Abstract
The human immunodeficiency virus type-1 (HIV-1) accessory protein Vif serves to neutralize the human antiviral proteins apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G (APOBEC3G [A3G]) and A3F. As such, the therapeutic blockade of Vif function represents a logical objective for rational drug design. To facilitate such endeavors, we have employed molecular genetics to define features of A3G that are required for its interaction with Vif. Using alanine-scanning mutations and multiple different substitutions at key residues, we confirm the central role played by the aspartic acid at position 128 and identify proline 129 and aspartic acid 130 as important contributory residues. The overall negative charge of this 3-amino-acid motif appears critical for recognition by Vif, as single lysine substitutions are particularly deleterious and a double alanine substitution at positions 128 and 130 is far more inhibitory than single-residue mutations at either position. Our analyses also reveal that the immediately adjacent 4 amino acids, residues 124 to 127, are important for the packaging of A3G into HIV-1 particles. Most important are tyrosine 124 and tryptophan 127, and mutations at these positions can ablate virion incorporation, as well as the capacity to inhibit virus infection. Thus, while pharmacologic agents that target the acidic motif at residues 128 to 130 have the potential to rescue A3G expression by occluding recognition by Vif, care will have to be taken not to perturb the contributions of the neighboring 124-to-127 region to packaging if such agents are to have therapeutic benefit by promoting A3G incorporation into progeny virions.
Proteins of the vertebrate apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3 (APOBEC3) family protect cells against invasion by a broad range of viruses and mobile genetic elements, such as retroviruses, hepadnaviruses, endogenous retroviruses, and retrotransposons (3, 5, 8, 16, 30, 42, 46, 47, 52, 56). These proteins belong to a larger family of proteins that contain one or two copies of the signature histidine/cysteine-X-glutamic acid-X23-28-proline-cysteine-X2-cysteine motif that is characteristic of cytidine and adenosine deaminases found in species ranging from bacteria to vertebrates (7, 20, 22, 47). Expression of APOBEC3 proteins can lead to their encapsidation into progeny virions through recruitment to substrate viral or transposon capsid structures, and this appears to involve interactions with both Gag (or Gag-like) proteins and RNA (6, 12, 14, 28, 43, 54, 63).
Following reverse transcription of the retroelement genomic RNA into single-stranded DNA, the APOBEC proteins can deaminate cytidines to uridines, causing deleterious mutations that result in the loss of genetic integrity and protein function, a process that is commonly referred to as hypermutation (19, 30, 64). More recently, it has emerged that hypermutation is not the only means by which APOBEC3 proteins can inhibit infectivity or transposition. Specifically, it has been reported that inhibition of retrotransposons and hepatitis B virus occurs in the absence of detectable hypermutation (5, 36, 42, 52, 56) and that mutant versions of the human APOBEC3G (A3G) and A3F proteins with inactive deaminase catalytic domains can still suppress human immunodeficiency virus type 1 (HIV-1) infection (2, 21, 38). In the latter case, the inhibition of infection correlates well with reduced accumulations of HIV-1 reverse transcripts in challenged cells (2, 21). The A3G and A3F proteins both contain duplications of the cytidine deaminase (CDA) catalytic motif, but they are not equivalent, as only the C-terminal CDA motif can mediate cytidine deamination (21, 38): the molecular basis for this domain difference remains to be defined.
Lentiviruses, such as HIV-1, encode a virion infectivity factor (Vif) protein that prevents the antiviral effects of APOBEC3 family members, in particular, A3G and A3F (3, 47, 58, 65). The mode of action by which HIV-1 Vif counters A3G-mediated antiviral function has been elucidated in some detail. HIV-1 Vif interacts with human A3G and thereby recruits a cullin 5-elongin B/C E3 ubiquitin ligase so that A3G is polyubiquitinated and targeted to the proteasome for degradation (9, 32-34, 48, 61, 62). The elimination of A3G at the time of virus production consequently prevents its encapsidation, although Vif has also been reported to occlude A3G encapsidation directly (48) and to reduce A3G translation (53). The interactions of Vif with components of this E3 ligase complex are mediated by a histidine-cysteine-cysteine-histidine zinc binding motif that provides a binding site for cullin 5 (35, 59) and a BC box that binds to elongin C (62).
Our molecular understanding of the Vif-A3G interaction has been advanced by the observation that a single-amino-acid difference between the A3G proteins of humans and African green monkeys (AGMs) at position 128 modulates the species-specific interaction with the Vif proteins of HIV-1 or AGM-derived simian immunodeficiency virus (SIVAGM) (4, 31, 44, 60). Most notably, replacement of the aspartic acid residue that naturally occurs at this position (D128) in human A3G with the lysine found at the corresponding position in AGM A3G abrogates the interaction with HIV-1 Vif but confers sensitivity to the SIVAGM Vif protein. The A3G-Vif interaction is of considerable interest, as it provides a compelling target for novel therapeutic strategies for treating HIV-1 infections. Specifically, successful disruption of the Vif-A3G interaction is predicted to rescue A3G expression, induce virion packaging, and therefore stimulate natural antiviral activity. Similarly, pharmacologic suppression of proteasome-mediated A3G degradation has already been shown to enhance A3G expression and thereby inhibit HIV-1 infectivity (61). The rational design of inhibitors of the Vif-A3G interaction will be facilitated by detailed molecular descriptions of the determinants present in both Vif and A3G. Accordingly, and with the current absence of structural information, we have undertaken a molecular genetic approach to address this question.
