [Google Scholar]
- 24.Mozzherin, D. J., M. McConnell, M. V. Jasko, A. A. Krayevsky, C.-K. Tan, K. M. Downey, and P. A. Fisher. 1996. Proliferating cell nuclear antigen promotes misincorporation catalyzed by calf thymus DNA polymerase delta. J. Biol. Chem. 271:31711-31717. [DOI] [PubMed] [Google Scholar]
- 25.Mozzherin, D. J., S. Shibutani, C.-K. Tan, K. M. Downey, and P. A. Fisher. 1997. Proliferating cell nuclear antigen promotes DNA synthesis past template lesions by mammalian DNA polymerase delta. Proc. Natl. Acad. Sci. USA 94:6126-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Day, C., P. M. J. Burgers, and J.-S. Taylor. 1992. PCNA-induced DNA synthesis past cis-syn and trans-syn-l thymine dimers by calf thymus DNA polymerase δ in vitro. Nucleic Acids Res. 20:5403-5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parris, D. S., A. Cross, L. Haarr, A. Orr, M. C. Frame, M. Murphy, D. J. McGeoch, and H. S. Marsden. 1988. Identification of the gene encoding the 65-kilodalton DNA-binding protein of herpes simplex virus type 1. J. Virol. 62:818-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purifoy, D. J. M., R. B. Lewis, and K. L. Powell. 1977. Identification of the herpes simplex virus DNA polymerase gene. Nature 269:621-623. [DOI] [PubMed] [Google Scholar]
- 29.Randell, J. C., and D. M. Coen. 2004. The herpes simplex virus processivity factor, UL42, binds DNA as a monomer. J. Mol. Biol. 335:409-413. [DOI] [PubMed] [Google Scholar]
- 30.Randell, J. C., and D. M. Coen. 2001. Linear diffusion on DNA despite high-affinity binding by a DNA polymerase processivity factor. Mol. Cell 8:911-920. [DOI] [PubMed] [Google Scholar]
- 31.Randell, J. C. W., G. Komazin, C. Jiang, C. B. C. Hwang, and D. M. Coen. 2005. Effects of substitutions of arginine residues on the basic surface of herpes simplex virus UL42 support a role for DNA binding in processive DNA synthesis. J. Virol. 79:12025-12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, L., M. Chaudhuri, C. W. Knopf, and D. S. Parris. 2004. Contribution of the 3′- to 5′-exonuclease activity of herpes simplex virus type 1 DNA polymerase to the fidelity of DNA synthesis. J. Biol. Chem. 279:18535-18543. [DOI] [PubMed] [Google Scholar]
- 33.Taddeo, B., A. Esclatine, and B. Roizman. 2002. The patterns of accumulation of cellular RNAs in cells infected with a wild-type and a mutant herpes simplex virus 1 lacking the virion host shutoff gene. Proc. Natl. Acad. Sci. USA 99:17031-17036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trego, K. S., and D. S. Parris. 2003. Functional interaction between the herpes simplex virus type 1 polymerase processivity factor and origin-binding proteins: enhancement of UL9 helicase activity. J. Virol. 77:12646-12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Umar, A., A. B. Buermeyer, J. A. Simon, D. C. Thomas, A. B. Clark, R. M. Liskay, and T. A. Kunkel. 1996. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell 87:65-73. [DOI] [PubMed] [Google Scholar]
- 37.Weisshart, K., C. S. Chow, and D. M. Coen. 1999. Herpes simplex virus processivity factor UL42 imparts increased DNA-binding specificity to the viral DNA polymerase and decreased dissociation from primer-template without reducing the elongation rate. J. Virol. 73:55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weller, S. K. 2006. HSV-1 DNA replication, p. 85-104. In R. M. Sandri-Goldin (ed.), Alphaherpesviruses: pathogenesis, molecular biology and infection control. Caister Academic Press, Norfolk, United Kingdom.
- 39.Wyman, C., D. Ristic, and R. Kanaar. 2004. Homologous recombination-mediated double-strand break repair. DNA Repair 3:827-833. [DOI] [PubMed] [Google Scholar]
- 40.Zhu, Y., K. S. Trego, L. Song, and D. S. Parris. 2003. 3′ to 5′ exonuclease activity of herpes simplex virus type 1 DNA polymerase modulates its strand displacement activity. J. Virol. 77:10147-10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuccola, H. J., D. J. Filman, D. M. Coen, and J. M. Hogle. 2000. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell 5:267-278. [DOI] [PubMed] [Google Scholar]