Abstract
Conserved among all coronaviruses are four structural proteins: the matrix (M), small envelope (E), and spike (S) proteins that are embedded in the viral membrane and the nucleocapsid phosphoprotein (N), which exists in a ribonucleoprotein complex in the lumen. The N-terminal domain of coronaviral N proteins (N-NTD) provides a scaffold for RNA binding, while the C-terminal domain (N-CTD) mainly acts as oligomerization modules during assembly. The C terminus of the N protein anchors it to the viral membrane by associating with M protein. We characterized the structures of N-NTD from severe acute respiratory syndrome coronavirus (SARS-CoV) in two crystal forms, at 1.17 Å (monoclinic) and at 1.85 Å (cubic), respectively, resolved by molecular replacement using the homologous avian infectious bronchitis virus (IBV) structure. Flexible loops in the solution structure of SARS-CoV N-NTD are now shown to be well ordered around the β-sheet core. The functionally important positively charged β-hairpin protrudes out of the core, is oriented similarly to that in the IBV N-NTD, and is involved in crystal packing in the monoclinic form. In the cubic form, the monomers form trimeric units that stack in a helical array. Comparison of crystal packing of SARS-CoV and IBV N-NTDs suggests a common mode of RNA recognition, but they probably associate differently in vivo during the formation of the ribonucleoprotein complex. Electrostatic potential distribution on the surface of homology models of related coronaviral N-NTDs suggests that they use different modes of both RNA recognition and oligomeric assembly, perhaps explaining why their nucleocapsids have different morphologies.
Infection by severe acute respiratory syndrome coronavirus (SARS-CoV) is initiated by the recognition of ACE-2 receptor on the surface of respiratory epithelial cells by the “spike” glycoprotein present on the viral surface (27, 29, 34). Subsequent progression of infection involves a series of complex, tightly regulated processes that begin by the entry of genomic RNA into the cytosol and culminate with the budding of infectious progeny (14, 15). These mature, fully formed virions are functionally as well as morphologically indistinguishable from their parents and have a quasi-fluid-like, pleomorphic, bilipid envelope whose surface is studded with three main structural transmembrane proteins: the matrix (M), the small envelope (E), and the trimeric spike (S) glycoproteins (16, 40, 54). The envelopes of these particles encase the ∼29-kb genomic RNA that is thought to be organized as a helical filamentous ribonucleoprotein (RNP) complex. Several copies of the N protein self-associate and form a template for binding RNA during nucleocapsid formation (13, 16, 18, 35, 61). As noted in studies done using murine hepatitis virus (MHV), the initial steps of virus assembly, including the formation of the RNP complex and its eventual packaging into the virion lumen, occurs in a temporally regulated manner, mainly at the endoplasmic reticulum-Golgi intermediate compartments just prior to budding (1, 8, 22, 55). Successful targeting of the RNP into the virion lumen is thought to be facilitated by its anchoring onto the membrane-embedded M protein by specific interaction between their respective C-terminal tails (10, 23, 32, 39, 56). Despite extensive studies on several model coronaviruses spanning 25 years, our structural understanding of these assembly events remains sketchy (5, 7, 8, 15, 24, 34).
SARS-CoV N protein is translated from the smallest of the eight subgenomic RNAs (the bicistronic sg-mRNA 9) (15, 26, 54) that spans the genomic 3′-most open reading frame, ORF9a (Fig. 1a). Coronaviral N proteins are typically ca. 45 to 50 kDa, very basic (with typical pIs of ∼10), prone to aggregate into large homopolymers (16), phosphorylated at multiple sites (3, 50, 58), and extremely labile to proteolytic degradation (39, 57, 61). These characteristics have hindered in vitro structural studies on full-length N. The N-terminal domains of coronaviral N proteins (N-NTDs) typically share about 30 to 40% sequence identity (Fig. 1c). As in most nidoviruses, the full-length SARS-CoV N protein (430 residues) has three main protein domains: an N-terminal RNA-binding domain (i.e., the N-NTD), a poorly structured central serine-rich region that is thought to house the primary sites of phosphorylation (33, 58), and a C-terminal domain (N-CTD [52]) that is mainly involved in oligomerization and self-association (4; Fig. 1b). In addition, a few coronaviruses have about 20 residues upstream of the NTD that are rich in serine, glycine, and arginine (SRG motif; Fig. 1b). N protein is also known to undergo sumoylation (28). Several other ancillary functions have been ascribed to coronaviral N proteins. In MHV as well as infectious bronchitis virus (IBV), N not only binds to genomic RNA but to the six subgenomic RNAs as well (62). It is involved in cell signaling (19, 20) and is known to interact with several human proteins, including human cyclophilin A (31) and human RNP A1. Anti-N monoclonal antibodies protect mice from lethal coronaviral infection (43). SARS-CoV N is known to elicit a well-defined immunological response, as evidenced by its peptides binding to human lymphocyte antigens with nanomolar affinities (2, 53), which underscores the importance of N as a potential target in neutralizing SARS infection (30). The structure of a highly conserved nine-residue peptide corresponding to the region 362KTFPPTEPK370 has been resolved in complex with a class I major histocompatibility complex molecule (2, 53).
FIG. 1.
