Abstract
Nanoviruses, multicomponent single-stranded DNA plant viruses, encode a unique cell cycle link protein, Clink, that interacts with retinoblastoma-related proteins (RBR). We have established transgenic Arabidopsis thaliana lines that conditionally express Clink or a Clink variant deficient in RBR binding. By controlled induction of Clink expression, we demonstrated the capacity of the Clink protein to alter RBR function in vivo. We showed that transcription of both S-phase-specific and G2/M-phase-specific genes was up-regulated depending on the RBR-binding proficiency of Clink. Concomitantly, ploidy levels increased in a substantial fraction of leaf cell nuclei. Also, leaf epidermis cells of transgenic plants producing Clink were smaller and more numerous, indicating additional cell divisions in this tissue. Furthermore, cytogenetic analyses following induction of Clink expression in mature leaves revealed the presence of metaphasic and anaphasic nuclei, clear evidence that Clink-mediated RBR inactivation is sufficient to induce quiescent cells to reenter cell cycle progression and, for at least a fraction of them, to pass through mitosis. Expression of Clink had no effect on genes transcribed by RNA polymerases I and III, suggesting that, in contrast to its mammalian homologue, A. thaliana RBR is not involved in the repression of polymerase I and polymerase III transcription. The results of these in vivo analyses firmly establish Clink as a member of the diverse class of multifunctional cell cycle modulator proteins encoded by small DNA viruses.
Due to their restricted genome size, small DNA viruses do not encode polymerases and other enzymes of the DNA synthesis machinery. Instead, they exploit host DNA replication to multiply their genomes (19). This is a general feature of mammalian tumor viruses, e.g., simian virus 40 (SV40) or the papillomaviruses, which encode multifunctional regulatory proteins that cause the host cell to enter S phase, thereby making the host's DNA synthesis machinery available for virus DNA replication. Key regulators of cell cycle progression are the members of the retinoblastoma protein (RB) family, which sequester E2F/DP transcription factors in inactive complexes, thereby preventing them from gene activation (13, 53). The RB-controlled block of cell cycle progression is released in various ways, frequently by the binding of other proteins to RB and the subsequent release of the previously sequestered transcription factors. Various cellular or viral proteins bind to RB or otherwise prevent it—by hyperphosphorylation (48) or degradation (8)—from complexing S-phase relevant transcription factors. Among the best-studied examples are the SV40 large T antigen (T-ag), human papillomavirus E7, and adenovirus E1A protein, all of which bind to the pocket domain of RB through a sequence containing the conserved amino acid motif LxCxE (11, 16).
In mammals, RB also acts as a general repressor of transcription by RNA polymerase III (PolIII) and PolI, potentially to control cell growth (reviewed in reference 29). Repression of PolI transcription is mediated through interaction between RB and the RNA PolI transcription factor UBF (10), while the different classes of PolIII-transcribed genes appear to be down-regulated via distinct mechanisms involving the interaction of RB with different transcription factors (21, 22, 47, 54). These effects require the pocket domain of RB, and LxCxE-containing viral proteins such as E1A, T-ag, and E7 are able to relieve the repressive effects of RB on PolIII and PolI transcription (29, 54).
Plant DNA viruses include members of the families Geminiviridae and Nanoviridae (43, 52). These viruses possess small single-stranded DNA (ssDNA) genomes, as opposed to the double-stranded genomes of the mammalian tumor viruses cited above, but still show striking similarities with them in the way they induce host cells to enter S phase or trigger progress beyond the G1/S checkpoint (20).
The multifunctional replication initiator protein Rep (or AL1) of geminiviruses has been shown to bind to the plant homologues of RB, retinoblastoma-related proteins (RBR), and release the block imposed by RBR on cell cycle progression (18, 26). The binding of geminivirus Rep proteins to RBR is not always mediated by an LxCxE motif; expression of Rep proteins from viruses of the genus Begomovirus which lack this sequence systematically leads to the induction of cell cycle progression and S-phase-relevant genes, such as the proliferating cell nuclear antigen gene (PCNA) (18, 35, 36).
Members of the second plant virus family with ssDNA genomes, the nanoviruses, encode a multitude of replication initiator proteins, but only one of them, the master Rep (M-Rep), is essential for nanovirus genome multiplication (49). In addition, all nanoviruses encode a small (20-kDa) protein named Clink (for “cell cycle link”) that has been shown to bind human RB and plant RBR in vitro through its LxCxE motif (5). Furthermore, Clink is an F-box protein that also interacts with Skp1 protein, a member of the Skp1/cullin/F-box protein complex targeting proteins for ubiquitin-dependent degradation by the 26S proteasome. Clink acts as a replication enhancer during infection by a nanovirus (5). The stimulating effect of Clink on DNA replication is not nanovirus specific; geminivirus DNA replication is equally enhanced (3). Although present in the genomes of all known nanoviruses, Clink is not essential for the replication of faba bean necrotic yellows virus (FBNYV) DNA or for the infectivity and symptom expression of this nanovirus (50).
The in vitro interaction of Clink with RBR and its function as a replication enhancer in vivo suggested that Clink might interact with RBR in planta and trigger cell cycle progression in a manner similar to that of geminivirus Rep proteins or, for instance, SV40 T-ag. Therefore, we studied here the influence of Clink on cell cycle regulation in vivo by using transgenic Arabidopsis thaliana plants carrying clink gene constructs, conditionally inducible by the glucocorticoid hormone dexamethasone (Dex) (2).
MATERIALS AND METHODS
Recombinant DNA plasmids and plant transformation.
The binary transformation vector pTA7002, containing the complete two-component glucocorticoid-inducible system (2), was cleaved with XhoI and SpeI. The DNA sequence coding either for wild-type Clink or for the mutated ClinkC112R E114A (Clinkmut) protein (5) was introduced into the pTA7002 vector as an XhoI-SpeI fragment to create pTA7002-Clink and pTA7002-Clinkmut, respectively. The correctness of all constructs was verified by sequencing using an automated capillary sequencer (CEQ2000) and the DTCS sequencing kit (both from Beckman Coulter).
A. thaliana Col4 plants were transformed with the pTA7002-Clink or -Clinkmut construct as previously described (12). Transgenic T1 seedlings were selected after 2 to 3 weeks of growth under hygromycin selection (20 μg/ml), and several independent transgenic plant lines were produced. A preliminary screening of eight clink and six clinkmut independent lines was performed to confirm that the expression of the viral transgenes was dependent on the presence of Dex (see Fig. S1 in the supplemental material). E2F-controlled PCNA gene expression was also tested and found to be up-regulated in response to Dex treatment for the eight transgenic clink lines, while Dex treatment had no effect on PCNA expression in the six clinkmut lines (see Fig. S1 in the supplemental material). Therefore, for each construct, one representative line that segregated for a single T-DNA insertion locus according to the hygromycin resistance segregation pattern was selected and established at the homozygous state. These two transgenic clink and clinkmut plant lines display roughly similar transgene expression under glucocorticoid induction and were used for the whole set of experiments described.
