Abstract
Gliomas are often resistant to the induction of apoptotic cell death as a result of the development of survival mechanisms during astrocyte malignant transformation. In particular, the overexpression of Bcl-2-family members interferes with apoptosis initiation by DNA-damaging agents (e.g., cisplatin) or soluble death ligands (e.g., TRAIL). Using low-passage-number cultures of glioma cells, we have shown that parvovirus H-1 is able to induce death in cells resistant to TRAIL, cisplatin, or both, even when Bcl-2 is overexpressed. Parvovirus H-1 triggers cell death through both the accumulation of lysosomal cathepsins B and L in the cytosol of infected cells and the reduction of the levels of cystatin B and C, two cathepsin inhibitors. The impairment of either of these effects protects glioma cells from the viral lytic effect. In normal human astrocytes, parvovirus H-1 fails to induce a killing mechanism. In vivo, parvovirus H-1 infection of rat glioma cells intracranially implanted into recipient animals triggers cathepsin B activation as well. This report identifies for the first time cellular effectors of the killing activity of parvovirus H-1 against malignant brain cells and opens up a therapeutic approach which circumvents their frequent resistance to other death inducers.
The action of successful anticancer drugs depends largely on their ability to trigger cell death, and particularly apoptosis, in tumor cells. Their efficacy is often impaired by death escape mechanisms resulting from the accumulation of genetic alterations during the malignant transformation process (12). Drugs inducing apoptosis fall into two classes according to their ability to activate either the extrinsic receptor-dependent apoptotic pathway, as do TRAIL and tumor necrosis factor alpha (TNF-α) (22), or the intrinsic pathway, as do cisplatin and other DNA-damaging agents (3). The extrinsic pathway relies on binding of a ligand to its death receptor at the cell surface and on activation of procaspase 8 by protein complexes associated with the intracellular domain of the receptor (31). The intrinsic pathway can be stimulated by genotoxic stress or by activation of the extrinsic pathway; such stimulation results in the release from injured mitochondria of proapoptotic molecules (e.g., cytochrome c) that are components of the apoptosome, which cleaves the cytosolic procaspase 9 (18). The extrinsic and intrinsic pathways converge in the activation of downstream effector caspases (e.g., caspase 3) by cleaved caspase 8 and 9 (18, 31). Multiple mechanisms have been identified in cancer cells that prevent these pathways from being activated. Most anticancer drugs are efficiently neutralized in tumor cells before they can induce DNA damage (24). Down-regulation of death receptors or surface (over)expression of receptors lacking their cytoplasmic tails prevents the extrinsic pathway from being activated (30). In addition, tumor cells overexpress antiapoptotic molecules (e.g., Bcl-2 family members, Flip, IAP) that prevent procaspase cleavage by activating complexes (3).
Gliomas are the most common brain cancers, and the life expectancy of newly diagnosed patients is often less than a year (25). These tumors are particularly resistant to conventional cancer therapy, and novel approaches are eagerly sought. Tumor progression and resistance to clinical treatment may be due in part to a defective apoptotic program and to overexpression of antiapoptotic molecules such as Bcl-2 or PEA15 (phosphoprotein enriched in astrocyte 15) (13). Investigators have proposed novel therapeutic strategies based on targeting of the antiapoptotic pathways to restore apoptotic cell death, but the multiplicity of genetic alterations occurring in tumor cells jeopardizes treatment efficacy (48).
Oncolytic viruses, and particularly some rodent parvoviruses, can interfere with the survival of low-passage-number and established cultures of human glioma cells (15). Parvoviruses can induce death in a number of tumor cells while being innocuous to healthy tissues (37). The exact mechanism of cell death triggered by these viruses is still unclear. Rodent parvovirus infections can induce either necrosis or apoptosis, depending on the tumor model considered. For example, after infection with the rat parvovirus H-1 (H-1PV), human monoblastic leukemia cells (U937) and several hepatocarcinoma cell lines die from apoptosis (26, 34) whereas transformed rat fibroblasts and human keratinocytes show signs of necrosis (32). In the present study we have investigated whether human gliomas that have acquired resistance to death-inducing drugs can still be killed by parvovirus H-1. We have used low-passage-number cultures of glioma cells isolated from cancer patients to show that parvovirus H-1 can induce nonapoptotic cell death irrespective of the responsiveness of tumor cells to apoptotic stimuli. This mechanism is dependent on both the accumulation of cathepsin B and L in the cytosol and the down-regulation of cystatins, the physiologic inhibitors of cathepsins. It is insensitive to Bcl-2 overexpression and results in the killing of tumor cells that have mechanisms enabling them to survive conventional treatments.
MATERIALS AND METHODS
Cell cultures and reagents.
Human glioma cell lines (U373MG and U138MG) were provided by Tumorbank (DKFZ, Heidelberg, Germany). Low-passage-number cell cultures were prepared at the Neurosurgery Department of Heidelberg University Hospital from glioblastomas (NCH82, NCH89, NCH125, NCH149) and one gliosarcoma (NCH37) or from samples from epileptic patients (normal astrocytes). Tumor primary cultures were characterized as previously described (14). Cells were cultured in Dulbecco's modified Eagle medium (Sigma, Schnelldorf, Germany) supplemented with 10% (vol/vol) fetal calf serum (Biochrom, Berlin) and kanamycin (100 μg/ml)-gentamicin (50 μg/ml) (GIBCO, Karlsruhe, Germany). Recombinant human TRAIL and cisplatin (Platinex) were obtained from PeproTech EC Ltd. (London, United Kingdom) and Bristol-Myers Squibb (New York, NY), respectively. Caspase 3 inhibitor (DEVD-CHO; cell permeable) and cathepsin B inhibitor (Ca-074 Me) were purchased from Calbiochem (Darmstadt, Germany) and resuspended in dimethyl sulfoxide (DMSO) (Sigma).
Infection and transfection.
H-1PV was amplified in human NBK cells and purified on iodixanol gradients as previously described (42). H-1PV titers were determined on NBK indicator cells by plaque assay and further used at various multiplicities of infection (expressed in PFU per cell). Cells were infected with H-1PV suspensions in medium without serum or antibiotics for 2 h at 37°C. Retroviruses were produced by transfection of helper-free ecotropic virus-producing BOSC cells by use of FuGENE 6 (Roche, Mannheim, Germany) according to the manufacturer's instructions. Cells were cotransfected with the retroviral vector and the pseudotyping vector pVSVg (Invitrogen, Karlsruhe, Germany) at a ratio of 3:1 (wt/wt), and supernatants were collected 48 h posttransfection. Recipient cells were incubated for 4 h with 3 ml inoculum supplemented with Polybrene (6 μg/ml; Sigma). Retrovirus-infected cells were selected with puromycine (2.5 μg/ml; Sigma) and collected when control uninfected cells were killed. Small interfering RNA (siRNA) oligonucleotides targeting hCathB (5′-GCU UGG AAC UUC UGG ACA ATT-3′) or hCathL (5′-GAA CAU GAA GAU GAU UGA ATT-3′) transcripts were synthesized at MWG (Ebersberg, Germany) and transiently transfected using X-tremeGENE (Roche) according to the manufacturer's instructions. Luc (5′-AAC GUA CGC GGA AUA CUU CGA-3′) control oligonucleotide was purchased from Xeragon (Zürich, Switzerland). The cystatin B expression vector was a gift from E. Spiess (DKFZ, Heidelberg, Germany). NF-κB activity was measured after cell transfection with the Translucent reporter vector (Panomics, Heidelberg, Germany) by use of a luciferase assay system (Promega, Mannheim, Germany) and a luminometer (Fluoroskan & FL; Thermolabsystem, Dreieich, Germany).
Cell fractionation and protease activity measurements.
