Abstract
All gammaretroviruses, including murine leukemia viruses (MuLVs), feline leukemia viruses, and gibbon-ape leukemia virus, encode an alternate, glycosylated form of Gag polyprotein (glyco-Gag or gPr80gag) in addition to the polyprotein precursor of the viral capsid proteins (Pr65gag). gPr80gag is translated from an upstream in-frame CUG initiation codon, in contrast to the AUG codon used for Pr65gag. The role of glyco-Gag in MuLV replication has been unclear, since gPr80gag-negative Moloney MuLV (M-MuLV) mutants are replication competent in vitro and pathogenic in vivo. However, reversion to the wild type is frequently observed in vivo. In these experiments, in vivo inoculation of a gPr80gag mutant, Ab-X-M-MuLV, showed substantially lower (2 log) initial infectivity in newborn NIH Swiss mice than that of wild-type virus, and revertants to the wild type could be detected by PCR cloning and DNA sequencing as early as 15 days postinfection. Atomic force microscopy of Ab-X-M-MuLV-infected producer cells or of the PA317 amphotropic MuLV-based vector packaging line (also gPr80gag negative) revealed the presence of tube-like viral structures on the cell surface. In contrast, wild-type virus-infected cells showed the typical spherical, 145-nm particles observed previously. Expression of gPr80gag in PA317 cells converted the tube-like structures to typical spherical particles. PA317 cells expressing gPr80gag produced 5- to 10-fold more infectious vector or viral particles as well. Metabolic labeling studies indicated that this reflected enhanced virus particle release rather than increased viral protein synthesis. These results indicate that gPr80gag is important for M-MuLV replication in vivo and in vitro and that the protein may be involved in a late step in viral budding or release.
Moloney murine leukemia virus (M-MuLV) is a well-studied replication-competent oncogenic retrovirus of the gammaretrovirus genus. Gammaretroviruses are unique among retroviruses in that they encode an alternate, glycosylated form of Gag polyprotein (glyco-Gag or gPr80gag for M-MuLV) in addition to the polyprotein precursor of the internal capsid proteins (Pr65gag for M-MuLV) (2, 5). Glyco-Gag was first recognized as a cell surface antigen present on AKR MuLV-induced tumor cells (16, 28), but it subsequently became apparent that all MuLVs carry gPr80gag (5, 7, 20, 25) and that gPr80gag differs from Pr65gag in that it has additional peptide sequences at the amino terminus (6). gPr80gag is translated from the same unspliced full-length viral mRNA as that for Pr65gag, by way of an upstream in-frame CUG initiation codon relative to the AUG codon used for Pr65gag (23). The N-terminal extension in gPr80gag comprises 88 amino acids, including a signal peptide that targets gPr80gag to the rough endoplasmic reticulum for glycosylation and export to the cell surface. At the cell surface, gPr80gag is cleaved in the CA domain (5, 16, 22), and the amino-terminal fragment adopts a type II integral membrane configuration in which the N terminus is inserted back into the cytosol (11). While large amounts of glyco-Gag (gPr80gag or its cleavage fragments) are not present in virions (1, 5), small amounts can be detected with glyco-Gag-specific antisera (9). In infected cells, translation of gPr80gag and Pr65gag is approximately equimolar (5).
Despite its conservation in all gammaretroviruses, the function of glyco-Gag has been unclear. M-MuLV mutants defective in expression of gPr80gag are replication competent in vitro (3, 8, 12, 26), although there was some suggestion of physical differences in gPr80gag-negative virions (8). In vivo, infection of gPr80gag-negative M-MuLV results in the appearance of wild-type revertants in tumors (3, 4) or at preleukemic times (4). For the neuropathic MuLV recombinant FrCasE, glyco-Gag has been shown to be a major pathogenic determinant associated with early efficient infection (10, 21, 22). Rapid in vivo reversion of glyco-Gag-negative FrCasE was also observed (4). Thus, glyco-Gag is important for MuLV replication in vivo, although it may not be absolutely required.
In these experiments, we have continued our in vivo and in vitro investigations on the role of gPr80gag in M-MuLV replication. The results confirm the importance of gPr80gag for efficient replication in vivo, and they indicate that this protein is involved in a late step in viral budding or assembly.
MATERIALS AND METHODS
Cell culture and viruses.
Ab-X-M-MuLV is a mutant M-MuLV containing a stop codon in the gPr80gag reading frame (UAU→UAG at nucleotide [nt] 608) between the start codons for gPr80gag and Pr65gag (8). 43D and 17-5 cells are NIH 3T3 fibroblasts stably infected with wild-type M-MuLV and Ab-X-M-MuLV, respectively. They were maintained in Dulbecco modified Eagle's medium (DMEM) supplemented with 10% calf serum and antibiotics. PA317/BAG cells are PA317 amphotropic MuLV-based packaging cells (18) stably transfected with an M-MuLV-based vector expressing bacterial beta-galactosidase and the bacterial neomycin resistance gene (BAG vector) (24). They were maintained in DMEM containing 10% fetal bovine serum (FBS), antibiotics, and 400 μg/ml G418. PA317/BAG 8065-2 and PA317/BAG ΔMCS cells are PA317/BAG cells stably transfected with the p8065-2 and pZeoSV ΔMCS plasmids, respectively. These cells were maintained in the same medium as that for PA317/BAG, but with the addition of 25 μg/ml Zeocin to select for stable expression of the pZeoSV-based plasmids.
