Abstract
In the efforts of viruses to dominate and control critical cellular pathways, viruses generate considerable intracellular stress within their hosts. In particular, the capacity of resident endoplasmic reticulum (ER) chaperones to properly process the acute increase in client protein load is significantly challenged. Such alterations typically induce the unfolded protein response, one component of which acts through IRE1 to restore ER homeostasis by expanding the folding capabilities, whereas the other arm activates the eIF-2α (α subunit of eukaryotic initiation factor 2) kinase PERK to transiently arrest production of new polypeptide clientele. Viruses, such as herpes simplex virus type 1 (HSV-1), however, go to great lengths to prevent the inhibition of translation resulting from eIF-2α phosphorylation. Here, we establish that PERK, but not IRE1, resists activation by acute ER stress in HSV-1-infected cells. This requires the ER luminal domain of PERK, which associates with the viral glycoprotein gB. Strikingly, gB regulates viral protein accumulation in a PERK-dependent manner. This is the first description of a virus-encoded PERK-specific effector and defines a new strategy by which viruses are able to maintain ER homeostasis.
By virtue of the intrinsic ability of stress-sensing molecules to monitor the concentration of misfolded proteins within the endoplasmic reticulum (ER) lumen, a set of stress-sensing molecules ensures that the load of ER client proteins does not overwhelm the capacity of the organelle to correctly fold them. Once activated, one arm of the unfolded protein response (UPR) represented by IRE1 and ATF-6 activates a gene expression program that extends the folding capability of the ER, whereas the other branch, corresponding to PERK, causes a transient translational arrest (reviewed in reference 14).Together, the components of this system control homeostasis within the ER lumen in the face of a diverse assortment of physiological and pharmacological insults, one of which is viral infection.
Many aspects germane to viral replication result in significant intracellular stress and may irreversibly perturb this homeostasis. Virus replication on internal membranes, retention of major histocompatibility complex (MHC) class I chains within the ER, or high-level production of secreted viral proteins and particles tax the capability of the ER to properly fold and process its client proteins, potentially triggering the UPR (22). In particular, activation of PERK, an eIF-2α (α subunit of eukaryotic initiation factor 2) kinase that spans the ER membrane, by misfolded proteins within the lumen of this organelle (reviewed in reference 14) is poised to negatively impinge upon viral replication. Phosphorylation of eIF-2 on its α subunit inhibits the exchange of GDP for GTP, preventing the recycling reaction necessary to maintain supplies of active eIF-2, a critical translation initiation factor that chaperones the initiator tRNA to the 40S ribosomal subunit. Unopposed, the action of an activated eIF-2α kinase, such as PERK, can rapidly inhibit protein synthesis (reviewed in reference 26).
Besides PERK, mammalian cells contain three additional eIF-2α kinases, each of which inhibits translation in response to a different physiological stress (reviewed in reference 26). One of these is the double-stranded RNA (dsRNA)-dependent interferon-induced protein kinase PKR, an important mediator of innate defenses in virus-infected cells. Intensive investigation in many different viral systems has demonstrated that viral countermeasures to limit PKR activity and effectively prevent the accumulation of phosphorylated eIF-2α are important components of viral pathogenesis (reviewed in reference 26). Indeed, herpes simplex virus type 1 (HSV-1) encodes two different functions, each of which is expressed at a discrete point in the viral developmental program. While the Us11 gene product specifically antagonizes the eIF-2α kinase PKR, the γ134.5 gene encodes a GADD34-related subunit that targets the protein phosphatase 1α (PP1α) catalytic activity to phosphorylated eIF-2α (16, 36). Significantly, irrespective of the fact that the γ134.5 phosphatase holoenzyme can counteract multiple eIF-2α kinases by virtue of its intrinsic ability to remove phosphate from phosphorylated eIF-2α, recent work has established that HSV-1 mutants deficient in both Us11 and γ134.5 are still able to resist the effects of acute ER stress (30).
Presently, relatively little is known regarding how viruses counter the actions of eIF-2α kinases other than PKR. Indeed, in some cases, infected cells ultimately succumb to the effects of PERK-mediated apoptosis (10, 21, 23, 34, 49). However, not all of the consequences of the UPR are detrimental to viral replication. Human cytomegalovirus (HCMV) appears to activate the aspects of the UPR beneficial to viral replication, such as chaperone induction to extend the folding capacity of the ER, while suppressing the particularly harsh consequences resulting from eIF-2α phosphorylation by an unknown mechanism (19, 20). In other cases, effectors that operate downstream of eIF-2α phosphorylation, such as pseudosubstrates specified by hepatitis C virus (HCV) (39, 40, 50) along with vaccinia virus (46) or the HSV-1 γ134.5 phosphatase component (6) have been reported to counter the effects of activated PERK. While in some respects, it is not at all surprising that the latter class of eIF-2α pseudosubstrates and phosphatases are proficient to counter multiple eIF-2α kinases, kinase-specific antagonists for eIF-2α kinases other than PKR have yet to be described.
To address this issue, HSV-1 derivatives deficient in all characterized functions known to regulate eIF-2α phosphorylation were shown to effectively resist acute ER stress (30). This does not involve the cellular eIF-2α kinase PKR, nor does it involve induction of the cellular GADD34 phosphatase component, the principal means by which uninfected cells recover from the UPR. Instead, the resistance of HSV-1 mRNA translation to ER stress requires the expression of viral genes other than γ134.5 and Us11 (30). Herein, we not only establish that both PERK and IRE1 remain unactivated in HSV-1-infected cells, but PERK is selectively resistant to activation by acute ER stress. The luminal domain of PERK specifically associates with glycoprotein B (gB), one of several virus-specified glycoproteins, in infected cells. Moreover, the accumulation of normal quantities of viral polypeptides in infected cells requires a wild-type (WT) glycoprotein B allele. Strikingly, regulation of viral protein abundance by glycoprotein B is obviated in cells with a homozygous PERK deficiency. These genetic and physical interactions between glycoprotein B and PERK suggest that HSV-1 glycoprotein B interacts with PERK to maintain ER homeostasis, defining a new strategy by which viral functions can subvert a cellular ER stress sensor.
MATERIALS AND METHODS
Cell culture and viruses.
Vero cells were propagated in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% calf serum (CS), 2 mM glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin. U373 cells were maintained similarly to Vero cells except that in place of CS, a mixture of 5% CS and 5% fetal bovine serum (FBS) was used. Primary normal human diploid fibroblasts (Clonetics, Walkersville, MD), were cultured in DMEM plus 5% fetal bovine serum. Mouse 3T3 cells were obtained from the American Type Culture Collection and maintained in DMEM supplemented with 10% CS. PKR−/− and PKR−/− PERK−/− cells (4) were maintained in DMEM containing 10% FBS. The HT22Fv2ePERK cells were a kind gift from D. Ron and P. Lu (New York University School of Medicine) and maintained in DMEM plus 10% FBS. The HSV-1 gB mutant KO82 (a gift from Prashad Desai and Stanley Person, John Hopkins University School of Medicine [5]) was propagated in D6 cells. The HSV-1 gH mutant (gift of Duncan Wilson, Albert Einstein College of Medicine, and Tony Minson, University of Cambridge, United Kingdom; 11) was propagated in F6 cells. Both D6 and F6 cells were maintained in DMEM plus 10% FBS. The ICP6-deficient HSV-1 mutant (ΔICP6) was a kind gift of Sandra Weller (University of Connecticut Health Science Center, Farmington, CT; 13).
Antibodies and chemicals.
The monoclonal antibody that detects total eIF-2α was originally produced by the late Edward Henshaw and was a gift from Mike Clemens (44) The anti-phospho-PERK (used exclusively for immunoprecipitations) and anti-total PERK polyclonal antibodies along with AP20187 (available from Ariad Pharmaceuticals and used with permission) were kind gifts from D. Ron and H. Harding. Antibodies recognizing the following HSV-1 proteins were graciously provided by the listed investigators: anti-gC (R. Eisenberg and G. Cohen, University of Pennsylvania School of Dental Medicine), anti-ICP27 (S. Silverstein, Columbia University College of Physicians & Surgeons), and anti-TK (B. Roizman, University of Chicago). The following antibodies were purchased commercially: anti-eIF-2α phospho-Ser51-specific antibody (StressGen), anti-phospho-PERK antibody for immunoblotting (Cell Signaling), anti-myc (Sigma), and anti-eIF4E (BD Transduction Laboratories, San Diego, CA). Dimethyl sulfoxide (DMSO), actinomycin D, thapsigargin (Tg), and tunicamycin were purchased from Sigma. Phosphonoacetic acid (PAA) and dithiothreitol were purchased from Calbiochem.
Analysis of PERK activation by immunoprecipitation.
Immunoprecipitations for PERK were performed as described previously (2). Briefly, confluent 10-cm dishes of mouse 3T3 cells were washed twice in phosphate-buffered saline (PBS) and once in PBS with 1 mM EDTA. The cells were solubilized in 1% Triton X-100 lysis buffer supplemented with protease and phosphatase inhibitors. PERK was then immunoprecipitated using 0.5 μl of both anti-phospho-PERK antibody (from D. Ron) and anti-total-PERK antibody. The samples were incubated overnight at 4°C with rocking. The immune complexes were washed three times with 1 ml lysis buffer supplemented with phosphatase inhibitors, once with 1 ml PBS, and boiled in sodium dodecyl sulfate (SDS) sample buffer. The entire sample was used for immunoblotting.
Isolation of PERK-associated proteins by coimmunoprecipitation.
Ten-centimeter dishes of confluent Vero cells were either left uninfected or infected (multiplicity of infection [MOI] of 10) with HSV-1 (ΔICP6). At 8 h postinfection (hpi), the cells were labeled with 0.5 mCi of [35S]cysteine and [35S]methionine for 2 h. Cell-free lysates were subsequently prepared without phosphatase inhibitors as described in the preceding immunoprecipitation section. Supernatants were precleared by incubation with 30 μl normal rabbit serum for 1 h at 4°C, which was collected by three consecutive incubations with 100 μl settled bed volume (SBV) pansorbin (catalog no. 507858; Calbiochem, CA). Following transfer of the precleared extract to fresh microcentrifuge tubes containing protein A Sepharose (10-μl SBV) previously bound to either 2 μl of the appropriate preimmune sera or to a mixture of 0.5 μl anti-PERK together with 0.5 μl anti-phospho-PERK polyclonal antiserum, the reaction mixtures were incubated on a rocking platform overnight at 4°C. Last, the beads were collected by brief centrifugation and washed three times in 1 ml radioimmunoprecipitation assay (RIPA) buffer (2). Following fractionation of immune complexes by SDS-polyacrylamide gel electrophoresis (PAGE), the gels were fixed, impregnated with a fluorophore, dried, and exposed to Kodak XAR film.
