Abstract
Glycoprotein B (gB), along with gD, gH, and gL, is essential for herpes simplex virus (HSV) entry. The crystal structure of the gB ectodomain revealed it to be an elongated multidomain trimer. We generated and characterized a panel of 67 monoclonal antibodies (MAbs). Eleven of the MAbs had virus-neutralizing activity. To organize gB into functional regions within these domains, we localized the epitopes recognized by the entire panel of MAbs and mapped them onto the crystal structure of gB. Most of the MAbs were directed to continuous or discontinuous epitopes, but several recognized discontinuous epitopes that showed some resistance to denaturation, and we refer to them as pseudo-continuous. Each category contained some MAbs with neutralizing activity. To map continuous epitopes, we used overlapping peptides that spanned the gB ectodomain and measured binding by enzyme-linked immunosorbent assay. To identify discontinuous and pseudocontinuous epitopes, a purified form of the ectodomain of gB, gB(730t), was cleaved by α-chymotrypsin into two major fragments comprising amino acids 98 to 472 (domains I and II) and amino acids 473 to 730 (major parts of domains III, IV, and V). We also constructed a series of gB truncations to augment the other mapping strategies. Finally, we used biosensor analysis to assign the MAbs to competition groups. Together, our results identified four functional regions: (i) one formed by residues within domain I and amino acids 697 to 725 of domain V; (ii) a second formed by residues 391 to 410, residues 454 to 475, and a less-defined region within domain II; (iii) a region containing residues of domain IV that lie close to domain III; and (iv) the first 12 residues of the N terminus that were not resolved in the crystal structure. Our data suggest that multiple domains are critical for gB function.
Herpes simplex virus (HSV) is a neurotropic agent responsible for episodic cold sores and genital legions. After primary infection of mucosal epithelial cells, the virus establishes lifelong latency in sensory neurons, from which it periodically reactivates. After reactivation, the virus migrates along the axons and infects cells at the site of primary infection, causing painful blisters on the surface of the lips in the case of HSV type 1 (HSV-1) or of the genital mucosa for the closely related HSV-2 (48).
A critical event in the life cycle resides in the entry of the virus into target cells. Recent progress has been made in understanding the mechanism governing this process (reviewed in references 21 and 38). Of the 12 different glycoproteins of the viral envelope, 4 have essential functions for entry, namely, glycoprotein B (gB), gD, gH, and gL. First, the virion attaches by interaction of gC and gB with cell surface heparan sulfate proteoglycans (42). Although not essential for entry, this step provides stable interactions between the virion and the cell that favor the next steps. These include the association of gD with one of its three identified receptors—HVEM, nectin-1, and 3-O-sulfated proteoglycans—and that of gB with an as-yet-unidentified receptor(s) (3, 11, 28, 41). Ultimately the viral envelope and the cell plasma or endosomal membrane fuse, a phenomenon that is still poorly understood except that it requires the cooperative function of gB, the heterodimer gH/gL, as well as gD and a gD receptor (36, 44).
The crystal structures of gD alone and bound to HVEM greatly enhanced our understanding of glycoprotein functions during entry (7, 22). Binding of gD to HVEM induces conformational changes in gD whereby the C terminus of the ectodomain is displaced and the N terminus is exposed to the receptor (22). Additional changes in gD are presumed to occur that favor interactions with the fusion core constituted by gB and gH/gL. The mechanism by which the latter proteins effect fusion is not understood, but evidence suggests the formation of a complex of all four essential glycoproteins (12-14, 35).
Recently, the crystal structure of gB from HSV-1 was solved, opening a new era of study (15). The molecule forms a five-domain elongated trimer with notable similarities to the structure of the fusion glycoprotein G of the vesicular stomatitis virus (VSV) (39). This observation strongly suggests that gB is the fusion protein in HSV (43). Considering the high degree of conservation, it is also reasonable to think that gB functions similarly among all members of the herpesvirus family (15).
Several investigators have used mutants and monoclonal antibodies (MAbs) to identify functional domains of HSV gB (16, 25, 33). Functional regions (FRs) were identified in parallel, and previously we attempted to reconcile the two resulting domain classifications (29). Taking advantage of the solved gB structure, we generated new MAbs to gB and characterized their epitopes on the structure. We also compared our MAbs with those that had been generated and characterized in other laboratories. Our panel of MAbs was directed to both continuous and discontinuous epitopes. Interestingly, several that bind to different regions on the surface of gB had virus-neutralizing activity, suggesting that functional sites are dispersed on the structure. More precisely, we identified three overlapping functional epitopes within domain II, one at the interface of domain III and domain IV, one at the tip of domain V, and one in domain I. The domain nomenclature is as reported previously (15). These results constitute a detailed map of FRs in gB and show that multiple domains play important roles in gB function.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney (HEK) 293T cells and mutant mouse fibroblast Gro2C cells derived from L cells, which are deficient in heparan sulfate proteoglycan synthesis (2), were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin (Invitrogen). African green monkey Vero cells were cultured the same way but supplemented with only 5% FCS. B78-H1-derived C10 cells engineered to express nectin-1 (27) were cultured in DMEM with 10% FCS, antibiotic, and 500 μg of G418/ml. Hybridoma cells were grown in Kennett's Hy complete medium, consisting of DMEM containing 10% NCTC-109 medium and 1× Opi (both from Gibco) supplemented with 20% FCS, 50 μM hypoxanthine, 8 μM thymidine, 2 mM l-glutamine, and 3% hybridoma cloning factor (Bio Veris). HSV-1 strain KOS and HSV-1 (KOS)tk12 that carries the β-galactosidase gene under the control of the HSV-1 immediate-early ICP-4 promoter (46) were both purified on sucrose gradients as described previously (14). The Gro2C cells were generously provided by B. Banfield and F. Tufaro. C10 cells and HSV-1 (KOS)tk12 were kindly provided by P. Spear.
Antibodies and reagents.
MAbs DL16 and DL21 were selected among a panel of antibodies generated against BHK cell extracts infected by HSV-1 and 2. DL16 recognizes trimeric gB (3). Mice were also injected with wild-type gB that was purified from lysates of HSV-1- and HSV-2-infected BHK cells by using a MAb DL16 immunosorbent column. The injected antigenic protein mix contained native, as well as alkylated and reduced, proteins. Hybridoma cells were obtained by fusion of SP2/0 cells with splenocytes of immunized mice and then injected into mice to generate ascites. Immunoglobulin G (IgG) was purified from ascites fluids with a Montage protein G spin column (Millipore), dialyzed against phosphate-buffered saline (PBS) and concentrated. Two series of MAbs were obtained, with the initials SS and RC, followed by a number. MAbs A22, C226, and 61.1.1 and MAb series BD were provided by Becton Dickinson. R69 serum was obtained from a rabbit immunized against purified and denatured HSV-1 gB (18). Other MAbs to gB, including the series B1 to B9, 3S to 109S, and H1781 to H1838 (listed in Fig. 1) were described elsewhere and were kindly provided by J. Glorioso, M. Zweig, and L. Pereira, respectively (26, 33, 40). An anti-tetrahistidine tag MAb was purchased from QIAGEN. Protein A, goat anti-mouse, and anti-rabbit antibodies coupled to horseradish peroxidase (HRP) were purchased from Kirkegaard and Perry Laboratories.
FIG. 1.