To prepare virus or vector stocks, 5 × 105 17-5 or 43D cells or 1 × 106 PA317/BAG-based cells were seeded on 10-cm plates in growth medium. Twenty-four hours later, the medium was changed. Twenty-four hours after the medium change, supernatants were collected and passed through a 0.45-μm filter. They were then divided into aliquots, frozen, and stored at −80°C.
Plasmids.
The gPr80gag expression plasmid p8065-2 was generated by first cloning M-MuLV gag sequences from nt 357 to 2235 (between restriction sites for Asp718 and EcoRI) into the pZeoSV expression plasmid (Invitrogen, Carlsbad, CA). The CUG start codon for gPr80gag was replaced by an AUG start codon by site-directed mutagenesis, resulting in the p8065-2 plasmid. pZeoSV ΔMCS is the pZeoSV plasmid from which the multiple cloning site has been removed.
Titrations.
Titrations of wild-type M-MuLV and Ab-X-M-MuLV stocks were performed as described previously (27), using a focal immunofluorescence assay. Briefly, 7 × 104 NIH 3T3 cells were seeded per 6-cm plate 24 h prior to infection. Dilutions of the viral stocks were then adsorbed to the cells in the presence of 2 μg/ml Polybrene, followed by the addition of growth medium. At 4 days postinfection, plates were incubated for 1 h at 4°C with the 548 Gag monoclonal antibody, washed with phosphate-buffered saline (PBS) supplemented with 2% FBS, and then stained for 30 min at 4°C with a secondary fluorescein-conjugated goat anti-mouse immunoglobulin G (IgG) antibody (1:200 dilution in 2% FBS-PBS; ICN). Plates were washed three times, and foci of immunofluorescence were counted with a UV microscope.
For titration of BAG vector stocks, NIH 3T3 cells were plated 24 h prior to infection at 2 × 105 cells/6-cm plate. Dilutions of the stocks were adsorbed to the cells as described above, and the plates were stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 2 days postinfection. For X-Gal staining, cultures were fixed with 0.05% glutaraldehyde in PBS for 5 min and incubated overnight at 25°C in PBS containing 400 μg/ml X-Gal, and colonies of blue cells were counted under the microscope.
Inoculation of animals.
Neonatal NIH Swiss mice were inoculated intraperitoneally with 0.2 ml of virus (ca. 105 IU) as described previously (15). Animals were sacrificed at different days postinfection, and cells from the bone marrow, spleen, and thymus were harvested. Single-cell suspensions in PBS were obtained and used in infectious center assays. Briefly, different dilutions of the cell suspensions were cocultured with NIH 3T3 cell cultures for 24 h, after which the hematopoietic cells were removed. Focal areas of infection were identified by immunofluorescence 4 days later, as described above.
DNA sequencing.
To test for reversion of gPr80gag-negative virus in vivo, PCR amplification was carried out on DNAs from cells obtained from animals at different times postinfection. The PCR primers flanked the region of the nonsense mutation in Ab-X-M-MuLV. The forward primer corresponded to nt 100 to 121 (GGTCTCCTCTGAGTGATTGACT), and the reverse primer corresponded to nt 656 to 635 (GGTCAAACTTAAGGGAGTGGTA). The sequencing reaction was done on 500 ng of DNA, using an ABI BigDye Terminator kit.
Western blots.
To quantify virus release, whole-cell lysates of PA317/BAG or PA317/BAG 8065-2 cells grown on 6-cm dishes for 24 h were harvested in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 50 mM Tris, pH 8.0). Prior to cell harvesting, 5 ml of tissue culture supernatant from these cells was subjected to ultracentrifugation at 45,000 rpm for 1 h in an SW41 rotor, and the pellets were resuspended in RIPA buffer. Samples representing equal amounts of starting cells were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to a nitrocellulose membrane. The membrane was incubated with a primary rabbit polyclonal antibody for M-MuLV p30 (CA) protein (1:3,333 dilution) (19), followed by washing and incubation with a horseradish peroxidase-conjugated secondary antibody (donkey anti-rabbit IgG; Amersham). Bands were detected with a Pico detection kit (Pierce) and X-ray film.
Metabolic labeling.
Cells were seeded at 50% confluence in 10-cm dishes the night before metabolic labeling. On the day of labeling, the growth medium was aspirated and replaced with DMEM lacking methionine and cysteine (Sigma), followed by incubation for 10 min at 37°C. The cultures were then aspirated, and 2 ml of methionine- and cysteine-free DMEM containing 300 μCi of [35S]methionine-[35S]cysteine protein labeling mix (NEN) was added, followed by incubation at 37°C for 15 or 60 min. At the end of the pulse period, the radioactive medium was removed, the cells were washed with 5 ml Tris-buffered saline, regular growth medium was then added, and incubation was continued at 37°C. At 0, 1, 2, and 4 h of chase, the released virus was harvested from one dish per time point, and cell lysates were also prepared. For released virus, growth medium (cell supernatants) was collected, passed through a 0.45-μm filter, and concentrated by ultracentrifugation in a Beckman SW41 rotor at 25,000 rpm for 1 h at 4°C. Pelleted virus was resuspended in 200 μl lysis buffer (1% Triton X-100, 1% sodium deoxycholate, 30 mM NaCl, 25 mM Tris-HCl, pH 8.0). For cell lysates, cells were washed with 5 ml cold Tris-buffered saline, followed by the addition of 1 ml lysis buffer. After incubation for 5 min at room temperature, the lysate was harvested and nuclei were removed by centrifugation (10 min in a microcentrifuge).
Continuous labeling was performed similarly to pulse-chase labeling, except that the labeling medium consisted of methionine- and cysteine-free DMEM containing 10% complete DMEM. Harvest of released virus and cell lysates was done as for the pulse-chase experiments.