(a) Organization of SARS-CoV genome. Locations of the open reading frames (ORFs) are indicated. The boundaries of the 16 nonstructural proteins (nsp1 to nsp16) that result from proteolytic processing of the replicase polyprotein (PP1ab) by PL-Protease (green) and 3CLpro (black) are marked by vertical lines. (b) Domain organization of coronaviral N proteins. The four domains labeled are as follows: SGRD, serine-glycine-arginine-rich domain; NTD, N-terminal domain; SRD, serine-rich domain; and CTD, C-terminal domain. (c) Multiple sequence alignment of NTD domains. The region for which structural coverage is provided in this study is marked by vertical lines. Hydrophobic residues are shown in yellow. Secondary structures observed for SARS-CoV N-NTD are shown above the alignment as arrows (strand) and cylinders (helix). Positively charged residues that have been implicated in RNA binding are indicated by asterisks above the sequence. The ICTV acronyms used for each viral sequence and their corresponding database accession numbers were as follows: HCoV-229E, human coronavirus 229E (NP_073556); TCoV-NC95, turkey coronavirus NC95 strain (gi32129798); BCoV-Lun, bovine coronavirus (AAL57313); HEV-VW572, porcine hemagglutinating encephalomyelitis virus (YP_459957); TGEV-Purdue, transmissible gastroenteritis virus Purdue strain (NP_058428); HCoV-NL63 human coronavirus NL63 (YP_003771); PEDV-CV777, porcine epidemic diarrhea virus CV777 strain (NP_598314); FCoV-79-1146, feline coronavirus (YP_239358); SARS-CoV-Tor2, severe acute respiratory syndrome coronavirus-Tor2 strain (AAP41047); MHV-JHM, murine hepatitis virus JHM strain (YP_209238); HCoV-OC43, human coronavirus OC43 (NP_937954); HCoV-HKU1, human coronavirus HKU-1 (YP_173242); RCoV, Rat coronavirus (AAD33104); HECoV-4408, human enteric coronavirus 4408 (AAQ67202); CCoV, canine coronavirus; ECoV-NC99, equine coronavirus NC99 (Q9DQX6); PgCoV, pigeon coronavirus (gi58416203); PCoV, puffinosis coronavirus, gi28460530; and IBV-Beaudette, avian infectious bronchitis virus (NP_040838).
The structure of the NTD of N protein from SARS-CoV has been determined by using nuclear magnetic resonance (NMR) spectroscopy, which revealed that the solvent exposed loops are flexible, existing in multiple conformers in the absence of RNA, a feature that probably helps in its primary biological function as the scaffolding agent in viral genomic RNA packaging (19). The crystal structure of its homologue from avian IBV has been reported recently in two crystal forms showing a similar domain architecture centered around a five-stranded β-sheet core (11, 21). The C-terminal oligomerization domain of N protein has also been structurally characterized both from SARS-CoV and IBV (4, 21, 60).
In the present study, we report the crystallographic characterization of SARS-CoV N-NTD spanning residues 47 to 175 in two crystal forms that have been solved at 1.17 Å and 1.85 Å, respectively. The structures have been phased by molecular replacement using the IBV homolog. Comparison of the two crystal forms versus the solution conformation of this domain (residues 45 to 181 [19]) and comparison with the two published IBV N-NTD structures (residues 29 to 160 [21]) shows several commonalities, as well as many subtle structural differences. The crystal packing noticed in the cubic form of SARS-CoV N-NTD and in the C2 lattice of its IBV homolog suggests that the two viruses probably employ different modes of oligomeric self-association during the RNP core formation. Modeling studies on this domain from related coronaviruses suggest that not only is the assembly of RNA around the helical N protein polymer likely to be different but the manner in which the N proteins recognize RNA is likely to be different as well. These observations might explain why the fully packaged nucleocapsids of different nidoviruses often exhibit different morphologies as observed in cryo-electron microscopy (cryoEM) studies (15).
MATERIALS AND METHODS
Cloning, expression, and purification.
Multiple constructs were designed covering different regions of ORF9a from Tor2 strain of SARS-CoV as part of the structural and functional proteomics of SARS-CoV (FSPS) project (http://sars.scripps.edu). Domain boundaries were arrived at based on secondary structure predictions, earlier observations made in the literature regarding proteolytic susceptibility, and sequence conservation characteristics. The sequence of the construct reported here corresponds to the N-terminal domain that covers residues 47 to 175 of the ORF9a gene (NP_828858.1, gi:29836503). The gene was amplified by PCR from genomic cDNA of the SARS-CoV Tor2 strain using Taq polymerase and primer pairs encoding the predicted 5′ and 3′ ends (forward, 5′-ATGCCCAATAATACTGCGTCTTGGTAGGGCCGGCCGGG-3′; reverse, 5′-CTCTGCGTAGAAGCCTTTTGGCCCCGGCCGGCCCTA-3′). The PCR product was cloned into plasmid pMH1f that encodes an N-terminal purification tag (MGSDKIHHHHHH). Protein was expressed from a sequence verified clone in 2×YT media. Bacteria were lysed by sonication in buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 10% glycerol) containing two Roche protease inhibitor tablets and 0.5 mg of lysozyme. After ultracentrifugation at 45,000 rpm for 20 min at 4°C, the soluble fraction was applied on a metal chelate column (Talon resin charged with cobalt; Clontech), washed in 20 mM Tris (pH 7.8)-300 mM NaCl-10% glycerol-5 mM imidazole, and eluted with 25 mM Tris (pH 7.8)-15 mM NaCl-150 mM imidazole. The resultant protein was further purified using anion-exchange chromatography on Poros HQ column with elution buffer containing 25 mM Tris (pH 8.0) and 1 M NaCl. The pure fractions of the protein were pooled, and buffer was exchanged into crystallization buffer (10 mM Tris [pH 7.8], 150 mM NaCl) and concentrated by ultrafiltration to a final concentration of 1.8 mM. It was either flash frozen or used immediately for crystallization trials.