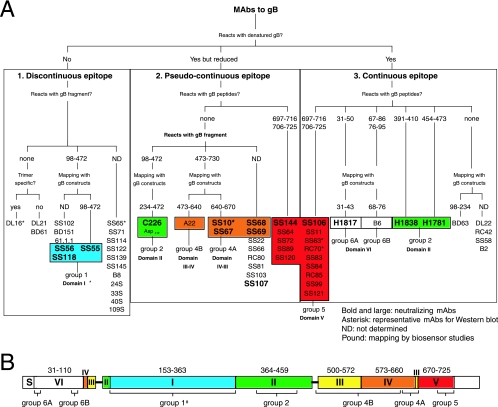
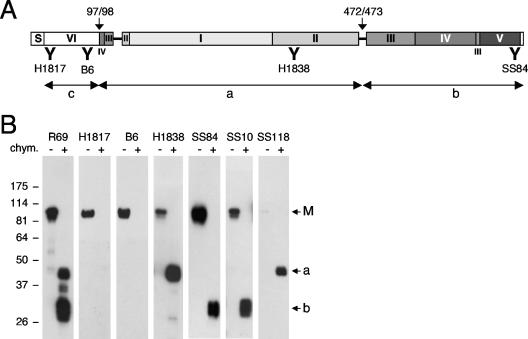
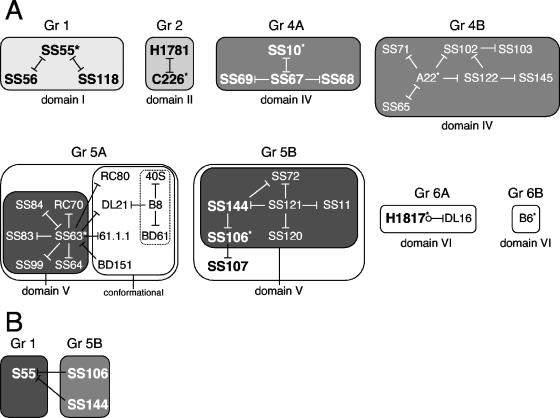
Diagrammatic representation of HSV gB antibodies. (A) MAbs were organized according to several criteria. The first criterion was reactivity of MAbs with denatured gB(730t) by Western blotting. MAbs that did not react with gB under these conditions were placed in box 1 (discontinuous epitope). MAbs reacting with denatured gB but more strongly with native gB were placed in box 2 (pseudocontinuous epitope). If reactivity was similar with both native and denatured gB, MAbs were placed in box 3 (continuous epitope). The MAbs in box 1 were further divided according to their reactivity with proteolytic fragments of gB: MAbs reacted either with gB proteolytic fragment 98-472 or with no fragment (none) or were not characterized by this criterion (ND). When MAbs did not react with fragments, they were shown to be either trimer specific (yes) or not (no). MAb SS55 that did react with proteolytic fragment 98-472 was also characterized by using gB deletion constructs expressed in mammalian cells. The shortest protein positive for this MAb was 98-472. All other MAbs positive for fragment 98-472 were not characterized using gB mutant proteins (ND). MAbs of boxes 2 and 3 were reacted against gB-derived peptides. MAbs were either negative for all overlapping gB peptides (none) or positive, in which case specific peptide sequences are listed. In box 2, MAbs negative for all peptides were characterized with gB fragments as in box 1. MAbs reacted either with fragment 98-472 or 473-730 or were not characterized by this criterion (ND). When a fragment was identified, MAbs were further mapped on the structure by using gB deletion proteins. The minimal sequences required for binding are indicated at branch points (234-472, 473-640, and 640-670), and the specific MAbs are listed below. Similar criteria were used to separate the MAbs of box 3. In each of three categories, MAbs that mapped to an identified domain (domain I to VI) in the crystal structure of gB are grouped (groups 1 to 6; the colors correspond to the ones used to indicate structural domains of gB in Fig. 1B). MAbs with virus-neutralizing activity are shown in boldface (Table 1). MAbs marked with an asterisk are representative of a distinctive pattern seen by Western blotting. (B) The domain architecture of gB as observed in the crystal structure is shown schematically. Amino acid numbers are indicated above the largest segments. Unresolved amino acids 31 to 110 in the crystal structure are referred to as domain VI. S, signal sequence. The location of binding sites for groups of MAbs is shown underneath the corresponding domains. The schematic organization of the gB domains and the color codes are as described previously (15).
Construction of gB mutants.
The gB1 and gB2 mutants truncated at the C terminus were created by changing a specific amino acid codon into a stop codon that also created a BclI restriction site in the gB codon sequence by using the QuikChange site-directed mutagenesis kit (Stratagene) as previously described (8). The QuikChange procedure was also used to generate deletions at the gB N terminus. For these, 30 nucleotide chimeric primers were designed so that the first 15 nucleotides of the primer matched the flanking 5′ end and the last 15 matched the 3′ flanking end of the gB sequence to be deleted. The templates used were plasmids pPEP98 and pTC580 that encode wild-type gB1 (36) and gB2 (5), respectively. A secreted version of the N terminus mutants was also engineered by changing the codon for Met731 of gB1 into a stop codon. The entire gB gene of all constructs was sequenced to ensure against PCR errors. The plasmid pPEP98 and the control empty vector pCAGGS/MCS (30) were kindly provided by P. Spear.
Purification of HSV-1 glycoproteins.
The ectodomain of gB truncated after His724 and terminated by an additional 5 histidine tag [gB(724tHis)] was engineered, expressed from recombinant baculovirus-infected insect cells, and purified essentially as described for gB(730t) (4). The construction procedure differed only by the use of a downstream primer that incorporated the codons for the histidine tag.
Virus neutralization assays.
Serial dilutions of IgGs were mixed with 50 PFU of purified HSV-1 (KOS) in Vero cell culture medium supplemented with heat-inactivated FCS. The MAb-virus mixture was incubated for 1 h at 37°C before being added to a monolayer of Vero cells grown in 48-well plates. Cells were incubated at 37°C for an additional 24 h, fixed with methanol and acetone (2:1 [vol/vol]) for 30 min, and air dried. Plaques were stained by using a cocktail of polyclonal antibody (PAb) to gB, gC, gD, and gH/gL, followed by protein A-HRP and substrate, and then counted (47). The neutralization titer was measured as a 50% plaque reduction. Virus-neutralizing activity was also measured in Gro2C cells by using a β-galactosidase entry assay as previously described (3).
Expression of gB proteins in infected or transfected cells.
B78-C10 cells seeded in T75 flasks were infected with 5 PFU/cell of either HSV-1 (KOS strain) or HSV-2 (333 strain) for 12 h. Cells were washed with PBS, and proteins were extracted with 4 ml of a 50 mM Tris buffer (pH 7.4) containing 150 mM NaCl, 1% Triton X-100, and complete protease inhibitors (Roche). The nuclei were removed by centrifugation at 500 × g for 5 min. Proteins from cleared detergent extracts were then Western blotted and probed with R69 or with MAbs to gB. HEK-293T cells grown in six-well plates (80% confluent) were transfected with 2 μg of endotoxin-free purified plasmid (QIAGEN) encoding gB1, gB2, or plasmids encoding gB mutants by using GenePorter, according to the manufacturer's recommendations (Gene Therapy Systems). After 48 h, cell supernatants were collected, the cells were washed with PBS, the proteins were extracted with 400 μl of Triton X-100-containing buffer, and nuclei were pelleted as described above. Proteins from cleared detergent extracts or from 293T cell supernatants were then either directly Western blotted or else first immunoprecipitated prior to electrophoresis, Western blotted, and probed with antibodies to gB.
Western blotting and immunoprecipitation.
Purified gB(730t) from baculovirus-infected cells, proteins from cell extracts, or proteins from cell supernatants were mixed with an equal volume of polyacrylamide gel electrophoresis (PAGE) sample buffer containing either no reducing agent and 0.2% sodium dodecyl sulfate (SDS) (“native” conditions) or with 200 mM dithiothreitol and 2% SDS (“denaturing” conditions). Proteins from denatured samples were also boiled for 5 min at 100°C before electrophoresis. Proteins were resolved by SDS-PAGE. After transfer to nitrocellulose, strips of membranes were blocked and incubated with PAb R69 or with one of our panel of MAbs to gB at a concentration of 1 μg/ml. Bound IgGs were visualized with secondary antibodies, enhanced chemiluminescence (Amersham), and exposure to X-ray film.