Glucocorticoid treatments and plant growing conditions.
Dex (Sigma) was dissolved in 98% ethanol at 30 mM Dex (stock solution) and stored at −20°C in a light-tight vial. clink and clinkmut plants were grown in the greenhouse for 6 to 7 weeks. For detached-leaf inductions, fully expanded mature leaves were cut from each rosette and incubated either in a solution of distilled water including Dex at a final concentration of 30 μM in 0.1% ethanol or in 0.1% ethanol as a negative (mock) control. At 0, 2, 5, 24, 48, or 72 h after induction, the leaves were collected and used for total-RNA extraction. In addition, leaves were used for total-protein extraction after 24 h of induction.
Plants were germinated in vitro on standard 0.5× Murashige and Skoog (MS) medium (M-5519; Sigma) supplemented with 1% sucrose and 0.8% Bacto agar at 21°C under growth conditions of 16 h of light and 8 h of darkness. For inductions lasting 3 to 4 weeks, 3-day-old seedlings were transferred either to fresh 0.5× MS medium-1% sucrose plates containing 30 μM Dex or to control plates containing 0.1% ethanol and were maintained under the same growth conditions. After the respective induction period, the third and fourth rosette leaves and the first cauline leaf were harvested for flow cytometry or scanning electron microscopy (SEM) analysis. Whole plants were harvested for protein extraction and immunoblot analysis. Alternatively, fully expanded mature leaves from greenhouse plants were incubated in 0.1% ethanol in the presence or absence (mock control) of 30 μM Dex and were analyzed by flow cytometry and immunoblotting after 4 days of treatment.
Infection of plants by FBNYV.
A. thaliana plants were either infected with FBNYV by viruliferous Aphis craccivora aphids or mock treated with nonviruliferous aphids reared as described elsewhere (51). Two-week-old A. thaliana plants were exposed for an inoculation access period of 3 to 5 days, and aphids were killed with 0.2% Dedevap (Bayer). Healthy and infected plants as well as nonviruliferous and viruliferous aphids were maintained at 25°C and 50% humidity with 16 h of light in cages inside growth chambers in a restricted-access S3 confinement facility.
Cytogenetics.
Plant leaf material was obtained as described for detached-leaf inductions (see above). Following fixation of the leaves in ethanol-acetic acid (3:1), nuclei were spread on a slide according to reference 40. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI).
Flow cytometry.
One to two square centimeters of fresh leaves was finely chopped in 600 μl of Galbraith buffer (17) supplemented with 0.1% Triton X-100 and 1% polyvinylpyrrolidone 10,000 with a sharp razor blade. The extract was filtered on 48-μm mesh, treated with 20 μg/ml of RNase A (Roche) for 10 min on ice, and stained with 20 μg/ml propidium iodide. The samples were analyzed on an EPICS Elite ESP cytometer (Coulter, Hialeah, FL). Excitation of the fluorochrome was performed by an air-cooled argon laser (Uniphase) at 488 nm and 20 mW.
Total protein extraction and immunoblot analysis.
A. thaliana leaves or whole plants were harvested after Dex or ethanol treatment (see above). Total-protein extracts were prepared as described elsewhere (32). Leaf tissue (fresh weight, 100 mg) was ground to a fine powder using an MM301 mixer mill (Retsch) and resuspended at a 1:2 (wt/vol) ratio in 2× Laemmli buffer. The extracts were centrifuged at 15,000 × g for 15 min at 4°C, and 1/20 (10 μl) of the total proteins was fractionated by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis and analyzed by immunoblotting using polyclonal anti-Clink antibodies (1:500 dilution) as primary antibodies as described previously (4). Antigen was detected using an ECL detection kit (Amersham).
SEM.
The leaf epidermal surface was visualized by SEM with a Hitachi S-3000N microscope under the environmental secondary electron detector mode. Samples were slowly frozen at −18°C under a partial vacuum (90 Pa) on the Peltier stage prior to observation. The adaxial lamina surfaces of the third and fourth expanded leaves of 4-week-old plants grown in the presence of Dex or ethanol were visualized. Cell areas were measured using ImageJ 1.28u software.
Total-RNA isolation, Northern blot analysis, quantification, and cell cycle gene-specific probes.
Total RNA was extracted as described elsewhere (31). For Northern blot analysis, RNA samples were denatured and separated on 1% agarose-formaldehyde gels (39). The RNAs were transferred to Hybond-N+ membranes (Amersham-Pharmacia Biotech.) by capillary blotting and fixed by UV cross-linking. Prehybridizations and hybridizations were carried out in 50% formamide-5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-1% SDS-1× Denhardt solution-100 μg/ml of sheared, denatured herring sperm DNA-10% (wt/vol) dextran sulfate at 42°C. Membranes were washed with 1× SSC-1% SDS for 15 min at 42°C and then for 15 min at 65°C, followed by two washes with 0.1× SSC-1% SDS for 15 min at 65°C. Signals were visualized by autoradiography or by using a phosphorimager (Molecular Imager FX; Bio-Rad) for quantification.
For the detection of cell cycle-regulated gene transcripts, probes corresponding to parts of the PCNA-2 (At2g29570), CDC6a (At2g29680), CDKB2;1 (At1g76540), CycB1;1 (At4g37490), and CycB1;4 (At2g26760) cDNAs were PCR amplified (primers available on request).
RESULTS
Production of stably transformed A. thaliana plants with a glucocorticoid-inducible clink gene.