Crystallization and data collection.
Crystals were grown by the nano-volume sitting-drop method. Typically, 100 nl of protein was mixed with 100 nl of well solution. Monoclinic crystals grew in solution containing 0.2 M sodium bromide, 0.1 M sodium acetate (pH 5.5), and 25% polyethylene glycol 2000 MME. The crystal that was used for data collection contained BCIP (5-bromo-4-chloro-3-indolylphosphate) as an additive. Cubic crystal form grew in 40% methyl pentanediol and 0.1 M Tris (pH 8.0), typically within 2 weeks. These were cryoprotected in a solution containing mother liquor and 15% glycerol and flash frozen in liquid nitrogen. Crystal screening and data collection were done by using the BLU-ICE (36) interface at the remote facility at the Stanford Synchrotron Radiation Laboratory Beamline-11.1, and all diffraction data were processed using HKL2000 (41).
Phasing and refinement.
Initial phases for the monoclinic crystal form were obtained by molecular replacement using a full-atom model of the corresponding domain of the IBV nucleocapsid (PDBId 2BTL) by using the program Phaser (44) with data from 20.0 to 3.0 Å. Rigid body refinement using Refmac5 revealed a clearly interpretable electron density map. Both phases and the model were further improved by one round of automated model building cycle followed by a solvent atom search in Arp/wARP (25). The resulting model was improved by subsequent rounds of manual model building in Coot (9) alternated with restrained refinement in Refmac5 (37) of CCP4 using anisotropic B factor refinement. Optimum TLS parameters were analyzed at the TLSMD Web server (42), and 11 TLS groups covering 139 residues were used during refinement. Similarly, the structure of the cubic crystal form was solved by using the IBV structure of N-NTD as the query model and searched based on data from 20.0 to 3.0 Å, followed by a rigid body refinement and finally by alternating manual model building and restrained refinement cycles using Coot and Refmac5 (37), respectively. The final model statistics, validation, and stereochemical quality for the two structures are reported in Table 1.
TABLE 1.
Data collection and refinement statisticsa
| Data collection | Monoclinic | Cubic |
|---|---|---|
| Space group | P21 | I213 |
| Unit cell (size [Å], angle [°]) | a = 36.57, α = 90 | a = 110.01, α = 90 |
| b = 36.44, β = 94.7 | b = 110.01, β = 90 | |
| c = 47.73, γ = 90 | c = 110.01, γ = 90 | |
| Wavelength (Å) | 0.9793 | 0.9797 |
| Resolution range (Å) | 40-1.17 (1.21-1.17) | 40.0-1.85 (1.89-1.85) |
| Total no. of observations | 351,234 | 73,926 |
| Unique no. of reflections | 40,016 (2,882) | 18,018 (1,337) |
| Completeness score (%) | 97.4 (99.7) | 99.78 (100.0) |
| Redundancy | 3.8 (3.2) | 4.1 (1.9) |
| Mean I (σ) | 22.19 (2.25) | 29.33 (1.91) |
| Rsym on I | 0.059 (0.526) | 0.068 (0.446) |
| Refinement statistics | ||
| Resolution range (Å) | 40-1.17 | 40-1.85 |
| Rcryst | 0.168 (0.205) | 0.189 (0.307) |
| Rfree | 0.199 (0.229) | 0.243 (0.372) |
| Bond length (Å) (RMSD) | 0.009 | 0.011 |
| Bond angle (°) (RMSD) | 1.26 | 1.45 |
| Avg isotropic B value (Å2) | 12.1 | 25.1 |
Rsym = Σhkl[(Σj|Ij − 〈I〉|)/Σj|Ij|]. Rcryst = Σhkl|Fo − Fc|/Σhkl |Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. Five percent of randomly chosen reflections were used to calculate Rfree. Values in parentheses are for data corresponding to the outermost reflection shell.
Homology modeling and electrostatic calculations.
Comparative molecular models of N-NTDs from related coronaviruses were built using the full structure of SARS-CoV N-NTD as a template. The program Modeler was used (automodel class) with default parameters. Regions of coronaviral N proteins that aligned with the sequence of SARS-CoV N protein boundaries described in the present study were used for homology modeling. Electrostatic calculations were done using the Adaptive Poisson-Boltzmann Solver (APBS) module using PYMOL (6).
Protein structure accession numbers.
The structure factors and coordinates of SARS-CoV N-NTD in the two crystal forms have been deposited in PDB under accession numbers 2OFZ (monoclinic form) and 2OG3 (cubic form), respectively.
RESULTS AND DISCUSSION
Structure of the SARS-CoV N-NTD.
The structure of N-NTD (residues 47 to 175) was determined in monoclinic and cubic lattices to 1.17 and 1.85 Å, respectively. As anticipated, SARS-CoV N-NTD, with its single-domain β-sheet core (with the exception of a single short 310 helix) and large loops on the outside (Fig. 2a), is similar in overall topology and surface electrostatic profiles (Fig. 2b) to both its solution structure (residues 45 to 181 [19]) and to the two IBV crystal structures (residues 29 to 160 [11, 21]).