Proteins were also immunoprecipitated with MAbs to gB. Proteins from cell extracts or supernatants were incubated for 1 h at 4°C with 5 μg of IgG and then for an additional hour with protein A-agarose beads (Sigma). Beads were extensively washed with cold PBS, and the proteins were eluted with PAGE sample buffer, boiled, and electrophoresed. After transfer to membranes, the proteins were probed with PAb R69.
Peptide mapping.
Epitope mapping by enzyme-linked immunosorbent assay (ELISA) using peptides was performed as previously described (1, 6) with some modifications. Synthetic 20-mer peptides with biotin at the N terminus were purchased from Mimotopes (Australia). Peptides, numbered 1 to 82, spanned the entire extra cellular portion of gB (amino acids 31 to 773) and overlapped one another by 11 amino acids, except that peptide 82 overlapped peptide 81 by 17 amino acids. Each peptide was dissolved in 5% acetic acid-40% (vol/vol) acetonitrile in water and diluted in PBS to a working concentration of 1 μM. Streptavidin-coated 96-well plates (Pierce) were exposed to 100 pmol of each peptide for 1 h, blocked with 5% milk in PBS-0.1% Tween 20 for 30 min, and incubated with 20 μg of IgG/ml for 1 h. Wells were washed, and bound IgGs were revealed with anti-mouse-HRP. ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid); Moss, Inc.] was added as a substrate, and the absorbance was read at 405 nm.
Chymotrypsin digestion of gB.
Chymotrypsin treatment of gB was carried out as previously described for trypsin (15). Purified gB(730t) from baculovirus-infected cells (4) was further purified by gel filtration chromatography (Superdex S200; GE Healthcare) using 10 mM Tris-HCl (pH 7.6), 100 mM NaCl, and 1 mM EDTA as a buffer and concentrated by using a Millipore Ultra-4 centrifugal filter unit (molecular mass cutoff of 50 kDa). The purified protein was incubated with chymotrypsin at a ratio of 100:1 (wt/wt) for 3 h at room temperature. The reaction was stopped by passing the digested mix over a benzamidine Sepharose column (GE Healthcare). The flowthrough and the wash were concentrated and passed over a Superdex S200 gel filtration column that had been equilibrated in 10 mM Tris-HCl (pH 7.6) and 100 mM NaCl. The eluted fractions containing the chymotrypsin-trimmed core of gB, residues Arg98 to Ala730, were concentrated to 4 to 5 mg/ml by using Millipore Ultra-4 (molecular mass cutoff of 50 kDa). Chymotrypsin cleaves gB between residues Leu97/Arg98 and Gln472/Ser473, as determined by N-terminal sequencing. Cleavage effectively removed residues Ala31 to Leu97, yielding a stable homogeneous product. The chymotryptic core of gB remained a trimer.
IgG blocking studies.
MAb binding studies were carried out on a BIACore X optical biosensor as previously described (1, 20). Briefly, an anti-tetrahistidine MAb was covalently attached to the surface of the biosensor chip type CM5. Next, 250 resonance units of gB(724tHis) was captured via its C-terminal His tag. Purified IgGs (20 μg/ml) were then injected, and association was monitored for 3 min. Blocking studies were performed by injecting a second MAb and recording its association for 3 min.
RESULTS
Eleven of the MAbs to gB have virus-neutralizing activity.
MAbs with virus-neutralizing activity likely interfere with gB function as an essential glycoprotein for HSV entry. We used a plaque reduction assay on Vero cells to test the virus-neutralizing activity of the MAbs. Of the 67 new antibodies tested, 11 reduced the plaque number by 50% at concentrations ranging from 1.2 to 5 μg of IgG/ml (Table 1) . These MAbs also all blocked entry of a virus carrying a β-galactosidase reporter gene (not shown). Among these 11 MAbs, 4 were tested for their ability to block binding of soluble gB to cells (3). SS10, SS55, and SS118 blocked binding, whereas C226 had no effect in this assay (Table 1). To obtain information about gB's FRs, we characterized the epitopes recognized by these and other MAbs and mapped them to the crystal structure. Various criteria were used as described below. Figure 1 summarizes our findings and serves as a guide.
TABLE 1.
MAbs to gB with virus-neutralizing activity
| MAb | Neutralizationa | Blockingb | Categorye | Residues | Group | Domain | FRf |
|---|---|---|---|---|---|---|---|
| SS55 | 2 | +c | DE | 98-472 | 1 | ||
| SS56 | 5 | NDd | DE | 98-472 | 1 | I/II | |
| SS118 | 5 | + | DE | 98-472 | 1 | 1 | |
| SS106 | 5 | ND | CE | 697-725 | 5B | ||
| SS107 | 5 | ND | PE | ND | 5B | V | |
| SS144 | 5 | ND | PE | 697-725 | 5B | ||
| C226 | 1.2 | − | PE | 234-472 | |||
| H1838 | Yes | ND | CE | 391-410 | 2 | II | 2 |
| H1781 | Yes | ND | CE | 454-473 | |||
| SS10 | 5 | + | PE | 640-670 | |||
| SS67 | 2 | ND | PE | 640-670 | 4A | III/IV | 3 |
| SS68 | 1.2 | ND | PE | ND | |||
| SS69 | 1.2 | ND | PE | ND | |||
| H1817 | Yes | ND | CE | 31-43 | 6A | VI | 4 |
| A22 | No | − | PE | 473-640 | 4B | III/IV | None |
That is, the concentration of IgG in μg/ml required to reduce plaque number by >50% on Vero cells. Yes, reported previously (33).
That is, the ability of IgG to block the binding of gB(730t) to Gro2C cells by >50% with a fivefold excess (wt/wt) of IgG to gB.
+, reported previously (3).
ND, not determined.
DE, discontinuous epitope; PE, pseudocontinuous epitope; CE, continuous epitope.
FR, functional region.
MAbs to gB show three distinct patterns by Western blotting.
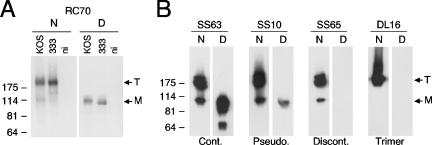
Antibodies were first characterized by Western blotting against extracts of HSV-1- or HSV-2-infected cells or a recombinant form of gB, gB(730t) (Fig. 2). Prior to electrophoresis, samples were either heat denatured and reduced (denatured) or not boiled and not reduced (“native”). All antibodies reacted with gB expressed in lysates of cells infected by either HSV-1 or HSV-2, as well as with gB(730t) (data not shown). This indicates that epitopes were conserved in the different expression systems, that they were conserved between HSV type 1 and 2, and that they were all in the extracellular portion of gB, as exemplified by RC70 in Fig. 2A. Finally, all MAbs also recognized gB expressed on the surface of infected cells by using a cell-based ELISA (data not shown). For these reasons, further characterization of the antibodies was done using gB(730t) or other baculovirus-derived gB fragments.
FIG. 2.
Characterization of MAbs according to their reactivity with gB under different Western blot conditions. (A) Proteins from cells that were infected by HSV-1 (KOS strain) or HSV-2 (333 strain) or not infected (ni) were extracted and separated by SDS-PAGE under native (N) or denaturing (D) conditions. After transfer to nitrocellulose, proteins were probed with RC70 as a representative MAb to gB. The migration positions of the gB trimer (T) and monomer (M) are indicated on the right (arrows). Molecular size markers are shown on the left in kilodaltons. (B) Recombinant gB(730t) was resolved by SDS-PAGE under native (N) or denaturing (D) conditions and transferred to nitrocellulose membrane. The membrane was blocked, and proteins were probed with MAbs (indicated at the top) to gB. The MAbs were representative of the typical patterns seen with any MAbs and used to assemble all MAbs to the three categories listed below. Abbreviations: Cont., continuous epitope; Pseudo., pseudocontinuous epitope; Cont., continuous epitope; Trimer, continuous epitope formed by gB trimer.