The amount of radioactivity in released virus or intracellular Gag protein was determined by immunoprecipitation with a rabbit polyclonal antiserum raised against M-MuLV CA (5), as described previously. Half of the cell lysates or virus supernatants from each time point were incubated with diluted antiserum for 2 h at 4°C in a total volume of 500 μl, with rocking. Protein A and G agarose beads (Roche Diagnostics) were diluted 1:20 in lysis buffer, and 100 μl of the diluted beads was added to each sample, followed by incubation at 4°C for 2 h on a rocker. The beads were then washed three times with lysis buffer, dried, and resuspended in SDS sample buffer. The samples were boiled for 10 min, the beads were removed, and the samples were then loaded onto 10% polyacrylamide gels in SDS. SDS-PAGE was carried out, followed by drying and autoradiography of the gels.
AFM.
Atomic force microscopy (AFM) of virus-infected or vector-producing cells was performed as described previously (13). Cells were grown on glass coverslips. They were then fixed with 0.05% glutaraldehyde in PBS and postfixed with a 1% solution of osmium tetroxide in double-distilled H2O. The samples were dehydrated and imaged as previously described (13).
RESULTS
Requirement of glyco-Gag for efficient M-MuLV infection in vivo.
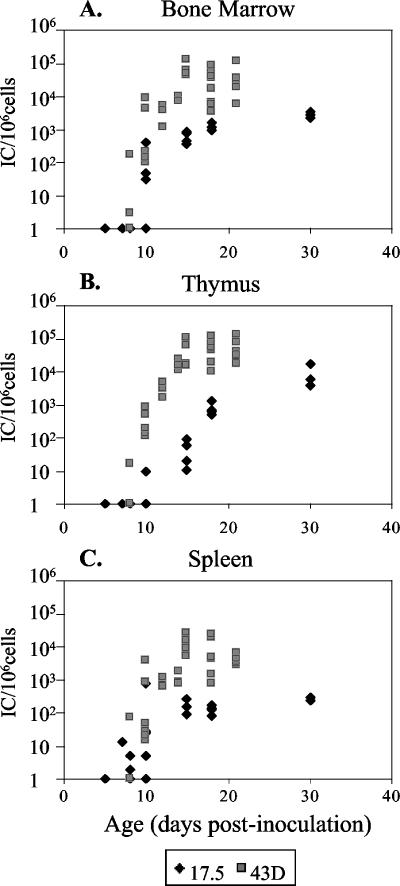
To test the biological importance of gPr80gag for M-MuLV infection in vivo, we employed a gPr80gag mutant that we previously generated, Ab-X-M-MuLV (Fig. 1) (8). Infectious stocks of wild-type and Ab-X-M-MuLV (titrated in vitro) virus were inoculated intraperitoneally into newborn NIH Swiss mice, and the levels of infection in the bone marrow cells at different times postinoculation were determined by infectious center assays as described previously (27). As shown in Fig. 2, animals inoculated with wild-type M-MuLV established maximal levels of infection in the bone marrow, spleen, and thymus by 15 days (ca. 105 infected cells per 106 cells assayed), as described previously (15). In contrast, animals inoculated with equivalent amounts of Ab-X-M-MuLV showed substantially lower levels of bone marrow infection (ca. 100-fold less) during the same time frame, and the levels of in vivo infection never reached the levels observed with wild-type M-MuLV (e.g., at 30 days). These results indicate that glyco-Gag is important for establishment of efficient infection in vivo by M-MuLV, and they confirm previous findings on the role of glyco-Gag in early in vivo infection by the neuropathic FrCasE MuLV (21).
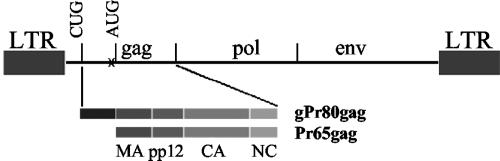
FIG. 1.
gPr80gag and Ab-X-M-MuLV. The relationship of the coding sequences for Pr65gag and gPr80gag is shown. gPr80gag is translated from the same reading frame as Pr65gag, but from an upstream CUG initiation codon. The location of the stop codon present in the genome of Ab-X-M-MuLV is shown (X); it is upstream from the Pr65gag AUG codon.
FIG. 2.
Establishment of in vivo infection by Ab-X-M-MuLV. Neonatal NIH Swiss mice were inoculated intraperitoneally with wild-type M-MuLV or Ab-X-M-MuLV. At different days postinoculation, the animals were sacrificed, and single-cell suspensions were prepared from bone marrow, thymus, and spleen. The levels of infection (number of infectious centers [ICs]/106 cells plated) are shown. Each point represents the result for one animal. Shaded boxes, wild-type M-MuLV (43D cells); solid diamonds, Ab-X-M-MuLV (17-5 cells). (A) Bone marrow; (B) thymus; (C) spleen.