Cultures (106 cells per plate) were collected in phosphate-buffered saline (PBS), pelleted by centrifugation (500 × g, 10 min, 4°C), and resuspended in 1.5 ml hypotonic buffer (0.25 M sucrose, 10 mM HEPES-NaOH [pH 7.4], 1 mM EDTA) for the measurement of cathepsin B activities or in 0.5 ml PBS for caspase activities. For cathepsin B activity, cells were further homogenized in a cell cracker (bead diameter of 8.006 mm; 10 strokes). Nuclei and heavy mitochondria were pelleted by centrifugation at 2,500 × g for 10 min at 4°C. An aliquot (200 μl) of the supernatant (postnuclear supernatant) was kept and used to determine enzyme latency. The light mitochondrial fraction (LMF) was obtained by centrifugation of 1 ml postnuclear supernatant at 17,000 × g for 20 min at 4°C. The supernatant was kept as a cytosolic extract, and the LMF pellet was either resuspended in 1 to 1.3 ml hypotonic buffer for the determination of enzyme activity or further fractionated. Lysosomes were purified from the LMF (1.3 ml) on a self-generated gradient of iodixanol (OptiPrep; Axis-Shield, Heidelberg, Germany) by adding 0.7 ml of 50% iodixanol in 1 mM EDTA-10 mM HEPES-NaOH (pH 7.4)-42 mM sucrose and centrifuging at 380,000 × g for 1 h 30 min at 4°C in a TLV100 rotor. Twelve 150-μl fractions were collected and analyzed by immunoblotting. Cathepsin B activities were determined by adding 25 μl of whole-cell extract, cytosolic extract, or LMF subfractions to a reaction mixture consisting of 50 mM morpholineethanesulfonic acid (MES) (pH 6.0), 0.25 M sucrose, 1 mM EDTA, and 2 mM N-acetyl-L-cysteine. After a 10-min incubation, the substrate Z-Arg-Arg-AMC (Calbiochem) was added (1 mM), and the reaction was monitored for 1 h on a Fluoroskan & FL luminometer (Thermolabsystem) for measurement of emission at a wavelength of 455 nm after excitation at a wavelength of 360 nm. Caspase activity was determined by adding 25 μl of whole-cell extract to caspase buffer (50 mM HEPES [pH 7.4], 150 mM NaCl, 0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 0.1% Triton X-100, 1 mM EDTA, 10% glycerol, 10 mM dithiothreitol) supplemented with 1 mM caspase 3 (Ac-Asp-Glu-Val-Asp-AFC), 8 (Z-Ile-Glu-Thr-Asp-AFC), or 9 (Ac-Leu-Glu-His-Asp-AFC) substrate (Calbiochem). The reaction was monitored for 1 h on a Fluoroskan & FL luminometer (Thermolabsystem) for measurement of emission at a wavelength of 510 nm with the excitation wavelength set at 390 nm.
Immunoblot analyses.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Protran; Schleicher & Schuell, Relliehausen, Germany) by use of a semidry blotting apparatus (Bio-Rad Laboratories, Munich, Germany). The primary antibodies used for immunostaining were directed against human cathepsin B (clone CB 59-4B11), cathepsin L (clone CPLH 33/2), cystatin B (clone RJMW 2E7), β-tubulin (Sigma), cystatin C, PTEN (Upstate Biotech, Hamburg, Germany), green fluorescent protein (GFP), Lamp2 (Santa Cruz, Heidelberg, Germany), Bcl-2 (Calbiochem), Flip (Apotech, Epalinges, Switzerland), NS1 (rabbit antiserum αSP8), or VP (rabbit antiserum polypeptide; region of nucleotides 3610 to 4310 of the H-1PV genome). Peroxidase-conjugated secondary antibodies were all purchased from Santa Cruz. Immunodetection was performed with Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences, Rodgau-Jügesheim, Germany).
Immunoprecipitation analyses.
Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate, protease inhibitor cocktail), and 200 μg total protein extract was precleaned with 40 μl protein G Sepharose bead slurry (80%) for 1 h at 4°C. Precleaned extracts were subsequently incubated for 2 h at 4°C with 1 μg of p53 (Ab-6)- or mutp53 (Ab-3)-specific antibodies bound to 50 μl protein G Sepharose bead slurry (80%). Proteins were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted, and immunostained as described above, using antibodies directed against p53 (Ab-7), biotin-conjugated anti-sheep antibodies, and peroxidase-conjugated streptavidin. Antibodies were all purchased from Calbiochem, protease inhibitor cocktail from Roche, and protein G Sepharose from Upstate Biotech.
Southern blot analysis.
Low-molecular-weight DNA was extracted from H-1PV- and mock-infected cell pellets by the Hirt procedure (15a). DNA was fractionated by agarose gel electrophoresis and blotted onto nylon membranes. Membrane-bound DNA was hybridized to an NS-1 gene-specific DNA probe (42).
PCR analysis.
Genomic DNA was extracted from cultured cells by the standard phenol-chloroform-proteinase K protocol (38). The primer sequences to determine the integrity of the INK4 locus were as follows: for p16 exon 2, 5′-CAA TTA CCA CAT TCT GCG CTT-3′ (sense) and 5′-CAG GCA GAG AGC ACT GTG AG-3′ (antisense); for p19 exon 2, 5′-GGA AAC TGA GGC GAA CAG AG-3′ (sense) and 5′-TAT CTC ACG GGT CCT CCA CT-3′ (antisense); and for INK4 exon 2, 5′-TGG ACG TGC CGC GGA TGC-3′ (sense) and 5′-GGA AGC TCT CAG GGT ACA AAT TC-3′ (antisense).
Assessment of cell viability and lysis.
Viability was determined using hexosaminidase assays performed according to a previously well-established protocol. Briefly, cultures in 96-well plates (3,000 cells per well) were washed twice in PBS. Reaction buffer (3.75 mM p-nitrophenol-N-acetyl-β-d-glucosaminide in 0.05 mM citrate buffer [pH 5.0]-0.25% Triton X-100) was added (100 μl per well). After a 3-h incubation at 37°C, the assay was stopped and the color developed by adding a 1.5 vol of 50 mM glycine-5 mM EDTA (pH 10.4). The absorbance was read at 405 nm. Cell lysis was determined by measuring the release of glucose 6-phosphate dehydrogenase into culture medium by use of a Vybrant cytotoxicity assay kit (Molecular Probes) according to the manufacturer's instructions.
Animal treatment.
Rat glioma (RG2) cells were intracerebrally implanted (3,000 cells/animal) into Wistar Kyoto inbred rats, and tumor development was monitored by magnetic resonance imaging. On day 11 postimplantation, both the tumor and its corresponding portion in the tumor-free hemisphere were stereotactically injected with H-1PV (1 PFU/cell) or mock treated. Animals were sacrificed on day 3 postinfection, and protein extracts from resections of tumors and healthy brains were prepared for measurement of cathepsin B activity.
RESULTS
Low-passage-number cultures of glioma cells are resistant to death induced by genotoxic stress and soluble death ligands.