MAbs were initially placed in three categories according to their reactivity with gB under different Western blot conditions (Fig. 1A), and representative examples are shown (Fig. 2B). Antibodies in category 1, such as SS63, reacted equally well with denatured and native samples. When gB was electrophoresed under native conditions, these MAbs reacted with both the trimeric and the monomeric forms of gB. When gB was electrophoresed under denaturing conditions, these MAbs reacted with the monomer, as well as with smaller bands resulting from partial degradation. Thus, these antibodies recognize continuous or linear epitopes in the extracellular portion of gB. Antibodies in category 2, such as SS10, also reacted with native and denatured gB but differed from SS63 in two ways. First, the reactivity with denatured gB was always weaker than with the native form of the protein (Fig. 2B). Second, degradation fragments were barely detectable or undetectable. The results indicate that these antibodies recognize a discontinuous epitope or, as termed here, a “pseudocontinuous” epitope that was either partially resistant to denaturation or that reformed during Western blotting. Antibodies in category 3, such as SS65, reacted only with native gB. Most MAbs of this group recognized both monomer and trimer. Thus, these antibodies each recognize a discontinuous or “conformational” epitope present in gB that is sensitive to heat denaturation. One MAb in this family, DL16, was of particular interest since it reacted only with the multimeric form of gB under native conditions (Fig. 2B) (3). We believe that this particular antibody specifically recognizes an epitope that is present only in trimeric gB.
MAbs with neutralizing activity include ones that are directed at continuous epitopes (SS106), pseudocontinuous epitopes (C226, SS10, SS67, SS68, SS69, SS107, and SS144), or discontinuous epitopes (SS55, SS56, and SS118) (Fig. 1A and Table 1). Two strategies were used to further characterize these diverse antibodies. Continuous and pseudocontinuous epitopes were mapped by using a panel of overlapping gB-derived peptides. MAbs to pseudocontinuous epitopes and discontinuous epitopes were tested against gB fragments generated with proteases or against protein constructs with portions of gB deleted.
MAbs to continuous and pseudocontinuous epitopes are mapped by using gB peptides.
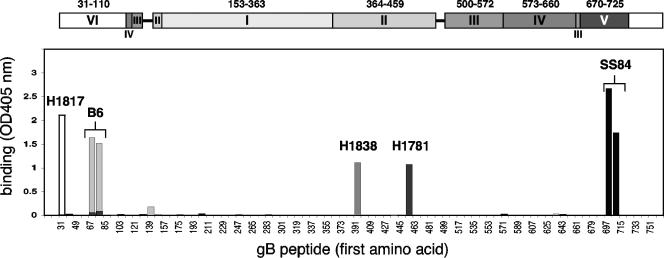
To identify specific epitopes, we used ELISA to screen the reactivity of MAbs to a panel of peptides based on the gB1 sequence. Peptides were 20 amino acids long, spanned the entire ectodomain of gB (amino acids 30 to 773), and overlapped one another by 11 amino acids. Peptides had biotin coupled to their N terminus so that they could be immobilized on streptavidin-coated ELISA plates (Fig. 3). Fourteen MAbs reacted with the same pair of overlapping peptides spanning residues 697 to 725 (Fig. 1A, group 5), as exemplified by SS84 (Fig. 3). These included nine MAbs to continuous epitopes and five MAbs to pseudocontinuous epitopes. Interestingly, of this group, SS106 and SS144 also have virus-neutralizing activity (Table 1) (15). No other linear peptide was recognized by any of the antibodies in our new collection. To validate our method and also to identify other linear epitopes, we screened antibodies generated in other laboratories (Fig. 1A, third box, and Fig. 3). As previously reported (33), antibody H1817 recognized a single peptide spanning residues 31 to 50. In addition, we found that antibodies H1838 and H1781 reacted with single peptides comprising residues 391 to 410 and residues 454 to 473, respectively (15). These results slightly differ with the initial mapping of H1838 and H1781, which localized the epitope of H1838 within a larger fragment comprising amino acids 31 to 220 and the epitope for H1781 to residues 470 to 487 (37). Antibody B6, a complement-dependent neutralizing MAb, recognized a pair of consecutive peptides spanning amino acids 67 to 95. This last result is in agreement with the existence of a monoclonal antibody-resistant (mar) mutation to B6 at position 85 (17). Taken together, these results identify five continuous epitopes spanning amino acids 31 to 50, 67 to 95, 391 to 410, 454 to 473, and 697 to 725 of gB (Fig. 1A, third box). With the exception of the region from amino acids 67 to 95, each of these sequences might be involved in an essential function for virus entry because they bind neutralizing MAbs. Interestingly, residues 391 to 410, 454 to 473, and 697 to 725 are located on the surface of the protein (15). Amino acids 391 to 410 and 454 to 473 are located in domain II, and amino acids 697 to 725 are located in domain V (see Fig. 9). The epitopes for H1817 and B6 map to amino acids 31 to 110, which are missing from the crystal structure. We refer to that region as a putative domain VI. Thus, peptide mapping identified epitopes in three domains of gB. To map MAbs that did not react with peptides, we used partial proteolysis and gB deletion protein constructs.
FIG. 3.
Peptide mapping of MAb to gB by ELISA. Biotin-coupled overlapping peptides mimicking the gB sequence were immobilized on a 96-well plate coated with streptavidin and incubated with 20 μg of purified IgG/ml. Bound IgG was revealed with anti-mouse peroxidase and substrate (ABTS). The absorbance was read at 405 nm. For simplicity, the graph is a composite result of separate experiments with the four MAbs indicated. On the x axis, the numbers refer to the first amino acid of the peptide sequence, and every other peptide is indicated. At the top, the domain architecture of gB is shown in the scale of the x axis. Bar graph binding profiles for MAb H1817 (white), B6 (light gray), H1838 (gray), H1781 (dark gray), and SS84 (black) are presented.
FIG. 9.
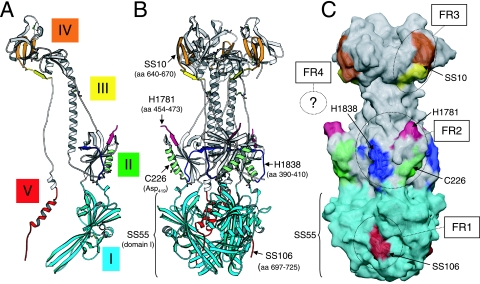
Location of the epitopes of neutralizing MAbs and of FRs on the surface of the crystal structure of gB. (A) Ribbon diagram of a gB protomer showing in color the epitopes of representative neutralizing MAbs. The epitopes of SS10 and SS67 (amino acids 640 to 670) include β-strands 33 to 35 from domain IV (orange) and β-strand 36 from domain III (yellow). The H1838 (blue) and H1781 (magenta) epitopes (residues 391 to 410 and residues 454 to 473, respectively) are linear stretches within domain II. Part of the H1781 epitope is not visible since it is located in a disordered portion of gB (residues 459 to 473). The C226 epitope includes Asp419 located in α-helix B (green) also of domain II. The conformational epitope for SS55 includes unidentified amino acids within domain I (cyan). Finally, the epitope of MAb SS106 (amino acids 697 to 725) is located at the C terminus of domain V (red). Disulfide bonds are shown in ball-and-stick representation. (B) Ribbon diagram of a gB trimer. Color code is as in panel A. (C) Epitopes are rendered on an accessible surface area representation of the gB trimer. FR1 to FR3 are indicated with circles. Since FR4 is present in an unresolved region of gB, it is symbolically represented with a question mark. Colors are as defined in panels A and B.