Extensive site-directed mutagenesis of human A3G has enabled us to map a 3-amino-acid motif comprising aspartic acid-proline-aspartic acid at positions 128 to 130 as being crucial for the interaction with HIV-1 Vif. Further substitutions in this region with amino acids carrying side chains with different properties revealed that the negative charges at positions 128 and 130 play a central role in the HIV-1 Vif interaction but that there is redundancy between the two positions. In addition, a 4-amino-acid region of A3G immediately N-terminal to residue 128 is essential for virion packaging and hence for antiviral efficacy. In developing therapeutic moieties that interfere with the Vif-A3G interaction, consideration will have to be given to retaining efficient A3G packaging.
MATERIALS AND METHODS
Plasmids and cloning.
The wild-type (parental) human A3G cDNA was inserted into pCMV4-HA (1) as PCR-amplified HindIII-XbaI fragments to create (i) pA3G, in which the natural stop codon of the A3G gene preceded the triple hemagglutinin (HA) tag, or (ii) pA3G-HA, in which the HA tag was fused in frame to the C terminus of hA3G. An overlapping PCR-based strategy was used to generate derivatives that encoded mutated A3G proteins. Briefly, for each mutation, N- and C-terminal fragments of A3G containing the desired point mutation in an ∼20-nucleotide region that was common to both fragments were generated by PCR. These were used as the template for a second PCR in which only the outer primers were present. The resulting mutated cDNA was then inserted into pCMV4-HA to yield vectors expressing untagged or tagged proteins, depending upon the 3′ primer that was used. Bacterial A3G expression vectors were produced by subcloning A3G cDNAs into pTrc99A as Asp718-SmaI fragments (2). The wild-type and vif-deficient (Δvif) HIV-1 proviruses, pIIIB and pIIIB/Δvif; the HIV-1 Vif expression vector, pcVIF; and its corresponding control, pcΔVIF, have all been described previously (48, 50).
Single-cycle infectivity assays.
Stocks of wild-type or Δvif HIV-1 were prepared by cotransfection of 35-mm-diameter monolayers of 293T cells with 0.5 μg of pA3G expression vector and 2.0 μg of pIIIB or pIIIB/Δvif using polyethylenimine (13). After 24 h, the supernatants were harvested and virus levels were quantified by p24gag enzyme-linked immunosorbent assay (Perkin-Elmer). The producer cells were lysed in sodium dodecyl sulfate (SDS) containing loading dye for the analysis of protein expression. Viral stocks were stored at −80°C, and volumes corresponding to 5 ng p24gag were used to infect 105 TZM-bl indicator cells (11, 40, 57). The induced expression of β-galactosidase in whole-cell lysates was measured 24 h after the initiation of infection using the Galacto-Star system (Applied Biosystems).
Analysis of protein expression by immunoblotting.
Whole-cell lysates prepared from virus-producing cells were resolved by SDS-polyacrylamide gel electrophoresis and analyzed by immunoblotting using primary antibodies specific for A3G (rabbit polyclonal [38] or Hsp90 [Invitrogen], horseradish peroxidase-conjugated secondary antibodies) and enhanced chemiluminescence (Pierce). In experiments examining the effects of Vif on A3G accumulation, 35-mm cultures of 293T cells were transfected with 0.2 μg pA3G (or a mutant derivative) and 0.2 μg pcVIF or pcΔVIF. After 24 h, the cells were lysed in loading buffer and analyzed by immunoblotting using primary antibodies specific for A3G, Hsp90, or Vif (319 mouse monoclonal antibody) (50).
Coimmunoprecipitation assays.
293T cells were transfected with 2 μg pA3G-HA (or a mutant derivative) and 2 μg pcVIF or pcΔVIF in 35-mm cultures. After 24 h, the cells were washed and lysed in 1 ml lysis buffer (1% Triton X-100, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and complete protease inhibitor cocktail [Roche]). The lysates were cleared by centrifugation in a bench-top centrifuge at 21,000 × g for 10 min, and 850 μl of each was incubated with the 12CA5 antibody and protein A-agarose (Invitrogen) for 2 h at 4°C. The agarose beads were washed three times with lysis buffer and resuspended in 50 μl loading dye; 10 μl of the immunoprecipitated samples, as well as 10 μl of the cleared lysate, was resolved by SDS-polyacrylamide gel electrophoresis (11% gel) and analyzed by immunoblotting using primary antibodies specific for HA, Vif, or Hsp90.
Packaging assays.
Virus stocks containing 20 ng p24gag were spun in a bench-top centrifuge at 21,000 × g for 2 h at 4°C through a 20% (wt/vol) sucrose cushion (500 μl) in a 2-ml Eppendorf tube. The viral pellets were resuspended in loading dye and analyzed by immunoblotting using antibodies specific for A3G or p24gag (mouse monoclonal 24-2 [49]). Whole-cell lysates from the corresponding producer cells were assessed for A3G expression.
E. coli mutation assay.
The KL16 strain of Escherichia coli was transformed with pTrc99A-based, IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible A3G expression vectors or the empty vector. Individual colonies were picked and grown to saturation in LB medium containing 100 μg/ml ampicillin and 1 mM IPTG. Appropriate dilutions were spread onto agar plates containing either 100 μg/ml ampicillin or 100 μg/ml rifampin and incubated overnight at 37°C. Mutation frequencies were recorded as the number of rifampin-resistant colonies per 109 viable cells (which were enumerated using the ampicillin-containing plates) (38, 39).