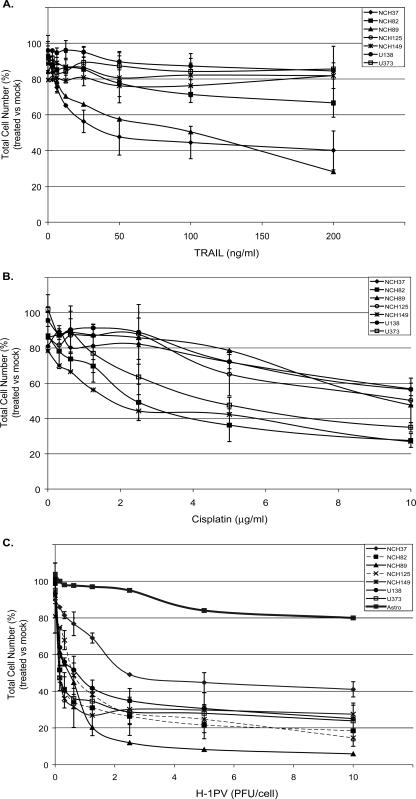
The resistance of gliomas to anticancer drugs is attributed to inefficient crossing of the blood-brain barrier and/or to inefficient killing of cancer cells (20). Our first step in the present work was to analyze the sensitivity of low-passage-number cultures of human glioma cells to a DNA-damaging agent and a soluble death ligand that are both known to induce apoptosis. Since preexposure of cells to clinical treatment could lead to selection of resistant variants, the cultures were derived from tumor resections from patients who had not been exposed to antitumor therapy prior to surgery. Low-passage-number cultures and two established glioma cell lines were incubated with increasing concentrations of cisplatin or TRAIL, which induce cell death via the intrinsic or extrinsic apoptotic pathways, respectively. After a 30-h exposure, the hexosaminidase assay was used to compare numbers of living cells in treated and control cultures (Fig. 1). Out of seven cultures tested, only two were sensitive to TRAIL (Fig. 1A) and three to cisplatin (Fig. 1B), while the others showed little alteration of their proliferation and/or viability even at high doses of these agents. It is noteworthy that primary cultures of normal human astrocytes proved sensitive to cisplatin (data not shown), indicating that in the in vitro system tested the drug had no specificity for cancer cells. The different cultures displayed a heterogeneous pattern of oncogene and antioncogene mutations, reflecting the known high genetic diversity of human gliomas (Table 1) (25, 48). This cell collection thus appeared to be representative of human gliomas with regard to both its genetic alterations and frequent resistance to conventional treatments. It therefore appeared suitable for evaluating the antiglioma efficacy of novel therapeutic agents.
FIG. 1.
H-1PV-induced killing of glioma cells resistant to genotoxic agents and/or death ligands. Cell survival was measured by means of the hexosaminidase assay, using low-passage-number cultures (NCH37, NCH82, NCH89, NCH125, NCH149) or established lines (U138MG, U373MG) of human glioma cells and primary cultures of human astrocytes. Cells were analyzed 30 h after treatment with TRAIL (A) or cisplatin (B) or 48 h after infection with parvovirus H-1 (C). Cell survival is expressed as a percentage of vital staining (i.e., total living cells) in treated versus mock-treated cultures. Average values and standard deviation bars were derived from the results of three independent experiments, each performed in triplicate.
TABLE 1.
Genetic alterations in the human gliomas testeda
| Cell culture | p53 statusb | PTEN expressionc | INK4 integrityc |
|---|---|---|---|
| Astrocytes | wt | H | H |
| NCH37 | mut | LOH | H |
| NCH82 | wt | Del | LOH |
| NCH89 | mut | H | LOH |
| NCH125 | mut | LOH | LOH |
| NCH149 | mut | LOH | H |
| U138MG | mut | Del | H |
| U373MG | mut | LOH | LOH |
Low-passage-number glioma cell cultures (NCH37, NCH82, NCH89, NCH125, NCH149), established glioma cell lines (U138MG, U373MG), and astrocyte primary cultures were screened for p53 mutations, PTEN expression, and the integrity of the INK4 locus by immunoprecipitation, immunoblot, and PCR analysis, respectively.
wt, wild type; mut, mutated.
Del, deleted; H, heterozygosis; LOH, loss of heterozygosis.
Glioma cells resistant to inducers of apoptotic death are sensitive to the oncolytic activity of H-1PV.
The resistance of many gliomas to anticancer drugs has been attributed to the suppression of apoptotic pathways in those cells. In this context it is noteworthy that parvoviruses have been shown to induce the death of certain cells by activating caspase 3 via a TNF-dependent and/or a Bcl-2-sensitive pathway (17, 34). This prompted us to test whether H-1PV could still bypass the resistance to cisplatin or TRAIL in the glioma cell collection described above. Cultures of glioma cells and of normal human astrocytes were inoculated with H-1PV at increasing multiplicities of infection (i.e., numbers of particles per cell), and their subsequent growth was compared with that of mock-treated cultures. All cultures tested, apart from normal astrocyte cultures, proved sensitive to H-1PV, as shown by a 60% to 80% reduction in the number of living cells counted at 48 h postinfection (Fig. 1C). This inhibition of glioma cell growth was due, at least in part, to the ability of H-1PV to induce lysis of these cells, resulting in a 50% release of glucose-6-phosphate dehydrogenase (G6PD) into the medium by 48 h postinfection (data not shown). In contrast, the growth of cultured H-1PV-infected normal astrocytes was only slightly reduced (Fig. 1C). This slight reduction was not accompanied by detectable cell lysis and was transient (data not shown). H-1PV can thus efficiently kill cultured glioma cells that are resistant to cisplatin, TRAIL, or both.
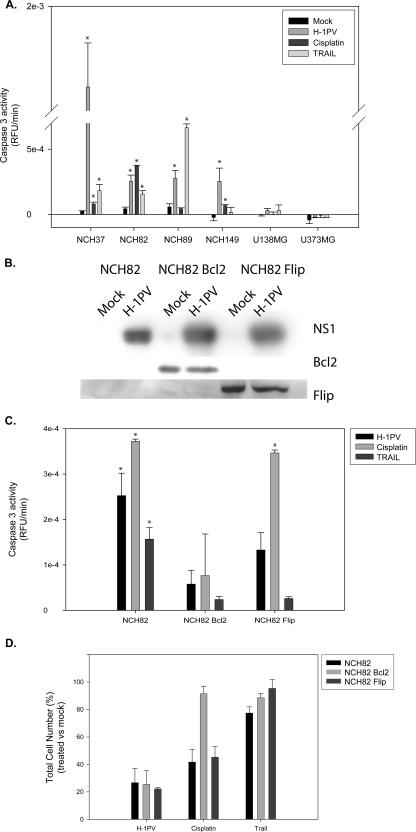
Given the above-mentioned apoptotic activity of H-1PV in certain cell systems, the question arose whether this virus would still trigger apoptosis in cells that resist treatment with conventional inducers. The ability of H-1PV to trigger apoptotic cell death in glioma cells was tested by measuring the induction of caspase 3 activity in infected cells in comparison to the results seen with cultures treated with cisplatin or TRAIL (Fig. 2A). We observed a correlation between the ability of cisplatin or TRAIL to cause cell death (see Fig. 1) and the induction of caspase 3 activity, except for the U373MG line, in which cell death was detected after treatment with cisplatin despite the absence of caspase 3 activity. In cells sensitive to either cisplatin (NCH82, NCH149) or TRAIL (NCH37, NCH89), H-1PV infection led to caspase 3 activation. As expected, no caspase 3 activation was detected after cisplatin or TRAIL treatment of glioma cells resistant to these drugs (U138MG). Interestingly, H-1PV also failed to cause caspase 3 activation in these cells, although it did kill them. It thus appears that H-1PV cannot bypass the resistance mechanism(s) preventing cisplatin and TRAIL from activating the apoptotic machinery in some glioma cells, although it does jeopardize the survival of these cells through another death pathway.
FIG. 2.
H-1PV-induced glioma cell killing in the absence of apoptosis. (A and C) H-1PV (5 PFU/cell), cisplatin (2.5 μg/ml), and TRAIL (100 ng/ml) were compared for their ability to activate caspase 3 in glioma cells of various origins (A) and in NCH82 cells transfected or not transfected to overexpress Bcl-2 or Flip (C). Caspase 3 activity is expressed in relative fluorescence units per minute. Statistically significant differences between treated and control samples (P < 0.05) are indicated by a star. (B) Accumulation of Bcl-2, Flip, and the parvoviral cytotoxic protein NS1, as measured by Western blotting after H-1PV or mock infection of NCH82 cells and stable NCH82 transductants expressing Bcl-2 or Flip from a recombinant retrovirus. (D) Effect of TRAIL, cisplatin, and H-1PV on the survival of these cells, expressed as a percentage of vital staining (i.e., total living cells) in treated versus control cultures. Average values and standard deviation bars were derived from the results of three independent experiments, each performed in triplicate (A, C, and D).