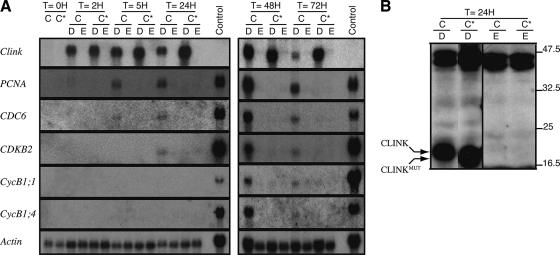
In vitro and yeast two-hybrid assays both suggested that Clink is able to interact with RB and potentially modulate the host cell cycle (5). Because we failed to establish Arabidopsis lines containing a clink transgene under the control of the strong, constitutive cauliflower mosaic virus 35S promoter, we took advantage of the tightly regulated glucocorticoid-inducible system (2). With this system, we established transgenic A. thaliana lines with conditional expression of Clink or of a mutated version of Clink (Clinkmut). The Clinkmut protein differs from wild-type Clink by 2 amino acids that convert the LxCxE motif to LxRxA and render it incompetent for binding to the A/B pocket domain of RBR (5). Based on a preliminary screening, representative homozygous lines expressing Clink or Clinkmut were selected for further studies (see Materials and Methods; also Fig. S1 in the supplemental material). Following Dex treatment for 2 to 72 h, efficient expression of the respective clink transgenes was obtained, leading to accumulation of comparable levels of clink-specific transcripts (Fig. 1A). Transcript levels were correlated with the accumulation of Clink or Clinkmut protein in leaf tissue as revealed by Western blot analysis using Clink-specific antibodies (Fig. 1B; see also Fig. 3D).
FIG. 1.
Expression of viral sequences in transgenic plants, showing the impact of Clink on cell cycle-regulated gene expression. Following Dex (D) or ethanol (E) treatment of detached, fully expanded leaves, total RNAs (panel A) or proteins (panel B) were extracted and analyzed. (A) Six micrograms of total RNAs extracted from the expanded rosette leaves was used for Northern blot analysis. The time (T) of the treatment and the identity of the transgenic plant line (C, Clink; C*, Clinkmut) are given above the panels. The two membranes were hybridized successively with probes specific for the classes of genes given on the left (see Materials and Methods). The transcript level of actin was used as a control for quantification. Six micrograms of total RNA extracted from cells from an actively growing A. thaliana T87 suspension culture (6) was also loaded and served as a positive control for hybridization with the cell cycle-specific probes. (B) Typical immunoblot result obtained with the anti-Clink antibodies. Molecular masses of protein markers (in kilodaltons) are given on the right. As previously reported (5), the Clinkmut protein migrates slightly faster than the Clink protein. Equivalent amounts of proteins were loaded in each lane, as confirmed by the similar intensities of the ≅45-kDa bands resulting from cross-reactions to endogenous proteins.
FIG. 3.
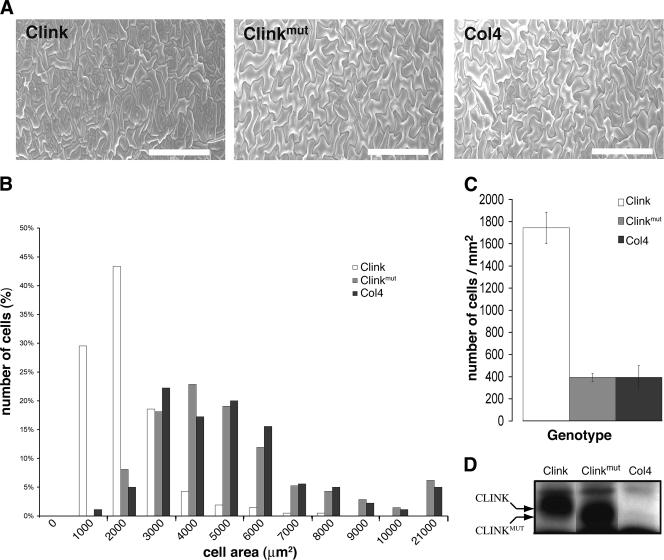
Effect of Clink expression on cell size. (A) SEM of the leaf adaxial epidermis of Clink-expressing, Clinkmut-expressing, and nontransgenic Col4 plants. Bars, 200 μm. (B) Cell area distribution of Clink-expressing (open bars), Clinkmut-expressing (shaded bars), and nontransgenic (solid bars) Col4 plants. (C) Number of cells per square millimeter of leaf epidermis of Clink-expressing (open bar), Clinkmut -expressing (shaded bar), and nontransgenic (solid bar) Col4 plants. (D) Immunoblot analysis of total proteins from Col4 plants and from the two transgenic plant lines using Clink-specific antibodies. The identity of the plant line is given above the panel. As explained for Fig. 1B, the Clinkmut protein migrates faster than the Clink protein. Plants for which results are shown in panels A and D were grown and induced under the same conditions.
Clink-specific induction of cell cycle-regulated genes.
To assess the cellular impact of Clink, we studied the expression of two well-known E2F-regulated genes, encoding PCNA and the chromosomal DNA replication origin protein (CDC6), in relation to Clink induction. Expression of Clink, but not Clinkmut, in detached, expanded 6- to 7-week-old rosette leaves was clearly associated with the transcriptional activation of both genes (Fig. 1A). PCNA- and CDC6-specific transcripts were already detectable 5 h after the beginning of Dex treatment, and a longer induction time with Clink correlated consistently with PCNA and CDC6 gene expression. No impact on PCNA or CDC6 expression was observed for Dex-treated Clinkmut plants or ethanol-treated Clink or Clinkmut plants. To analyze whether Clink expression could impact the transcription of cell cycle genes other than E2F-regulated S-phase genes, the same Northern blots were hybridized with probes specific for B2-type cyclin-dependent kinases (CDKB2) and the B1-type cyclins CycB1;1 and CycB1;4. In plants, CDKB2 proteins are thought to control entry and progression through the M phase, and this class of genes (CDKB2;1 and CDKB2;2) is expressed only during the G2 and M phases (24, 27, 34). Similarly, accumulation of transcripts of plant mitotic cyclins of the B1 class is limited to the G2 and M phases of dividing tissues (23, 37). For all three G2/M markers, specific up-regulation was detected only when wild-type Clink was expressed (Fig. 1A). In contrast to that of PCNA and CDC6, transcriptional expression of the CDKB2, CycB1;1 and CycB1;4 genes became clearly detectable at 48 h after Clink induction (Fig. 1A). When whole plants were treated with Dex, analysis of mature leaves showed results similar to those reported in Fig. 1 (data not shown). Therefore, for short-term induction experiments, we decided to work on detached mature leaves.