To identify viral proteins bound to an epitope-tagged version of PERK, 10-cm dishes of 293 cells were transfected with 7 μg of mammalian expression plasmids encoding myc-tagged versions of PERK (kindly provided by H. Harding) using 17.5 μl of Lipofectamine 2000 (Life Technologies) per dish. At 16 h after transfection, the cells were infected with WT HSV-1 and at 8 h postinfection labeled with 0.5 mCi of [35S]cysteine and [35S]methionine for 2 h. The cells were then processed as described above, and an anti-myc monoclonal antibody (Sigma) was added to precipitate myc-tagged PERK. The complexes were then separated via 7% SDS-PAGE, and the gels were fixed, impregnated with a fluorophore, dried, and exposed to film.
Glycoprotein synthesis.
Triplicate sets of 60-mm dishes of Vero cells were either left untreated or treated with 300 μg/ml PAA for 1 h. The cells were then either mock infected or infected with WT HSV-1. At 15 h postinfection, cultures were overlaid with 1 ml DMEM lacking methionine and cysteine but supplemented with 50 to 70 μCi/ml 35S-labeled Express (commercial mixture of methionine and cysteine from Perkin-Elmer) for 15 min. The cells were then processed as described under immunoprecipitations (except no phosphatase inhibitors were included in the lysis buffer), and 150 μg of protein from each sample was incubated for 2 h at 4°C with 10 μl (SBV) of concanavalin A (ConA)-conjugated agarose beads (Calbiochem) which was previously blocked for 1 h in a 5-mg/ml fraction V bovine serum albumin solution. The beads were washed three times in RIPA buffer and then boiled in SDS sample buffer for 5 min. The denatured proteins released from the ConA beads were diluted 1:25 in fresh 1% Triton X-100 lysis buffer supplemented with protease inhibitors, 7.5 μl (SBV) of bovine serum albumin-blocked ConA beads was added, and the mixture was incubated at 4°C for 2 h for a second round of glycoprotein purification. The beads were then washed three times in RIPA buffer and boiled in 40 μl SDS sample buffer. Ten microliters was then counted in a liquid scintillation counter to assess the amount of labeled glycoproteins recovered from each sample.
Endoglycosidase digests.
[35S]cysteine- and [35S]methionine-labeled myc-tagged PERK immune complexes solubilized in sample buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 5% β-mercaptoethanol, 10% glycerol) were diluted 1:6 in 1× N-glycosidase F (PNGase F) or endoglycosidase H (endo H) reaction buffer supplemented with either 16.7 U/μl PNGase F or 33.4 U/μl endo H (New England Biolabs) and incubated at 37°C for 1 h. The reactions were terminated by adding an equal volume of 2× sample buffer and boiling for 3 min.
XBP-1 splicing.
Total RNA was isolated and subjected to reverse transcription-PCR using primers MXBP1.3S and MXBP1.12AS as described previously (25). The spliced product was 448 bp long, while the unspliced precursor was 473 bp long.
Construction of targeting plasmids. (i) pXN1-ΔNot.
pXN1 (32) was digested with NotI, and the overhanging ends were filled in with the Klenow fragment of DNA polymerase I and self-ligated with T4 DNA ligase to remove the unique NotI site in the pXN1 vector backbone.
(ii) pXN1-BGHpA-LoxP.
A PCR product containing the bovine growth hormone (BGH) polyadenylation signal was amplified from the plasmid pcDNA4 (Invitrogen) using Pfu polymerase (Stratagene) with the following primers: Pf-BGHpA (5′-TCCCCACAAGATGGCTGTGCCTTCTAGTTGCCAGCCATCTGTTGTTT-3′) and NI-Not-LoxP-BGHpA (5′-TGCCCTAGCACAGGGCGGCCGCATAACTTCGTATAATGTATGCTATACGAAGTTATCCATAGAGCCCACC-3′). The resulting 294-bp product is flanked by PflMI and EcoNI sites and contains internal loxP and NotI sites. After digestion with PflMI and EcoNI, the fragment was ligated into EcoNI- and PflMI-digested, shrimp alkaline phosphatase-treated pXN1-ΔNot to create pXN1-BGHpA-LoxP. The region isolated by PCR in the plasmid was sequenced to confirm that no mutations were introduced by Pfu polymerase.
(iii) pXN1-Bac.
pBelo-Bac11 (New England Biolabs) was digested with NotI, and the 6,876-bp fragment was ligated into NotI-digested, shrimp alkaline phosphatase-treated pXN1-BGHpA-LoxP to create pXN1-Bac.
Construction of the Δ34.5 bacterial artificial chromosome (BAC).
pXN1-Bac (10 μg) was linearized by NsiI digestion and subsequently cotransfected with 50 μg Δ34.5ΔUs11 viral DNA (33) into Vero cells using the calcium phosphate method as described previously (32). After 5 days, cell-free lysates were prepared by freeze-thawing, diluted, and used to infect confluent monolayers of U373 cells. U373 cells do not support the growth of the parental Δ34.5ΔUs11 virus and can thus be used to select for recombinants expressing Us11 as an immediate early (IE) protein (32). Once significant cytopathic effect was observed, cell-free lysates were prepared by freeze-thawing, diluted, and used to infect fresh, confluent U373 cell monolayers in order to further amplify the recombinant virus and obtain enriched stocks. Each enriched, cell-free lysate derived from independent transfections was used to infect one 10-cm dish of confluent Vero cells in the presence of 100 μg/ml cycloheximide for 3 h at 37°C and 5% CO2. Circular viral DNA was isolated as described previously (17). Briefly, the infected Vero cells were washed with ice cold PBS, transferred to microcentrifuge tubes, and resuspended in 10 mM EDTA, pH 8.0. SDS and NaCl were then added to 0.6% and 1 M, respectively, and incubated on ice overnight. DNA was then phenol-chloroform extracted and ethanol precipitated. The isolated DNA was dissolved in H2O supplemented with 50 μg/ml RNase A. Ten percent of the isolated DNA sample was subsequently electroporated into Escherichia coli DH10B cells, spread onto LB agar supplemented with chloramphenicol, and incubated at 37°C to select for transformants. Infectious virus was generated by isolating BAC DNA from 1.5 ml of saturated LB culture inoculated with DH10B cells harboring Δ34.5ΔSUPΔUs10-BGHpA-Bac by standard alkaline lysis. The entire DNA preparation was subsequently transfected into Vero cells by using the calcium phosphate method. Plaques were clearly discernible by 4 days posttransfection.
Construction of Δ34.5ΔgB recombinant virus.
Allelic replacement was performed as described previously (9). Briefly, pKD42 (9) encoding the λ red protein recombination machinery was introduced into E. coli DH10B cells maintaining Δ34.5ΔSUPΔUs10-BGHpA-Bac by electroporation and selection at 30°C on LB agar supplemented with chloramphenicol and ampicillin. A PCR product containing the kanamycin resistance (Kanr) gene flanked on each side by 42 nucleotides homologous to sequences in the UL27 (gB) gene was then generated using pKD13 as the template and the following primers: UL27pKD13P1-55710 (5′-TCGGCGGCTCCGAGTTCCCCCGGCACGCCTGGGGTCGCGGCCGCGGTGTAGGCTGGAGCTGCTTC-3′) and UL27pKD13P4-55621 (5′-GGCGGGCGGCGCCGGAGTGGCAGGGCCCCCGTTCGCCGCCTGGGTATTCCGGGGATCCGTCGACC-3′).
This PCR product was then electroporated into pKD42-transformed Δ34.5ΔSUPΔUs10-BGHpA-Bac E. coli DH10B cells, and recombinants were selected on LB agar supplemented with kanamycin. Proper integration of the Kanr cassette was determined by PCR using the following primers: UL27DnSt-55451 (5′-ACGTAAAAGTTTGCATCGGTG-3′), UL27UpST-55766 (5′-GCCGGTGGTTCGTCGTATGG-3′), K1 (5′-CAGTCATAGCCGAATAGCCT-3′), K2 (5′-CGGTGCCCTGAATGAACTGC-3′), UL10-DnSt-24792 (5′-ACAATGTTCTGTATACGGTC-3′), and UL10-UpSt-23525 (5′-TATGCCGTGGTCGGCGCCGTG-3′).
Infectious ΔΔΔBAC-B2-UL27::Kanr virus was generated as described above by transfecting viral DNA into D6 cells which contain an integrated copy of the HSV-1 gB gene under transcriptional control of the HSV-1 gD promoter. Southern analysis was performed as described previously (32).
RESULTS
Cross-reaction of an HSV-1 polypeptide with phospho-PERK-specific antisera.
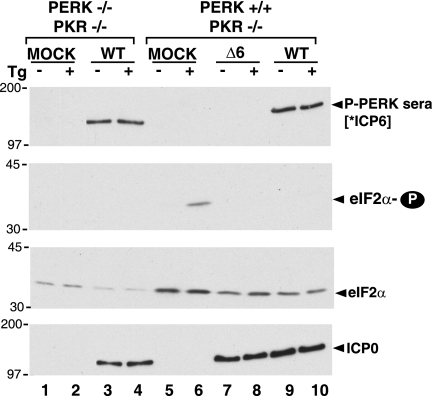
The ER-resident eIF-2α kinase PERK is not an abundant polypeptide in most cells, making it difficult to detect by simply fractionating total cellular protein by SDS-PAGE, followed by immunoblotting. Ascertaining the activation status of PERK by immunoblotting total cellular protein in conjunction with commercially available antisera specific for phospho-PERK is similarly compromised due to the paltry concentration of the target antigen in extracts prepared from the vast majority of cells. An immunoprecipitation enrichment step is typically required in such protocols to sufficiently concentrate PERK such that it can be detected by immunoblotting (15). Moreover, when combined with gel electrophoresis in low percentage gels for lengthy time periods, this procedure resolves activated, phosphorylated PERK from unphosphorylated PERK. While a recent study claims that PERK is activated in HSV-1-infected cells, all of the data in support of this conclusion were surprisingly derived by simple SDS-PAGE fractionation of total cellular protein, followed by immunoblotting using a commercially available phospho-specific anti-PERK antisera (6). To ascertain the precise nature of the immunochemical signal generated using phospho-specific PERK antiserum to probe immunoblots of HSV-1-infected cell lysates fractionated by SDS-PAGE, we performed a similar experiment in PKR-deficient and PERK-deficient cells.