A mechanism developed by tumor cells to prevent caspase activation is the overexpression of polypeptides that directly inactivate caspases or their activating complexes. In particular, Flip interacts with the cytoplasmic domain of death receptors, preventing cleavage and activation of caspase 8 (43). Bcl-2, on the other hand, prevents mitochondrial membrane depolarization and relocation of cytochrome c to the cytosol. Bcl-2 has been shown to interfere with either the intrinsic or the extrinsic apoptotic pathway, depending on the cell type (8, 39). To test whether Flip or Bcl-2 can modulate H-1PV-induced cell death, each of these factors was overexpressed in NCH82 cells. This low-passage-number glioma culture was chosen for its sensitivity to H-1PV, cisplatin, and, to some extent, TRAIL (Fig. 1). Expression of Flip or Bcl-2 was confirmed by immunoblotting and was unaffected by cell infection with H-1PV (Fig. 2B). Bcl-2 overexpression prevented caspase 3 activation upon exposure of the cells to H-1PV, cisplatin, or TRAIL. As expected, Flip overexpression inhibited the activation of caspase 3 following treatment with TRAIL but not treatment with cisplatin. On the other hand, Flip interfered with H-1PV-induced caspase 3 activation to only a small extent (Fig. 2C), suggesting that in infected cells, this process may depend mostly on the loss of mitochondrial membrane potential. Interestingly, although overexpression of Bcl-2 blocked caspase 3 activation in cells treated with cisplatin, TRAIL, or H-1PV, it protected the cells only against the cytotoxic action of cisplatin and TRAIL but not against H-1PV-induced killing (Fig. 2D). This result, along with that of the absence of caspase 3 activation in infected U138MG and U373MG cultures, shows that parvovirus H-1 can promote procaspase 3 cleavage but that this process is not essential for H-1PV-induced cell death.
Lysosomal cathepsins accumulate in the cytosol of H-1PV-infected cells.
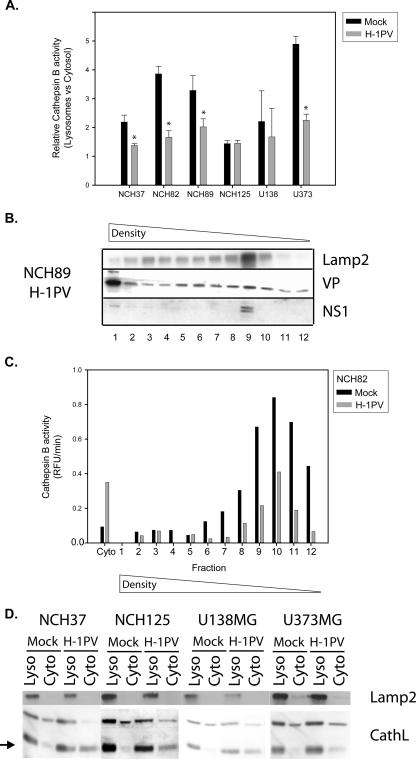
The above-described observations led us to hypothesize that caspase 3 cleavage may be a side effect of H-1PV infection and reflect the activation of proteases in response to the viral infection of glioma cells. In keeping with this possibility, caspase 3 was previously reported to be a target for active cathepsins (16, 19). Cathepsins are normally located inside lysosomes, and their targeted transport from the endoplasmic reticulum to these organelles minimizes their impact on the cytosol. Under stress, e.g., exposure to lysosomotropic agents or starvation, the integrity of the lysosomal membrane is lost, and the release of cathepsins into the cytosol induces cell necrosis (11). In H-1PV-infected glioma cells, a twofold increase in the cytosolic fraction of total cathepsin B activity was observed (Fig. 3A). This parvovirus-induced imbalance of cytosolic versus lysosomal cathepsin was further confirmed by cell fractionation. Postnuclear extracts were fractionated by density gradient centrifugation, and the lysosomal fractions were identified by detection of the lysosome-associated protein Lamp2. The enzymatic activity of cathepsin B decreased in the Lamp2-positive fractions from infected cells, which showed a much higher level of cathepsin B in the cytosolic fraction compared to mock-treated cells (Fig. 3B and C). Similarly, the cytosol of infected cells became richer in the active cathepsin L form (Fig. 3D). Altogether, these data lead to the conclusion that H-1PV infection results in an alteration of the distribution of lysosomal proteases, an enhanced proportion of these enzymes being present and active in the cytosol. How H-1PV induces cathepsin accumulation in the cytosol of infected cells remains an open question. Alteration of cathepsin distribution and concomitant death of glioma cells were not observed after treatment with empty (genome-free) capsids or UV-inactivated virus or after infection with full virions and cotreatment with bromodeoxyuridine (BrdU; 30 μM) or aphidicolin (20 μM), conditions allowing viral entry but preventing the onset of genome replication (21, 36) (data not shown). Thus, H-1PV DNA replication, gene expression, or both appear to be required for these viral effects. Viral structural (VP) and nonstructural (NS) proteins were found mostly in the cytosol and nucleus of infected cells, and yet a significant minor fraction of both products was associated with organelles, and in particular with the Lamp2-positive lysosomal fraction (Fig. 3B). Viral proteins may thus be present in the lysosomes and participate directly in the location of cathepsins in the cytosol. Yet parvoviral products, particularly the NS proteins, have multiple effects on both virus and host cell physiology (41) which might indirectly cause the observed change in the fate of cathepsins.
FIG. 3.
Cytosolic accumulation of cathepsin B and L in H-1PV-infected glioma cells. Cultures were infected or with H-1PV (5 PFU/cell) or left uninfected and examined after 24 h to determine the intracellular distribution of cathepsin B and L. (A) Cathepsin B activity in mock- and H-1PV-treated glioma cells, expressed as a ratio of lysosomal versus cytosolic activity. Statistically significant differences between infected and control samples (P < 0.05) are indicated by a star. Average values and standard deviation bars were derived from the results of three independent experiments, each performed in triplicate. (B) Distribution of the lysosomal transmembrane protein Lamp2 and parvoviral structural (VP) and nonstructural (NS) proteins in individual vesicle fractions obtained from H-1PV-infected NCH89 cells, after separation by iodixanol gradient centrifugation. (C) Distribution of cathepsin B activity between the cytosol and vesicle fractions (see panel B) obtained from H-1PV- and mock-treated NCH82 cells, as measured by a fluorescence-based enzymatic assay (RFU, relative fluorescence units). (D) Western blotting detection of the proteins Lamp2 and cathepsin L in the cytosolic and lysosomal fractions of the indicated glioma cells, after mock or H-1PV infection. The cathepsin L active form is indicated (lower band, black arrow).
Cathepsin B inhibition protects glioma cells from H-1PV-induced oncolysis.