FIG. 2.
(a) Structural representation of N-NTD monomer. The structure is colored from the N terminus (blue) to the C terminus (red). (b) Distribution of electrostatic potential on the surface of N-NTD. The potential distribution was calculated by using APBS module in Pymol (6). The values range from −5 kT (red) to 0 (white) and to +5 kT (blue), where k is the Boltzmann constant and T is the temperature. The orientation of the molecule is about 180° rotation along y axis of panel a. (c) The crystal structure of the monoclinic form of SARS-CoV N-NTD over the average coordinates of the NMR structure of the same domain as reported by Huang et al. (19). The four regions along the polypeptide that differ the most between the two structures are indicated by L1 to L4. Loop L1 is colored cyan for the NMR structure and blue for the crystal structure. (d) Stereo diagram showing the Cα trace of superimposed structures of SARS-CoV N-NTD and IBV N-NTD. The cubic and monoclinic forms of SARS-CoV N-NTD are shown in green and blue, respectively, while the structure from IBV is traced in red.
However, a number of differences between these structures help elucidate the function of full-length N protein and its interaction with the viral RNA. These differences were not only restricted to the loops that show high disorder in the solution structure (19) but also were present in the functionally important β-hairpin and a few residues in the conserved β-sheet core. A superimposition of the monoclinic form of SARS-CoV N-NTD with the average solution structure is shown in Fig. 2c. The structural similarity only appears to be at the overall “fold” level. The root mean square deviation (RMSD) between the solution structure coordinates (average structure, PDB ID 1SSK) and the crystal structure of the monoclinic form are 2.6 Å over 112 superimposed Cα atoms. These might account for the failure of attempts at phasing the two datasets using the solution structure coordinates (either the average, or the ensemble in toto, as well as individually) as the query model. Similarly, Fan et al. reported large RMSDs between the IBV crystal structure and the NMR spectroscopy structure of SARS-CoV N-NTD as a possible cause of failure in phasing the IBV structure using the SARS-CoV NMR coordinates (14). The most dramatic differences are in the loops L1 (residues 91 to 100) and L3 (residues 118 to 123), which show a concerted inward shift by as much as 4.2 Å in the X-ray structure (Fig. 2c). This movement, combined with a corresponding outward hinge motion of the β-hairpin L2 (Glu 106 and Gly 107) and the loop L4 (residues 127 to 134), results in the RNA-binding cleft (discussed below) being both narrow and shallow in the X-ray structure compared to the solution structure.
The Cα traces of the superimposed structures of the two crystal forms of SARS-CoV N-NTD and the corresponding domain of IBV are shown in (Fig. 2d). As one would anticipate (given the success of molecular replacement), the crystal forms of IBV and SARS-CoV N-NTDs are quite similar (RMSD = 1.22 Å for 110 superimposed Cα atoms). The two SARS-CoV N-NTD crystal forms themselves superimpose quite well with an RMSD of 0.3 Å over 111 Cα residues. They differ in the side chain rotamers of a few residues. The most important difference is in the positively charged β-hairpin (residues 57 to 72), implicated in RNA binding, which is disordered in the cubic form (Fig. 2d).
Putative RNA binding surface.
Solution studies by NMR spectroscopy of SARS-CoV N-NTD and in vitro RNA binding studies in IBV clearly indicate that this domain binds to viral RNA corresponding to a highly conserved region at the genomic 3′ end (61). We also noticed that this specific region encompassed by N-NTD construct binds single-stranded RNA, double-stranded RNA, single-stranded DNA, and double-stranded DNA (in decreasing order of affinity) in gel shift assays (data not shown). As observed in the previous structures, there is a clear segregation of positive and negative charges in the crystal structure of SARS-CoV N-NTD. The positively charged residues are largely confined to a groove that includes the β-hairpin, and the cleft whereas the negatively charged residues are clustered around the β-sheet core (Fig. 2b). It is therefore likely that the model of RNA-N interaction that was proposed for IBV (a group III coronavirus) with the phosphate backbone of RNA stacking against the conserved arginine and lysine residues of the β-hairpin due to favorable electrostatics is likely to be true for SARS-CoV as well. The two tyrosine residues of IBV N-NTD (Y92 and Y94) that have been proposed to potentially stack with consecutive nucleotide bases are conserved in SARS-CoV N-NTD (Y76 and Y78) and lie at the base of the RNA binding groove. Similar modes of RNA-protein interactions (mostly in mRNA cap recognition) have been reported in unrelated RNA-binding viral proteins such as VP40 of Ebola virus (16) and vaccinia virus VP39 (22, 51). The positively charged β-hairpin was reported to be flexible due to weaker than expected nuclear Overhauser effects in the heteronuclear NMR experiment and higher-than-average B factors observed in the IBV crystal structure. This hairpin is completely disordered in the one of the crystal forms (cubic form) reported here. However, it is clearly ordered in the high-resolution structure of the monoclinic form. It is oriented in a similar conformation as one of the IBV structures and is almost perpendicular to the central core domain (Fig. 2a).
Although N-NTD binds to all four forms of nucleic acid polymers (single- and double-stranded RNA and single- and double-stranded DNA), specificity for RNA over DNA is probably provided more by context (localization to the replicase complex) rather than biochemical selectivity. Whether coronaviral N protein traffics to the nucleus (20, 59) or, more specifically, to the nucleolus (17) has been the subject of debate over many years. Given the shallow nature of the RNA binding groove of N-NTD, it is possible that full-length SARS N might bind to one or both RNA forms (single and double stranded) within infected cells.