It is well established that treatment with thapsigargin, by depleting Ca2+ stores from the ER, inhibits the activity of resident Ca2+-dependent chaperones and thereby induces the unfolded protein response (51). One component of this response involves activation of the eIF-2α kinase PERK. Indeed, phosphorylated eIF-2α accumulates in mock-infected PKR−/− cells treated with Tg (Fig. 1). The increase in eIF-2α phosphorylation is PERK dependent, as it is not observed in Tg-treated PKR−/− PERK−/− cells (Fig. 1). Notably, an immunoreactive signal from mock-infected PKR−/− lysates is not detected using a phospho-PERK-specific antibody, despite the fact that PERK-dependent eIF-2α phosphorylation is easily observed. However, a prominent 140-kDa band immunoreactive with phospho-PERK-specific sera is detected in PKR−/− cells infected with WT HSV-1 (Fig. 1). The intensity of this signal is unchanged by Tg treatment. Surprisingly, this band is present at a similar intensity in HSV-1-infected PKR−/− PERK−/− cells. Thus, although the high-molecular-weight antigen that cross-reacts with phospho-PERK-specific antisera is found in HSV-1-infected cells, it does not require a wild-type PERK allele and therefore is not likely to be the phosphorylated product of the cellular PERK gene. Instead, we hypothesized that it might instead reflect the fortuitous reaction of the commercially available phospho-specific PERK antisera with an HSV-1-encoded protein. It is likely that the band immunoreactive with phospho-PERK antisera in HSV-1-infected cells described by Cheng and colleagues is produced relatively early in the viral reproductive cycle, as its detection requires viral protein synthesis but is unaffected by phosphonoacetic acid treatment, an inhibitor of viral DNA replication and late gene expression (6). Examination of the HSV-1 proteome for 140-kDa antigens expressed with this particular profile revealed that the viral ICP6 gene was a likely candidate. Parallel experiments carried out with an ICP6 null mutant confirmed that the phospho-PERK-specific antisera immunoreactive signal detected in HSV-1-infected cells absolutely required the viral ICP6 gene and not the product of the cellular PERK gene. Thus, simple SDS-PAGE fractionation of total protein from HSV-1-infected cells followed by immunoblotting using a commercially available phospho-specific anti-PERK antisera cannot be used to draw conclusions regarding PERK activation. We thus set out to carefully characterize the effect of HSV-1 infection on PERK.
FIG. 1.
Cross-reaction of phospho-PERK antiserum with the HSV-1 ICP6 polypeptide. Immortalized mouse embryonic fibroblasts (PERK−/− PKR−/− or PERK+/+ PKR−/−) were mock infected (MOCK) or infected with HSV-1 (MOI of 5) (wild-type [WT] or an ICP6-deficient strain [Δ6]). At 9 hpi, cultures were treated with thapsigargin (Tg) (+) or DMSO (−) for 30 min. Total protein was subsequently isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera (P-PERK sera, antigen affinity-purified phospho-PERK sera; eIF2α-P, phospho-specific eIF-2α sera; eIF2α, total eIF-2α sera; ICP0, anti-ICP0 monoclonal antibody as a marker for viral infection). [*ICP6] indicates that the P-PERK sera cross-reacts with the HSV-1 ICP6 gene product. The migration positions of molecular mass standards (in kilodaltons) appear to the left of the individual membrane strips.
Resistance of PERK to activation by acute ER stress in HSV-1-infected cells.
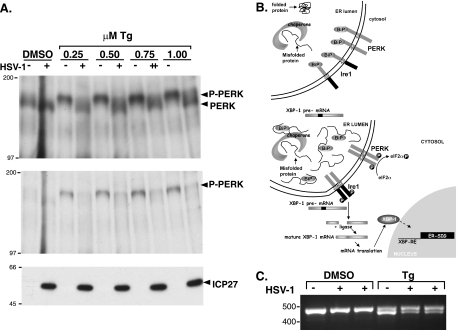
To evaluate the extent to which HSV-1 infection activates PERK, it was absolutely necessary to first immunoprecipitate PERK. Not only does this remove the viral contaminant that cross-reacts with phospho-PERK antisera on an immunoblot, it also facilitates the detection of PERK on immunoblots by concentrating the antigen. Immune complexes were subsequently fractionated by electrophoresis in 7% polyacrylamide gels for 3 h at 100 V, such that the 66-kDa molecular mass standard migrated to the bottom of the gel. Following this extensive electrophoresis regimen, which is sufficient to resolve the activated and unactivated forms of PERK, proteins were transferred to nitrocellulose and immunoblotted using the indicated antisera (Fig. 2A). A further advantage of this technique is that the ratio of activated to unactivated PERK can be visualized, as they can be detected with the same antisera. Under these conditions, we were unable to observe an increase in the abundance of activated PERK in HSV-1-infected cells. However, an increase in the abundance of a slower-migrating form of PERK together with a corresponding increase in phospho-PERK was easily detected in Tg-treated, uninfected cells (Fig. 2A). Strikingly, under conditions where Tg treatment effectively shifted 100% of the detectable PERK into a slower-migrating, activated form in uninfected cells, the vast majority of PERK remained unactivated in HSV-1-infected cultures (Fig. 2A). Even at the highest Tg concentration tested, most of the PERK in HSV-1-infected cells was refractory to activation. Essentially the same result was achieved when anti-PERK immune complexes were fractionated by SDS-PAGE and immunoblotted using phospho-PERK-specific sera (Fig. 2A). Thus, PERK not only remains unactivated in HSV-1-infected cells, it is remarkably resistant to activation by acute ER stress.
FIG. 2.
Resistance of PERK to activation in HSV-1-infected cells. (A) Murine 3T3 cells were mock infected (−) or infected (MOI of 5) with wild-type HSV-1 (+). At 7 hpi, the cultures were exposed to solvent (DMSO) or the indicated concentrations (0.25, 0.5, 0.75, and 1.0 μM) of Tg for 30 min. Total protein was subsequently isolated and subjected to immunoprecipitation using anti-PERK polyclonal antisera. Immune complexes were fractionated by SDS-PAGE and analyzed by immunoblotting using total anti-PERK sera (top gel) or phospho-PERK-specific antisera (middle gel). Total protein in initial lysates (prior to immunoprecipitation) was directly fractionated by SDS-PAGE and analyzed by immunoblotting using antisera directed against the viral ICP27 protein as a marker for viral infection (bottom gel). The migration positions of molecular mass markers (in kilodaltons) appear to the left of the gels. (B) The top panel illustrates a nonstressed ER under conditions of chaperone sufficiency. The stress sensors IRE1 and PERK are both inactive, bound to the chaperone BiP in the ER lumen. The XBP-1 precursor mRNA (pre-mRNA) remains unprocessed, unable to direct XBP-1 protein synthesis. In the bottom panel, ER stress, as measured by chaperone insufficiency, results in the release of BiP from the luminal domains of both IRE1 and PERK, allowing them to form homodimers within the plane of the ER membrane. Once each subunit of the dimer phosphorylates the other, activated PERK phosphorylates eIF-2α, whereas the activated endoribonuclease activity of IRE1 cleaves the XBP-1 pre-mRNA to initiate proper processing. Once processing is completed by an RNA ligase, the mature XBP-1 mRNA is translated and the protein product translocates to the nucleus where it activates expression of ER stress-induced genes (SIG). (C) Processing of XBP-1 mRNA in HSV-1-infected cells exposed to ER stress (0.5 μM Tg). This was performed as in panel A, except that total RNA was isolated and subjected to reverse transcription-PCR using primers specific for the XBP mRNA. PCR products were fractionated by electrophoresis on a 2.2% agarose gel and visualized under UV illumination following ethidium bromide staining. The product derived from mature XBP-1 mRNA is approximately 26 bp smaller and migrates faster.
To discern whether the resistance of PERK to activation by ER stress in HSV-1-infected cells reflected a global impairment of all UPR stress sensors, the ability of IRE1 to respond to acute ER stress in virus-infected cells was evaluated. Like PERK, IRE1 is a resident ER kinase with a luminal sensor domain joined to an intracellular kinase domain (Fig. 2B). Upon activation by malfolded proteins within the ER lumen, IRE1 dimerizes, and each subunit phosphorylates the other. Subsequent allosteric changes in IRE1 structure activate a latent endoribonuclease which initiates processing of the stable, cytosolic XBP-1 mRNA precursor (Fig. 2B). Once correctly processed, the XBP mRNA directs production of a transcription factor that in turn induces transcription of stress-responsive genes (reviewed in reference 14). Whereas IRE1 activation is difficult to reproducibly measure by direct assessment of IRE1 phosphorylation, processing of XBP precursor mRNA into mature mRNA provides a convenient, accepted measure of IRE1 activation (20, 25). Like PERK, IRE1 is not activated in HSV-1-infected cells (Fig. 2C). However, processing of XBP precursor mRNA into mature mRNA is not precluded in HSV-1-infected cells exposed to acute ER stress (Fig. 2C). Similar results were obtained at either 7 or 12 hpi (not shown). Thus, activation of IRE1 by acute ER stress is not prevented in HSV-1-infected cells. This establishes that the observed resistance of PERK to activation by ER stress in HSV-1-infected cells likely reflects the selective targeting of PERK, as opposed to both PERK and IRE1, by a virus-specified function.
Suppression of PERK activation by HSV-1 requires the PERK ER luminal domain.