Identifying pseudocontinuous and discontinuous epitopes: proteolytic digestion of gB shows two major fragments.
The strategy for identifying gB domains involved in the binding of MAbs to pseudocontinuous and conformational epitopes was to use gB fragments generated by proteolysis. Because of its compact structure, gB is mostly resistant to proteases such as trypsin (15). Nevertheless, α-chymotrypsin cleaves gB(730t) between Leu97 and Arg98 and between Gln472 and Ser473 (Fig. 4A). The three resulting fragments comprise (i) domain VI (residues 31 to 110); (ii) domain I (residues 154 to 363) and domain II (residues 364 to 459); and (iii) the major parts of domain III (residues 500 to 572 and residues 661 to 669), domain IV (residues 573 to 660), and domain V (residues 670 to 725), respectively. We analyzed the chymotrypsin degradation products by Western blotting using a PAb and MAbs (Fig. 4B). As expected, uncleaved gB(730t) migrated mostly as a single band of 95 kDa under denaturing conditions as revealed by Western blotting with R69, a PAb to gB (3) (Fig. 4B). When gB(730t) was treated with α-chymotrypsin, a doublet of bands of about 45 kDa (Fig. 4B, band a) and a triplet of about 30 kDa (Fig. 4B, band b) were detected with this PAb. Based on N-terminal sequencing of the individual fragments and of the peptide mapping data for H1838 and SS84 (Fig. 3), we deduced that the 45-kDa fragment corresponds to residues 98 to 472 and that the 30-kDa fragment corresponds to residues 473 to 730. Migration as multiple bands was probably the result of a difference in the state of glycosylation of individual fragments. The short fragment containing the N-terminal domain VI, residues 31 to 97, was likely further degraded because it was not detected by SDS-PAGE and H1817 (Fig. 4A and data not shown).
FIG. 4.
Characterization of gB proteolytic fragments. (A) The different fragments generated by α-chymotrypsin digestion are presented graphically, and MAbs used to identify each fragment are shown underneath. N-terminal sequencing revealed a cleavage site (arrow) between residues Leu97 and Arg98 and another cleavage site between Gln472 and Ser473. The four MAbs used for the identification of each fragment (see panel B) and the position of their binding sites (indicated by a bold “Y”) are shown. (B) Baculovirus expressed gB(730t) was digested (+) or not (−) with α-chymotrypsin and then resolved by SDS-PAGE under denaturing conditions. After transfer to nitrocellulose, fragments were probed with PAb R69 or MAbs to gB (indicated on top). The migration positions of molecular mass markers are shown (left, in kilodaltons). Arrows point to the undigested full-length gB (M) and two prominent cleavage products of 45 (a) and 30 kDa (b). The N-terminal derived fragment (c) was further degraded and thus was not detected.
The two major proteolytic fragments were used to map antibodies that could not be mapped with peptides. MAb SS10 (Fig. 4B) and A22 (not shown) reacted specifically with band b (C terminus). Since SS10 neutralizes virus (Table 1), we conclude that the C-terminal proteolytic fragment of the gB ectodomain contains epitopes that are important for gB function.
Two other neutralizing antibodies, SS55 and SS118, reacted with the 45-kDa band a (N terminus) (shown for SS118 in Fig. 4B). However, detection of this band required a longer film exposure time. This result was particularly interesting for two reasons. First, these MAbs identified an important FR within residues 98 to 472. Second, although SS55 and SS118 recognized the “a” fragment under denaturing conditions, neither MAb recognized undigested gB under these conditions. On the crystal structure, fragment “a” mainly consists of an independently folded module comprising domains I and II. We hypothesize that either they refold after denaturation during the course of electrophoresis or they resist denaturation.
MAbs lose reactivity to gB mammalian proteins truncated at the C terminus or N terminus.
To confirm mapping of the MAbs to linear epitopes to narrow down the mapping or to identify new epitopes, we introduced stop codons into the gB open reading frame to generate additional gB truncations [named according to the format gB(xt), where “x” is the last amino acid before the truncation] at positions suggested by the crystal structure. These mammalian-cell-derived proteins and their expected properties are described in Fig. 5A and Table 2. We also made two sets of gB protein constructs lacking different portions of the N terminus [named according to the format gB(Δy-z), where “y” and “z” refer to the first and last deleted (Δ) amino acids, respectively] in either a full-length molecule or a truncated form. Finally, a gB protein construct from HSV-2 was generated [gB2(727t)] that was truncated at position 727. Constructs were expressed in 293T cells and the secreted proteins or those in extracts of transfected cells were detected by Western blotting with a PAb (R69) and MAbs (Fig. 5 to 7).
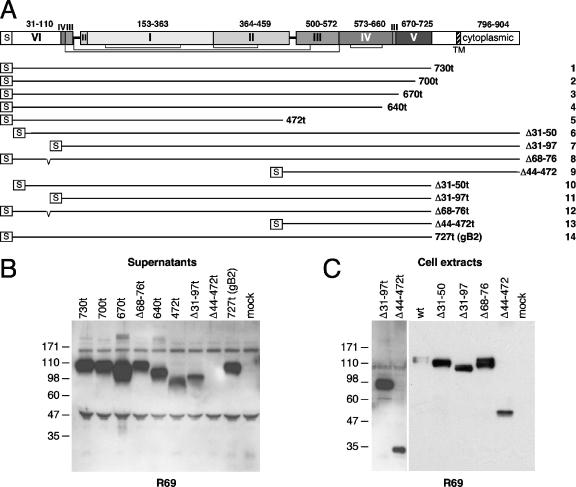
FIG. 5.
Generation and characterization of gB deletion mutants. (A) The portions of gB expressed by deletion mutants are represented by lines (numbered 1 to 14 on the right) of various lengths and are aligned with a diagram of the sequence of the schematic domain architecture of full-length gB. Disulfide bonds are represented schematically underneath by brackets. Each mutant starts with the natural gB signal sequence (S). Mutants 1 to 5 are C-terminal truncations at the positions indicated on the right. The next four mutants (mutants 6 to 9) contain a deletion (Δ) in the N-terminal region indicated on the right. Mutants 10 to 13 have N-terminal deletions indicated on the right and are truncated at position 730. Mutant 14 is derived from gB2 and is truncated after residue Ala727. This residue corresponds to Ala730 of gB type 1 (15). (B) C-terminal truncation mutants indicated on the top were expressed in 293T cells. Secretion of gB proteins in the cell supernatant was analyzed by Western blotting with PAb R69. (C) C- or N-terminal deletion mutants of gB (indicated on the top) were expressed in 293T cells. Proteins from cell extracts were resolved by SDS-PAGE, followed by Western blotting and probing with R69. Migration positions of molecular size markers are indicated on the left in kilodaltons.
TABLE 2.
Description of gB deletion proteins
| Terminus and construct | Residues | Deleted sequence/comments |
|---|---|---|
| C terminus | ||
| gB(730t) | 31-730 | Membrane-proximal, transmembrane, and cytoplasmic domains |
| gB(700t) | 31-700 | Group 5 epitope |
| gB(670t) | 31-670 | Domain V |
| gB(640t) | 31-640 | Shortest deletion keeping all disulfite bonds |
| gB(472t) | 31-472 | C-terminal fragment generated by α-chymotrypsin |
| gB2(727t) | 23-727 | Protein is the type 2 equivalent of gB(730t) |
| N terminus | ||
| gB(Δ31-50) | 51-904 | H1817 epitope (33) |
| gB(Δ31-97) | 97-904 | Short N-terminal fragment cleaved off by α-chymotrypsin |
| gB(Δ68-76) | 31-67 + 77-904 | Potential heparan sulfate-binding site (23) |
| gB(Δ44-472) | 31-43 + 473-904 | Large N-terminal fragment generated by α-chymotrypsin |
| N-term/730t | ||
| gB(Δ31-50t) | 51-730 | Same as gB(Δ31-50) but with truncation at 730 |
| gB(Δ31-97t) | 97-730 | Same as gB(Δ31-97) but with truncation at 730 |
| gB(Δ68-76t) | 31-67 + 77-730 | Same as gB(Δ68-76) but with truncation at 730 |
| gB(Δ44-472t) | 31-43 + 473-730 | Same as gB(Δ44-472) but with truncation at 730 |
FIG. 7.