Packing of SARS-CoV N-NTD monomers in the two crystal forms.
We observed distinctly different modes of packing in the two crystal forms of N-NTD monomers. The asymmetric units of both crystal forms contain one monomer each. In the monoclinic crystal form, the symmetry mates pack in a linear three-dimensional array as head-to-head dimers, with most of the interfacial interactions being made by residues of the positively charged β-hairpin (Fig. 3a and b). In the cubic form, the N-NTD monomers pack as helical tubules. Here, individual monomers are organized as trimeric units (Fig. 3c), where two consecutive trimers exhibit a right-handed twist, coiling around a pseudohelical axis. Three trimers arch around this axis and form one full turn of the helix (total of nine monomers per turn; Fig. 3d). A trimeric form of nucleocapsid has been observed for full-length N proteins of MHV A59 strain in RNA protein overlay blot assay experiments on a nonreduced preparation of purified virions (46). The relationship between these in vivo observations and the crystal packing we observed, however, remains unclear in the absence of bound RNA in our structure. Nonetheless, the possibility that this helical arrangement might be of physiological relevance cannot be ignored in light of the observation of a similar tubular mode of crystal packing highlighted in the IBV N-NTD structure by Jayaram et al. (21).
FIG. 3.
Crystal packing in the two crystal forms of SARS-CoV N-NTD. (a) Side-on view of three crystallographically related monomers showing stacking interactions in the monoclinic form. (b) Larger end-on view of the same crystal showing two primary modes of packing between the β-sheet cores (green and blue monomers on the left) and the protruding hairpins of adjacent monomers (yellow and brown monomers in the middle). (c and d) Three symmetry related monomers viewed along a threefold axis of the cubic crystal form (c) and a zoomed-out stereo view showing one turn along the helical axis of the cubic form (d). Equivalent trimers are labeled A, B, and C and colored green, red, and blue.
Modeling of related coronaviral N-NTDs.
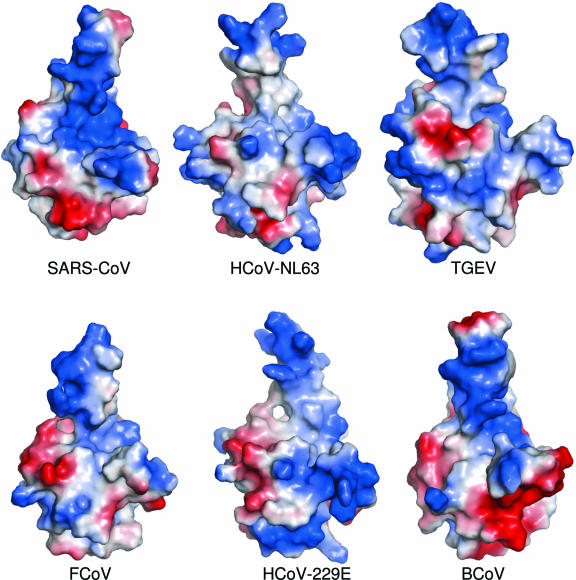
The two crystal forms each of N-NTD from SARS-CoV reported here and the two forms of IBV N-NTDs (11, 21) allowed us to generate homology models of N-NTDs of related coronaviruses with high accuracy. The distribution of the electrostatic potential on the surfaces of these models is shown in Fig. 4. Although these models retain a similar overall organization of β-strands within the core, the models markedly differ in their surface charge distribution patterns, despite having short stretches of locally conserved sequences with similar electrostatic sequence profiles. While speculative, it is likely that the RNA interacting residues and, therefore, the mode of interaction in these coronaviruses, is likely to be different compared to SARS-CoV (group II) and IBV (group III). This is not surprising given that RNP core itself is packaged differently among the various morphologically distinct nidoviruses (reviewed in reference 15). For example, in transmissible gastroenteritis virus (TGEV), a group I coronavirus, cryoEM studies of detergent-treated virions clearly show that the RNP cores are organized as almost regular icosahedrons (47). This is also true for arteriviruses, which have icosahedral RNPs of ca. 20 to 30 nm (58). Roniviruses, on the other hand, encase a rod-like RNP core within elongated lipidic envelopes (48). Finally, nucleocapsids of toro- and coronaviruses (17, 46) exhibit rather disperse fluid-like structures with a beaded appearance.
FIG. 4.
Electrostatic charge distribution on the surfaces of homology models of coronaviral N-NTDs. As in Fig. 1, the values range from −5 kT (red) to 0 (white) and to +5 kT (blue), where k is the Boltzmann constant and T is the temperature. The sequences and database accession numbers that were used as templates are the same as in Fig. 1. The boundaries of the model correspond to the regions that align with that of the SARS construct as shown in Fig. 1c.
Implications for nucleocapsid formation.