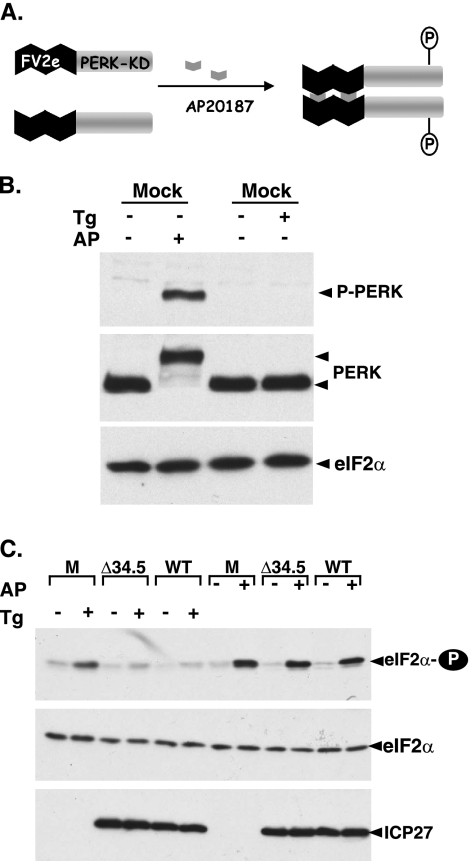
As a resident type I ER membrane protein, PERK contains both a cytosolic eIF-2α kinase domain fused to an ER luminal regulatory domain. To determine whether the subcellular localization of PERK to the ER was required for HSV-1 to prevent kinase activation, HT22-Fv2e cells that express a cytosolic PERK derivative were employed (24). In addition to relatively small quantities of endogenous PERK, these cells overexpress a synthetic, hybrid PERK derivative where the PERK eIF-2α kinase domain is fused to two Fv2e-binding domains, which promotes dimer formation in the presence of a synthetic small molecule, AP20187 (Fig. 3A). Unlike natural PERK, Fv2e-PERK is an abundant cytosolic protein (24). Thus, application of AP20187 to HT22-Fv2e cells promotes Fv2e-PERK dimer formation, kinase activation, and subsequent eIF-2α phosphorylation without activating endogenous, natural PERK (24). As this synthetic PERK derivative is abundantly expressed, its activation state can be easily examined by immunoblotting using total PERK antisera, as opposed to phospho-specific PERK antisera, without prior immunoprecipitation (Fig. 3B). Significantly, whereas both WT HSV-1 and a Δ34.5 mutant derivative are able to effectively prevent the accumulation of phosphorylated eIF-2α in response to Tg-mediated activation of endogenous PERK, both viruses are unable to prevent the accumulation of phosphorylated eIF-2α in response to AP20187 (Fig. 3C). Thus, the suppression of PERK activation observed in HSV-1-infected cells requires the luminal regulatory domain of PERK, as activation of the PERK eIF-2α kinase activity by a small molecule that promotes dimerization of the kinase domain is unimpaired in virus-infected cells. Surprisingly, phosphorylated eIF-2α accumulates to the same extent regardless of the state of the viral γ134.5 gene (Fig. 3C). This further suggests that the proper subcellular localization of PERK to the ER is necessary for the γ134.5-PP1α holoenzyme to effectively counteract PERK eIF-2α kinase activity. These observations raise the possibility that the luminal domain of PERK might somehow be modified in HSV-1-infected cells.
FIG. 3.
Suppression of PERK activation in HSV-1-infected cells depends upon the luminal domain of PERK. (A) Conditional activation of a synthetic PERK derivative by a small molecule. FV2e PERK is comprised of two modified FK506-binding domains, termed FV2e segments, fused to the cytosolic catalytic PERK kinase domain. In the presence of AP20187, this fully cytoplasmic protein forms active dimers when each subunit phosphorylates the other. The activated kinase can then phosphorylate eIF-2α. (B) HT22 cells expressing FV2e-PERK were exposed to either DMSO (−), 0.5 μM Tg, or 1 nM alkaline phosphatase (AP) for 30 min. Total protein was subsequently isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting with the antisera specific for PERK, total eIF-2α, or phospho-PERK (P-PERK). Due to the abundance of the FV2e-PERK fusion protein in these cells, the commercially available phospho-PERK-specific antisera easily detects PERK in unfractionated lysates. (C) As in panel B except mock-infected (M), Δ34.5, and wild-type virus-infected (WT) cultures were used. Following electrophoresis, proteins were analyzed by immunoblotting with the indicated antisera. ICP27 serves as a marker for viral infection.
Association of a 105-kDa protein with the luminal domain of PERK in HSV-1-infected cells.
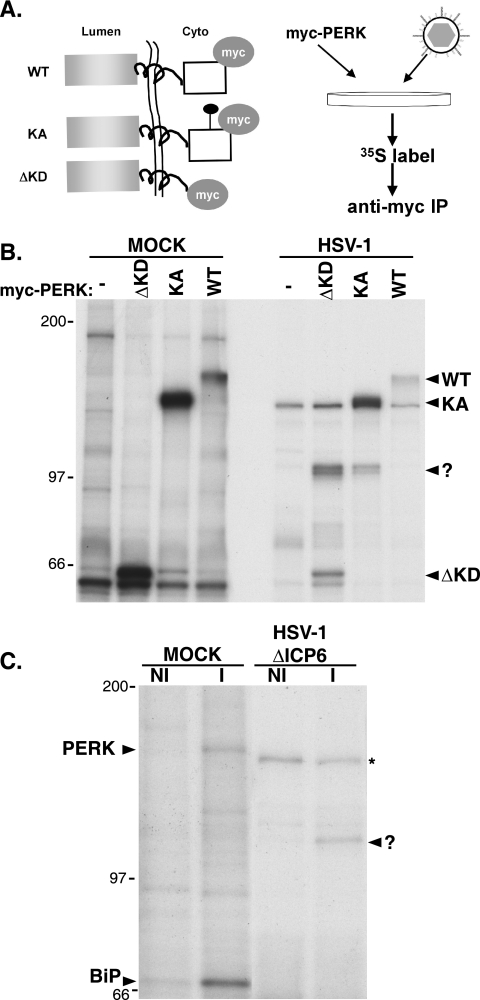
To investigate the possibility that a virus-specified or virus-induced protein might associate with PERK in infected cells, we made use of a series of epitope-tagged PERK derivatives, as limited quantities of endogenous PERK are present in most cultured cell lines. In addition, the use of an epitope-tagged derivative allows for immunoprecipitation to be executed with a monoclonal antibody specific for the tag, eliminating many spurious nonspecific background bands. Plasmids expressing either myc-tagged WT PERK, a luminal domain derivative that lacked a kinase domain (ΔKD) or a catalytically inactive protein (KA) harboring a Lys-to-Arg substitution at residue 618 (15) were transfected into 293 cells, and the cells were subsequently mock infected or infected with WT HSV-1 (Fig. 4A). After radiolabeling with 35S-labeled amino acids, cell-free lysates were prepared under nondenaturing conditions and subjected to immunoprecipitation with an anti-myc monoclonal antibody. Following several washes to remove nonspecifically bound material, the immune complexes were fractionated by SDS-PAGE, and the radiolabeled proteins were visualized by autoradiography. In mock-infected cells, myc-tagged polypeptides of the expected size were recovered following transfection of plasmids expressing the WT, ΔKD, or KA PERK derivative (Fig. 4B). However, the WT myc-tagged protein is activated shortly after transfection, resulting in its retarded migration relative to the full-length catalytically inactive KA derivative (Fig. 4B). This is in accord with published observations from other groups (15). Due to this transfection-mediated activation of WT myc-tagged PERK, which of course promotes eIF-2α phosphorylation and the inhibition of protein synthesis, the WT myc-tagged protein is labeled poorly compared to the catalytically inactive KA mutant (Fig. 4B) and is less abundant (unpublished observation). In addition to the myc-tagged PERK derivatives, a radiolabeled antigen migrating with an apparent molecular mass of 105 kDa is present exclusively in immune complexes isolated from cells transfected with ΔKD or KA myc-tagged PERK derivatives and subsequently infected with HSV-1 (Fig. 4B). The 105-kDa protein was not present in immune complexes isolated from mock-transfected cells, nor was it detected in complexes prepared from cells transfected with myc-tagged WT PERK. This likely reflects the global inhibition of protein synthesis resulting from activation of myc-tagged WT PERK in transfected cells prior to infection (Fig. 4B), preventing any subsequent incorporation of 35S-labeled amino acids into viral proteins.
FIG. 4.
Association of a 105-kDa protein with PERK in HSV-1-infected cells. (A) Illustration of myc-tagged protein derivatives and protocol for detecting PERK-associated proteins in infected cells. All PERK derivatives contain the myc epitope tag on their cytosolic side. WT is wild-type PERK, KA has a single amino acid substitution at residue 618 within the cytosolic kinase domain and that is kinase negative, and ΔKD lacks the cytosolic kinase domain and fuses the luminal and transmembrane segments directly to the myc tag. Following transfection of myc-tagged PERK expression plasmids, 293 cells were infected with HSV-1. At 8 hpi, cultures were radiolabeled with 35S-labeled amino acids for 2 additional hours, and detergent lysates were subjected to immunoprecipitation using an anti-myc monoclonal antibody. After the immunoprecipitates were washed to remove unbound proteins, immune complexes were fractionated by SDS-PAGE, and the bound proteins were visualized by autoradiography. (B) Cultures transfected with plasmids expressing the indicated myc-tagged PERK derivatives or untransfected counterparts (−) were either mock infected or infected with HSV-1. Lysates were prepared and analyzed as described above for panel A. The migration positions of the different myc-tagged PERK derivatives are indicated to the right of the gel. The reduced radiolabeling of the myc-tagged derivatives in infected cells reflects the potent suppression of host protein synthesis in HSV-1-infected cells. The mobility of the 105-kDa PERK-associated protein is indicated with a question mark. The migration positions of molecular mass standards (in kilodaltons) are indicated to the left of the gel. (C) Vero cells were either mock infected (MOCK) or infected with an HSV-1 ICP6-deficient mutant (HSV-1 ΔICP6). Following a 2-h incubation with 35S-labeled amino acids at 8 hpi, detergent lysates were prepared and subjected to immunoprecipitation with either normal, nonimmune rabbit sera (NI) or anti-PERK immune rabbit sera (I). Immune complexes were fractionated by SDS-PAGE and analyzed as described above for panel A. The migration positions of PERK and BiP in immune complexes isolated from mock-infected cells are indicated to the left of the gel. The migration of the 105-kDa PERK-associated protein in immune complexes derived from HSV-1-infected cells is indicated with a question mark to the right of the panel. The asterisk indicates a nonspecific background band found in immune complexes isolated from HSV-1-infected cells using both NI or I sera. The migration positions of molecular mass standards (in kilodaltons) appear to the left of the gel.
To establish that an association between endogenous PERK and the 105-kDa protein could occur in infected cells, detergent extracts prepared from mock-infected or HSV-1-infected radiolabeled cultures were immunoprecipitated using either nonimmune or anti-PERK sera. Both PERK and its more abundant chaperone partner BiP are components of immune complexes isolated from mock-infected cells using anti-PERK, but not nonimmune sera (Fig. 4C). This not only validates the specificity of the anti-PERK immune sera but also demonstrates that our experimental conditions are adequate to detect the well-characterized PERK-associated protein BiP. Whereas the strong inhibition of host protein synthesis in HSV-1-infected cells eliminates the radiolabeled signal from PERK and BiP, the 105-kDa PERK-associated protein is clearly visible only in immune complexes isolated using anti-PERK sera (Fig. 4C). Detection of the 105-kDa protein in association with endogenous, natural PERK in HSV-1-infected cells not only establishes that the 105-kDa polypeptide interacts with wild-type PERK but that the myc tag did not contribute to this association in the initial transfection experiments. Taken together, the simplest interpretation of these results is that a virus-encoded or virus-induced protein associates with the luminal domain of PERK in HSV-1-infected cells. Given that the data using the ΔKD derivative suggests that the luminal domain of PERK is required for this association, we focused our attention on viral proteins known to reside within this subcellular compartment.