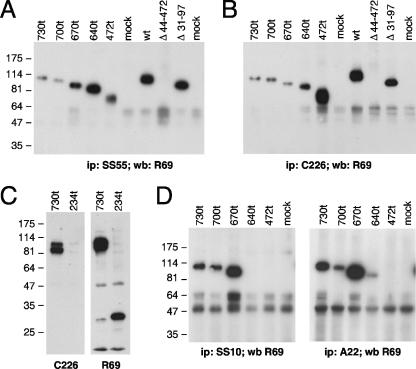
Epitope mapping of MAbs to gB by immunoprecipitation and Western blotting using gB deletion mutants. Mutant proteins were expressed in 293T cells, and their reactivity with MAbs was analyzed. Proteins were either directly probed on a Western blot (C) or were first immunoprecipitated (A, B, and D) with an MAb to gB and then detected by Western blotting with PAb R69. Mutants are labeled at the top of each blot. The MAb used for Western blotting (wb) or immunoprecipitation (ip) is indicated at the bottom of each blot.
As expected, gB proteins truncated at positions 730t, 700t, 670t, 640t, and 472t were detected in the cell supernatant with R69 and displayed the expected sizes by denaturing Western blot (Fig. 5B). Extracts of transfected cells containing full-length gB with deletion Δ31-50, Δ31-97, Δ68-76, or Δ44-472 also displayed the expected SDS-PAGE migration size (Fig. 5C). When a stop codon at position 730 was further engineered into these mutants, all but gB(Δ44-472t) were readily secreted (Fig. 5B). The latter protein was expressed and retained in the cell (Fig. 5C).
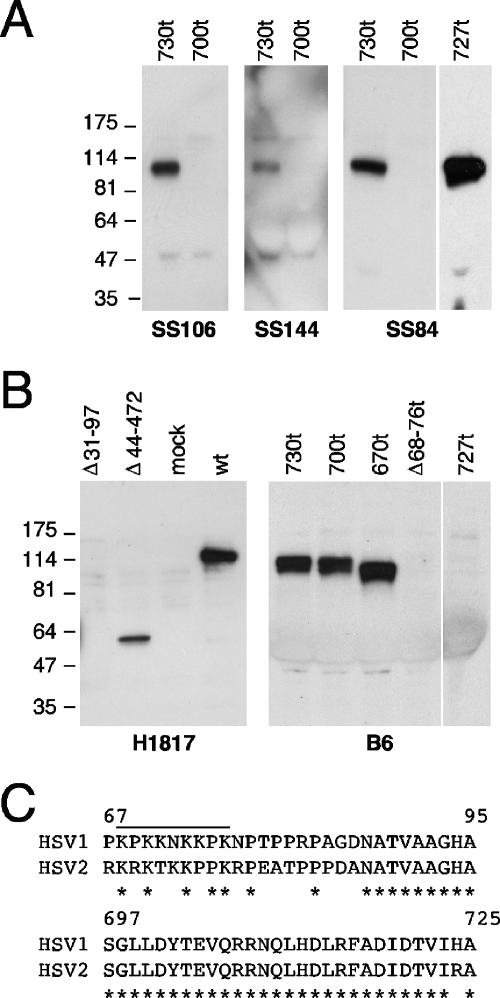
To analyze epitopes at the C terminus, each protein was tested with MAbs SS84, SS106, and SS144 (Fig. 6A) and MAbs SS11 and SS63 (results not shown). These results agree with peptide mapping of these MAbs to amino acids 697 to 725. In addition, SS84 also reacted with gB2(727t), a finding consistent with the high amino acid sequence identity in this stretch of gB1 and gB2 (amino acids 697 to 725, Fig. 6C). MAb H1817 (peptide mapping to residues 31 to 50) failed to react with gB(Δ31-50) or gB(Δ31-97) but did react with gB(Δ44-472) (Fig. 6B), and MAb B6 (peptide mapping to residues 67 to 95) reacted with all proteins except for gB(Δ68-76t) (Fig. 6B) and gB(Δ68-76) (not shown). The predicted heparan sulfate-binding region (23) had been deleted from these last two proteins. B6 was also negative for gB2(727t), a finding consistent with the divergent sequences of gB1 and gB2 in this epitope (Fig. 6C). These results specifically identify the epitopes for H1817 as residues 31 to 43 and demonstrate that residues 68 to 76 were essential for B6 binding. Together, these results indicated that protein constructs had the expected size, reacted appropriately with antibodies to linear epitopes, and were secreted appropriately.
FIG. 6.
Characterization of gB deletion mutants using MAbs to linear epitopes. (A) gB1 and gB2 mutants, truncated at the C terminus, were transiently expressed in 293T cells. Proteins from the cell supernatants were resolved by SDS-PAGE and probed after Western blot with MAbs to linear epitopes of gB. The mutant proteins used are indicated at the top of each blot. The MAbs are indicated at the bottom. (B) Mutants with N-terminal deletions of gB, with either a wild-type C terminus or truncated after residue A730, were expressed in 293T cells. Proteins from cell extracts (probed with MAb H1817) or from cell supernatants (probed with B6) were separated by SDS-PAGE, followed by Western blotting. (C) The sequences of gB types 1 and 2 are aligned to highlight the divergence between the two within residues 67 to 95 and the identity within residues 697 to 725. A line indicate the position of the lysine-rich putative heparan sulfate-binding motif of gB1 (23).
gB protein constructs allow the identification of gB FRs.
To verify proper conformation and identify gB surface epitopes, MAbs were used to detect the gB proteins by immunoprecipitation or Western blotting (Table 3). Priority was given, but not restricted, to neutralizing MAbs to identify functional epitopes, and representatives are shown in Fig. 7. As one example, all of the C-terminal truncation mutants were precipitated by the neutralizing MAb SS55 (Fig. 7A). SS55 also recognized gB(Δ31-97) but not gB(Δ44-472). Thus, the epitope for SS55 is located within residues 98 to 472 (Fig. 1). The results using constructs expressed in mammalian cells agree completely with those using proteolytic fragments of gB expressed in insect cells. Moreover, the results also show that all C-terminal truncation mutants were properly folded.
TABLE 3.