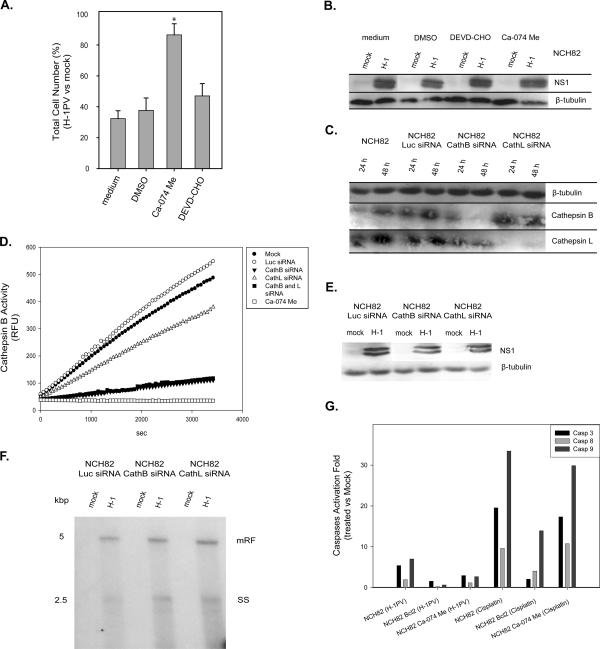
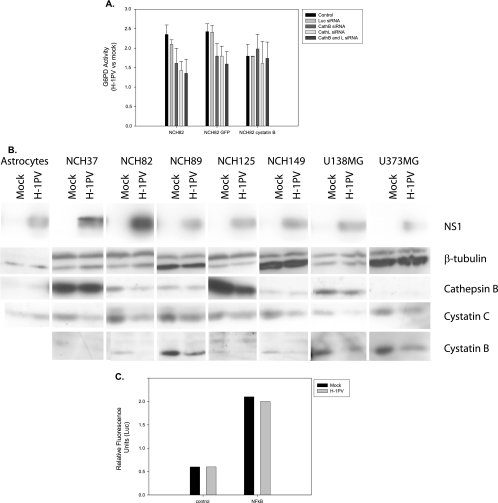
To further confirm the role of cytosolic cathepsins in the death of H-1PV-infected glioma cells, we tested the ability of the cathepsin B inhibitor Ca-074 Me to protect these cells from H-1PV-induced killing. NCH82 cells were infected with H-1PV (5 PFU/cell) and cultured under standard growth conditions or in medium supplemented with Ca-074 Me (10 μM) or the caspase 3 inhibitor DEVD-CHO (20 μM). As shown in Fig. 4A, Ca-074 Me exerted almost full protection, indicating that cathepsin B is an indispensable component of virus-induced glioma cell death. This drug is likely to interfere directly with H-1PV-triggered death pathway rather than with production of cytotoxic viral intermediates, as no change in H-1PV protein synthesis was detected in the presence of Ca-074 Me (Fig. 4B). The role of lysosomal proteases in H-1PV-triggered induction of glioma cell death was further substantiated by siRNA knockdown experiments. Down-regulation of cathepsin B expression in siRNA-treated NCH82 cells (Fig. 4C) was found to correlate with a reduced ability of cell protein extracts to cleave a cathepsin B substrate (Fig. 4D). NCH82 cells transfected with cathepsin-specific siRNAs proved less sensitive to H-1PV-induced killing than the corresponding parental cells (Fig. 5A), confirming the involvement of cathepsins in viral cytopathic effects. As with the above-mentioned inhibitors, the presence of cathepsin-specific siRNA had no detectable effect on virus replication or expression (Fig. 4E and F).
FIG. 4.
Role of cathepsins in H-1PV-induced killing of glioma cells. (A) NCH82 cell rescue from viral infection was determined 48 h postinfection (5 PFU/cell) by means of the hexosaminidase assay. Infected cells were incubated in medium alone or in medium supplemented with Ca-074 Me, DEVD-CHO, or 28 μM DMSO (used as control). Cell survival is expressed as a percentage of vital staining (i.e., total number of living cells) in H-1PV-infected versus mock-treated cultures. Statistically significant differences (P < 0.05) are indicated by a star. Average values and standard deviation bars were derived from the results of three independent experiments, each performed in triplicate. (B and E) Western blotting detection of NS1 protein at 48 h after H-1PV infection (5 PFU/cell) of NCH82 cells that were either incubated in medium supplemented with Ca-074 Me, DEVD-CHO, or DMSO (B) or transfected with luciferase, cathepsin B, or cathepsin L siRNAs (E). β-Tubulin was used as a standard for protein load matching. (C) Cathepsin B and L production in glioma cells was determined by Western blotting at 24 or 48 h after mock treatment or transfection with nonspecific (Luc) or specific siRNAs. (D) Cathepsin B activity was measured in H-1PV-infected (5 PFU/cell) NCH82 cells that were either mock treated, treated with a cathepsin-specific or control (Luc) siRNA, or treated with the cathepsin inhibitor Ca-074 Me (10 μM). Protein extracts were prepared and incubated for the indicated times with a fluorescent cathepsin B substrate, and emitted fluorescence (RFU, relative fluorescence units) was monitored for 1 h. (F) Southern blotting analysis of viral DNA replication in NCH82 cells transfected with nonspecific (Luc) or specific siRNAs 48 h after mock treatment or H-1PV infection (5 PFU/cell). mRF, monomer replicative form; SS, single-stranded DNA. (G) Caspase activation in NCH82 cells after H-1PV infection (5 PFU/cell) or cisplatin treatment (2.5 μg/ml) in the presence or absence of Ca-075 Me (10 μM) or Bcl-2 overexpression. Specific substrates for caspases 3, 8, and 9 were used. Caspase activity levels are expressed relative to control culture levels.
FIG. 5.
Interrelation of H-1PV infection and expression of cystatins in glioma cells. (A) Cell lysis was detected by measuring the release of G6PD into the medium after H-1PV infection (5 PFU/cell) of NCH82 cells or NCH82 derivatives overexpressing either cystatin B or GFP (as a control). The cells were transfected with cathepsin B-specific, cathepsin L-specific, or control (Luc) siRNAs or left untreated. Data are expressed as total G6PD activity in H-1PV- versus mock-infected cultures normalized with respect to a fully lysed positive control. (B) Steady-state levels of cathepsin B and of its inhibitors cystatin B and C were measured by Western blotting performed with H-1PV- versus mock-infected normal astrocytes and indicated glioma cells. β-Tubulin was used to control protein load matching. The parvoviral NS1 protein served as a marker of H-1PV infection. (C) NF-κB activity was determined in NCH82 glioma cells after H-1PV or mock infection. Transient expression assays were carried out, using a reporter Luc gene under the control of a promoter containing multiple copies of a NF-κB-responsive element. A minimal cytomegalovirus promoter lacking these elements was used as a control. Gene expression is expressed in relative fluorescence units.
As shown in Fig. 4G, infection of low-passage-number glioma cell cultures with H-1PV resulted in weak, but significant, Bcl-2-sensitive activation of caspase 3 (see also Fig. 2) and caspase 9. By preventing the release of cytochrome c from mitochondria, Bcl-2 prevents it from contributing to the formation of the caspase-9/caspase-3-activating complex. Previous reports have shown that cathepsins can participate in caspase 3 activation and, as a result, promote cell death (16, 19, 28). Treatment of glioma cells with the cathepsin B inhibitor Ca-074 Me reduced H-1PV-triggered caspase 3 and caspase 9 activation while having no effect on the induction of this activation by cisplatin, which does not lead to cathepsin activation. These results suggest that caspases are indeed direct or indirect targets of the cathepsins accumulating in the cytosol of H-1PV-infected cells. Yet this cathepsin-mediated activation of caspases is not essential for the killing effect of the virus, since neither the suppression of caspase activation by Bcl-2 overexpression nor coincubation with the caspase 3 inhibitor DEVD-CHO protected NCH82 cells from H-1PV-induced death (Fig. 2D and 4A).
Physiological inhibitors of cathepsins are down-regulated in H-1PV-infected glioma cells.