The 105-kDa PERK-associated protein is a virus-encoded glycoprotein.
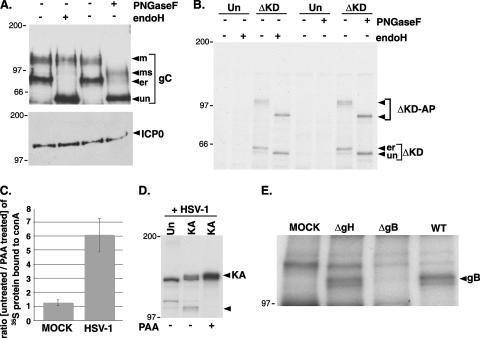
Proteins that are translocated into the ER lumen are frequently glycosylated. To investigate whether the PERK-associated protein found in HSV-1-infected cells was glycosylated, its sensitivity to cleavage by endo H and PNGase F was examined. While both of these enzymes cleave the high-mannose sugars covalently attached to proteins in the ER lumen, fully mature glycoproteins that have successfully moved through the Golgi apparatus are resistant to endo H but remain sensitive to PNGase F. For a control, we verified the specificity of these enzymes in HSV-1-infected cells by evaluating the mobility of glycoprotein C following SDS-PAGE and immunoblotting. Multiple bands representing immature and mature glycosylated forms of glycoprotein C are clearly evident in HSV-1-infected cells; however, only the immature high-mannose form is sensitive to endo H cleavage, whereas both forms are sensitive to PNGase F (Fig. 5A). ICP0, on the other hand, is not translocated into the ER and is completely resistant to the action of both of these enzymes (Fig. 5A). Having verified the specificity of endo H and PNGase F with these control experiments in our system, the sensitivity of the 105-kDa PERK-associated protein to these enzymes was subsequently evaluated. Figure 5B clearly shows that the myc-tagged PERK derivative ΔKD and the 105-kDa PERK-associated protein are sensitive to both endo H and PNGase F. Identical results were obtained using the larger, myc-tagged KA derivative (data not shown). Thus, the 105-kDa PERK-associated protein identified in HSV-1-infected cells is glycosylated, containing the high-mannose form of sugars characteristic of proteins resident within the lumen of the ER.
FIG. 5.
Identification of the 105-kDa PERK-associated protein as an HSV-1-encoded glycoprotein. (A) Specificity of endoglycosidases for viral glycoproteins. Detergent extracts from HSV-1-infected 293 cells were prepared at 10 hpi and either mock treated (−) or digested with endoglycosidase H (endoH) or PNGase F. At the conclusion of the digestion period, lysates were fractionated by SDS-PAGE and analyzed by immunoblotting using antisera specific for glycoprotein C or ICP0. The different glycosylated forms of gC are indicated to the right of the gel (un, unglycosylated; er, high-mannose form produced in the ER lumen; ms, mature post-Golgi form sensitive to PNGase F; m, fully mature glycosylated form). The migration positions of molecular mass standards (in kilodaltons) are indicated to the left of the gel. (B) 293 cells (untransfected [Un] or transfected with the ΔKD myc-tagged PERK expression plasmid) were infected with HSV-1 at high MOI. Following a 2-h incubation with 35S-labeled amino acids at 8 hpi, detergent lysates were prepared, immunoprecipitated with an anti-myc monoclonal antibody, and analyzed by SDS-PAGE as described in the legend to Fig. 4. Prior to electrophoresis, the immune complexes were incubated in the absence (−) or presence (+) of the indicated endoglycosidase. The migration positions of the immunoprecipitated proteins are indicated to the right of the gel (ΔKD-AP, 105-kDa protein associated with the myc-tagged ΔKD-PERK). The migration positions of molecular mass standards (in kilodaltons) appear to the left of the gel. (C) Mock-infected or HSV-1-infected Vero cells either treated or untreated with PAA were radiolabeled with 35S-labeled amino acids for 15 min at 15 hpi. Detergent lysates were incubated with concanavalin A Sepharose, and after extensive washing, the bound proteins were eluted and quantified by counting in liquid scintillant. Results are expressed as the ratio of proteins synthesized in untreated cells compared to PAA-treated cells. (D) 293 cells (untransfected [Un] or transfected with the KA myc-tagged PERK expression plasmid [KA]) were infected with HSV-1 in the presence (+) or absence (−) of PAA. After radiolabeling with 35S-labeled amino acids for 2 h at 8 hpi, detergent lysates were prepared, immunoprecipitated with anti-myc antibody, and analyzed as described in the legend to Fig. 4. The migration positions of KA and the 105-kDa protein-associated protein are indicated by arrowheads to the right of the gel. The migration positions of molecular mass standards (in kilodaltons) appear to the left of the gel. (E) 293 cells transfected with the KA myc-tagged PERK derivative were mock infected (MOCK) or infected with a gH-deficient mutant (ΔgH), a gB-deficient mutant (ΔgB), or wild-type virus (WT). Following a 2-h incubation with 35S-labeled amino acids at 8 hpi, detergent lysates were prepared, immunoprecipitated with an anti-myc antibody, and analyzed as described in the legend to Fig. 3. The migration position of the 105-kDa glycoprotein B protein is indicated to the right of the gel.
Many of the proteins within the ER lumen in HSV-1-infected cells are in fact different viral glycoproteins that perform a variety of characterized roles in cell entry and spread (reviewed in reference 47). Indeed, the vast majority of glycoprotein synthesis in HSV-1-infected cells evaluated by retention of radiolabeled proteins on concanavalin A Sepharose beads is sensitive to phosphonoacetic acid, a DNA replication inhibitor that prevents entry into the late phase of the viral developmental cycle (Fig. 5C). Thus, the virus-imposed load on the ER in terms of new glycoprotein synthesis late in the productive growth cycle is considerably greater than the burden at earlier times. In addition, the virus-induced impairment of host protein synthesis, an early event requiring HSV-1 gene expression together with components deposited in the cytosol by incoming virions (reviewed in reference 27), likely contributes to the reduction in ER clients early in the life cycle as well. Significantly, the association of the 105-kDa glycoprotein with PERK is likewise sensitive to PAA and thereby correlates with the time of maximal virus-imposed ER load (Fig. 5D).
A search of the HSV-1 glycoprotein proteome for 105-kDa proteins suggested that the myc-tagged PERK-associated protein might in fact be glycoprotein B. To test this hypothesis, cells transfected with a myc-tagged KA PERK expression plasmid were mock infected or infected with either WT HSV-1, a glycoprotein B-deficient virus (ΔgB), or a glycoprotein H-deficient virus (ΔgH) as a specificity control that produces gB but is unable to synthesize a different virus-encoded glycoprotein (gH) with a similar molecular weight. After radiolabeling with 35S-labeled amino acids, myc-tagged immune complexes were isolated, and the proteins bound were fractionated by SDS-PAGE and visualized by autoradiography. While the 105-kDa myc-tagged PERK-associated protein is observed only in WT and ΔgH virus-infected cells, it is not detected in mock-infected cells and in cells infected with the gB-deficient virus, ΔgB (Fig. 5E). Thus, detection of the 105-kDa myc-tagged PERK-associated protein present in HSV-1-infected cells requires an intact gB gene, suggesting that the gB gene product can physically associate with PERK in virus-infected cells. In addition, the 105-kDa protein that coimmunoprecipitates with myc-tagged PERK is immunoreactive with anti-gB antisera (data not shown).
Regulation of viral protein accumulation by a genetic interaction between PERK and glycoprotein B.
A critical function of ER stress sensors is to maintain ER homeostasis (reviewed in reference 14). This is achieved in effect by evaluating the capacity of ER chaperones to fold client proteins and adjusting protein accumulation accordingly. To determine whether the physical association between gB and PERK has the functional capability to regulate protein accumulation, viral polypeptide abundance was evaluated in the presence and absence of PERK using recombinant viruses either proficient or deficient in gB synthesis. This required the construction of an HSV-1 strain lacking gB together with both γ134.5 genes, as the γ134.5 gene product, through its interaction with protein phosphatase 1α also regulates eIF-2α phosphorylation by multiple eIF-2α kinases, including PERK (6). The use of a γ134.5-deficient strain, combined with PKR−/− and PKR−/− PERK−/− deficient cells, affords us the unique opportunity to directly evaluate whether gB and PERK contribute to the accumulation of viral proteins in infected cells.