The Ab-X-M-MuLV mutant differs from wild-type M-MuLV by the presence of a single base change leading to the presence of a nonsense codon in the gPr80gag reading frame upstream from the initiation codon for Pr65gag (8). Thus, it seemed possible that some or all of the infectious viruses detected in the Ab-X-M-MuLV-infected mice might represent revertants to wild-type gPr80gag-positive virus, since a single base substitution could restore the gPr80gag reading frame. Therefore, DNAs were prepared from bone marrow, splenocytes, or thymocytes from Ab-X-M-MuLV-infected animals at different days postinoculation, and the 5′ noncoding region and part of gag were PCR amplified with primers specific for M-MuLV. PCR products from independent amplifications were then sequenced, and the results are shown in Table 1. In three out of four animals analyzed, revertants to the wild-type gPr80gag codon were observed in at least one tissue, and for two of the animals, reversion was extensive. These results supported the importance of gPr80gag for in vivo infectivity, since selection for wild-type revertants of Ab-X-M-MuLV was observed as soon as 15 days postinfection. These results also suggested that the differences in infectivity in vivo between wild-type MuLV and Ab-X-M-MuLV were even greater than those apparent in Fig. 2, since the infectious center assays with the Ab-X-M-MuLV-infected animals would have scored both the mutant virus and wild-type revertants. Indeed, of the four animals analyzed for Table 1, the animal that showed only input virus and no revertants in any tissue (animal 3) also had the lowest levels of infection. Rapid in vivo reversion of gPr80gag-negative FrCasE (14) and F-MuLV (4) mutants has also been described previously.
TABLE 1.
Appearance of revertant gPr80gag-positive viruses in Ab-X-M-MuLV-infected micea
| Animal | Age (days) | Tissue | Level of infection (IC/106 cells)b | Virusc |
|---|---|---|---|---|
| 1 | 15 | Bone marrow | 3,000 | R, R |
| Thymus | 50 | I, I | ||
| Spleen | 580 | R, R, R | ||
| 2 | 15 | Bone marrow | 90 | R, R, R |
| Thymus | 3,100 | I, I, I | ||
| Spleen | 4,400 | R, R, R | ||
| 3 | 18 | Bone marrow | 5 | I, I, I |
| Thymus | 40 | I, I, I | ||
| Spleen | 60 | I, I, I | ||
| 4 | 18 | Bone marrow | 4 | I, R/I, I |
| Thymus | 380 | I, I, I | ||
| Spleen | 4,100 | I, I, I |
Neonatal NIH Swiss mice were inoculated intraperitoneally with Ab-X-M-MuLV as described in Materials and Methods. On different days of age, single-cell suspensions of bone marrow, thymuses, and spleens were prepared.
The levels of infection (number of infectious centers [IC]/106 cells) were determined by infectious center assays.
DNA was extracted from the harvested cells, and the region surrounding the nonsense mutation in Ab-X-M-MuLV was PCR amplified and sequenced. I, input sequence; R, sequence showing reversion of the stop codon, resulting in reversion to gPr80gag-positive virus. Multiple entries indicate the sequencing results for independent PCR amplifications. In one case, both input and revertant sequences were detected (R/I).
Glyco-Gag is involved in a late step in virus release.
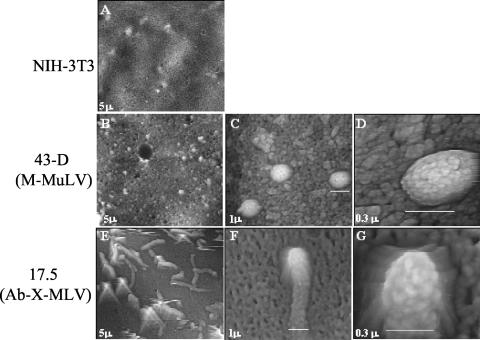
While it is clear that gPr80gag-negative viruses such as Ab-X-M-MuLV are infectious in vitro and in vivo, our previous studies suggested that these virions might differ somewhat from wild-type M-MuLV in that they have a slightly decreased (fivefold) specific infectivity and a slightly lower buoyant density (8). We imaged NIH 3T3 cells productively infected with wild-type M-MuLV (43D cells) or with Ab-X-M-MuLV (17-5 cells) by AFM as described previously (13). AFM is similar to scanning electron microscopy in that surface contours of objects (e.g., cells) are visualized, but the harsh fixation and shadowing techniques necessary for scanning electron microscopy are not employed. As shown in Fig. 3A, uninfected NIH 3T3 cells showed relatively smooth cell surfaces. In contrast, 43D cells showed characteristic spherical particles on the cell surface of ca. 145 nm (Fig. 3B to D), and we have previously established that these represent M-MuLV virions associated with the infected cell (13, 14). In contrast, some Ab-X-M-MuLV-infected 17-5 cells showed the presence of long tube-like structures on the cell surface. The tube-like structures had the same approximate width as a wild-type M-MuLV virion (ca. 145 nm), and the substructure of the tips of the tubes closely resembled that of the spherical M-MuLV particle (Fig. 3 E to G) (13). This suggested that when gPr80gag is not expressed in M-MuLV-infected cells, there may be a partial defect in a late step in virus release, resulting in a temporal delay in “pinching off” of virions as they bud from the surface. Alternatively, glycosylated Gag in the plasma membrane might enhance membrane deformation to allow the formation of spherical particles. Also, incorporation of small numbers of glyco-Gag proteins into virus particles (9) might be important for proper assembly of Gag and Gag-Pol polyproteins into spherical particles.
FIG. 3.
AFM of infected cells. AFM was performed on NIH 3T3 cells infected with wild-type M-MuLV (43D) or Ab-X-M-MuLV (17-5). (A) Uninfected NIH 3T3 cell; (B to D) wild-type M-MuLV-infected 43D cell at three magnifications; (E to G) Ab-X-M-MuLV-infected 17-5 cell at three magnifications. The widths of the fields shown are indicated in micrometers. The white bars indicate the widths of the particles. The surface viral structures are typical of infected cells and absent from uninfected NIH 3T3 cells, as described previously (13).
The PA317 packaging line shows the late viral release defect.