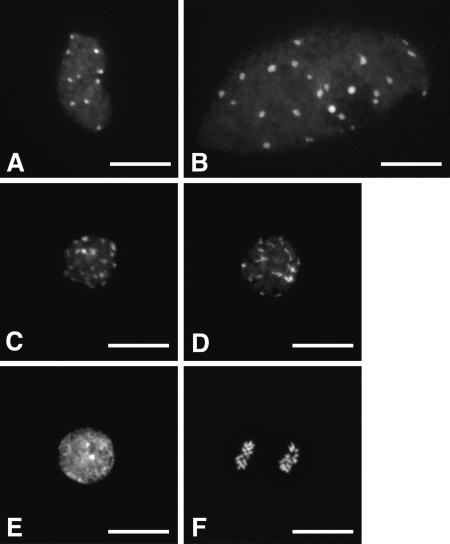
To further investigate the impact of Clink on the cell cycle, nuclei were isolated from detached, expanded rosette leaves of 6- to 7-week-old Clink and Clinkmut transgenic plants that had been treated for 4 days with Dex or ethanol, spread, and stained with DAPI. Ethanol-treated tissues and Dex-treated Clinkmut plant tissues displayed similar distributions of only two types of nuclei (Fig. 2A and B), a situation typical of mature A. thaliana leaves (41). About 95% of these nuclei showed a mostly oval shape with irregular contours and exhibited about 10 intensely DAPI stained regions termed chromocenters (CCs), which reflect the classical organization of the centromeric and nucleolus organizer heterochromatic regions (Fig. 2A). The other 5% of nuclei (Fig. 2B) appeared larger, with a higher number of CCs, a characteristic associated with a high (usually 16C to 32C) ploidy level (41). In Clink-expressing tissues, these two types of nuclei were also observed, but the proportion of the major fraction was drastically reduced, from 95% to about 33%, while the proportion of the minor fraction was increased from 5% to 10%, and new nucleus configurations became visible. Among these, a large fraction of nuclei clearly displayed some of the morphological changes associated with young, dividing cells, such as nuclear rounding and smaller nuclei (Fig. 2C, D, E, and F). For 19% of the nuclei, this phenotype also correlated with the presence of more than 10 CCs, suggesting increased levels of ploidy (Fig. 2C and D). Seven percent of the nuclei displayed an overall intense DAPI staining indicative of highly condensed euchromatin (Fig. 2E). Moreover, 1% of the nuclei were unambiguously identified as metaphasic (not shown) or anaphasic (Fig. 2F), thus providing direct evidence of mitotic activity in the Clink-expressing leaf tissue.
FIG. 2.
Representative DAPI-stained nuclei isolated after 4 days of Dex treatment on detached, expanded 6- to 7-week-old rosette leaves from Clink-transgenic plants. The relative proportions of these different types of nuclei in Clink-expressing tissues were as follows: 33% for oval nuclei with irregular contours and ∼10 CCs (A), 10% for larger nuclei with more CCs (B), 19% for smaller, rounded nuclei with >10 CCs (C and D), 7% for nuclei with highly condensed euchromatin (E), and 1% for dividing (e.g. anaphasic) nuclei (F). For Dex-treated Clinkmut plant leaves and ethanol-treated Clink or Clinkmut plant leaves, only nuclei of the types shown in panels A and B were observed, in proportions of about 95% and 5%, respectively. In each case, the relative proportions are based on the observation of 800 to 1,100 nuclei. Bars, 15 μm.
Phenotypic effects of Clink expression in A. thaliana plants.
Three-day-old A. thaliana seedlings germinated in the absence of Dex were transferred to new plates containing 30 μM Dex or control plates containing 0.1% ethanol and kept at 21°C with a photoperiod of 16 h of light. Under these conditions, no visible differences in the growth or morphology of leaves or roots between A. thaliana plants expressing Clink or Clinkmut and wild-type Col4 plants were observed (data not shown). The accumulation of Clink or Clinkmut protein in leaves was confirmed by Western blot analysis using Clink-specific antibodies (Fig. 3D). The adaxial lamina surfaces of the third and fourth expanded rosette leaves of 4-week-old plants grown in the presence of Dex or ethanol were analyzed by SEM. The wild-type A. thaliana leaf epidermis contains three cell types: pavement cells in a jigsaw puzzle-like pattern, stomatal cells, and trichomes. Rosette leaves of Clink-expressing plants showed clusters of small pavement cells surrounded by almost normal puzzle-shaped cells (Fig. 3A). Ethanol-treated plants (data not shown) or Dex-treated nontransgenic Col4 and Clinkmut plants (Fig. 3A) showed epidermal pavement cells of a normal puzzle shape and size. Comparison of nontransgenic Col4 plants and Clink-expressing plants revealed a decrease in cell size in the Clink-expressing plants (Fig. 3A and B). Cell size was quantified using SEM images of the third rosette leaves of each genotype. The results showed a significant (P < 0.05 by Student's t test) decrease in the mean cell surface area (∼65%) for Clink-expressing plants (1,633 μm2 for 210 cells measured) compared to wild-type Col4 plants (4,612 μm2 for 180 cells measured). No significant change (P < 0.1) in the mean cell surface area was observed for Clinkmut-expressing plants (4,769 μm2 for 210 cells measured) compared to nontransgenic Col4 plants (4,612 μm2 for 180 cells measured). The reduction in the size of pavement cells for Clink-expressing plants was accompanied by a 4.5-fold increase in the number of pavement cells (Fig. 3C), while the numbers of pavement cells in leaves of Clinkmut-expressing plants and nontransgenic control plants were similar. In addition, the number of stomatal cells was 2.5-fold higher in Clink-expressing plants than in Clinkmut-expressing and nontransgenic control plants (Fig. 3A). These results suggest that the arrest of cell division of epidermal leaf cells in Clink-expressing plants was less synchronous than that in nontransgenic or Clinkmut expressing plants.
Effect of Clink expression on ploidy levels.
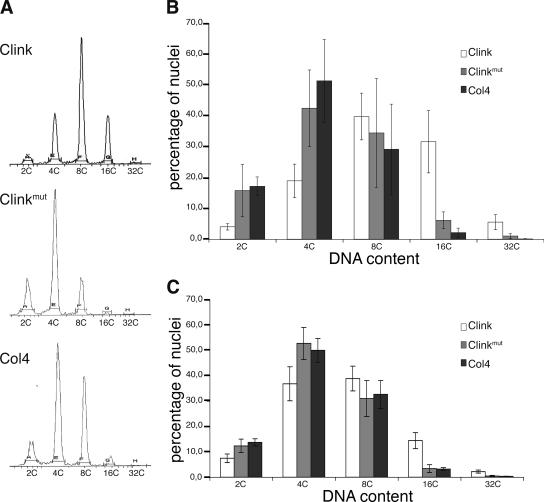
To analyze the impact of Clink expression on endoreduplication cycles, the ploidy levels of leaves were measured by flow cytometry. The third and fourth expanded rosette leaves from 4-week-old plants of the three genotypes, grown on plates and subjected to the same induction treatment as that for plants analyzed by SEM, were used (Fig. 4A and B). Six replicates were analyzed in three independent experiments. The ploidy level of ethanol-treated plants (data not shown) and those of Dex-treated nontransgenic Col4 and Clinkmut-expressing plants did not show significant differences. For Clink-expressing plants, however, the fractions of 2C (P < 0.001 by Student's t test) and 4C (P < 0.01) ploidy level nuclei were lower, and the fractions of 16C (P < 0.001) and 32C (P < 0.01) nuclei were drastically higher, than those for nontransgenic Col4 plants. Similar results were obtained using detached leaves of 6-week-old greenhouse-grown plants when the leaves were induced for 4 days in liquid Dex medium (Fig. 4C). Collectively, these results indicate that leaf cells in Clink-expressing plants had undergone more endoreduplication cycles than those in nontransgenic Col4 and Clinkmut-expressing plants. This induction of the endoreduplication cycles clearly depended on an intact RBR-binding domain (LxCxE) of Clink; hence, interaction with RBR is required.