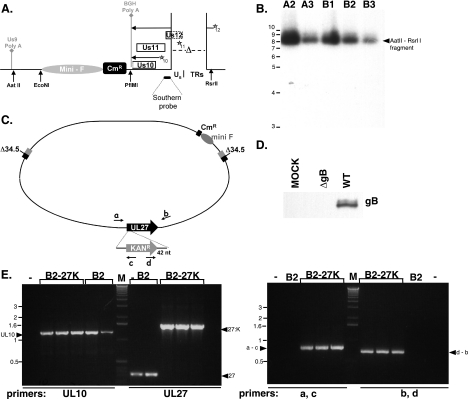
To create an HSV-1 strain doubly deficient in both γ134.5 and gB, we constructed a new bacterial artificial chromosome derivative to overcome some of the growth limitations imposed by the γ134.5 deficiency and simultaneously facilitate the isolation of a gB mutant. It has been well established that γ134.5 mutants replicate poorly in a variety of established cell lines due to activation of PKR, the accumulation of phosphorylated eIF-2α, and the resulting inhibition of protein synthesis (7). We therefore sought to link the incorporation of BAC genetic elements with a compensatory suppressor allele that substantially improves the replication capacity of a γ134.5-deficient virus. In this scheme, the BAC mini-F cis elements required to ensure maintenance in bacteria are present within a cassette that also directs the expression of the Us11 gene from an immediate-early promoter (Fig. 6A). Normally, Us11 is expressed as a true-late gene and functions to antagonize PKR. However, we have previously demonstrated that its expression as an IE gene allows Δ34.5 mutants to replicate in nonpermissive cells (29, 32). In addition, replication of Δ34.5 mutants in restrictive cells can be used as a powerful selection to enrich for desired Δ34.5 recombinants (28, 29). Thus, cells were cotransfected with HSV-1 DNA (prepared from a Δ34.5ΔUs11 mutant strain) together with the mini-F-IE Us11 expression plasmid (Fig. 6A). Once plaques appeared, lysates were prepared by freeze-thawing, and Δ34.5 recombinants that expressed Us11 as an IE protein were enriched by passage in nonpermissive U373 cells. After two cycles of enrichment, viral DNA was isolated and introduced into E. coli, and chloramphenicol-resistant colonies were isolated. BAC DNA prepared from these isolates was subsequently transfected into permissive Vero cells to prepare clonal viral stocks. This functionally established that a bacterial DNA element could establish an infection when introduced into a mammalian host cell (unpublished observation). Southern analysis of viral DNA confirmed the proper integration of the mini-F-IE US11 expression cassette into the Us-TRs locus for all of the independent isolates analyzed (Fig. 6B). Moreover, examination of the BamHI cleavage pattern of multiple BAC DNA isolates following fractionation by agarose gel electrophoresis and ethidium bromide staining revealed that gross DNA rearrangements were not detectable (data not shown). One of these isolates, B2 BAC, was chosen to be the parent strain for all subsequent modifications. To introduce the gB mutant allele into the Δ34.5 mutant BAC genetic background, a kanamycin resistance cassette flanked by sequences within the gB gene was introduced into bacteria that harbored the phage λ red recombination machinery (9) together with the B2 BAC (Fig. 6C). Kanr colonies were isolated, and the disruption of the gB gene was verified by PCR (Fig. 6E). Insertion of the Kanr element after the first nucleotide of the codon for residue 43 disrupts the gB gene at a site identical to the well-characterized KO82 mutation (5). The KO82 allele essentially does not produce any detectable gB protein (5). As gB is essential for virus growth in cultured cells (5), BAC DNA was isolated and transfected into gB-expressing Vero cells to prepare infectious viral stocks which are effectively pseudotyped with gB. Analysis of proteins produced in cells infected with the Δ34.5 gB-deficient BAC-derived virus by immunoblotting established that the gB protein was not detected (Fig. 6D).
FIG. 6.
Construction of a γ134.5 gB doubly deficient BAC. (A) Map of the Us-TRs junction resulting from integration of the mini-F-IE Us11 expression cassette. In addition to eliminating much of the Us12 gene (deleted portion of open reading frame rectangle shown as a broken line), which encodes an immunomodulatory protein and is nonessential for growth in cultured cells, deletion of a 585-bp sequence spanning the Us-TRs junction (Δ) removes the late Us11 promoter (represented by a star subscript 11). This allows for the expression of the Us11 protein from an immediate-early (IE) transcript initiating from the Us12 promoter (represented by a star subscript 12). The resulting deletion creates a suppressor allele that has been shown to allow γ134.5-deficient mutants to replicate in nonpermissive cells (29, 32). Transcripts initiating from the Us12 and Us10 promoters are polyadenylated at an ectopic bovine growth hormone (BGH) poly(A+) site. Mini-F cis elements required for propagation and maintenance in bacteria, adjacent to a chloramphenicol resistance (Cmr) gene for selection in bacteria, are shown inserted into Us10 coding sequences. The endogenous Us9 polyadenylation site (Us9 Poly A) is shown on the left. Select restriction enzyme cleavage sites, along with the small fragment used for the Southern analysis, appear below the diagram. (B) Multiple, independent isolates of BAC DNA were prepared from bacteria and digested with AatII and RsrII. DNA were subsequently fractionated by agarose gel electrophoresis, transferred to nitrocellulose, and probed with a 32P-labeled 100-bp fragment encompassing the 3′ portion of the Us12 gene (Southern probe depicted in panel A). After the filter was washed, it was exposed to X-ray film. The migration positions of molecular size standards (in kilobases) are shown to the left of the gel. The parental AatII-RsrII fragment lacking the mini-F cassette runs at 1,956 bp (M. Mulvey and I. Mohr, unpublished observation). (C) Illustration of the targeted recombination procedure used to isolate a UL27 (glycoprotein B)-deficient HSV-1 γ134.5 mutant BAC. A PCR fragment containing the Kanr gene flanked by 42-bp sequences designed to facilitate insertion in a homologous segment of the UL27 gene was electroporated into bacteria containing the γ134.5-deficient (Δ34.5) B2 BAC. BAC DNA from Kanr Cmr colonies was isolated and analyzed by PCR using the indicated primers (a, b, c, and d). nt, nucleotides. (D) The BAC-derived Δ34.5 gB-deficient recombinant virus is unable to produce detectable gB. PKR−/− cells were mock infected (MOCK) or infected (MOI of 1) with either a γ134.5 gB-deficient virus (ΔgB) or its γ134.5-deficient parent (WT gB). At 13 hpi, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting using antisera directed against gB. (E) In the left panel, BAC DNA from the parental B2 BAC (B2) or the B2 BAC with a disrupted UL27 (glycoprotein B) gene (B2-27K) was analyzed by PCR using primers specific for a control, unrecombined genome region (UL10) or primers specific for the UL27 gene. The migration positions of PCR products are indicated by the arrowheads (UL10, control product from unrearranged UL10 gene; 27, product from the wild-type UL27 gene; 27:K, expected size of products from UL27 genes that contain the insertion of a kanamycin resistance gene). The migration positions of molecular size standards (lane M) (in kilobases) appear to the left of the gel. The right panel is the same as the left panel, except that the primer pairs illustrated in panel C were used. PCR products spanning the UL27-Kanr junction were detected only in B2-27K BACs. Arrowheads denote the migration of PCR products using primers a and c or primers d and b.
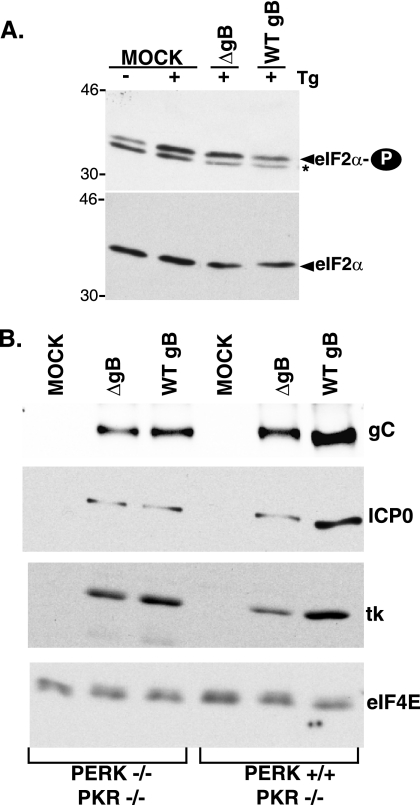
Similar levels of phospho-eIF-2α were routinely detected in normal human diploid fibroblasts or Vero cells infected with the HSV-1 Δ34.5 derivatives irrespective of the gB allele (data not shown). However, the abundance of phospho-eIF-2α was slightly elevated when cells infected with a Δ34.5-gB-deficient virus were exposed to ER stress (Fig. 7A). While this increase in phospho-eIF-2α was not sufficient to completely inhibit protein synthesis (data not shown), we reasoned that it might in fact impact upon viral polypeptide accumulation. To evaluate how the eIF-2α kinase PERK might contribute to viral protein abundance and to define a genetic interaction between gB and PERK, the accumulation of viral proteins was compared in the presence and absence of PERK using viruses proficient or deficient in gB. All the cells utilized in this experiment were PKR deficient, and all the viruses were γ134.5 deficient to rule out any possible involvement of a virus eIF-2α-phosphatase component or the cellular eIF-2α kinase PKR. Cells (PKR−/− PERK+/+ or PKR−/−PERK−/−) were infected with an HSV-1 Δ34.5 derivative containing a WT gB or ΔgB gene. At 13 h postinfection, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting with the indicated antisera (Fig. 7B). In all cases, three representative viral proteins (ICP0, thymidine kinase, and glycoprotein C) accumulated to equivalent levels regardless of the gB allele in PERK-deficient cells. Strikingly, in PERK+/+ cells, the overall accumulation of representative viral proteins was reduced between 2- and 2.5-fold in the absence of gB. Thus, the accumulation of wild-type levels of viral polypeptides in PERK+/+ cells requires gB; in addition, the gB dependence for protein accumulation is obviated in PERK-deficient cells. This establishes a genetic interaction between PERK and the gB gene product which regulates viral protein accumulation and ER homeostasis in infected cells. However, we were unable to prevent PERK activation by ectopically expressing gB in transiently transfected cells (unpublished observation). Thus, whereas gB may not be sufficient to suppress PERK activation in uninfected cells, its effects on protein accumulation likely requires additional virus-encoded or virus-induced components present in infected cells.
FIG. 7.
Regulation of HSV-1 protein accumulation by gB in a PERK-dependent manner. (A) Normal human diploid fibroblasts were infected (MOI of 5) with either a γ134.5 gB-deficient virus (ΔgB) or its γ134.5-deficient parent (WT gB). At 16 hpi, cultures were treated with Tg (+), and total protein was subsequently isolated. Polypeptides were fractionated by SDS-PAGE and analyzed by immunoblotting with antibodies directed against phosphorylated eIF-2α (eIF2α-P) or total eIF-2α. Extracts from mock-infected (MOCK) cells treated (+) or untreated (−) with Tg are shown for comparison. The asterisk to the right of the top gel denotes a nonspecific band detected in some preparations that migrates faster than phospho-eIF-2α does. The migration positions of molecular mass markers (in kilodaltons) are shown to the left of the gel. (B) Cells (PERK+/+ PKR−/− or PERK−/− PKR−/−) were mock infected (MOCK) or infected (MOI of 1) with either a γ134.5 gB-deficient virus (ΔgB) or its γ134.5-deficient parent (WT gB). At 13 hpi, total protein was isolated, fractionated by SDS-PAGE, and analyzed by immunoblotting using antisera directed against the indicated viral proteins (ICP0, thymidine kinase [tk], or gC). The abundance of the cellular translation initiation factor eIF4E serves as a control.