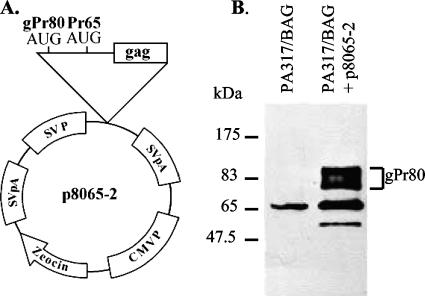
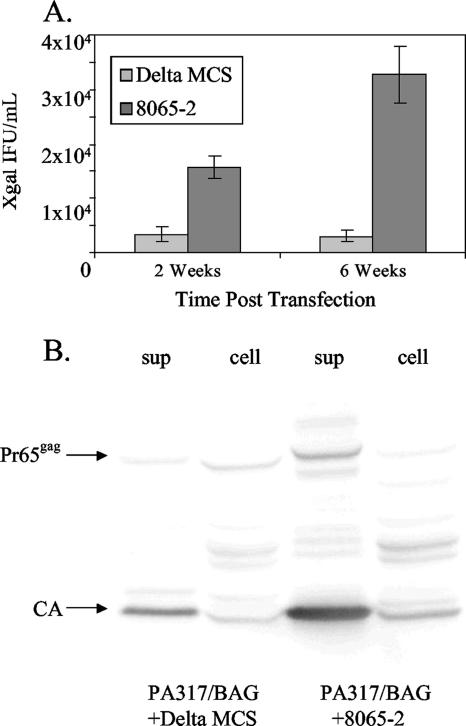
One of the challenges in these experiments was that not all of the 17-5 cells showed the tube-like virus structures, and the relative frequency of cells showing this phenotype varied depending on the experiment (5 to 50%). Therefore, we sought a gPr80gag-negative MuLV producer cell that showed the tube phenotype more consistently. Virtually all MuLV-based retroviral vector packaging cell lines express Pr65gag (for production of the capsid proteins) but not gPr80gag, since the gPr80gag coding sequences begin in the region containing the RNA packaging (Ψ) sequences (17). If these packaging cells express a transfected MuLV-based vector that does not itself encode gPr80gag, then the packaging cells will produce the vector particles in the absence of gPr80gag. We found that the amphotropic MuLV vector packaging line PA317 (18) consistently showed the tube phenotype. Figure 4A to C show AFM of PA317 cells stably expressing an M-MuLV-based vector expressing the bacterial β-galactosidase gene (BAG vector) (24), PA317/BAG. The surfaces of these cells were uniformly covered with viral “tubes,” and few spherical particles were evident. To test if the tube phenotype was related to the lack of gPr80gag expression in the PA317 cells, the PA317/BAG cells were stably transfected with an expression plasmid for gPr80gag, p8065-2 (Fig. 5A). In p8065-2, the CUG initiation codon for gPr80gag was converted to a more efficient AUG codon, and expression was driven from a simian virus 40 early viral promoter from the backbone expression plasmid pZeoSV. This leads to high-level expression of gPr80gag in transfected cells (Fig. 5B). When PA317/BAG cells were transfected with p8065-2 and selected for stable transfection by growth in Zeocin, the resulting PA317/BAG-8065-2 cells consistently showed typical 145-nm spherical virus particles on the surface instead of the tubes (Fig. 4D to F). These experiments strongly suggest that expression of gPr80gag in PA317/BAG cells converted the viral tube-like structures to spherical particles, consistent with a role for gPr80gag in a late step in viral budding or release.
FIG. 4.
AFM of PA317/BAG cells. AFM of the Am-MuLV-based retroviral vector packaging cell line PA317 is shown. These cells were stably transfected with the BAG vector plasmid and produced infectious BAG vector. (A to C) Surface of a PA317/BAG cell at three magnifications. (D to F) Surface of a PA317/BAG cell expressing gPr80gag (PA317/BAG 8065-2) at three magnifications. Note the presence of spherical particles instead of tubes.
FIG. 5.
gPr80gag expression plasmid. (A) Organization of the gPr80gag expression plasmid p8065-2. The CUG initiation codon has been converted to an AUG codon; the backbone expression plasmid is pZeoSV. (B) Western blotting of PA317/BAG versus PA317/BAG 8065-2, using two antibodies raised against the N terminus of gPr80gag (a gift of John Portis) (9). The location of gPr80gag is indicated.
gPr80gag increases virus release.
We also tested if gPr80gag results in enhanced virus release. PA317/BAG cells were transfected in parallel with p8065-2 or the backbone vector pZeoSV ΔMCS, and stably transfected cells were selected by growth in medium containing Zeocin. At the end of selection (2 weeks), supernatants from equal numbers of transfected cells were harvested, and the amounts of infectious BAG vector released were measured by titration of the supernatants on NIH 3T3 cells, followed by X-Gal staining for β-galactosidase (Fig. 6A). The p8065-2-transfected packaging cells produced about five times as much infectious BAG vector as those transfected with the backbone vector. The transfected cultures were passaged for an additional 4 weeks, at which time the amounts of vector released were again titrated. In the passaged cultures, the difference in infectious vector production between gPr80gag-positive packaging cells was 10-fold. The number of virions released from the PA317/BAG cells at 6 weeks was also assessed by SDS-PAGE and Western blotting (Fig. 6B). Consistent with the infectivity measurements, the gPr80gag-expressing PA317/BAG cells released more virus particles than did the nonexpressing cells. While more viral protein was produced from the PA317/BAG-8065-2 cells, the difference did not appear to be quite as much as the 10-fold difference evident in the infectivity assay. This might have been influenced by the fact that packaging cells such as PA317 cells are continually releasing viral particles regardless of whether they are packaging a vector RNA or not. In sum, these results supported the conclusions from the AFM experiments that gPr80gag facilitates a late stage in MuLV budding or release.