Cathepsin activity is tightly regulated by pH and by polypeptides of the cystatin family. Among these, cystatin C has a signal peptide for extracellular transport and controls secreted cathepsin activity, while cystatin B is intracellularly located and inhibits cytosolic cathepsins accidentally released from or not delivered to the lysosomes (33). To investigate a possible involvement of cystatin B in H-1PV-induced killing, NCH82 cells were infected with a recombinant retrovirus driving overexpression of either a cystatin B-GFP fusion protein or GFP alone (as a control) or were left untreated. Cells overexpressing cystatin B were less sensitive than control cells to H-1PV-induced killing and could not be further protected from the viral toxic effects by means of cathepsin-specific RNA interference (Fig. 5A).
Since cystatin B overexpression reduced H-1PV-induced cell death, we measured the levels of cathepsin B and cystatins in H-1PV- and mock-treated glioma cells. While cathepsin B levels were either unchanged or only slightly reduced, the expression of both cystatin B and C was clearly down-regulated in infected versus mock-treated cells (Fig. 5B). It has been reported that NF-κB-responsive genes such as Spi2A can also inhibit cathepsin B activation (23). This prompted us to examine whether H-1PV infection might modulate NF-κB activity in NCH82 cells. To this end, cultures were transfected with a NF-κB-driven Luc reporter gene, and the relative fluorescence levels were compared between mock- and H-1PV-infected cells. Data were normalized using a vector expressing GFP under the control of the cytomegalovirus promoter. No significant change in Luc activity was observed upon infection (Fig. 5C). Altogether, these data indicate that H-1PV-induced activation of cathepsins is due, at least in part, to reduced cathepsin inhibitor expression and that this effect is not mediated by NF-κB.
H-1PV-induced tumor regression is associated with cathepsin B activation in vivo.
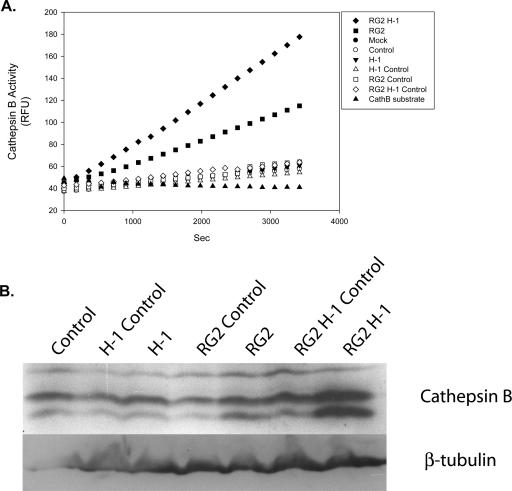
In a rat syngeneic glioma model based on intracerebrally implanted rat glioma (RG2) cells, the presence of H-1PV results in striking and irreversible tumor regression (K. Geletneky, M. Herreroy Calle, L. Deleu, C. Sommer, R. Koch, J. Rommelaere, and J. R. Schlehofer, unpublished data). This prompted us to determine the effect of the intracerebral injection of H-1PV on cathepsin B activity in healthy brains and gliomas (Fig. 6A). In the healthy brains, cathepsin B activity was very low and not significantly affected by H-1PV injection. In contrast, H-1PV-induced glioma regression in vivo was found to be associated with a striking enhancement of cathepsin B activity. This induction could already be detected in mock-treated tumors, and it was further increased as a result of their infection with the virus. While H-1PV-dependent activation of cathepsins in human glioma cell cultures took place in the absence of a significant change in enzyme concentrations (see above), rat glioma in vivo infection with parvovirus H-1 resulted in a clear increase in the total amount of tumor-associated cathepsin B (Fig. 6B). Altogether, these data argue for the possibility that the accumulation of cathepsin B inside infected tumor cells and its release in the extracellular space after cell death may contribute to glioma regression and the observed formation of a vitreous cavity in the place of the tumor.
FIG. 6.
In vivo activation of cathepsin B after H-1PV infection of tumor cells. Protein extracts were prepared from brain resections performed on control and tumor-bearing rats 3 days after mock or H-1PV (1 PFU/cell) intracerebral treatment. Samples were incubated for the indicated time with a fluorescent cathepsin B substrate and monitored for 1 h (emitted fluorescence expressed in relative fluorescence units [RFU]) (A) or analyzed by Western blotting for cathepsin B and β-tubulin expression (B). Protein amounts were normalized according to tissue masses and extract volumes. RG2 H-1, tumor-implanted rat, virus-infected tumor-bearing hemisphere; RG2 H-1 Control, tumor-implanted rat, virus-infected tumor-free hemisphere; RG2, tumor-implanted rat, mock-treated tumor-bearing hemisphere; RG2 Control, tumor-implanted rat, mock-treated tumor-free hemisphere; H-1, control rat, virus-infected hemisphere; H-1 Control, control rat, virus-free hemisphere; Mock, control rat, mock-treated hemisphere; Control, control rat, untreated hemisphere.
DISCUSSION
Cancer cells have developed numerous mechanisms to escape from induction of death by anticancer drugs. Here we show that the oncolytic rodent parvovirus H-1 can circumvent glioma cell-acquired resistance to inducers of both the intrinsic and extrinsic apoptotic pathways by activating a nonapoptotic type of death. Investigation of the interaction of H-1PV with gliomas may thus help to unravel novel facets of cell death mechanisms and to develop new therapeutics likely to be effective against tumors that resist conventional anticancer treatments.
Parvovirus H-1 was found to kill glioma cell death via a nonapoptotic cell mechanism mediated by cathepsins. Lysosomal membrane permeabilization (LMP) and the resulting release of lysosomal enzymes, and in particular cathepsins, into the cytosol have been already shown to mediate cell death (7, 10). A unique feature of H-1PV-induced glioma cell killing lies in the fact that it requires the cytosol to both accumulate cathepsins and lose cathepsin inhibitors. Experimental conditions interfering with either of these changes in apoptosis-competent cells result in a reduction of the ability of H-1PV to exert a cytotoxic activity and process effector caspases. Yet cathepsin-dependent caspase activation is not essential to virus-induced glioma cell death, since Bcl-2 suppresses the former but not the latter. Further studies are thus needed to identify the cellular targets of cytosolic cathepsins mediating H-1PV cytotoxicity.
Since cathepsins are normally confined to the lysosomes, their accumulation in an active form in the cytosol of infected cells implies that H-1PV either inhibits their translocation into the organelles or causes their release by LMP. Several reports indicate that LMP is triggered by various inducers of p53-dependent and -independent death, including TNF and proapoptotic drugs, in rodent fibroblasts (7), colon cancer cells (5), and myeloid leukemic cells (44). H-1PV-induced redistribution of lysosomal cathepsins is not mediated by TNF, since all cells tested in this study were TNF resistant. Furthermore, H-1PV cytotoxicity does not appear to be associated with the induction of detectable TNF production (34). TRAIL is also unlikely to mediate redistribution, since this ligand has recently been shown to promote cell death through caspase 8-induced cathepsin B activation (28). Generally speaking, the cytosolic accumulation of cathepsins in H-1PV-infected glioma cells can occur under conditions in which effector caspases are not activated. Permeabilization of vesicles may result from reactive oxygen species production (2) or from the activation of cellular phospholipases (47). H-1PV has been shown to block the generation of reactive oxygen species in HeLa cells (32), and inhibition of cPLA2 by chemical agents has no effect on H-1PV-induced cell killing in gliomas (data not shown). Yet a domain displaying PLA2 activity has been identified within the capsid protein VP1. Under acidic conditions (e.g., in endosomes), the capsid structure is modified, resulting in exposure of this domain and enabling the virus to exit from the acidic vesicles (6, 40, 45). Should neosynthesized VP proteins or progeny particles transit through endolysosomal vesicles, VP1 might conceivably affect the stability of vesicle membranes and, as a result, alter the trafficking of cathepsins between the cytosol and the organelles. This hypothesis is consistent with the detection of viral proteins in the vesicles of H-1PV-infected glioma cells, particularly in the Lamp2-positive fraction. The degree of LMP was proposed to determine the balance between apoptotic and necrotic death, with partial lysosomal permeabilization favoring the former and extensive lysosomal destabilization leading to the latter (11). This does not appear to always hold true, since our results indicate that limited and very selective permeabilization of lysosomes is enough to trigger a necrotic type of death in H-1PV-infected glioma cells. The dominance of necrosis in this system may be due, at least in part, to the presence of antiapoptotic signals in glioma cells. Other cells, such as U937 cells derived from a myeloid leukemia, have indeed been shown to die of apoptosis upon H-1PV infection (34). It is noteworthy that U937 cell variants surviving H-1PV infection are also resistant to death induction by TNF (34) and that TNF-induced killing of U937 cells depends on the expression of cathepsins (4). Thus, cathepsin activation appears to be a key element in parvoviral cytotoxicity, although the type of ensuing cell death may differ according to other cellular determinants. Inhibitors of NAD-consuming enzymes have also been shown to affect the balance between apoptosis and necrosis after cell infection with H-1PV (32). This may be due to the role of lysosomes in parvovirus-induced cell death, since the NAD+/NADH system is involved in electron transport and acidification in these organelles (9).