FIG. 4.
Effect of Clink expression on endoreduplication in rosette and cauline leaves. (A) Flow cytometry profiles of the DNA contents of nuclei from third and fourth expanded rosette leaves of 4-week-old Clink-expressing, Clinkmut-expressing, and nontransgenic Col4 plants grown on plates in the presence of Dex. (B) Comparison of the DNA contents in nuclei of rosette leaves derived from Clink-expressing (open bars), Clinkmut-expressing (shaded bars), and nontransgenic (solid bars) Col4 plants. (C) Comparison of the DNA contents in nuclei of detached cauline leaves following 4 days of Dex treatment. For panels B and C, three independent experiments with six replicates were performed; representative results for one replicate are shown in panel A.
Ploidy levels in FBNYV-infected tissue.
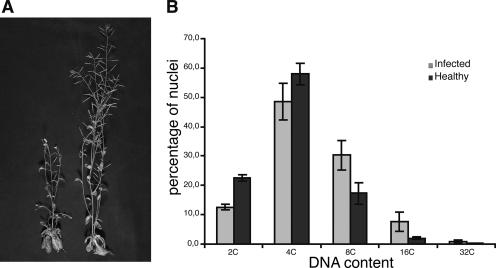
Infection of several ecotypes of A. thaliana with FBNYV by means of viruliferous Aphis craccivora has been established previously (J. Vega-Arreguin et al., unpublished data). Figure 5A shows the symptoms of FBNYV infection in a Col4 plant 3 weeks after inoculation by viruliferous aphids. Infected plants were stunted compared to plants that had been exposed to nonviruliferous aphids (Fig. 5A). FBNYV levels were measured by an enzyme-linked immunosorbent assay (kindly provided by H. J. Vetten). The virus accumulated to high levels in newly developed tissues (data not shown). As in the case of Clink expression, infection with FBNYV resulted in a significant increase in ploidy levels in the leaf cells (Fig. 5B). Infected cauline leaves showed lower levels of 2C (P < 0.001) and 4C (P < 0.05) nuclei and higher levels of 8C (P < 0.01), 16C (P < 0.05), and 32C (P < 0.01) nuclei than leaves exposed to nonviruliferous aphids. The shift of ploidy to higher levels in the nuclei of FBNYV-infected cauline leaves suggests that the nuclei with elevated ploidy levels underwent virus-induced endoreduplication that paralleled viral DNA replication.
FIG. 5.
FBNYV infection of A. thaliana plants. (A) Symptoms of an infected plant (left) compared to a healthy plant (right). (B) Ploidy levels in cauline leaves of FBNYV-infected plants (shaded bars) or healthy control plants (solid bars) 29 days postinfection.
Clink expression does not influence expression of PolIII- and PolI-transcribed genes.
In mammals, RB also acts as a general repressor of transcription by PolIII and PolI, potentially to control cell growth, and interacting viral proteins such as T-ag, E7, and E1A are able to neutralize this repression (29). To address the question whether the same may occur in plants following the Clink-RBR interaction, we used the same RNA samples analyzed in Fig. 1A and performed polyacrylamide gel electrophoresis and several hybridizations using probes specific for (i) the PolI-transcribed 5.8S rRNA gene, (ii) the PolIII-transcribed class-1 5S rRNA gene and class-2 tRNAala gene (controlled by different intragenic promoter elements), and (iii) the PolIII-transcribed class-3 7SL, U6, and U3 genes (relying solely on extragenic promoter elements) (data not shown). For the 5.8S rRNA gene and all PolIII-transcribed genes tested, induction of Clink did not correlate with a significant change in the accumulation of the respective RNA. A similar result was observed for the PolI-transcribed 18S rRNA gene (data not shown). Therefore, these results indicate that expression of Clink is not accompanied by a significant increase in gene transcription by PolIII or PolI, suggesting a difference in the pathways of cell growth regulation by RB and RBR in mammals and plants.
DISCUSSION
Having established that the nanovirus protein Clink binds the key cell cycle regulator protein RBR and enhances the replication of FBNYV DNA (5), we studied here the influence of Clink on cell cycle regulation. For that purpose, transgenic A. thaliana lines were constructed in which the expression of Clink or the RBR-binding-deficient variant Clinkmut could be controlled by induction with the glucocorticoid hormone Dex (2).
Depending on the expression of a functional Clink protein, transcription of the E2F/DP-controlled S-phase-specific genes encoding the PCNA and CDC6 proteins was readily induced, whereas expression of Clinkmut, an RBR-binding-deficient Clink variant, did not cause any induction of PCNA- and CDC6-specific transcripts. This is consistent with the assumption that via interaction with Arabidopsis RBR, Clink triggers a cell cycle progression beyond the RBR-controlled checkpoint. Similar effects on the S-phase transition were reported for another plant DNA virus RBR-binding protein, the tomato golden mosaic virus Rep (or AL1) protein (15, 26, 36). Clink expression also caused increased transcription of G2-M-specific genes encoding the B2-type cyclin-dependent kinases and the B1-type cyclins CycB1;1 and CycB1;4 (Fig. 1A). This is suggestive evidence that not only DNA synthesis (during S phase) but also, at least to some extent, mitoses and cell divisions might be triggered by the induced expression of Clink. These findings were further supported by in situ fluorescence analyses of DAPI-stained nuclei derived from Clink- and Clinkmut-transgenic plant leaves (Fig. 2). Following expression of Clink, but not Clinkmut, the fraction of leaf nuclei displaying more than 10 CCs increased from 5% to 29% (Fig. 2B, C, and D); moreover, about 1% of metaphasic or anaphasic nuclei (Fig. 2F) were clearly identified among Clink-expressing cells, a situation unexpected in mature Arabidopsis leaves. These results provide clear evidence of Clink-induced cell cycle reactivation in mature leaf tissues associated with enhanced endoreduplication and some mitotic activity. Recently, the geminivirus RBR-binding RepA protein was also shown to affect the G2-M transition in transgenic Arabidopsis plants (14). Following disruption of RBR function by RepA in late-developing rosette leaves, a general increase in the ploidy levels of leaf nuclei was observed, and fully expanded pavement cells displayed the capacity to engage in a single division event. In our case, these epidermal cells are also likely to account for at least a fraction of the mitotic events observed in response to Clink induction in fully differentiated rosette leaves from 6- to 7-week-old seedlings.