DISCUSSION
Given the magnitude of the burden that replicating viruses impose upon the ER, their potential to produce intracellular stress by challenging the folding capacity of the ER and activating the UPR is not at all surprising. Altering ER homeostasis triggers activation of PERK, an ER-resident kinase that inactivates the critical translation initiation factor eIF-2 by phosphorylating its alpha subunit (reviewed in reference 14). Thus, inhibiting new client protein production alleviates the ER protein load (14). Viral protein synthesis, however, requires active eIF-2, and viruses go to extraordinary lengths to prevent eIF-2α phosphorylation (reviewed in reference 23). Indeed, failure to do so would severely compromise their virulence. Of the four known eIF-2α kinases, the role of the interferon-induced, dsRNA-dependent eIF-2α kinase PKR in virus-infected cells has received the most scrutiny (reviewed in reference 26). Much less attention, however, has been directed at how other eIF-2α kinases, such as PERK, react to the stresses generated during viral infection. Recently, we established that HSV-1 encodes yet another function, distinct from those required to counteract PKR, to allow continued translation in the presence of acute ER stress (30). Herein, we show that PERK not only remains inactive upon HSV-1 infection but also is dramatically resistant to activation by acute ER stress compared with IRE1. Resistance of PERK to activation by ER stress in HSV-1-infected cells requires the luminal domain of PERK. Strikingly, we specifically find glycoprotein B associated with the luminal domain of PERK in virus-infected cells. Moreover, we identify a genetic interaction between PERK and gB that regulates viral protein accumulation in infected cells, delineating a new strategy whereby viruses regulate ER homeostasis.
Although PERK remains resistant to activation by acute ER stress in HSV-1-infected cells, this does not reflect a global attenuation of all ER stress sensors. Indeed, IRE1 activation, as evidenced by the processing of XBP mRNA is easily detected when infected cells are exposed to ER stress. This selective modulation of the UPR is similar to what has been reported in HCMV-infected cells (20). This preserves the beneficial responses, such as the transcriptional induction of stress response genes mediated by XBP-1, but curtails the potentially deleterious ones, such as the accumulation of phosphorylated eIF-2α and the global inhibition of translation. While this comparison is theoretically appealing, its potential relevance in HSV-1-infected cells is uncertain, as the inhibition of host mRNA translation might blunt the synthesis of XBP-1-induced gene products, which include ER chaperones. It is, however, certainly more likely to occur in HCMV-infected cells where host mRNA translation is not significantly impaired (48, 52).
While the ER lumen provides an environment for secreted proteins to fold, it clearly has a finite capacity to deal with client proteins at a given moment in time. Any physiological or pharmacological insult that perturbs this capacity has the potential to trigger the UPR, which is activated when the ER client protein load exceeds the folding capacity of chaperones (reviewed in reference 14). Physiological sources of ER stress disturb the homeostasis between chaperones and client proteins and therefore reflect a state of chaperone insufficiency (14). Indeed, as the UPR stress sensors PERK and IRE1 remain inactive in HSV-1-infected cells, the virus is clearly able to maintain homeostasis within the ER. This is somewhat surprising, given the acute demands the virus places on the ER, including the accumulation of cellular MHC class I molecules and a wide variety of abundant virus-specified glycoproteins (reviewed in references 42 and 47). While it is unlikely that HSV-1 glycoproteins fold independently of cellular chaperones, as they have been found physically associated with calnexin along with calreticulin (54), perhaps they can achieve their final conformation even when chaperone concentrations become somewhat limiting. Although we are unable to completely rule this out, another attractive alternative is that the virus uses a variety of independent mechanisms to maintain ER homeostasis and chaperone sufficiency. Indeed, both HCMV and HSV-1 go to great lengths to ensure that MHC class I molecules bereft of their peptide ligand are promptly dislocated from the ER and destroyed (18, 53). In a similar fashion, the rapid, global inhibition of host gene expression profoundly limits the load of cellular proteins entering the ER early in the HSV-1 life cycle. This requires the concerted action of multiple, independent viral gene products which reduce host mRNA transcription, inhibit mRNA splicing, and destabilize host mRNA (27). As host cell shutoff occurs very early in the viral life cycle, it is likely that this event is responsible for the initial resistance of HSV-1-infected cells to acute ER stress (30). Eliminating the cellular ER clientele early on in the viral developmental program together with the relatively light burden of viral clients produced early in the productive growth cycle (as shown in Fig. 5C) thus contributes to ER homeostasis by promoting a state of chaperone sufficiency.
The contribution of an initial reduction in ER client proteins through a complex host shutoff mechanism along with the relatively light virus-imposed client load early in the life cycle, however, does not in any way ensure that chaperone sufficiency will be maintained and viral protein synthesis sustained for the duration of the productive growth cycle. This requires other functions, specified by the viral GADD34 homolog, γ134.5, together with the PERK-associated viral glycoprotein gB. Whereas viral protein accumulation is unaffected in PERK−/− cells infected with a γ134.5 gB-deficient virus, HSV-1 proteins accumulate to reduced levels in PERK+/+ cells. Strikingly, this parallels what is observed following exposure of uninfected GADD34-deficient cells to ER stress. Since these cells are unable to induce a functional phosphatase to dephosphorylate eIF-2α in response to PERK activation, they simply preserve ER homeostasis by producing less protein (25, 37). Nevertheless, γ134.5 gB-deficient viruses are still able to maintain some degree of ER homeostasis in PERK+/+ cells. While their ability to impair host protein synthesis likely contributes to an initial, early state of chaperone sufficiency, it does not rule out the existence of additional mechanisms designed to posttranslationally and/or cotranslationally extract proteins from the ER, the latter of which involves the cytosolic cochaperone P58IPK (3, 12, 38).
The resistance of PERK to activation by ER stress in HSV-1-infected cells requires the luminal domain of PERK. Significantly, this same luminal domain of PERK physically associates with glycoprotein B. Furthermore, PERK regulates viral polypeptide accumulation in a gB-dependent manner. On the basis of these physical and genetic interactions, we suggest that gB binding to PERK modifies the kinase, making it more resistant to activation by ER stress, should it arise in virus-infected cells. Whereas other viral functions have been reported to counteract activated PERK, all of them are in fact broad-spectrum eIF-2α kinase inhibitors, such as the eIF-2α pseudosubstrates exemplified by vaccinia virus K3L (46) along with HCV E2 (39) and HSV-1 γ134.5 phosphatase component (6). Moreover, while the HCV E2 glycoprotein interacts with the cytosolic PERK kinase domain, gB represents the first example of a viral glycoprotein that specifically targets the luminal domain of PERK. As the luminal domains of PERK together with IRE1 are related and interchangeable (2, 15), it is likely that both sense unfolded proteins in the ER by a similar mechanism involving oligomer formation among their respective luminal domains. Recent structural data demonstrates that IRE1 luminal domain dimers generate a groove similar to the peptide-binding domain found in major histocompatibility complexes important for recognizing unfolded proteins (8). Remarkably, gB contains an MHC class II invariant chain homologous sequence that reportedly binds in the peptide-binding groove of HLA-DR molecules to disrupt the MHC class II processing pathway in infected cells (35, 45). Perhaps by interacting with the PERK luminal domain, a similar segment of gB lodges within the MHC-related fold responsible for sensing unfolded proteins and interferes with its ability to perceive ER stress in infected cells.
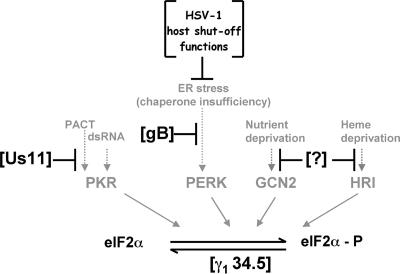
Targeting the ER stress sensor PERK in conjunction with the PP1α phosphatase catalytic component potentially allows the pathway to be tightly controlled at restriction points both upstream and downstream of the eIF-2α phosphorylation event. In the event that ER burden is not sufficiently reduced by early acting viral host shutoff functions, or gB is unable to effectively complex all of the PERK luminal domains, the γ134.5-protein phosphatase 1 holoenzyme lies ready to counterbalance any activated PERK should the need arise. The coordinate operation of these multiple overlapping pathways allows the infected cell to adapt and respond to ER stress-inducing stimuli (Fig. 8). A strikingly similar paradigm operates to control eIF-2α phosphorylation directed by PKR. Here, the Us11 gene product physically interacts with PKR and prevents its activation in response to either dsRNA or PACT protein ligands (41, 43). Once again, the upstream kinase-specific antagonist works in conjunction with the γ134.5 phosphatase component to achieve extraordinarily tight control over eIF-2α phosphorylation (31, 33). Indeed, the diverse strategies employed by HSV-1 to maintain proper supplies of active eIF-2 at all costs reflects its importance in the proper production of viral proteins and pathogenesis. Conceivably, this may lead to the discovery of yet other viral functions dedicated to controlling the impact of the two remaining eIF-2α kinases present in mammalian hosts (1).
FIG. 8.
Multiple HSV-1-specified functions interact with host eIF-2α kinases to regulate viral protein accumulation. The four known mammalian kinases, PKR, PERK, GCN2, and HRI, capable of phosphorylating eIF-2α (eIF-2α-P) are shown along with their respective activating stimuli. The HSV-1 γ134.5 protein is a GADD-4-related polypeptide that binds to PP1α to maintain adequate supplies of phosphorylated, active eIF-2α. By virtue of acting downstream of eIF-2α phosphorylation, it has the potential to counteract many eIF-2α kinases. Us11 is a PKR-specific antagonist that physically associates with PKR to prevent its activation by dsRNA and PACT. It also physically associates with dsRNA and PACT. The powerful impairment of host gene expression in HSV-1-infected cells results from the combined action of virion components together with immediate-early genes (collectively termed HSV-1 host shutoff functions). This eliminates the majority of cellular ER clients, and combined with the limited load of viral ER clients early in the productive life cycle, ensures chaperone sufficiency and thereby does not produce ER stress. As most viral glycoprotein production ramps up later in the life cycle, glycoprotein B physically associates with PERK to regulate viral protein accumulation in a PERK-dependent manner, thereby maintaining ER homeostasis. Hypothetical functions targeting the remaining eIF-2α kinases GCN2 and HRI are indicated by a question mark (?).
Acknowledgments
We are most grateful to Roz Eisenberg, Gary Cohen, Gus Kousoulas, Stan Person, Sandy Weller, Duncan Wilson, Tony Minson, and Bernard Roizman for their generous gifts of mutant viruses and antisera; Heather Harding and Phoebe Lu along with David Ron for knockout cells, antisera, and helpful discussions; Wade Breshnahan and Dom Tortorella for their helpful suggestions; Andrew Darwin for reagents, strains, and expertise in constructing targeted mutations in bacterial chromosomes; Derek Walsh, Carolina Arias, and Cesar Perez for critically reading the manuscript; and members of the Mohr lab for their insights and critical comments as this work progressed. Finally, we especially thank Paola Vallejo for her efforts in assisting with the BAC manipulations.