Reactivity of MAbs with gB deletion proteinsa
| Terminus and construct | Reaction (antibody)a
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R69b
|
Gr. 1, DE (SS55) | Group 2
|
Group 4A
|
Group 4B
|
Group 5A
|
Group 6A
|
None, CE (BD63) | |||||||||
| SN | E | CE (H1781) | PE (C226) | PE (SS10) | PE (SS67) | PE (A22) | DE (SS65) | DE (SS102) | DE (SS145) | CE (SS84) | DE (DL21) | CE (H1817) | Trimer (DL16) | |||
| C terminus | ||||||||||||||||
| 730t | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| 700t | + | + | + | + | + | + | + | + | + | + | + | − | + | + | − | + |
| 670t | + | + | + | + | + | + | + | + | + | + | + | − | − | + | − | + |
| 640t | + | + | + | + | − | − | + | − | − | − | − | − | − | + | ||
| 472t | + | + | + | + | − | − | − | − | − | − | − | − | − | + | ||
| 234t | + | − | − | − | − | − | + | |||||||||
| gB2-727t | + | + | + | + | + | +/− | + | +/− | +/− | |||||||
| N terminus | ||||||||||||||||
| Δ31-51 | + | |||||||||||||||
| Δ31-97 | − | + | + | + | + | + | + | + | + | + | + | + | + | − | + | + |
| Δ68-76 | − | + | + | + | + | + | + | + | + | + | + | + | ||||
| Δ44-472 | − | + | − | − | + | + | + | − | ||||||||
| N-term/730t | ||||||||||||||||
| Δ68-76t | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| Δ44-472t | − | + | ||||||||||||||
| wt | ||||||||||||||||
| gB1 | − | + | + | + | + | + | + | + | + | + | ||||||
| gB2 | − | + | + | + | + | + | +/− | |||||||||
Abbreviations: CE, continuous epitope, PE, pseudocontinuous epitope, DE, discontinuous epitope. +, Positive reaction, −, no reaction; +/−, the reaction for gB2 was reduced compared to gB1.
Expression was tested by Western blotting in the cell supernatants (SN) or extracts (E) of transfected 293T cells using the R69 PAb.
As a second example, the neutralizing MAb C226 immunoprecipitated the same set of protein constructs as SS55 (Fig. 7B) and also recognized them by Western blotting (not shown). This MAb was the least sensitive to conformation in our collection of MAbs to pseudocontinuous epitopes (Fig. 7C). In addition, C226, in contrast to PAb R69, failed to recognize a frameshift mutant protein truncated at position 234. Thus, this neutralizing MAb bound to a surface pseudocontinuous epitope within amino acid residues 235 to 472.
Third, the neutralizing MAbs SS10 (Fig. 7D) and SS67 (results not shown) immunoprecipitated protein constructs 730t, 700t, and 670t but not the shorter forms of gB. These MAbs also recognized the same proteins on a Western blot (results not shown). Moreover, A22 immunoprecipitated protein construct 640t, although weakly. Thus, the epitopes for SS10 and SS67 are located within the sequence from positions 640 to 670, and that for A22 involves an adjacent stretch of residues from 473 to 640. Since SS10 and SS67 can neutralize virus infectivity, the 640-670 epitope that includes a β-strand of domain III and part of domain IV defines another FR of gB (Fig. 9). The other part of domain IV, including the epitope to the non-neutralizing MAb A22, might be less critical for function.
gB epitopes are analyzed by biosensor to define groups of MAbs.
To understand the spatial organization of the various gB epitopes, to locate FRs, and to assemble MAbs into groups, we carried out antibody blocking studies by biosensor (Fig. 8). A mutant protein consisting of the first 724 amino acids of the gB ectodomain followed by six histidines [gB(724tHis)] was captured by an anti-His antibody that was covalently linked to the surface of a CM5 biosensor chip. In this way, all of the proteins were oriented similarly and presented in a native structural state. A first MAb was then bound to the immobilized gB(724tHis). Next, a second MAb (test) was injected, and its binding pattern was monitored. We hypothesize that if the second MAb fails to bind to immobilized gB(724tHis), its epitope overlaps that of the first MAb or is hidden in the folded molecule. In contrast, if the second MAb binds efficiently to gB(724tHis), regardless of the first MAb, the epitopes for the two MAbs are spatially distinct and therefore independent (1).
FIG. 8.
Biosensor analysis of competitive MAb binding to gB. (A) Grouping of MAbs. MAbs were grouped based on binding competition as measured by a biosensor. Blocking is symbolized by a line going from the first MAb and ending with a bar facing the second MAb. Bars on both sides of the line indicate that the competition was reciprocal. The absence of a bar between the MAbs of a group means that competition was not studied. Circles indicate that competition was studied but that there was no competition. There was no competition between representative MAbs of each group. Group (Gr) numbers are indicated at the top, and gB domains in which epitope has been mapped are indicated at the bottom. Neutralizing MAbs are shown in boldface and in a larger typeface. MAbs marked with an asterisk are representative of each group. The MAbs of group 1 are neutralizing and conformational and bind to the N-terminal region from residues 98 to 472 (domain I plus domain II). The MAbs of group 2 are neutralizing and bind to a linear (H1838 and H1781) or pseudocontinuous (C226) epitope within domain II. The MAbs of group 4 bind to residues included in domain IV. On the basis of competition with SS10 (residues 640 to 670) and A22 (residues 473 to 640), the MAbs were subdivided into groups 4A (those that are related directly or indirectly to SS10, all neutralizing) and 4B (those related to A22, not neutralizing). A22 and SS10 do not compete for binding to gB. The MAbs of group 5 recognize the C-terminal epitope 697-725. Group 5 is divided into subgroup 5A, with SS63 being its representative, and subgroup 5B, which includes group 5 MAbs that are not blocked by SS63. Three MAbs of the last subgroup have neutralizing activity. Three MAbs (dotted line box) were indirectly associated to group 5A via competition with DL21 and B8. The MAbs of group 6 bind to the N-terminal domain VI. MAbs H1817 (residues 31 to 43) and B6 (residues 67 to 95) do not compete for binding to gB and thus define MAb subgroups 6A and 6B, respectively. (B) Competition experiments with MAbs of different groups. Competition between representative MAbs from different groups for binding to gB was studied by biosensor. The binding of SS55 (group 1) to gB was reduced by both SS106 and SS144 of group 5B.
Initially, each MAb was tested for its ability to bind to immobilized gB(724tHis). Most of the MAbs presented in Fig. 1 bound to captured gB(724tHis), therefore allowing their use in blocking experiments. One exception was BD63, the epitope for which is located between amino acids 98 and 234 (Fig. 1A, third box). This sequence is partially buried in the native molecule (15).
MAbs were preassigned by the above techniques to various groups, and pairs of MAbs were chosen in the blocking studies. MAbs competing for gB binding were assigned to six major competition groups, and the results for all of the biosensor competition data are illustrated graphically (Fig. 8). Below we describe two examples of these data in detail.
Group 1 and group 5B.
Our mapping studies identified the epitope for group 1 MAbs within a large gB fragment that consist mainly of domains I and II (positions 98 to 472). To gain more information on these MAbs, we tested the ability of SS55, as a representative, to block other MAbs from binding to gB. SS55 efficiently blocked the binding of both SS56 and SS118, two other neutralizing MAbs that recognize a conformational epitope within domains I and II (residues 98 to 472). Reciprocally, either MAb blocked the binding of SS55. The data suggest that these three MAbs (group 1) bind to overlapping epitopes within the domain I+II module and that this module is important for gB function.
To further analyze the group 1 MAbs, we did competition experiments with representative MAbs of the other groups. Interestingly, binding of SS55 (group 1) to gB was reduced by both SS106 and SS144, both in group 5B. MAbs of that last group recognize the C-terminal epitope 697-725 (Fig. 1A, domain V). This result suggests that the epitope for SS55 is near the C terminus of gB. Since domain I is closer to the C terminus than domain II, the competition data suggest that the epitope for SS55 is located in domain I (Fig. 9).
Group 2.
The MAbs binding to domain II (H1838, H1781, and C226) define group 2. Of these, the epitopes of H1838 (amino acids 391 to 410) and H1781 (amino acids 454 to 473) were precisely mapped with synthetic peptides. The epitope of C226 was mapped to amino acids 234 to 472. Interestingly, MAb C226 completely blocked the binding of H1781. Blocking was reciprocal. These results suggest that the C226 epitope is close to or overlaps amino acids 454 to 473. It also suggests that the epitope for C226 is located in domain II. Since all MAbs of group 2 have virus-neutralizing activity, we suggest that the middle portion of domain II contains an important FR.