FIG. 6.
Vector release from PA317/BAG cells. (A) PA317/BAG cells were transfected with the gPr80gag expression plasmid p8065-2 or the backbone plasmid pZeoSV ΔMCS, and the cells were grown in medium containing Zeocin to select for stable transfectants. After 2 weeks (when selection was complete), supernatants were collected from the cultures and assayed for production of infectious BAG vector as described in Materials and Methods. The cultures were passaged for an additional 4 weeks in the presence of Zeocin, and the amounts of infectious BAG vector released were again assayed. The titrations were performed with four replicates for each time point. (B) The amount of vector particles released from transfected cultures at the 6-week time point was determined by SDS-PAGE and Western blotting for viral CA (p30) protein. The amount of Gag protein in the cell extracts is also shown; samples representing equal amounts of cells were loaded in all lanes.
Virus particles released from the PA317/BAG-8065-2 cells also showed detectable levels of uncleaved Pr65gag, while virus particles released from PA317/BAG cells did not. This would be consistent with a more rapid release of virus when glyco-Gag is expressed, such that some of the Gag protein is in uncleaved form.
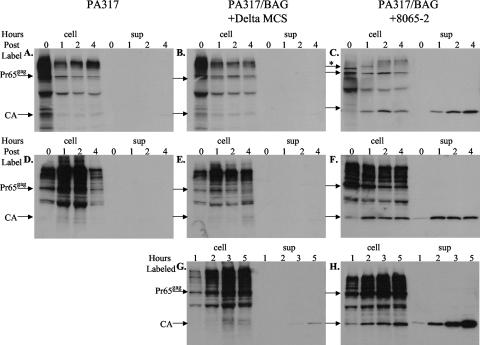
To more directly test if virus release from PA317/BAG cells was enhanced in the presence of glyco-Gag, PA317/BAG cells transfected with either p8065-2 or the backbone vector (Fig. 6) were pulse labeled with [35S]methionine and cysteine for 15 min, followed by a chase with unlabeled methionine and cysteine for periods of up to 4 h, and the amounts of labeled virus released were quantified by immunoprecipitation of viral pellets with anti-CA antibody, followed by SDS-PAGE and autoradiography. As shown in the top panels of Fig. 7, virus release (sup) from cells expressing glyco-Gag was detected (Fig. 7C), while the amount released from PA317 or PA317/BAG cells was not detectable. Even when the pulse period was increased to 1 hour (Fig. 7, D to F), the amount of virus released in the absence of glyco-Gag was below detection. Finally, cells were continuously radiolabeled (Fig. 7, G to H), and virus release from PA317/BAG cells could be detected (Fig. 7G), although it was much less (<10-fold) than that from cells expressing glyco-Gag (Fig. 7H). In addition, the relative labeling of Gag protein in the cells was similar in cells expressing or not expressing glyco-Gag, as evidenced from labeling of Pr65gag (left four lanes in each panel). These results confirmed that glyco-Gag increased the rate of virus release from PA317/BAG cells without altering the overall rate of Gag protein synthesis.
FIG. 7.
Virus release by metabolic labeling. PA317, PA317/BAG + ΔMCS, and PA317/BAG + 8065-2 cells, as described in the legend to Fig. 6, were labeled with [35S]methionine-cysteine by a 15-min pulse-chase (A to C), 60-min pulse-chase (D to F), or continuous labeling (G and H). One-half of the released virus or cell lysate for each sample was analyzed by SDS-PAGE. The lengths of the chases or continuous labels are indicated in hours at the top of each panel (0 to 4 h); cell lysates (cell) are shown in the left four lanes, and released viruses (sup) are shown in the right four lanes. The locations of Pr65gag and cleaved CA protein in released virus are indicated. The more slowly migrating radioactivity in the cell lysates is nonspecific binding; a partial proteolytic cleavage product of Pr65gag is evident in the cell lysates as well (6). gPr80gag is evident in the cell lysate from the 15-min pulse (*); during the chase, this protein was glycosylated to form higher-molecular-weight products (6). Autoradiographic exposures were done for 2 weeks at −80°C.
DISCUSSION
In this report, we further investigated the role of glyco-Gag in M-MuLV replication in vivo and in vitro. As described in the introduction, glyco-Gag is a conserved feature of gammaretroviruses, which strongly suggests that it plays a significant role in viral replication. On the other hand, glyco-Gag-negative MuLV mutants are replication competent in vitro and in vivo. For infection of newborn NIH Swiss mice, we found that the gPr80gag-negative M-MuLV mutant Ab-X-M-MuLV showed substantially (2 log) lower viral infection levels than those of wild-type M-MuLV in hematopoietic tissues at early times. Moreover, by 15 days, revertants of Ab-X-M-MuLV to wild-type gPr80gag-positive virus could be detected. These results indicated that gPr80gag is important for efficient in vivo infection by M-MuLV, and they confirm similar conclusions reached by other investigators (4, 21).
AFM of cells producing MuLV particles in the absence of gPr80gag (Ab-X-M-MuLV-infected NIH 3T3 cells [17-5 cells] and the Am-MuLV retroviral packaging line PA317) showed the presence of viral tube-like structures at the cell surface instead of spherical particles. The tube phenotype was due to the lack of gPr80gag as established by vectored expression of gPr80gag in PA317 cells, which resulted in the appearance of spherical particles at the surface instead of the tubes. This indicated that gPr80gag may be involved in a late step in viral budding or release—perhaps in “pinching off” of spherical virions during budding at the cell surface. A role for gPr80gag in virus budding or release was also supported by the finding that PA317 cells expressing gPr80gag produce 5 to 10 times more viral vector particles than do parental PA317 cells (measured either by infectivity or by viral protein release).