Accommodation of the exceptionally large (∼29-kb) SARS-CoV genome into newly formed virion spherules approximately 82 to 120 nm size (14, 45) necessitates an extremely well-packed, largely helical, supercoiling of the nucleic acid within the RNP core. Mature virions are thought to have about 50 to 100 copies of spike trimers and ca. 200 to 400 copies of N in the membrane-proximal region arranged in a paracrystalline lattice. Our recent cryoEM study on the supramolecular organization of the structural proteins on the coats of both SARS-CoV and feline-CoV showed that the RNP appears as punctuate electron-dense features that are clearly associated with M protein and organized as a linear S-M-RNP layer (outside to inside [45]). Our results and those reported by Risco et al. (47) suggest a two-layered organization wherein thread-like densities project from the inner face of the top S-M layer into the two-dimensionally ordered quasilattice of the RNP layer. If indeed the trimeric helical arrangement of N-NTD seen in the cubic form is one possibly physiologically relevant form of RNP, it is likely to face the other face of the quasilattice. This orientation would enable the terminal residue of the N-CTDs to adhere to M, thus anchoring the two layers. The absence of the structures of the other domains (or that of full-length N protein), especially the presence of the intervening serine-rich domain that is largely unstructured, preclude the development of molecular models that explain the higher-order organization of the RNP.
Virus assembly and maturation.
Enveloped viruses use one of three main mechanisms of assembly and budding (reviewed in references 14 and 15). Previously published studies have suggested a process that is independent of functional N protein (26, 38, 55). Experiments on tunicamycin-treated infected cells suggests that the role of spike in the budding process is also limited. Instead, assembly and budding of mature virions appear to be largely driven by correct folding and assembly of M and E proteins. Interference with M-N protein interaction has little effect on the correct incorporation of M protein into the envelope in the early stages of assembly leading to morphologically indistinguishable virions. An even less-understood process is viral closure or the pinching-off event (17). Nonetheless, it is becoming increasingly clear from multiple independent studies that an ordered lattice formation of the RNP in the immediate vicinity of the luminal face of virion envelope is integral to coronavirus budding. In SARS, MHV, and TGEV coronaviruses, the predominant forces at play in this region are those between the C-terminal tails of the M and N proteins, with the interacting residues of the M protein coming from its C-terminal luminal domain (residues 194 to 205 in the case of SARS-CoV [12]). Since the last few residues from N-CTD (the last residue being the most important) are thought to play the main anchoring role in the N-M layer, there is increasing consensus that, within the RNP, the CTDs of individual N monomers are oriented such that their C-terminal tails point toward the envelope (45). However, both the positioning and orientation of NTD remains nontrivial because of the fibrous organization of the helical RNP and the complex curved path that a fully assembled RNP traverses within the viral lumen. Further studies on the full-length N protein and complementation studies between the NTD and CTDs of N protein are needed to understand the interplay between these two domains within the N-M layer of coronaviruses.
Conclusion.
This study describes the high-resolution structures of two crystal forms of the N-terminal RNA-binding domain of SARS-CoV N protein. Structure analysis in the context of ribonucleocapsid assembly of SARS-CoV, IBV, and porcine reproductive and respiratory syndrome virus hints at both common features and differences in the ribonucleocapsid assembly of these three closely related Nidovirales members. The high degree of similarity of SARS-CoV N-NTD with other coronaviral N-NTDs compared to the IBV homolog has allowed the construction of accurate homology models. The lack of conserved electrostatic profiles in the RNA binding groove in these homology models suggests the use of disparate mechanisms for RNA recognition and RNP assembly by different coronaviruses. In conjunction with the structures of N-CTD oligomerization domains, these results are beginning to provide important insights into generic and unique aspects of coronaviral ribonucleocapsid assembly and set the stage for further structural studies on full-length N proteins by cryoEM and related techniques, which would hopefully shed further light on this very important aspect of coronaviral genome assembly and packaging.
Acknowledgments
We thank Q. Huang's group at Abbott Laboratories for providing the coordinates of the NMR ensemble for structure analysis; Tom Clayton, Anna Tang, Mark Griffith, and Kin Moy for help with protein expression and purification; Alexei Brooun and Jeffery Velasquez for cloning; and the staff scientists at SSRL for help with remote crystal screening and data collection support. SSRL Beamline BL-11.1 is supported by the National Institutes of Health (NIH) National Center for Research Resources, the NIH National Institutes of General Medical Sciences, the U.S. Department of Energy, the Office of Biological and Environmental Research, Stanford University, and The Scripps Research Institute. We also acknowledge the General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT) of Argonne National Laboratory, Chicago, IL, for help with initial crystal screening. GM/CA-CAT is supported by the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Sciences (Y1-GM-1104).
This study was supported by National Institutes of Allergy and Infectious Disease/NIH contract HHSN 266200400058C (Functional and Structural Proteomics of the SARS-CoV) to P.K.
Footnotes
Published ahead of print on 17 January 2007.
REFERENCES
- 1.Baric, R. S., G. W. Nelson, J. O. Fleming, R. J. Deans, J. G. Keck, N. Casteel, and S. A. Stohlman. 1988. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J. Virol. 62:4280-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blicher, T., J. S. Kastrup, S. Buus, and M. Gajhede. 2005. High-resolution structure of HLA-A*1101 in complex with SARS nucleocapsid peptide. Acta Crystallogr. D Biol. Crystallogr. 61:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvo, E., D. Escors, J. A. Lopez, J. M. Gonzalez, A. Alvarez, E. Arza, and L. Enjuanes. 2005. Phosphorylation and subcellular localization of transmissible gastroenteritis virus nucleocapsid protein in infected cells. J. Gen. Virol. 86:2255-2267. [DOI] [PubMed] [Google Scholar]
- 4.Chang, C. K., S. C. Sue, T. H. Yu, C. M. Hsieh, C. K. Tsai, Y. C. Chiang, S. J. Lee, H. H. Hsiao, W. J. Wu, C. F. Chang, and T. H. Huang. 2005. The dimer interface of the SARS coronavirus nucleocapsid protein adapts a porcine respiratory and reproductive syndrome virus-like structure. FEBS Lett. 579:5663-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, H. A., R. R. Dourmashkin, and R. MacNaughton. 1981. Ribonucleoprotein of avian infectious bronchitis virus. J. Gen. Virol. 53:67-74. [DOI] [PubMed] [Google Scholar]
- 6.DeLano, W. L. 2002. The PyMOL molecular graphics system. [Online.] http://pymol.sourceforge.net.