RESULTS
Modeling the A3G structure.
We previously exploited the availability of the crystal structures of some nucleotide cytidine deaminases to produce a model for APOBEC proteins by the combined use of structure prediction and sequence alignment (22). This analysis demonstrated that most structural features of cytidine deaminases can readily be assigned counterparts in APOBEC proteins. In particular, a mixed sheet composed of five β-strands and two α-helices that contain the catalytic site residues was readily predicted. One key difference that emerged was the presence of the additional α-helix 3 between β-strands 4 and 5 in APOBEC proteins, which is absent from the nucleotide cytidine deaminases. Based on the crystal structures of the human adenosine-to-inosine editing enzyme ADAR2 (29), the tRNA adenosine deaminases from Aquifex aeolicus (15, 25) and Staphylococcus aureus (27), Saccharomyces cerevisiae cytosine deaminase (23, 24), and APOBEC2 (an orphan APOBEC family member whose crystal structure was reported while this work was being reviewed) (41), it is now believed that α-helix 3 of APOBEC protein CDA cores crosses the β-sheet, resulting in a parallel positioning of β-strands 4 and 5. As a consequence, this helix is located toward the same face of the β-sheet as helices α1 and α2 (Fig. 1A). Because this conformation may limit the accessibility of α3 and the immediately adjacent residue D128 to interacting factors, such as Vif, we set out to analyze in detail the importance of the region between predicted β-strands 4 and 5 of the amino-terminal CDA domain of A3G using site-directed mutagenesis. In particular, we wished to probe the contributions of the residues surrounding D128 for involvement in regulation by HIV-1 Vif and to investigate the role of α3 in A3G function (Fig. 1B).
FIG. 1.
(A) Structure model of the N-terminal CDA domain of A3G. Zinc-coordinating residues are indicated by circles, α-helices are in yellow, and β-strands are in pink. The position of residue D128 is indicated in the predicted loop between β4 and α3. (B) The amino acid sequence of residues 114 to 153 of A3G. The positions of predicted α-helix (yellow) and β-strand (pink) regions are indicated, as are residues 119, 128, and 146. The motifs defined here as being important for HIV-1 packaging and Vif responsiveness are indicated.
Alanine-scanning mutagenesis of A3G.
We initially used site-directed mutagenesis to construct a series of A3G expression vectors encoding proteins with single alanine substitutions covering residues 119 to 146. This region contains two naturally occurring alanine residues, A121 and A134 (Fig. 1B), and they were not included in our analysis. Wild-type or vif-deficient (Δvif) HIV-1 stocks were produced using 293T cells in the presence of mutant or wild-type A3G proteins and used in single-cycle challenges of TZM-bl indicator cells to assay A3G-mediated antiviral activity. All mutants with alterations between residues 131 and 146 displayed antiviral properties comparable to those of the wild-type protein in that virus infection was efficiently suppressed in the absence of Vif but was restored in the presence of Vif to levels close to that of vector-only controls (data not shown). From this, we concluded that the region spanning residues 131 to 146 is not involved in the interaction with HIV-1 Vif and that the predicted helix α3 most likely plays a predominantly structural role. We did not examine this region further and instead focused our attention on residues 119 to 130. Mutations in this more N-terminal region gave rise to proteins with altered phenotypes that can be organized into two categories (Fig. 1B). First, alanine substitutions at positions 128 to 130 yielded proteins that showed increased capacities to inhibit wild-type HIV-1 infection, indicating resistance to the action of Vif. Second, various proteins with mutations between residues 119 and 127 display decreased abilities to inhibit HIV-1/Δvif, caused by reduced expression or packaging into virus particles. The further analyses of each class of mutated protein are discussed in turn below.
Analysis of A3G mutations affecting regulation by Vif.
The aspartic acid at position 128 plays a pivotal role in the responsiveness of A3G to HIV-1 Vif (4, 31, 44, 60). In addition to D128A, mutants P129A and D130A also emerged with Vif-resistant phenotypes (Fig. 2A). Though the disruption of the capacity of Vif to regulate A3G was not nearly as marked for the D128A mutant as it was for P129A, a similarly modest loss of Vif responsiveness was also seen for D130A. Since these three residues may constitute the core of a site required for the Vif interaction, we constructed additional mutant proteins in which the chemical natures of the three side chains were changed. The aspartic acids at positions 128 or 130 were altered to lysine, asparagine, glutamic acid, or serine to assess the consequences of replacing a negatively charged side chain with a side chain that was positively charged, basic, alternatively negatively charged, or polar. In addition, the proline at position 129 was changed to glycine to test whether the inherent flexibility of this amino acid might be able to compensate for the bend that would be imposed by the naturally occurring proline. Single-cycle infectivity assays were performed with this set of A3G mutants (as well as with the initial alanine substitutions) using wild-type or Δvif virus stocks, and protein expression was examined in whole-cell lysates by immunoblotting (Fig. 2A).
FIG. 2.