Interestingly, the present report shows for the first time that an increase in the cytosolic cathepsin activity can result not only from the enrichment of the cytosol in cathepsin molecules but also from the reduction of the content of its inhibitors (cystatin B and C). The combination of both of these effects presumably accounts for the striking increase in cathepsin activity that was observed in the cytosol of H-1PV-infected glioma cells and that was necessary for their killing. This finding points to a role of cystatin family proteins in the survival of cancer cells. In keeping with this, cystatin B and C are overexpressed in gliomas (46), accounting for the resistance of these tumors to conventional LMP-inducing agents. Successful H-1PV-induced glioma cell killing thus requires both the relocation of cathepsins and their relief from cystatin inhibition. It is noteworthy that highly malignant gliomas show lower cystatin C levels than astrocytomas (29), suggesting that the oncolytic activity of H-1PV may be even stronger against high-grade than against low-grade gliomas.
A main limitation of anticancer treatments lies in their side effects on healthy tissues. Previous data (15) and results presented here show that H-1PV infection does not jeopardize the survival of normal human astrocytes. This oncospecificity of H-1PV cytotoxic effects may be due to various factors, as normal human astrocytes differ from glioma cells by their lower permissiveness for parvovirus multiplication (1, 15), their lower level of cathepsin B, and their lack of cystatin B down-regulation upon infection.
In conclusion, H-1PV has developed an efficient strategy for killing human glioma cells through cytosolic activation of cathepsins. This parvovirus is indeed able to target lysosomal proteases from two sides, by inducing their cytosolic accumulation and suppressing their cytosolic inhibitors. Cathepsins are highly expressed in high-grade tumors and participate in normal tissue invasion by malignant cells (27, 35). A strategy taking advantage of this overexpression to induce killing of tumor cells seems particularly attractive. The evidence presented here suggests that parvoviruses could be used to this end and are especially relevant to the treatment of gliomas through their ability to efficiently hijack the lysosome-cathepsin pathway and kill tumor cells, whether or not these are resistant to conventional death inducers.
Acknowledgments
The work of M.D.P. was in part supported by a fellowship (Projects for International Mobility 2004) from Area di Ricerca Science Park, Trieste, Italy.
We are thankful to C. Cziepluch, L. Daeffler, A. Marchini, and J. Nuesch for critical reading of the manuscript.
We declare that no competing interests exist.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Abschuetz, A., T. Kehl, R. Geibig, B. Leuchs, J. Rommelaere, and A. Régnier-Vigouroux. 2006. Oncolytic murine autonomous parvovirus, a candidate vector for glioma gene therapy, is innocuous to normal and immunocompetent mouse glial cells. Cell Tissue Res. 325:423-436. [DOI] [PubMed] [Google Scholar]
- 2.Antunes, F., E. Cadenas, and U. T. Brunk. 2001. Apoptosis induced by exposure to a low steady-state concentration of H2O2 is a consequence of lysosomal rupture. Biochem. J. 356:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danial, N. N., and S. J. Korsmeyer. 2004. Cell death: critical control points. Cell 116:205-219. [DOI] [PubMed] [Google Scholar]
- 4.Deiss, L. P., H. Galinka, H. Berissi, O. Cohen, and A. Kimchi. 1996. Cathepsin D protease mediates programmed cell death induced by interferon-gamma, Fas/APO-1 and TNF-alpha. EMBO J. 15:3861-3870. [PMC free article] [PubMed] [Google Scholar]
- 5.Erdal, H., M. Berndtsson, J. Castro, U. Brunk, M. C. Shoshan, and S. Linder. 2005. Induction of lysosomal membrane permeabilization by compounds that activate p53-independent apoptosis. Proc. Natl. Acad. Sci. USA 102:192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farr, G. A., L. Zhang, and P. Tattersall. 2005. Parvoviral virions deploy a capsid-tethered lipolytic enzyme to breach the endosomal membrane during cell entry. Proc. Natl. Acad. Sci. USA 102:17148-17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foghsgaard, L., D. Wissing, D. Mauch, U. Lademann, L. Bastholm, M. Boes, F. Elling, M. Leist, and M. Jaattela. 2001. Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J. Cell Biol. 153:999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulda, S., E. Meyer, C. Friesen, S. A. Susin, G. Kroemer, and K. M. Debatin. 2001. Cell type specific involvement of death receptor and mitochondrial pathways in drug-induced apoptosis. Oncogene 20:1063-1075. [DOI] [PubMed] [Google Scholar]
- 9.Gille, L., and H. Nohl. 2000. The existence of a lysosomal redox chain and the role of ubiquinone. Arch. Biochem. Biophys. 375:347-354. [DOI] [PubMed] [Google Scholar]
- 10.Guicciardi, M. E., J. Deussing, H. Miyoshi, S. F. Bronk, P. A. Svingen, C. Peters, S. H. Kaufmann, and G. J. Gores. 2000. Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J. Clin. Investig. 106:1127-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guicciardi, M. E., M. Leist, and G. J. Gores. 2004. Lysosomes in cell death. Oncogene 23:2881-2890. [DOI] [PubMed] [Google Scholar]
- 12.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 13.Hao, C., F. Beguinot, G. Condorelli, A. Trencia, E. G. Van Meir, V. W. Yong, I. F. Parney, W. H. Roa, and K. C. Petruk. 2001. Induction and intracellular regulation of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) mediated apoptosis in human malignant glioma cells. Cancer Res. 61:1162-1170. [PubMed] [Google Scholar]
- 14.Herold-Mende, C., H. H. Steiner, T. Andl, D. Riede, A. Buttler, C. Reisser, N. E. Fusenig, and M. M. Mueller. 1999. Expression and functional significance of vascular endothelial growth factor receptors in human tumor cells. Lab. Investig. 79:1573-1582. [PubMed] [Google Scholar]
- 15.Herrero y Calle, M., J. J. Cornelis, C. Herold-Mende, J. Rommelaere, J. R. Schlehofer, and K. Geletneky. 2004. Parvovirus H-1 infection of human glioma cells leads to complete viral replication and efficient cell killing. Int. J. Cancer 109:76-84. [DOI] [PubMed] [Google Scholar]
- 15a.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 16.Hishita, T., S. Tada-Oikawa, K. Tohyama, Y. Miura, T. Nishihara, Y. Tohyama, Y. Yoshida, T. Uchiyama, and S. Kawanishi. 2001. Caspase-3 activation by lysosomal enzymes in cytochrome c-independent apoptosis in myelodysplastic syndrome-derived cell line P39. Cancer Res. 61:2878-2884. [PubMed] [Google Scholar]