The relative defect in virion release from M-MuLV-infected cells lacking gPr80gag appears to be more a kinetic effect or delay in release than an absolute block. We previously noted a somewhat different buoyant density of Ab-X-M-MuLV virions compared to wild-type particles (8), but on the other hand, the relative infectivities of the two viruses in in vitro plaque assays were similar. The fact that Ab-X-M-MuLV virions are infectious strongly implies that maturation and cleavage of the viral polyproteins (Pr65gag, Pr180gag-pol, and Pr80env) occur at some level, since cleavage is necessary for the generation of infectious virus. Indeed, cleaved CA protein is found in particles from gPr80gag-negative cells (e.g., see Fig. 6). AFM of Ab-X-M-MuLV virions released from 17-5 cells revealed standard 145-nm spherical particles, not tubes or multiple virions (Y. Kuznetsov and A. Low, unpublished data). Thus, virions released from gPr80gag-negative cells appear to be normal, although they are released less efficiently in the absence of the protein.
The viral release or budding defect in gPr80gag-negative MuLV mutants appears to be distinct from that of well-characterized late domain mutants (30). For M-MuLV, the L domain maps to the p12 domain of Pr65gag, not gPr80gag (31). The L domain (containing PPPY motifs for M-MuLV) (31) is responsible for binding of the Pr65gag polyprotein to components of the cellular vesicular export machinery, which is important for its delivery to the plasma membrane for assembly into virions. L domain mutants of M-MuLV also show a defect in virion release, but in this case the cell surfaces are covered with spherical particles that appear tethered to each other and to the cell (29) rather than the tubes observed in gPr80gag-negative infected cells. Moreover, M-MuLVs with L domain mutations are noninfectious, and they have immature cores indicative of uncleaved Gag and Gag-Pol polyproteins (31). An M-MuLV deleted of the entire p12 domain of Gag does show tube-like viral structures on the cell surface (29), but they differ from the tubes described here in their lack of infectivity.
The mechanism by which gPr80gag facilitates virion budding or release is currently unclear. gPr80gag is a type II integral membrane protein, with the amino-terminal gPr80gag unique domain exposed in the cytosol (11). Large amounts of gPr80gag (or its cleavage products) are not present in virions (5), although small amounts have been detected (9). One possible model could be that gPr80gag interacts with a cellular protein(s), localizing that protein to the plasma membrane, where it may facilitate pinching off or budding of nascent virions. It seems possible that gPr80gag could facilitate such a process but that in the absence of gPr80gag, the cellular protein could still function at some level. Depending on the relative abundance (or intracellular distribution) of such a cellular protein and the relative rate of viral production, the absence of gPr80gag might or might not have a visible effect on virus budding or release. Such a model could explain the variability of the “tube” phenotype in the AFM of Ab-X-M-MuLV-infected NIH 3T3 cells (17-5 cells). Indeed, in 17-5 cell cultures, if cells showed the tube phenotype, they had large numbers of tubes present (Y. Kuznetsov and S. Datta, unpublished data). In the future, it will be important to identify cellular proteins that interact with gPr80gag.
In these experiments, restoration of gPr80gag in PA317 cells with the p8065-2 expression vector was important in establishing the role of gPr80gag in viral budding or release. While it was possible to prepare stable p8065-2 transfectants of PA317 cells (or PA317 derivatives expressing retroviral vectors, such as the BAG vector), we could not generate stable p8065-2 transfectants of standard NIH 3T3 cells or the NIH 3T3-based M-MuLV vector packaging line Ψ2 (17). One possible explanation could be that high-level expression of gPr80gag from p8065-2 could be toxic in those cells due to sequestration of the putative cellular interacting protein, if that protein is essential for cell survival. The more moderate levels of gPr80gag expressed from the CUG initiation codon of genuine M-MuLV mRNA might not lead to this toxicity. The reasons why PA317 cells are not affected by p8065-2 are unclear, since PA317 cells are NIH 3T3-based cells (18).
Finally, it should be noted that essentially all MuLV-based vector packaging lines do not express glyco-Gag, for the reasons described in Results. It seems possible that the expression of gPr80gag in packaging lines could increase the yield of infectious vectors from these cells, as shown in these experiments for the BAG vector. This could be useful for future gene therapy experiments employing gammaretrovirus-based vectors.
Acknowledgments
This work was supported by NIH grant R01CA32455.