Mutational analysis of residues 128, 129, and 130 of A3G. (A) Analysis of the side chain requirements for Vif responsiveness at residues 128, 129, and 130. WT, wild type. (B) Simultaneous mutation of residues 128 and 130 to alanine increases the Vif resistance of A3G. Single-cycle infectivities of virus stocks produced in the presence of mutant or wild-type A3G (or the pCMV4 parental vector) were measured as relative luciferase units and are presented as percent infectivity relative to wild-type virus produced in the absence of A3G. The infectivities of wild-type HIV-1 stocks are shown as black bars and Δvif stocks as white bars. The data are the means of three infections performed with independently produced virus samples, and the error bars represent standard deviations. Below, immunoblots show the expression of A3G in the corresponding producer 293T cells for one set of transfections. The Hsp90 immunoblot is shown as a loading control.
The phenotypic trends for the mutations at residues 128 and 130 showed strong similarity, although subtle differences were also evident. In both cases, the strongest inhibition of wild-type HIV-1 infection (and hence resistance to Vif) was observed upon the introduction of lysine (mutants D128K and D130K), though the effect was greater at position 128. Among the other substitutions at this residue, the D128N mutation exhibited the most substantial loss of inhibition by Vif, whereas D128E and D128S behaved much like D128A. In comparison, at position 130, the introduction of asparagine, glutamic acid, or serine had negligible effects on susceptibility to Vif. The P129G mutant showed a phenotype very similar to that of P129A in that it inhibited wild-type HIV-1 infection very effectively, indicative of the loss of regulation by Vif. As anticipated, reductions in Vif responsiveness for this collection of mutants were accompanied by increased levels of protein accumulation in cell lysates (Fig. 2A, bottom). Together, these results suggest that the interaction between A3G and Vif is largely determined by electrostatic interactions involving residues 128 and 130, but with the dependency for aspartic acid at position 128 being more stringent.
The observation that reversing the negative charges on two closely positioned residues had marked effects on regulation by Vif but that the mere loss of one aspartic acid often had a relatively minor effect (e.g., the serine substitutions) implied that there may be redundancy between D128 and D130. To address this idea, a double-alanine-substitution mutant, D128A-D130A, was constructed and analyzed as described above (Fig. 2B). This protein was a considerably more potent inhibitor of wild-type HIV-1 infection than either single mutant, with infectivity being reduced by more than 90%. Consistent with this, examination of producer lysates revealed that Vif had little effect on its expression. Thus, while the removal of the aspartic acid residues does not completely abrogate responsiveness to Vif, these amino acids clearly play central and somewhat overlapping roles in the interaction of A3G with Vif.
The proline at position 129 also appears to be a key feature for recognition by Vif. As this could be due to either the structural or hydrophobic properties of the proline, we also analyzed an A3G mutant with valine at position 129, because the hydrophobicity of valine is similar to that of proline but it does not cause a bend in a peptide backbone. Though the infectivity of wild-type HIV-1 produced in the presence of the P129V mutant was mildly improved compared to that with P129A, the P129V protein was poorly expressed (data not shown). This suggests that the proline at position 129 is a key structural feature of A3G that contributes to correct protein folding, as well as being important for helping to establish an interaction with HIV-1 Vif.
Having examined the effects of Vif on the expression of A3G mutant proteins only in the context of virus-producing cells, we next ventured to verify the Vif-resistant properties of a subset of mutants by expressing them in the presence or absence of Vif in 293T cells independently of other viral proteins (Fig. 3A). This analysis confirmed the stability of the D128K, P129A, P129G, and D130K mutants in the presence of Vif. In contrast, the D128A and D130A mutants generally showed some Vif-induced degradation in this setting, confirming that these mutants have only moderately Vif-resistant phenotypes.
FIG. 3.
Vif-resistant A3G proteins do not interact with HIV-1 Vif. (A) Analysis of the stability of wild-type (WT) or mutant A3G proteins in the absence or presence of HIV-1 Vif. Proteins were detected by immunoblotting of cell lysates from transfected 293T cells using specific primary antibodies, and Hsp90 was measured as a loading control. (B) Coimmunoprecipitation of HIV-1 Vif with HA-tagged versions of wild-type or mutant A3G. The pCMV4 sample served as the negative control. Whole-cell lysates (top) were analyzed by immunoblotting using antibodies specific for A3G, HIV-1 Vif, or Hsp90 (loading control). The immunoprecipitates (IP) (bottom) were analyzed with antibodies specific for A3G or HIV-1 Vif. The band common to all coprecipitated Vif samples represents antibody light chains.
To this point, we have referred to the A3G-Vif interaction in only a genetic sense. To address whether the noted trends extended to biochemical interactions, the abilities of mutant A3G proteins to interact with Vif were assessed by coimmunoprecipitation (Fig. 3B). Cell lysates were prepared from coexpressing 293T cells and either examined directly by immunoblotting (upper three panels) or immunoprecipitated using an HA-specific antibody that recognized the C-terminally HA-tagged A3G proteins; the presence of A3G (fourth panel) or Vif (bottom panel) in the precipitates was then monitored by immunoblotting. We were able to coimmunoprecipitate Vif with wild-type A3G-HA and the D128A and D130A mutants, but not with the D128K, P129A, P129G, and D130K mutants. This confirmed that the partially Vif-resistant D128A and D130A proteins maintained an interaction with Vif (albeit at slightly reduced levels compared to wild-type A3G), but the profoundly Vif-resistant mutants did not. Thus, the capacity of altered A3G proteins to escape Vif-induced downregulation correlates well with their respective inabilities to form complexes with Vif.
Analysis of A3G mutants with reduced antiviral activities.