This work was supported by grants from the NIH and the Irma T. Hirschl Charitable Trust to I.M.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Berlanga, J. J., I. Ventoso, H. P. Harding, J. Deng, D. Ron, N. Sonenberg, L. Carasco, and C. de Haro. 2006. Antiviral effect of the mammalian translation initiation factor 2 alpha kinase GCN2 against RNA viruses. EMBO J. 25:1730-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 3.Besemer, J., H. Harant, S. Wang, B. Oberhauser, K. Marquardt, C. A. Foster, E. P. Schreiner, J. E. de Vries, C. Dascher-Nadel, and I. J. Lindley. 2005. Selective inhibition of cotranslational translocation of vascular cell adhesion molecule 1. Nature 436:290-293. [DOI] [PubMed] [Google Scholar]
- 4.Blais, J. D., V. Filipenko, M. Bi, H. P. Harding, D. Ron, C. Koumenis, B. G. Wouters, and J. C. Bell. 2004. Activating stress factor 4 is translationally regulated by hypoxic stress. Mol. Cell. Biol. 24:7469-7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, W. Z., S. Person, S. C. Warner, J. H. Zhou, and N. A. DeLuca. 1987. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J. Virol. 61:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, G., Z. Feng, and B. He. 2005. Herpes simplex virus 1 infection activates the endoplasmic reticulum resident kinase PERK and mediates eIF-2α dephosphorylation by the γ134.5 protein. J. Virol. 79:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ134.5− mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Credle, J. J., J. S. Finer-Moore, F. R. Papa, R. M. Stroud, and P. Walter. 2005. On the mechanism of sensing unfolded proteins in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 102:18773-18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimcheff, D. E., S. Askovic, A. H. Baker, C. Johnson-Fowler, and J. L. Portis. 2003. Endoplasmic reticulum stress is a determinant of retrovirus-induced spongiform neurodegeneration. J. Virol. 77:12617-12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrison, J. L., E. J. Kunkel, R. S. Hegde, and J. Taunton. 2005. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature 436:285-289. [DOI] [PubMed] [Google Scholar]
- 13.Goldstein, D. J., and S. K. Weller. 1988. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J. Virol. 62:196-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harding, H. P., M. Calfon, F. Urano, I. Novoa, and D. Ron. 2002. Transcriptional and translational control in the mammalian unfolded protein response. Annu. Rev. Cell Dev. Biol. 18:575-599. [DOI] [PubMed] [Google Scholar]
- 15.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 16.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1 alpha to dephosphorylate the alpha subunit of eukaryotic initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horsburgh, B. C., M. M. Hubinette, and F. Tufaro. 1999. Genetic manipulation of herpes simplex virus using bacterial artificial chromosomes. Methods Enzymol. 306:337-352. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, E. A., C. Hammond, and P. Cresswell. 1997. Misfolded major histocompatibility complex class I heavy chains are translocated into the cytoplasm and degraded by the proteasome. Proc. Natl. Acad. Sci. USA 94:1896-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isler, J. A., T. G. Maguire, and J. C. Alwine. 2005. Production of infectious human cytomegalovirus virions is inhibited by drugs that disrupt calcium homeostasis in the endoplasmic reticulum. J. Virol. 79:15388-15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isler, J. A., A. H. Skalet, and J. C. Alwine. 2005. Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 79:6890-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jordan, R., L. Wang, T. M. Graczyk, T. M. Block, and P. R. Romano. 2002. Replication of a cytopathic strain of bovine viral diarrhea virus activates PERK and induces endoplasmic reticulum stress-mediated apoptosis of MDBK cells. J. Virol. 76:9588-9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman, R. J. 2002. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 110:1389-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, N., X. Kuang, H. T. Kim, G. Stoica, W. Qiang, V. L. Scofield, and P. K. Wong. 2004. Possible involvement of both endoplasmic reticulum- and mitochondria-dependent pathways in MoMuLV-ts1-induced apoptosis in astrocytes. J. Neurovirol. 10:189-198. [DOI] [PubMed] [Google Scholar]
- 24.Lu, P. D., C. Jousse, S. J. Marciniak, Y. Zhang, I. Novoa, D. Scheuner, R. J. Kaufman, D. Ron, and H. P. Harding. 2004. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 23:169-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marciniak, S. J., C. Y. Yun, S. Oyadomari, I. Novoa, Y. Zhang, R. Jungreis, K. Nagata, H. P. Harding, and D. Ron. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18:3066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohr, I. 2006. Phosphorylation and dephosphorylation events that regulate viral mRNA translation. Virus Res. 119:89-99. [DOI] [PubMed] [Google Scholar]
- 27.Mohr, I. 2006. Hailing the arrival of the messenger: strategies for translational control in herpes simplex virus-infected cells, p. 105-120.In R. M. Sandri-Goldin (ed.), Alpha herpesviruses: molecular and cellular biology. Caister Academic Press, Norwich, United Kingdom.
- 28.Mohr, I., D. Sternberg, S. Ward, D. Leib, M. Mulvey, and Y. Gluzman. 2001. A herpes simplex virus type 1 γ34.5 second-site suppressor mutant that exhibits enhanced growth in cultured glioblastoma cells is severely attenuated in animals. J. Virol. 75:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohr, I., and Y. Gluzman. 1996. A herpesvirus genetic element which affects translation in the absence of the viral GADD34 function. EMBO J. 15:4759-4766. [PMC free article] [PubMed] [Google Scholar]
- 30.Mulvey, M., C. Arias, and I. Mohr. 2006. Resistance of mRNA translation to acute endoplasmic reticulum stress-inducing agents in herpes simplex virus type 1-infected cells requires multiple virus-encoded functions. J. Virol. 80:7354-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulvey, M., V. Camarena, and I. Mohr. 2004. Full resistance of herpes simplex virus type 1-infected primary human cells to alpha interferon require both the Us11 and γ134.5 gene products. J. Virol. 78:10193-10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mulvey, M., J. Poppers, A. Ladd, and I. Mohr. 1999. A herpesvirus ribosome-associated, RNA-binding protein confers a growth advantage upon mutants deficient in a GADD34-related function. J. Virol. 73:3375-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mulvey, M., J. Poppers, D. Sternberg, and I. Mohr. 2003. Regulation of eIF2α phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J. Virol. 77:10917-10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nanua, S., and F. K. Yoshimura. 2004. Mink epithelial cell killing by pathogenic murine leukemia viruses involves endoplasmic reticulum stress. J. Virol. 78:12071-12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann, J., A. M. Eis-Hubinger, and N. Koch. 2003. Herpes simplex virus type 1 targets the MHC class II processing pathway for immune evasion. J. Immunol. 171:3075-3083. [DOI] [PubMed] [Google Scholar]
- 36.Novoa, I., H. Zeng, H. P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2 alpha. J. Cell Biol. 153:1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novoa, I., Y. Zhang, H. Zeng, R. Jungreiss, H. P. Harding, and D. Ron. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22:1180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oyadomari, S., C. Yun, E. A. Fisher, N. Kreglinger, G. Kreibich, M. Oyadomari, H. P. Harding, A. G. Goodman, H. Harant, J. L. Garrison, J. Taunton, M. G. Katze, and D. Ron. 2006. Cotranslational degradation protects the stressed endoplasmic reticulum from protein overload. Cell 126:727-739. [DOI] [PubMed] [Google Scholar]
- 39.Pavio, N., P. R. Romano, T. M. Graczyk, S. M. Feinstone, and D. R. Taylor. 2003. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2α kinase PERK. J. Virol. 77:3578-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavio, N., D. R. Taylor, and M. M. C. Lai. 2002. Detection of a novel unglycosylated form of hepatitis C virus E2 envelope protein that is located in the cytosol and interacts with PKR. J. Virol. 76:1265-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peters, G. A., D. Khoo, I. Mohr, and G. C. Sen. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76:11054-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ploegh, H. L. 1998. Viral strategies of immune evasion. Science 280:248-253. [DOI] [PubMed] [Google Scholar]
- 43.Poppers, J., M. Mulvey, D. Khoo, and I. Mohr. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 74:11215-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scorsone, K. A., R. Pannierss, A. G. Rowlands, and E. C. Henshaw. 1987. Phosphorylation of eukaryotic initiation factor 2 during physiological stress which affect protein synthesis. J. Biol. Chem. 262:14538-14543. [PubMed] [Google Scholar]
- 45.Sievers, E., J. Neumann, M. Raftery, G. Schonrich, A. M. Eis-Hubinger, and N. Koch. 2002. Glycoprotein B from strain 17 of herpes simplex virus type I contains an invariant chain homologous sequence that binds to MHC class II molecules. Immunology 107:129-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sood, R., A. C. Porter, K. Ma, L. A. Quilliam, and R. C. Wek. 2000. Pancreatic eukaryotic initiation factor-2alpha kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to endoplasmic reticulum stress. Biochem. J. 346:281-293. [PMC free article] [PubMed] [Google Scholar]
- 47.Spear, P. G. 2004. Herpes simplex virus: receptors and ligands for entry. Cell. Microbiol. 6:401-410. [DOI] [PubMed] [Google Scholar]
- 48.Stinski, M. F. 1977. Synthesis of proteins and glycoproteins in cells infected with human cytomegalovirus. J. Virol. 23:751-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su, H., C. Liao, and Y. Li. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 76:4162-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 51.Treiman, M., C. Caspersen, and S. B. Christensen. 1998. A tool coming of age: thapsigargin as an inhibitor of sarco-endoplasmic reticulum Ca2+ ATPases. Trends Pharmacol. Sci. 19:131-135. [DOI] [PubMed] [Google Scholar]
- 52.Walsh, D., C. Perez, J. Notary, and I. Mohr. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 79:8057-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wiertz, E. J., D. Tortorella, M. Boygo, J. Yu, W. Mothes, T. R. Jones, T. A. Rapoport, and H. L. Ploegh. 1996. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature 384:432-438. [DOI] [PubMed] [Google Scholar]
- 54.Yamashita, Y. M. Yamada, T. Daikoku, H. Yamada, A. Tadauchi, T. Tsurumi, and Y. Nishiyama. 1996. Calnexin associates with the precursors of glycoproteins B, C, and D of herpes simplex virus type 1. Virology 225:216-222. [DOI] [PubMed] [Google Scholar]