We thank John Portis for the gift of antibodies. The support of the DNA sequencing shared resource of the Chao Family Comprehensive Cancer Center is acknowledged.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Aoki, T., R. B. Herberman, P. A. Johnson, M. Liu, and M. M. Sturm. 1972. Wild-type gross leukemia virus: classification of soluble antigens (GSA). J. Virol. 10:1208-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buetti, E., and H. Diggelmann. 1980. Murine leukemia virus proteins expressed on the surface of infected cells in culture. J. Virol. 33:936-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun, R., and H. Fan. 1994. Recovery of glycosylated gag virus from mice infected with a glycosylated gag-negative mutant of Moloney murine leukemia virus. J. Biomed. Sci. 1:218-223. [DOI] [PubMed] [Google Scholar]
- 4.Corbin, A., A. C. Prats, J. L. Darlix, and M. Sitbon. 1994. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J. Virol. 68:3857-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards, S. A., and H. Fan. 1979. Gag-related polyproteins of Moloney murine leukemia virus: evidence for independent synthesis of glycosylated and unglycosylated forms. J. Virol. 30:551-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards, S. A., and H. Fan. 1980. Sequence relationship of glycosylated and unglycosylated Gag polyproteins of Moloney murine leukemia virus. J. Virol. 35:41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, L. H., S. Dresler, and D. Kabat. 1977. Synthesis and glycosylation of polyprotein precursors to the internal core proteins of Friend murine leukemia virus. J. Virol. 24:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan, H., H. Chute, E. Chao, and M. Feuerman. 1983. Construction and characterization of Moloney murine leukemia virus mutants unable to synthesize glycosylated gag polyprotein. Proc. Natl. Acad. Sci. USA 80:5965-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujisawa, R., F. J. McAtee, C. Favara, S. F. Hayes, and J. L. Portis. 2001. N-terminal cleavage fragment of glycosylated Gag is incorporated into murine oncornavirus particles. J. Virol. 75:11239-11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujisawa, R., F. J. McAtee, K. Wehrly, and J. L. Portis. 1998. The neuroinvasiveness of a murine retrovirus is influenced by a dileucine-containing sequence in the cytoplasmic tail of glycosylated Gag. J. Virol. 72:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujisawa, R., F. J. McAtee, J. H. Zirbel, and J. L. Portis. 1997. Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: identification of differences in processing in vitro and in vivo. J. Virol. 71:5355-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goff, S. P., and L. I. Lobel. 1987. Mutants of murine leukemia viruses and retroviral replication. Biochim. Biophys. Acta 907:93-123. [DOI] [PubMed] [Google Scholar]
- 13.Kuznetsov, Y. G., S. Datta, N. H. Kothari, A. Greenwood, H. Fan, and A. McPherson. 2002. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV). Biophys. J. 83:3665-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuznetsov, Y. G., A. Low, H. Fan, and A. McPherson. 2005. Atomic force microscopy investigation of isolated virions of murine leukemia virus. J. Virol. 79:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lander, J. K., B. Chesebro, and H. Fan. 1999. Appearance of mink cell focus-inducing recombinants during in vivo infection by Moloney murine leukemia virus (M-MuLV) or the Mo+PyF101 M-MuLV enhancer variant: implications for sites of generation and roles in leukemogenesis. J. Virol. 73:5671-5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ledbetter, J., R. C. Nowinski, and S. Emery. 1977. Viral proteins expressed on the surface of murine leukemia cells. J. Virol. 22:65-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann, R., R. C. Mulligan, and D. Baltimore. 1983. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell 33:153-159. [DOI] [PubMed] [Google Scholar]
- 18.Miller, A. D., and C. Buttimore. 1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol. 6:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mueller-Lantzsch, N., and H. Fan. 1976. Monospecific immunoprecipitation of murine leukemia virus polyribosomes: identification of p30 protein-specific messenger RNA. Cell 9:579-588. [DOI] [PubMed] [Google Scholar]
- 20.Oroszlan, S., T. Copeland, M. Summers, and R. V. Gilden. 1972. Amino terminal sequences of mammalian type C RNA tumor virus group-specific antigens. Biochem. Biophys. Res. Commun. 48:1549-1555. [DOI] [PubMed] [Google Scholar]
- 21.Portis, J. L., R. Fujisawa, and F. J. McAtee. 1996. The glycosylated gag protein of MuLV is a determinant of neuroinvasiveness: analysis of second site revertants of a mutant MuLV lacking expression of this protein. Virology 226:384-392. [DOI] [PubMed] [Google Scholar]
- 22.Portis, J. L., G. J. Spangrude, and F. J. McAtee. 1994. Identification of a sequence in the unique 5′ open reading frame of the gene encoding glycosylated Gag which influences the incubation period of neurodegenerative disease induced by a murine retrovirus. J. Virol. 68:3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prats, A. C., G. De Billy, P. Wang, and J. L. Darlix. 1989. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J. Mol. Biol. 205:363-372. [DOI] [PubMed] [Google Scholar]
- 24.Price, J., D. Turner, and C. Cepko. 1987. Lineage analysis in the vertebrate nervous system by retrovirus-mediated gene transfer. Proc. Natl. Acad. Sci. USA 84:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schultz, A. M., E. H. Rabin, and S. Oroszlan. 1979. Post-translational modification of Rauscher leukemia virus precursor polyproteins encoded by the gag gene. J. Virol. 30:255-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartzberg, P., J. Colicelli, and S. P. Goff. 1983. Deletion mutants of Moloney murine leukemia virus which lack glycosylated Gag protein are replication competent. J. Virol. 46:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitbon, M., J. Nishio, K. Wehrly, D. Lodmell, and B. Chesebro. 1985. Use of a focal immunofluorescence assay on live cells for quantitation of retroviruses: distinction of host range classes in virus mixtures and biological cloning of dual-tropic murine leukemia viruses. Virology 141:110-118. [DOI] [PubMed] [Google Scholar]
- 28.Tung, J. S., T. Yoshiki, and E. Fleissner. 1976. A core polyprotein of murine leukemia virus on the surface of mouse leukemia cells. Cell 9:573-578. [DOI] [PubMed] [Google Scholar]
- 29.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan, B., A. Fassati, A. Yueh, and S. P. Goff. 2002. Characterization of Moloney murine leukemia virus p12 mutants blocked during early events of infection. J. Virol. 76:10801-10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]