In contrast to the small DNA viruses of mammals, which cause extended cell proliferation (tumors), such excessive cell proliferation has never been observed to be associated with infection of plants by ssDNA viruses. Only limited tissue proliferation or cell enlargement is caused by some geminivirus proteins (30, 44). It was therefore of interest to assess the effect of prolonged (3- to 4-week) expression of the cell cycle “deregulator” Clink on plant tissue. Clink-expressing as well as Clinkmut-expressing plants that had been grown for 4 weeks in the presence of Dex showed no obvious morphological differences. A closer inspection of the adaxial epidermis by SEM, however, revealed a clearly different phenotype for Clink-expressing plants than for Clinkmut-expressing plants or nontransgenic Col4 plants (Fig. 3). Clink-expressing plants had a significantly increased fraction of smaller epidermal cells (extrapolated from their smaller surface area), and this size reduction was accompanied by increases in the numbers of pavement and stomatal cells (Fig. 3). Similar phenotypes have been described for Arabidopsis plants overexpressing E2Fa-DPa (7), AtCDT1, or AtCDC6 (9) and for Arabidopsis plants subject to geminivirus RepA-induced RBR inactivation (14). This is in line with the assumption that due to prolonged expression of functional Clink, the RBR-imposed block of transcriptional activators of the E2F family remained suspended, and some mitotic cycles and subsequent epidermal cell divisions occurred. Concomitantly, the ploidy levels of nuclei from the third and fourth expanded rosette leaves and cauline leaves from the same plants (4 weeks of Dex induction), assessed by flow cytometry, revealed a clear shift toward 16C and 32C DNA contents (Fig. 4A and B). The beginning of such a shift toward a higher ploidy level could already be observed after 4 days of Clink induction (Fig. 4C). The Clink-induced increase in epidermal pavement cell and stomatal cell division and the concomitantly elevated level of ploidy of total nuclei from the corresponding leaves suggest a differential effect of Clink on the nuclear cycle in epidermal cells and total tissue nuclei. These changes are the consequence of transcriptional reprogramming suggested by the up-regulation of E2F target genes. In epidermal cells, no endoreduplication occurred beyond the already existing level (not determined), because larger epidermal cells, expected in the case of a higher degree of ploidy (see, for instance, references 28 and 33), were not observed. At the same time, in total cells from the same leaves, a fraction of nuclei existed that had undergone more endocycles. Since Clink clearly triggers transcription of S-phase-specific genes, e.g., PCNA and CDC6, as well as the mitotic cyclin cycB1;1, a dichotomy in the effect of Clink on cell cycle regulation in the nuclei of cells of different tissues is easily conceivable. Such a balanced dichotomy in E2F/DP-dependent effects on the cell cycle and cell fate has also been proposed by others (7, 9, 14). Indeed, the authors concluded that the RBR/E2F pathway regulates the balance between cell proliferation, endocycle program, and differentiation. They also concluded that the importance of this pathway depends on the cell type, the tissue, and the developmental stage.
A clear shift of the nuclear ploidy to elevated levels (8, 16, and 32C) was observed in cauline leaves of FBNYV-infected A. thaliana plants (Fig. 5). It is tempting to speculate that the endocycling nuclei are derived either from cells that are targeted by FBNYV for its replication or from cells that contain at least some FBNYV DNA components. Nanoviruses are assumed to be restricted to the phloem (52), but the impact on the overall ploidy level detected for FBNYV-infected cauline leaves suggests that this is not the case. No data are available on the multiplication of FBNYV in epidermal cells, and whether nanoviruses invade the mesophyll is not known. Attempts to determine the tropism of FBNYV in A. thaliana by immunolocalization with the available polyclonal antibodies against Clink, M-Rep, and FBNYV particles were unsuccessful, due to cross-reaction of these antibodies with host plant proteins. Nanoviruses (52), including FBNYV (B. C. Ramirez, unpublished results), code for two proteins involved in cell-to-cell movement. DNA-N codes for a nuclear shuttle protein and DNA-M for a movement protein. The movement protein and nuclear shuttle protein coordinate the movement of viral DNA as viral nucleoprotein complexes from the nucleus to the cytoplasm and from the cytoplasm to the neighboring cells. It is possible that nanoviruses have the capacity to invade nonphloem tissues, since this has been observed for some bipartite geminiviruses such as the begomovirus bean dwarf mosaic virus (38). On the other hand, it is also possible that some FBNYV DNA components are not limited to the phloem, at least in A. thaliana. Recent studies (42) with the nanovirus milk vetch dwarf virus (MDV) indicate that the expression of the individual DNA components is differentially regulated: the promoter of MDV-M is active not only in phloem and meristematic tissues but also in mesophyll and cortex cells. It therefore appears logical to assume that nuclei with an elevated ploidy level underwent virus-induced (i.e., Clink-triggered) endoreduplication that paralleled viral DNA replication. A similar situation was previously reported for infection of Digitaria setigera with the mastrevirus Digitaria streak virus, which was accompanied by an increase in the fraction of S-phase nuclei (1).
Since the effects of Clink on the expression of cell cycle-relevant genes involving interaction with RBR proteins are in many aspects comparable to those of the cell cycle regulators of small DNA tumor viruses such as SV40 T-ag or E7 of human papillomaviruses, we were interested in seeing whether Clink might also influence the transcription by PolI and PolIII of genes implicated in cell growth regulation (for a review, see reference 29). Northern blot analyses of the PolI-transcribed 5.8S rRNA gene and of five PolIII-transcribed genes, the 5S rRNA, tRNAala, 7SL, U6, and U3 genes, revealed no significant differences in transcript accumulation upon Clink induction (data not shown). Therefore, these classes of genes involved in cell growth regulation are not affected by Clink, in agreement with the observation that no gross increase in cell size was observed in Clink-expressing tissue. Also, for the RBR-interacting cell cycle modulators of other ssDNA plant viruses, e.g., the mastrevirus RepA protein and the begomovirus Rep (AL1) protein, no increase in cell size has been reported. The only protein known to cause an increase in cell size is the C4 protein of beet curly top virus (30). Hence, to regulate their cell growth (size), plants and animals have obviously evolved diverging molecular strategies (for reviews, see references 25, 45, and 46).