- 7.Doan, D. N., and T. Dokland. 2003. Structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Structure 11:1445-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubois-Dalcq, M., K. V. Holmes, and B. Rentier. 1984. Assembly of Coronaviridae, p. 100-119. In D. W. Kingsbury (ed.), Assembly of enveloped RNA viruses. Springer-Verlag, New York, NY.
- 9.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 10.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, H., A. Ooi, Y. W. Tan, S. Wang, S. Fang, D. X. Liu, and J. Lescar. 2005. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure 13:1859-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, X., L. L. Ye, K. A. Timani, S. Li, Y. Zen, M. Zhao, and H. Zheng, and Z. Wu. 2005. Peptide domain involved in the interaction between membrane protein and nucleocapsid protein of SARS-associated coronavirus. J. Biochem. Mol. Biol. 38:381-385. [DOI] [PubMed] [Google Scholar]
- 13.Fosmire, J. A., K. Hwang, and S. Makino. 1992. Identification and characterization of a coronavirus packaging signal. J. Virol. 66:3522-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, R., F. Dobie, M. Ballantine, A. Leeson, Y. Li, N. Bastien, T. Cutts, A. Andonov, J. Cao, T. F. Booth, F. A. Plummer, S. Tyler, L. Baker, and X. Li. 2004. Analysis of multimerization of the SARS coronavirus nucleocapsid protein. Biochem. Biophys. Res. Commun. 316:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiscox, J. A., T. Wurm, L. Wilson, P. Britton, D. Cavanagh, and G. Brooks. 2001. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J. Virol. 75:506-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsieh, P. K., S. C. Chang, C. C. Huang, T. T. Lee, C. W. Hsiao, Y. H. Kou, I. Y. Chen, C. K. Chang, T. H. Huang, and M. F. Chang. 2005. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 79:13848-13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, Q., L. Yu, A. M. Petros, A. Gunasekera, Z. Liu, N. Xu, P. Hajduk, J. Mack, S. W. Fesik, and E. T. Olejniczak. 2004. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry 43:6059-6063. [DOI] [PubMed] [Google Scholar]
- 20.Hurst, K. R., L. Kuo, C. A. Koetzner, R. Ye, B. Hsue, and P. S. Masters. 2005. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J. Virol. 79:13285-13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jayaram, H., H. Fan, B. R. Bowman, A. Ooi, J. Jayaram, E. W. Collisson, J. Lescar, and B. V. Prasad. 2006. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 80:6612-6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krijnse-Locker, J., M. Ericsson, P. J. Rottier, and G. Griffiths. 1994. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J. Cell Biol. 124:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamzin, V. S., and K. S. Wilson. 1997. Automated refinement for protein crystallography. Methods Enzymol. 277:269-305. [DOI] [PubMed] [Google Scholar]
- 26.Lau, Y. L., and J. S. Peiris. 2005. Pathogenesis of severe acute respiratory syndrome. Curr. Opin. Immunol. 17:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, F., W. Li, M. Farzan, and S. C. Harrison. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864-1868. [DOI] [PubMed] [Google Scholar]
- 28.Li, F. Q., H. Xiao, J. P. Tam, and D. X. Liu. 2005. Sumoylation of the nucleocapsid protein of severe acute respiratory syndrome associated coronavirus. FEBS Lett. 579:2387-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang, M. F., R. L. Du, J. Z. Liu, C. Li, Q. F. Zhang, L. L. Han, J. S. Yu, S. M. Duan, X. F. Wang, K. X. Wu, Z. H. Xiong, Q. Jin, and D. X. Li. 2005. SARS patients-derived human recombinant antibodies to S and M proteins efficiently neutralize SARS-coronavirus infectivity. Biomed. Environ. Sci. 18:363-374. [PubMed] [Google Scholar]
- 31.Luo, C., H. Luo, S. Zheng, C. Gui, L. Yue, C. Yu, T. Sun, P. He, J. Chen, J. Shen, X. Luo, Y. Li, H. Liu, D. Bai, J. Shen, Y. Yang, F. Li, J. Zuo, R. Hilgenfeld, G. Pei, K. Chen, X. Shen, and H. Jiang. 2004. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem. Biophys. Res. Commun. 321:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo, H., D. Wu, C. Shen, K. Chen, X. Shen, and H. Jiang. 2006. Severe acute respiratory syndrome coronavirus membrane protein interacts with nucleocapsid protein mostly through their carboxyl termini by electrostatic attraction. Int. J. Biochem. Cell. Biol. 38:589-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, H., F. Ye, K. Chen, X. Shen, and H. Jiang. 2005. SR-rich motif plays a pivotal role in recombinant SARS coronavirus nucleocapsid protein multimerization. Biochemistry 44:15351-15358. [DOI] [PubMed] [Google Scholar]
- 34.Masters, P. S. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macnaughton, M. R., and H. A. Davies. 1980. Two particle types of avian infectious bronchitis virus. J. Gen. Virol. 47:365-372. [DOI] [PubMed] [Google Scholar]