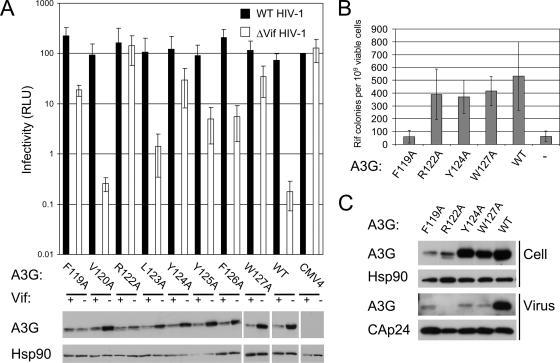
We next investigated the mutant proteins with changes between positions 119 and 127 that showed generalized reductions in antiviral activity (Fig. 4A). Most of the alanine-scanning mutants showed normal expression in producer cell lysates and still accumulated to lower levels in the presence of Vif. Mutants F119A and R122A, however, were expressed at lower levels, likely accounting for their respective losses of antiviral activity. In contrast, Y124A, Y125A, F126A, and W127A each showed levels of expression comparable to that of wild-type A3G, suggesting that their losses of antiviral activity, which were more pronounced for Y124A and W127A, may be caused by the disturbance of a functional moiety of A3G. The F119A, R122A, Y124A, and W127A mutants were therefore investigated to see whether editing activity (Fig. 4B) or virion packaging (Fig. 4C) had been compromised.
FIG. 4.
Alanine-scanning mutagensis of residues 119 to 127 of A3G. (A) Single-cycle infectivities of virus stocks produced in the presence of A3G mutant proteins. The experiments were performed, and are reported, as for Fig. 2. (B) Editing activities of wild-type (WT) and mutant A3G proteins as measured in a bacterial reporter assay. (C) Packaging of wild-type and mutant A3G into HIV-1 particles. Virions purified from supernatants of 293T cells cotransfected with Δvif and A3G expression vectors were analyzed by immunoblotting, together with corresponding producer cell lysates.
To begin to address whether the loss of antiviral function was due to impaired protein activity, we used an E. coli-based assay that measures the DNA-editing capability of APOBEC proteins. In this assay, APOBEC-driven mutation of the bacterial rpoB gene is registered by the emergence of rifampin-resistant colonies, which can be readily counted (38, 39). Mutants R122A, Y124A, and W127A all showed editing activities similar to that of wild-type A3G, but editing by the F119A mutant was reduced to background levels (Fig. 4B). Immunoblotting analysis showed that all proteins were expressed at similar levels (data not shown). The wild-type editing activities of R122A, Y124A, and W127A were perhaps not surprising, as previous studies had demonstrated that the editing activity of A3G is associated with the C-terminal CDA domain and not the N-terminal CDA domain in which these mutations are located. However, the inactivity of the F119A mutant protein was unexpected, suggesting that this mutation may have caused the loss of editing through misfolding of the protein in bacteria. In contrast, the R122A, Y124A, and W127A proteins presumably still adopted active conformations.
As A3G needs to be packaged into HIV-1 virions to exert its antiviral affect, we next produced Δvif virus particles in the presence of wild-type A3G or F119A, R122A, Y124A, or W127A and analyzed their encapsidation into virions by immunoblotting them (Fig. 4C). Consistent with their lower expression levels in cell lysates, only small amounts of the F119A and R122A proteins were detected in virus particles. In contrast, the Y124A and W127 mutant proteins were efficiently expressed but showed severe defects in packaging efficiency compared with wild-type A3G. Thus, the loss of antiviral activity of Y124A and W127A correlates well with the reduced packaging of these proteins.
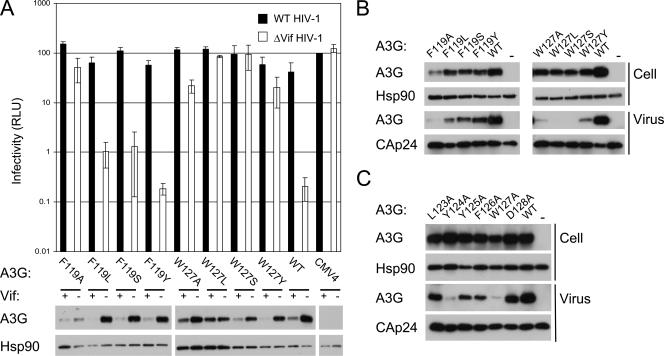
To investigate and compare further the expression and packaging phenotypes of mutant proteins with alterations at positions 119 or 127, we constructed another set of mutants in which these natural aromatic residues were replaced with amino acids carrying hydrophobic, polar, or alternative aromatic side chains (leucine, serine, or tyrosine, respectively). Single-cycle infectivity and protein expression assays were performed as described above (Fig. 5A). At position 119, all of these substitutions restored expression levels, Vif responsiveness, and inhibition of HIV-1/Δvif infectivity to levels close to those of wild-type A3G. In fact, the behavior of the F119Y protein that contains the alternative aromatic residue was indistinguishable from that of the parental protein in this experiment. On the other hand, none of these substitutions at position 127 restored the antiviral properties of the protein. Indeed, W127L and W127S both showed an even more pronounced loss of antiviral activity, as evidenced by their complete inability to inhibit infection by HIV-1/Δvif. Investigation of the packaging properties of these proteins revealed that all of the substitutions at position 119 restored encapsidation to close to wild-type levels but that the same substitutions at position 127 did not (Fig. 5B). At best, W127Y exhibited a marginal increase in packaging efficiency compared to W127A, but the presence of W127L or W127S in virions was undetectable. Thus, we conclude that the presence of tryptophan at position 127 is required for the correct packaging of A3G into HIV-1 particles.