In the present study, we have shown that the nanovirus cell cycle link protein Clink significantly interferes with the plant cell cycle in vivo, conceivably via interaction with cell RBR. An intriguing question still remains, however: what is the biological significance of Clink as an F-box protein, its binding to SKP1, and its connection to the SCF-regulated protein turnover pathway? A “transgenic” approach comparable to that described here, employing the tightly regulated conditional expression of Clink or mutant Clink, may be useful for shedding further light on the molecular details of other Clink functions and on nanovirus biology in general.
Supplementary Material
Acknowledgments
We thank Ingo Schubert and Olivier Mathieu for assistance in the analysis of the cytogenetics data, Julien Douet for help with the cytogenetics experiments, Séverine Domenichini for assistance with SEM, and Julio Vega-Arreguín for help with the aphid colonies. We thank the “Dynamique de la compartimentation cellulaire” laboratory for their help and valuable discussions with the immunolocalization and flow cytometry studies. We thank Susanne Bolte and Marie-Noëlle Soler for expert support with microscopy. We are grateful to Danielle Clérot for her valuable help concerning the measurements of the cell surfaces. We are also grateful to Nam-Hai Chua for the generous gift of the PTA7002 vector, to Eva Kondorosi for providing the CycB1;1 probe, and to H.-J. Vetten for the enzyme-linked immunosorbent assay and for antibodies to FBNYV. We thank Anne-Lise Haenni for valuable suggestions.
This work was partly supported by the CNRS (UMR 6547 BIOMOVE and GDR2157), by the Université Blaise Pascal, and by a grant from the European Commission as part of the RIBOREG EU FP6 project (LSHG-CT-2003503022). S. Lageix is the recipient of a French MNERT grant. The Imaging and Cell Biology facility of the IFR87 (FR-W2251) “La plante et son environnement” is supported by “Action de Soutien à la Technologie et la Recherche en Essonne,” Conseil Général de l'Essonne.
Footnotes
Published ahead of print on 31 January 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Accotto, G. P., P. M. Mullineaux, S. C. Brown, and D. Marie. 1993. Digitaria streak geminivirus replicative forms are abundant in S-phase nuclei of infected cells. Virology 195:257-259. [DOI] [PubMed] [Google Scholar]
- 2.Aoyama, T., and N. H. Chua. 1997. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11:605-612. [DOI] [PubMed] [Google Scholar]
- 3.Aronson, M. 2002. Ph.D. thesis. Institut National d'Agronomie Paris-Grignon, Paris, France.
- 4.Aronson, M. N., A. Complainville, D. Clerot, H. Alcalde, L. Katul, H. J. Vetten, B. Gronenborn, and T. Timchenko. 2002. In planta protein-protein interactions assessed using a nanovirus-based replication and expression system. Plant J. 31:767-775. [DOI] [PubMed] [Google Scholar]
- 5.Aronson, M. N., A. D. Meyer, J. Gyorgyey, L. Katul, H. J. Vetten, B. Gronenborn, and T. Timchenko. 2000. Clink, a nanovirus-encoded protein, binds both pRB and SKP1. J. Virol. 74:2967-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelos, M., C. Curie, L. Mazzolini, C. Bardet, and B. Lescure. 1992. A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell culture. Plant Physiol. Biochem. 30:1-5. [Google Scholar]
- 7.Boudolf, V., K. Vlieghe, G. T. Beemster, Z. Magyar, J. A. Torres Acosta, S. Maes, E. Van Der Schueren, D. Inze, and L. De Veylder. 2004. The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16:2683-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 9.Castellano Mdel, M., M. B. Boniotti, E. Caro, A. Schnittger, and C. Gutierrez. 2004. DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16:2380-2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavanaugh, A. H., W. M. Hempel, L. J. Taylor, V. Rogalsky, G. Todorov, and L. I. Rothblum. 1995. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature 374:177-180. [DOI] [PubMed] [Google Scholar]
- 11.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clough, S. J., and A. F. Bent. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16:735-743. [DOI] [PubMed] [Google Scholar]
- 13.de Jager, S. M., S. Maughan, W. Dewitte, S. Scofield, and J. A. Murray. 2005. The developmental context of cell-cycle control in plants. Semin. Cell Dev. Biol. 16:385-396. [DOI] [PubMed] [Google Scholar]
- 14.Desvoyes, B., E. Ramirez-Parra, Q. Xie, N. H. Chua, and C. Gutierrez. 2006. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 140:67-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egelkrout, E. M., D. Robertson, and L. Hanley-Bowdoin. 2001. Proliferating cell nuclear antigen transcription is repressed through an E2F consensus element and activated by geminivirus infection in mature leaves. Plant Cell 13:1437-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felsani, A., A. M. Mileo, and M. G. Paggi. 2006. Retinoblastoma family proteins as key targets of the small DNA virus oncoproteins. Oncogene 25:5277-5285. [DOI] [PubMed] [Google Scholar]
- 17.Galbraith, D. W., K. R. Harkins, J. R. Maddox, N. M. Ayres, D. P. Sharma, and E. Firoozabady. 1983. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220:1049-1051. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez, C. 2002. Strategies for geminivirus DNA replication and cell cycle interference. Physiol. Mol. Plant Pathol. 60:219-230. [Google Scholar]
- 19.Gutierrez, C., E. Ramirez-Parra, M. Mar Castellano, A. P. Sanz-Burgos, A. Luque, and R. Missich. 2004. Geminivirus DNA replication and cell cycle interactions. Vet. Microbiol. 98:111-119. [DOI] [PubMed] [Google Scholar]
- 20.Hanley-Bowdoin, L., S. B. Settlage, and D. Robertson. 2004. Reprogramming plant gene expression: a prerequisite to geminivirus DNA replication. Mol. Plant Pathol. 5:149-156. [DOI] [PubMed] [Google Scholar]
- 21.Hirsch, H. A., L. Gu, and R. W. Henry. 2000. The retinoblastoma tumor suppressor protein targets distinct general transcription factors to regulate RNA polymerase III gene expression. Mol. Cell. Biol. 20:9182-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirsch, H. A., G. W. Jawdekar, K. A. Lee, L. Gu, and R. W. Henry. 2004. Distinct mechanisms for repression of RNA polymerase III transcription by the retinoblastoma tumor suppressor protein. Mol. Cell. Biol. 24:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ito, M. 2000. Factors controlling cyclin B expression. Plant Mol. Biol. 43:677-690. [DOI] [PubMed] [Google Scholar]