- 17.Hsu, T. C., W. J. Wu, M. C. Chen, and G. J. Tsay. 2004. Human parvovirus B19 non-structural protein (NS1) induces apoptosis through mitochondria cell death pathway in COS-7 cells. Scand. J. Infect. Dis. 36:570-577. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, X., and X. Wang. 2004. Cytochrome C-mediated apoptosis. Annu. Rev. Biochem. 73:87-106. [DOI] [PubMed] [Google Scholar]
- 19.Johansson, A. C., H. Steen, K. Ollinger, and K. Roberg. 2003. Cathepsin D mediates cytochrome c release and caspase activation in human fibroblast apoptosis induced by staurosporine. Cell Death Differ. 10:1253-1259. [DOI] [PubMed] [Google Scholar]
- 20.Kartalou, M., and J. M. Essigmann. 2001. Mechanisms of resistance to cisplatin. Mutat. Res. 478:23-43. [DOI] [PubMed] [Google Scholar]
- 21.Kwant, M. M., and P. C. van der Vliet. 1980. Differential effect of aphidicolin on adenovirus DNA synthesis and cellular DNA synthesis. Nucleic Acids Res. 8:3993-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavrik, I., A. Golks, and P. H. Krammer. 2005. Death receptor signaling. J. Cell Sci. 118:265-267. [DOI] [PubMed] [Google Scholar]
- 23.Liu, N., S. M. Raja, F. Zazzeroni, S. S. Metkar, R. Shah, M. Zhang, Y. Wang, D. Bromme, W. A. Russin, J. C. Lee, M. E. Peter, C. J. Froelich, G. Franzoso, and P. G. Ashton-Rickardt. 2003. NF-κB protects from the lysosomal pathway of cell death. EMBO J. 22:5313-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longley, D. B., and P. G. Johnston. 2005. Molecular mechanisms of drug resistance. J. Pathol. 205:275-292. [DOI] [PubMed] [Google Scholar]
- 25.Maher, E. A., F. B. Furnari, R. M. Bachoo, D. H. Rowitch, D. N. Louis, W. K. Cavenee, and R. A. DePinho. 2001. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 15:1311-1333. [DOI] [PubMed] [Google Scholar]
- 26.Moehler, M., B. Blechacz, N. Weiskopf, M. Zeidler, W. Stremmel, J. Rommelaere, P. R. Galle, and J. J. Cornelis. 2001. Effective infection, apoptotic cell killing and gene transfer of human hepatoma cells but not primary hepatocytes by parvovirus H1 and derived vectors. Cancer Gene Ther. 8:158-167. [DOI] [PubMed] [Google Scholar]
- 27.Mohanam, S., S. L. Jasti, S. R. Kondraganti, N. Chandrasekar, S. S. Lakka, Y. Kin, G. N. Fuller, A. W. Yung, A. P. Kyritsis, D. H. Dinh, W. C. Olivero, M. Gujrati, F. Ali-Osman, and J. S. Rao. 2001. Down-regulation of cathepsin B expression impairs the invasive and tumorigenic potential of human glioblastoma cells. Oncogene 20:3665-3673. [DOI] [PubMed] [Google Scholar]
- 28.Nagaraj, N. S., N. Vigneswaran, and W. Zacharias. 2006. Cathepsin B mediates TRAIL-induced apoptosis in oral cancer cells. J. Cancer Res. Clin. Oncol. 132:171-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakabayashi, H., M. Hara, and K. Shimuzu. 2005. Clinicopathologic significance of cystatin C expression in gliomas. Hum. Pathol. 36:1008-1015. [DOI] [PubMed] [Google Scholar]
- 30.Pan, G., J. Ni, Y. F. Wei, G. Yu, R. Gentz, and V. M. Dixit. 1997. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277:815-818. [DOI] [PubMed] [Google Scholar]
- 31.Peter, M. E., and P. H. Krammer. 1998. Mechanisms of CD95 (APO-1/Fas)-mediated apoptosis. Curr. Opin. Immunol. 10:545-551. [DOI] [PubMed] [Google Scholar]
- 32.Ran, Z., B. Rayet, J. Rommelaere, and S. Faisst. 1999. Parvovirus H-1-induced cell death: influence of intracellular NAD consumption on the regulation of necrosis and apoptosis. Virus Res. 65:161-174. [DOI] [PubMed] [Google Scholar]
- 33.Rawlings, N. D., D. P. Tolle, and A. J. Barrett. 2004. Evolutionary families of peptidase inhibitors. Biochem. J. 378:705-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rayet, B., J. A. Lopez-Guerrero, J. Rommelaere, and C. Dinsart. 1998. Induction of programmed cell death by parvovirus H-1 in U937 cells: connection with the tumor necrosis factor alpha signalling pathway. J. Virol. 72:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rempel, S. A., M. L. Rosenblum, T. Mikkelsen, P. S. Yan, K. D. Ellis, W. A. Golembieski, M. Sameni, J. Rozhin, G. Ziegler, and B. F. Sloane. 1994. Cathepsin B expression and localization in glioma progression and invasion. Cancer Res. 54:6027-6031. [PubMed] [Google Scholar]
- 36.Rhode, S. L. 1974. Replication process of the Parvovirus H-1. III. Factors affecting H-1 RF DNA synthesis. J. Virol. 14:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rommelaere, J., and J. J. Cornelis. 1991. Antineoplastic activity of parvoviruses. J. Virol. Methods 33:233-251. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 39.Scaffidi, C., S. Fulda, A. Srinivasan, C. Friesen, F. Li, K. J. Tomaselli, K. M. Debatin, P. H. Krammer, and M. E. Peter. 1998. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suikkanen, S., M. Antila, A. Jaatinen, M. Vihinen-Ranta, and M. Vuento. 2003. Release of canine parvovirus from endocytic vesicles. Virology 316:267-280. [DOI] [PubMed] [Google Scholar]
- 41.Vanacker, J. M., and J. Rommelaere. 1995. Non-structural proteins of autonomous parvoviruses: from cellular effects to molecular mechanisms. Semin. Virol. 6:291-297. [Google Scholar]
- 42.Wrzesinski, C., L. Tesfay, N. Salome, J. C. Jauniaux, J. Rommelaere, J. Cornelis, and C. Dinsart. 2003. Chimeric and pseudotyped parvoviruses minimize the contamination of recombinant stocks with replication-competent viruses and identify a DNA sequence that restricts parvovirus H-1 in mouse cells. J. Virol. 77:3851-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao, C., B. F. Yang, N. Asadi, F. Beguinot, and C. Hao. 2002. Tumor necrosis factor-related apoptosis-inducing ligand-induced death-inducing signaling complex and its modulation by c-FLIP and PED/PEA-15 in glioma cells. J. Biol. Chem. 277:25020-25025. [DOI] [PubMed] [Google Scholar]
- 44.Yuan, X. M., W. Li, H. Dalen, J. Lotem, R. Kama, L. Sachs, and U. T. Brunk. 2002. Lysosomal destabilization in p53-induced apoptosis. Proc. Natl. Acad. Sci. USA 99:6286-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zádori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1:291-302. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, R., T. L. Tremblay, A. McDermid, P. Thibault, and D. Stanimirovic. 2003. Identification of differentially expressed proteins in human glioblastoma cell lines and tumors. Glia 42:194-208. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, M., U. T. Brunk, and J. W. Eaton. 2001. Delayed oxidant-induced cell death involves activation of phospholipase A2. FEBS Lett. 509:399-404. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, Y., and L. F. Parada. 2002. The molecular and genetic basis of neurological tumours. Nat. Rev. Cancer 2:616-626. [DOI] [PubMed] [Google Scholar]