- 36.McPhillips, T. M., S. E. McPhillips, H. J. Chiu, A. E. Cohen, A. M. Deacon, P. J. Ellis, E. Garman, A. Gonzalez, N. K. Sauter, R. P. Phizackerley, S. M. Soltis, and P. Kuhn. 2002. Blu-Ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9:401-406. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 38.Narayanan, K., C. J. Chen, J. Maeda, and S. Makino. 2003. Nucleocapsid-independent specific viral RNA packing via viral envelope protein and viral RNA signal. J. Virol. 77:2922-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuman, B. W., B. D. Adair, C. Yoshioka, J. D. Quispe, G. Orca, P. Kuhn, R. A. Milligan, M. Yeager, and M. Buchmeier. 2006. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J. Virol. 80:7918-7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 42.Painter, J., and E. A. Merritt. 2006. TLSMD web server for the generation of multi-group TLS models. Appl. Crystallogr. 39:109-111. [Google Scholar]
- 43.Peng, H., L. T. Yang, L. Y. Wang, J. Li, J. Huang, Z. Q. Lu, R. A. Koup, R. T. Bailer, and C. Y. Wu. 2006. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology 351:466-475. [DOI] [PMC free article] [PubMed]
- 44.Read, R. J. 2001. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D 57:1373-1382. [DOI] [PubMed] [Google Scholar]
- 45.Reed, M. L., B. K. Dove, R. M. Jackson, R. Collins, G. Brooks, and J. A. Hiscox. 2006. Delineation and modeling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic 7:833-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robbins, S. G., M. F. Frana, J. J. McGowan, J. F. Boyle, and K. V. Holmes. 1986. RNA-binding proteins of MHV: detection of monomeric and multimeric N protein with an RNA overlay protein blot assay. Virology 150:402-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Risco, C., I. M. Anton, L. Enjuanes, and J. L. Carrascosa. 1996. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J. Virol. 70:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spann, K. M., J. E. Vickers, and R. J. G. Lester. 1995. Lymphoid organ virus of Penaeus-Monodon from Australia. Dis. Aquat. Organisms 23:127-134. [Google Scholar]
- 49.Stohlman, S. A., R. S. Baric, G. N. Nelson, L. H. Soe, L. M. Welter, and R. J. Deans. 1988. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J. Virol. 62:4288-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surjit, M., R. Kumar, R. N. Mishra, M. K. Reddy, V. T. Chow, and S. K. Lal. 2005. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J. Virol. 79:11476-11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surjit, M., B. Liu, V. T. Chow, and S. K. Lal. 2006. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J. Biol. Chem. 281:10669-10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Surjit, M., B. Liu, P. Kumar, V. T. K. Chow, and S. K. Lal. 2004. The nucleocapsid protein of the SARS coronavirus is capable of self association through a C-terminal 209 amino acid interaction domain. Biochem. Biophys. Res. Commun. 317:1030-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sylvester-Hvid, C., M. Nielsen, K. Lamberth, G. Røder, S. Justesen, C. Lundegaard, P. Worning, H. Thomadsen, O. Lund, S. Brunak, and S. Buus. 2004. SARS CTL vaccine candidates: HLA supertype-, genome-wide scanning, and biochemical validation. Tissue Antigens 63:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan, Y. J., S. G. Lim, and W. Hong. 2005. Characterization of viral proteins encoded by the SARS-coronavirus genome. Antivir. Res. 65:69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by coexpression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verma, S., V. Bednar, A. Blount, and B. G. Hogue. 2006. Identification of functionally important negatively charged residues in the carboxy end of mouse hepatitis coronavirus A59 nucleocapsid protein. J. Virol. 80:4344-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Y., X. Wu, Y. Wang, B. Li, H. Zhou, G. Yuan, Y. Fu, and Y. Luo. 2004. Low stability of nucleocapsid protein in SARS virus. Biochemistry 43:11103-11108. [DOI] [PubMed] [Google Scholar]
- 58.Wootton, S. K., R. R. Rowland, and D. Yoo. 2002. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J. Virol. 76:10569-10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.You, J., B. K. Dove, L. Enjuanes, M. L. DeDiego, E. Alvarez, G. Howell, P. Heinen, M. Zambon, and J. A. Hiscox. 2005. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J. Gen. Virol. 86:3303-3310. [DOI] [PubMed] [Google Scholar]
- 60.Yu, I. M., M. L. Oldham, J. Zhang, and J. Chen. 2006. Crystal structure of the SARS coronavirus nucleocapsid protein dimerization domain reveals evolutionary linkage between Corona- and Arteriviridae. J. Biol. Chem. 281:17134-17139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou, M., A. K. Williams, S. I. Chung, L. Wang, and E. W. Collisson. 1996. The infectious bronchitis virus nucleocapsid protein binds RNA sequences in the 3′ terminus of the genome. Virology 217:191-199.8599203 [Google Scholar]
- 62.Zhou, M., and E. W. Collisson. 2000. The amino and carboxyl domains of the infectious bronchitis virus nucleocapsid protein interact with 3′ genomic RNA. Virus Res. 67:31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]