FIG. 5.
Mutational analysis of A3G residues 119 and 127. (A) Analysis of the side chain requirements for antiviral function at residues 119 and 127. Experiments were performed as for Fig. 2. (B) Roles of residues 119 and 127 in virion incorporation. Packaging was determined as for Fig. 4C. (C) Residues spanning positions 124 to 127 contribute to virion incorporation.
Finally, we sought to establish whether the reductions in antiviral activity associated with other mutations preceding residue 127 could also be explained by packaging defects. Accordingly, we analyzed the virion encapsidation of mutant proteins with amino acid substitutions at positions 123 to 128 (Fig. 5C). Consistent with their strong antiviral phenotypes, the L123A and D128A proteins were efficiently packaged. As in Fig. 4C, Y124A and W127A were packaged at very low levels, whereas the Y125A and F126A proteins showed intermediate levels of incorporation. In sum, the severity of the packaging defects for this set of mutant proteins correlated closely with the extent to which their abilities to inhibit HIV-1/Δvif had been impaired (Fig. 5A). Thus, our analysis identified the aromatic 4-amino-acid Y124-Y125-F126-W127 motif as being important for the incorporation of A3G into HIV-1 particles.
DISCUSSION
The primary intent of this structure-function study was to better understand molecular determinants of the interaction between A3G and the antagonistic HIV-1 Vif protein. A detailed knowledge of the interaction between these proteins is of particular interest for the rational design of future therapeutic approaches that aim to harness natural protection against HIV-1 infection by preventing the Vif-mediated degradation of A3G. We chose to mutate residues 119 to 146 of A3G because the region contains residue 128, which has previously been shown to be pivotal in facilitating the formation of A3G-Vif complexes (4, 31, 44, 60).
In addition to confirming the role of residue 128 in the interaction with Vif, we identified two additional amino acids, P129 and D130, whose alteration created proteins that displayed Vif-resistant phenotypes (Fig. 1B). More specifically, we found that insensitivity to Vif was conferred by mutating the aspartic acid residue at position 128 or 130 to the positively charged residue lysine. However, we did observe that residue 128 is more sensitive to mutation than 130, which can accommodate several amino acid changes and still behave like wild-type A3G, implying that D128 may play a more prominent role in the Vif interaction (Fig. 2A). Nevertheless, simultaneous mutation of residues D128 and D130 to alanine yielded a protein that was markedly more resistant to Vif than either singly mutated protein (Fig. 2B), suggesting a substantial degree of redundancy between these positions. In sum, these data indicate that the Vif-A3G interaction is dependent on electrostatic interactions, which is fully consistent with recent data demonstrating that mutation of positively charged residues in HIV-1 Vif can compensate for the D128K mutation in A3G (45). In addition to these electrostatic determinants, there also appears to be a rather specific structural requirement for the interaction, as evidenced by the strong Vif-resistant phenotypes of mutant A3G proteins carrying either alanine or glycine in place of proline at position 129 (Fig. 2A). Though it is most likely that Vif contacts A3G directly in this region, we emphasize that direct binding awaits formal demonstration. For this reason, we use the term “interaction” to encompass both direct and indirect contacts.
While our current study has provided novel insight into the interaction between HIV-1 Vif and A3G, the 3-amino-acid motif D128-P129-D130 is unlikely to represent a complete description of motifs within APOBEC proteins that are capable of interacting with HIV-1 Vif. More specifically, (i) one study reported that residues 54 to 124 of A3G are sufficient for coimmunoprecipitation with Vif, which would suggest the presence of an additional point (or points) of contact (9); (ii) the Vif proteins of HIV-2 and SIVmac are able to mediate the degradation of both wild-type A3G and the D128K mutant, indicating that these Vif proteins have different requirements for functional interactions with A3G (17, 60); and (iii) the interaction between HIV-1 Vif and human A3F is different from the interaction with A3G, as illustrated by the demonstration that amino acid differences in HIV-1 Vif can differentially affect the ability to neutralize A3F versus A3G (51, 55). Indeed, the human A3F protein contains an ERD motif at the position equivalent to the DPD motif of A3G (defined here), yet mutation of the glutamic acid in A3F does not affect sensitivity to Vif (10, 26); moreover, the naturally Vif-resistant human A3B protein also has this ERD motif at the equivalent position, again implying that an interaction with Vif requires more than just acidic amino acids at this position. Lastly, like A3G, A3F contains two CDA domains, and we noted that the C-terminal domain contains a DTD motif within the predicted β4-α3 loop. However, mutation of the first aspartic acid in this motif to lysine (D310K) did not affect downregulation by Vif (data not shown). Taken together, these assorted findings indicate that the DPD motif at residues 128 to 130 in human A3G represents a Vif interaction domain that is unique to this protein. A full understanding of the molecular determinants of this interaction will clearly depend on future structural analyses of Vif-A3G complexes. In this regard, it is interesting that the recently resolved structure of APOBEC2 (41) reveals that the corresponding glutamic acid-proline-glutamic acid motif (residues 159 to 161) also straddles the N terminus of helix α3 (Fig. 1A) and that the acidic side chains protrude outward from the helix, rendering them accessible for interactions with protein ligands.