- 24.Joubes, J., and C. Chevalier. 2000. Endoreduplication in higher plants. Plant Mol. Biol. 43:735-745. [DOI] [PubMed] [Google Scholar]
- 25.Kondorosi, E., F. Roudier, and E. Gendreau. 2000. Plant cell-size control: growing by ploidy? Curr. Opin. Plant Biol. 3:488-492. [DOI] [PubMed] [Google Scholar]
- 26.Kong, L. J., B. M. Orozco, J. L. Roe, S. Nagar, S. Ou, H. S. Feiler, T. Durfee, A. B. Miller, W. Gruissem, D. Robertson, and L. Hanley-Bowdoin. 2000. A geminivirus replication protein interacts with the retinoblastoma protein through a novel domain to determine symptoms and tissue specificity of infection in plants. EMBO J. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kono, A., C. Umeda-Hara, J. Lee, M. Ito, H. Uchimiya, and M. Umeda. 2003. Arabidopsis D-type cyclin CYCD4;1 is a novel cyclin partner of B2-type cyclin-dependent kinase. Plant Physiol. 132:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kudo, N., and Y. Kimura. 2002. Nuclear DNA endoreduplication during petal development in cabbage: relationship between ploidy levels and cell size. J. Exp. Bot. 53:1017-1023. [DOI] [PubMed] [Google Scholar]
- 29.Larminie, C. G., H. M. Alzuherri, C. A. Cairns, A. McLees, and R. J. White. 1998. Transcription by RNA polymerases I and III: a potential link between cell growth, protein synthesis and the retinoblastoma protein. J. Mol. Med. 76:94-103. [DOI] [PubMed] [Google Scholar]
- 30.Latham, J. R., K. Saunders, M. S. Pinner, and J. Stanley. 1997. Induction of plant cell division by beet curly top virus gene C4. Plant J. 11:1273-1283. [Google Scholar]
- 31.Logemann, J., J. Schell, and L. Willmitzer. 1987. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 163:16-20. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Garcia, J. F., E. Monte, and P. H. Quail. 1999. A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20:251-257. [DOI] [PubMed] [Google Scholar]
- 33.Melaragno, J. E., B. Mehrotra, and A. W. Coleman. 1993. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5:1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Menges, M., and J. A. Murray. 2002. Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J. 30:203-212. [DOI] [PubMed] [Google Scholar]
- 35.Nagar, S., L. Hanley-Bowdoin, and D. Robertson. 2002. Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant Cell 14:2995-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagar, S., T. J. Pedersen, K. M. Carrick, L. Hanley-Bowdoin, and D. Robertson. 1995. A geminivirus induces expression of a host DNA synthesis protein in terminally differentiated plant cells. Plant Cell 7:705-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potuschak, T., and P. Doerner. 2001. Cell cycle controls: genome-wide analysis in Arabidopsis. Curr. Opin. Plant Biol. 4:501-506. [DOI] [PubMed] [Google Scholar]
- 38.Rojas, M. R., C. Hagen, W. J. Lucas, and R. L. Gilbertson. 2005. Exploiting chinks in the plant's armor: evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 43:361-394. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Schubert, I., P. F. Fransz, J. Fuchs, and J. H. de Jong. 2001. Chromosome painting in plants. Methods Cell Sci. 23:57-69. [PubMed] [Google Scholar]
- 41.Schubert, V., M. Klatte, A. Pecinka, A. Meister, Z. Jasencakova, and I. Schubert. 2006. Sister chromatids are often incompletely aligned in meristematic and endopolyploid interphase nuclei of Arabidopsis thaliana. Genetics 172:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirasawa-Seo, N., Y. Sano, S. Nakamura, T. Murakami, S. Seo, Y. Ohashi, Y. Hashimoto, and T. Matsumoto. 2005. Characteristics of the promoters derived from the single-stranded DNA components of milk vetch dwarf virus in transgenic tobacco. J. Gen. Virol. 86:1851-1860. [DOI] [PubMed] [Google Scholar]
- 43.Stanley, J., D. M. Bisaro, R. W. Briddon, J. K. Brown, C. M. Fauquet, B. D. Harrison, E. P. Rybicki, and D. C. Stenger. 2005. Geminiviridae, p. 301-326. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 44.Stanley, J., and J. R. Latham. 1992. A symptom variant of beet curly top geminivirus produced by mutation of open reading frame C4. Virology 190:506-509. [DOI] [PubMed] [Google Scholar]
- 45.Stocker, H., and E. Hafen. 2000. Genetic control of cell size. Curr. Opin. Genet. Dev. 10:529-535. [DOI] [PubMed] [Google Scholar]
- 46.Sugimoto-Shirasu, K., and K. Roberts. 2003. “Big it up”: endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6:544-553. [DOI] [PubMed] [Google Scholar]
- 47.Sutcliffe, J. E., T. R. Brown, S. J. Allison, P. H. Scott, and R. J. White. 2000. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol. Cell. Biol. 20:9192-9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taya, Y. 1997. RB kinases and RB-binding proteins: new points of view. Trends Biochem. Sci. 22:14-17. [DOI] [PubMed] [Google Scholar]
- 49.Timchenko, T., F. de Kouchkovsky, L. Katul, C. David, H. J. Vetten, and B. Gronenborn. 1999. A single rep protein initiates replication of multiple genome components of faba bean necrotic yellows virus, a single-stranded DNA virus of plants. J. Virol. 73:10173-10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Timchenko, T., L. Katul, M. Aronson, J. C. Vega-Arreguin, B. C. Ramirez, H. J. Vetten, and B. Gronenborn. 2006. Infectivity of nanovirus DNAs: induction of disease by cloned genome components of Faba bean necrotic yellows virus. J. Gen. Virol. 87:1735-1743. [DOI] [PubMed] [Google Scholar]
- 51.Vega-Arreguin, J. C., T. Timchenko, B. Gronenborn, and B. C. Ramirez. 2005. A functional histidine-tagged replication initiator protein: implications for the study of single-stranded DNA virus replication in planta. J. Virol. 79:8422-8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vetten, H. J., P. W. G. Chu, J. L. Dale, R. Harding, J. Hu, L. Katul, M. Kojima, J. W. Randles, Y. Sano, and J. E. Thomas. 2005. Nanoviridae, p. 343-352. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom.
- 53.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 54.White, R. J., D. Trouche, K. Martin, S. P. Jackson, and T. Kouzarides. 1996. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature 382:88-90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.