In addition to defining this Vif interaction motif, our analyses revealed the existence of an adjacent motif that plays a central role in the packaging of A3G into virus particles (Fig. 1B). Alanine substitution mutations in this 4-amino-acid cluster spanning residues 124 to 127 generated proteins displaying reduced levels of antiviral activity against HIV-1/Δvif (Fig. 4A) that correlated with losses in virion incorporation (Fig. 4C and 5C). Specifically, mutants Y124A and W127A were almost entirely excluded from virions and had minimal effects on infectivity, whereas the Y125A and F126A proteins showed reduced levels of packaging and intermediate infectivity phenotypes. Further mutation analysis of residue 127 indicated that the presence of tryptophan at this position is particularly important, as all substitutions tested resulted in substantial diminutions of antiviral activity (Fig. 5A). A principal role for residues 124 and 127 in A3G function is also reflected in the strong conservation of these residues within A3G proteins from different species, whereas more variation was observed at the less important positions 125 and 126 (not shown).
It has been reported by several groups that incorporation of A3G into virus particles depends on a combination of its interaction with the NC domain of Gag and nonspecific RNA binding (6, 12, 14, 28, 43, 54, 63). Although the losses of antiviral activity for mutants in the 124-to-127 region correlated with defects in packaging, we found, using a previously described pull-down assay (12, 63) and lysates from transfected 293T cells, that proteins with mutations at positions 124, 125, and 126 interacted with HIV-1 Gag in an RNA-dependent manner but that the W127A mutant protein did not (data not shown). Thus, packaging capabilities cannot simply be ascribed to Gag-A3G-RNA interactions as monitored by this assay system. The reasons for these discrepancies are not yet evident and will require further investigation; however, one potential contributing factor to A3G packaging may be the requirement for spatial proximity between A3G and the cellular sites of viral particle assembly, and it is conceivable that this motif may participate in determining correct subcellular localization. A further possibility is that this motif may bind to an as-yet-unidentified molecular partner that may specify virus incorporation. In sum, further work is required to define the role of residues 124 to 127 in HIV-1 packaging and to determine how this region combines with other conserved elements of the N-terminal CDA of A3G that also contribute to encapsidation (18, 37, 38).
In this study, we demonstrated that crucial determinants for A3G interaction with Vif and its incorporation into HIV-1 particles are localized to a 7-amino-acid element spanning residues 124 to 130. Consistent with the notion that this region interacts with viral and cellular factors, structure predictions for A3G and comparisons with APOBEC2 place it in a loop region that connects β4 to α3 (Fig. 1). On the basis of our alanine scan, the residues that confer these two attributes appear to be distinct from each other. The W127L protein provides the only data that may dispute this: it is no longer packaged (Fig. 5B) yet has lost susceptibility to Vif-induced degradation (Fig. 5A). Since other mutations at position 127 remain Vif sensitive, it seems most likely that the W127L mutant is misfolded and consequently broadly nonfunctional (though it remains formally possible that this mutation genuinely influences both attributes). As discussed above, pharmacologic inhibition of the Vif-A3G interaction represents a potential therapeutic approach for suppressing HIV-1 replication. Should such an inhibitory agent interact with A3G at the DPD motif, it will be important to preserve the biological activities of the neighboring YYFW motif: failure to do so could lead to reduced packaging and antiviral efficacy. Further work defining the precise functions and structures of these motifs will inform rational drug design in this area.
Acknowledgments
The following reagent was obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl from John Kappes, Xiaoyun Wu, and Tranzyme Inc.
This work was supported in part by grant number 106637-38-RFHF from the American Foundation for AIDS Research (amfAR). Work in the Malim laboratory is supported by the United Kingdom Medical Research Council, and M.H.M. is an Elizabeth Glaser Scientist supported by the Elizabeth Glaser Pediatric AIDS Foundation.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Andersson, S., D. L. Davis, H. Dahlback, H. Jornvall, and D. W. Russell. 1989. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 264:8222-8229. [PubMed] [Google Scholar]
- 2.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 14:1392-1396. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., B. P. Doehle, H. L. Wiegand, and B. R. Cullen. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. USA 101:3770-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd, H. P., H. L. Wiegand, B. P. Doehle, K. K. Lueders, and B. R. Cullen. 2006. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 34:89-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 7.Chang, B. H., and L. Chan. 1998. Evolutionary analysis of RNA editing enzymes. Methods 15:41-50. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:15588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conticello, S. G., R. S. Harris, and M. S. Neuberger. 2003. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 13:2009-2013. [DOI] [PubMed] [Google Scholar]
- 10.Dang, Y., X. Wang, W. J. Esselman, and Y. H. Zheng. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 80:10522-10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doehle, B. P., A. Schafer, H. L. Wiegand, H. P. Bogerd, and B. R. Cullen. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 79:8201-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durocher, Y., S. Perret, and A. Kamen. 2002. High-level and high-throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elias, Y., and R. H. Huang. 2005. Biochemical and structural studies of A-to-I editing by tRNA:A34 deaminases at the wobble position of transfer RNA. Biochemistry 44:12057-12065. [DOI] [PubMed] [Google Scholar]
- 16.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 17.Gaddis, N. C., A. M. Sheehy, K. M. Ahmad, C. M. Swanson, K. N. Bishop, B. E. Beer, P. A. Marx, F. Gao, F. Bibollet-Ruche, B. H. Hahn, and M. H. Malim. 2004. Further investigation of simian immunodeficiency virus Vif function in human cells. J. Virol. 78:12041-12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakata, Y., and N. R. Landau. 2006. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J. Biol. Chem. 281:36624-36631. [DOI] [PubMed] [Google Scholar]
- 19.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 20.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 21.Holmes, R. K., F. A. Koning, K. N. Bishop, and M. H. Malim. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation: comparisons with APOBEC3G. J. Biol. Chem. 282:2587-2595. [DOI] [PubMed] [Google Scholar]
- 22.Huthoff, H., and M. H. Malim. 2005. Cytidine deamination and resistance to retroviral infection: towards a structural understanding of the APOBEC proteins. Virology 334:147-153. [DOI] [PubMed] [Google Scholar]
- 23.Ireton, G. C., M. E. Black, and B. L. Stoddard. 2003. The 1.14 Å crystal structure of yeast cytosine deaminase: evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure 11:961-972. [DOI] [PubMed] [Google Scholar]
- 24.Ko, T. P., J. J. Lin, C. Y. Hu, Y. H. Hsu, A. H. Wang, and S. H. Liaw. 2003. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J. Biol. Chem. 278:19111-19117. [DOI] [PubMed] [Google Scholar]
- 25.Kuratani, M., R. Ishii, Y. Bessho, R. Fukunaga, T. Sengoku, M. Shirouzu, S. Sekine, and S. Yokoyama. 2005. Crystal structure of tRNA adenosine deaminase (TadA) from Aquifex aeolicus. J. Biol. Chem. 280:16002-16008. [DOI] [PubMed] [Google Scholar]
- 26.Liu, B., P. T. Sarkis, K. Luo, Y. Yu, and X. F. Yu. 2005. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 79:9579-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Losey, H. C., A. J. Ruthenburg, and G. L. Verdine. 2006. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13:153-159. [DOI] [PubMed] [Google Scholar]
- 28.Luo, K., B. Liu, Z. Xiao, Y. Yu, X. Yu, R. Gorelick, and X. F. Yu. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macbeth, M. R., H. L. Schubert, A. P. Vandemark, A. T. Lingam, C. P. Hill, and B. L. Bass. 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309:1534-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 31.Mangeat, B., P. Turelli, S. Liao, and D. Trono. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481-14483. [DOI] [PubMed] [Google Scholar]
- 32.Mariani, R., D. Chen, B. Schrofelbauer, F. Navarro, R. Konig, B. Bollman, C. Munk, H. Nymark-McMahon, and N. R. Landau. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21-31. [DOI] [PubMed] [Google Scholar]
- 33.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 34.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 279:7792-7798. [DOI] [PubMed] [Google Scholar]
- 35.Mehle, A., E. R. Thomas, K. S. Rajendran, and D. Gabuzda. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 281:17259-17265. [DOI] [PubMed] [Google Scholar]
- 36.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 37.Navarro, F., B. Bollman, H. Chen, R. Konig, Q. Yu, K. Chiles, and N. R. Landau. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374-386. [DOI] [PubMed] [Google Scholar]
- 38.Newman, E. N., R. K. Holmes, H. M. Craig, K. C. Klein, J. R. Lingappa, M. H. Malim, and A. M. Sheehy. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166-170. [DOI] [PubMed] [Google Scholar]
- 39.Petersen-Mahrt, S. K., R. S. Harris, and M. S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature 418:99-103. [DOI] [PubMed] [Google Scholar]
- 40.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prochnow, C., R. Bransteitter, M. G. Klein, M. F. Goodman, and X. S. Chen. 2007. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature 445:447-451. [DOI] [PubMed] [Google Scholar]
- 42.Rosler, C., J. Kock, M. H. Malim, H. E. Blum, and F. von Weiszacker. 2004. Comment on “Inhibition of hepatitis B virus replication by APOBEC3G”. Science 305:1403. [DOI] [PubMed] [Google Scholar]
- 43.Schafer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163-168. [DOI] [PubMed] [Google Scholar]
- 44.Schrofelbauer, B., D. Chen, and N. R. Landau. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 101:3927-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrofelbauer, B., T. Senger, G. Manning, and N. R. Landau. 2006. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 80:5984-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. USA 102:9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 48.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 49.Simon, J. H., R. A. Fouchier, T. E. Southerling, C. B. Guerra, C. K. Grant, and M. H. Malim. 1997. The Vif and Gag proteins of human immunodeficiency virus type 1 colocalize in infected human T cells. J. Virol. 71:5259-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simon, J. H., T. E. Southerling, J. C. Peterson, B. E. Meyer, and M. H. Malim. 1995. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J. Virol. 69:4166-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 53.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591-601. [DOI] [PubMed] [Google Scholar]
- 54.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 55.Tian, C., X. Yu, W. Zhang, T. Wang, R. Xu, and X. F. Yu. 2006. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 80:3112-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 57.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 23:2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao, Z., E. Ehrlich, Y. Yu, K. Luo, T. Wang, C. Tian, and X. F. Yu. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349:290-299. [DOI] [PubMed] [Google Scholar]
- 60.Xu, H., E. S. Svarovskaia, R. Barr, Y. Zhang, M. A. Khan, K. Strebel, and V. K. Pathak. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. USA 101:5652-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056-1060. [DOI] [PubMed] [Google Scholar]
- 62.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78:6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]