In conclusion, epitope mapping and competition analysis suggest the existence of at least four FRs defined by the binding of neutralizing MAbs (Fig. 9). FR1 includes the epitopes of the MAbs of group 1 (SS55, SS56, and SS118) and group 5B (SS106 and SS144); it is formed by residues within domain I and the sequence from 697 to 725 of domain V. FR2 includes three overlapping epitopes within domain II; these epitopes correspond to MAbs of group 2 (MAbs H1838, H1781, and C226). FR3 is defined by the epitopes of MAbs of group 4A (MAbs SS10, SS67, SS68, and SS69); it includes residues of domain IV closest to domain III. Finally, FR4 consists of the first 12 residues of the N terminus that form the epitope of MAb H1817.
DISCUSSION
The recent determination of the crystal structure of HSV-1 gB represents a major advance in our understanding of herpesvirus entry (15). The high degree of structural homology with the fusion protein G of VSV (VSV-G) strongly suggests that gB is the corresponding fusion protein of HSV (15, 39, 43). Since gB is the most conserved entry glycoprotein among herpesviruses, gB proteins from other herpesviruses seem likely to function as fusion proteins as well. The molecule forms an elongated trimer with distinct domains whose individual contributions to gB function have yet to be defined. In an effort to identify the FR of gB in relation to its structure, we generated and characterized a panel of gB-specific MAbs.
Our study revealed four important FRs associated with domains I, II, IV, and V of the solved crystal structure, as well as a portion of the N-terminal region VI, missing from the structure. Thus, several domains of gB are required for its function in virus entry.
The MAbs used in the present study were generated against a mixture of full-length gB1 and gB2 purified from infected mammalian cells and tested for reactivity with gB. These antibodies reacted with gB in extracts of infected cells and bound the recombinant soluble form of gB [gB(730t)] expressed and purified from baculovirus-infected insect cells, which was used for crystallization. MAbs also recognized gB on the surface of HSV-infected cells. Many antibodies recognized conformational epitopes on gB, and some had neutralizing activity.
Antibodies with neutralizing activity might define a surface involved in interactions with other proteins or with a receptor, while others may bind in a way that would prevent gB from undergoing conformational changes during viral entry. Either would be important for function. Various regions in a protein serve the important functional purpose of maintaining the correctly folded structure. However, neutralizing MAbs do not bind to just any region on the surface of the protein. In this way, not just any region can be described as functional.
Seven out of a total of eleven neutralizing MAbs, including the representative MAbs of groups 2 (C226), 4A (SS10), 4B (A22), and 5B (SS144) define a class of epitope structure we refer to here as pseudocontinuous. These MAbs reacted with gB by Western blotting under denaturing and native conditions of electrophoresis, suggesting that they are directed to a continuous epitope. However, unlike classical MAbs to linear epitopes that react at least as well or better with denatured protein (e.g., SS63), the pseudocontinuous MAbs reacted more strongly with the native form of the protein, showing that conformation played a major role in the epitope structure (e.g., SS10). In addition, MAbs of this class failed to recognize or reacted weakly with synthetic peptides. For example, the crystal structure reveals that the epitope for SS10 (amino acids 640 to 670) encompasses four β-sheets (β33 to β36). That of SS144 (amino acids 697 to 725) contains the long α-helix of domain V (αF). Thus, sheets and helices might represent typical features of pseudocontinuous epitopes. This would explain the resistance of these epitopes to denaturation or their ability to reform after denaturation. More importantly, this type of epitope may represent critical elements for gB function(s).
Recently, we showed that antibodies SS10 (FR3, domains III and IV), SS55 and SS118 (FR1, domain I) inhibit attachment of gB(730t) to an unidentified nonproteoglycan receptor. These MAbs also block entry of HSV into heparan sulfate proteoglycan-deficient cells (3). Taken together, these results suggest that two of the FRs, FR1 and FR3, are important for attachment. These are located in domains I and III-IV, respectively.
FR1 is composed of residues of domain I and the C terminus of domain V.
Three of our most potent neutralizing MAbs—SS55, SS56, and SS118 (group 1)—have been mapped to domain I. Comparison of domain I of HSV gB and domain IV of VSV-G has revealed a significant structural homology (15, 39). In the case of G protein, this domain contains two fusion loops at the tip of an elongated four-strand β-sheet (9, 39). We hypothesize that the corresponding region in gB (15) is also involved in fusion and that binding of these antibodies to domain I interferes with the fusogenic activity of gB. Another remarkable property of domain I revealed by our MAb studies is its ability to either resist denaturation or refold after denaturation. Perhaps this stability may be critical for domain I function.
Fourteen MAbs that compose group 5 bind to a portion of domain V that forms an α-helix (αF) at the C terminus of the crystallized gB ectodomain. Only two MAbs of this group (SS106 and SS144) had virus-neutralizing activity. These two MAbs also competed with several group 1 MAbs, which bind to domain I. Unlike the rest of domain V, which is mostly hidden in the core of gB, the group 5 epitope is exposed. Specifically, the group 5 epitope of each protomer in gB is located between domains I of two neighboring protomers (Fig. 9). Our competition data suggest the existence of a unique FR at the base of gB that consists of residues present in helix αF of domain V and residues present in the neighboring domain I.
One possibility is that αF maintains domain I in close contact with the core of gB constituted mainly of domains III and V. In this conformation the fusion loops in domain I would not be exposed.
FR2 is composed of three overlapping epitopes within domain II.
Earlier we reported that MAbs H1838 and H1781 recognized amino acids 391 to 410 and amino acids 454 to 473, respectively, both within domain II (15, 39). Here we have additionally localized the epitope of the neutralizing MAb C226 to domain II of gB. C226 blocks the binding of H1781 to gB confirming that the two epitopes overlap on the surface of gB. In addition, we isolated a mar HSV-2 mutant to C226 that has a Glu414Lys mutation in gB (data not shown). The corresponding residue Asp419 in HSV-1 gB lies within the α-helix (αB) of domain II (Fig. 9). The amino acid sequence in this region of the protein is highly conserved between gB1 and gB2, and the three MAbs H1838, H1781, and C226 bind to gB from both HSV-1 and HSV-2. Together, these results demonstrate the critical role of domain II in the entry of HSV-1 and probably also HSV-2.
FR3 comprises amino acids located between domain III and domain IV.
Two groups of MAbs bind to domain IV of gB. MAbs of group 4A have virus-neutralizing activity and bind to an epitope within residues 640 to 670. This sequence is highly conserved among herpesviruses (15). Moreover, neutralizing MAbs to cytomegalovirus gB also map to domain IV (31, 45). Thus, domain IV may play an essential role in gB from different herpesviruses.
In contrast, group 4B MAbs do not neutralize virus and bind to less precisely defined epitopes within residues 473 to 640. Most of these epitopes are conformational and do not overlap the group 4A epitope. Thus, the group 4B epitope is located in a region that is likely not directly involved in the function of domain IV.
Antigenic analysis done by other laboratories and identification of FR4.
Anti-gB MAbs have been isolated in other laboratories (10, 26, 32, 34), and two groups undertook a systematic antigenic analysis of gB (17, 33). Domains were identified in parallel, and previously we attempted to reconcile the two resulting domain classifications (29). Among the Glorioso laboratory series of MAbs (B series) that define four distinct sites on gB (sites I to IV), only MAb B4 (site III) neutralizes virus in the absence of complement. It maps to a conformational epitope within residues 283 to 380, and a mar mutant Glu305Lys has been isolated (16, 17). Thus, the B4 epitope is located within domain I and might be a part of FR1. In contrast, MAbs B2 and B5 neutralize virus, but only in the presence of complement. Complement-dependent neutralization implies that the epitope is exposed but that it does not directly contribute to function. The epitope of B2 and B5 (amino acid 594 (16, 17) is located in domain IV and most likely corresponds to group 4B MAbs. Pereira's laboratory developed MAbs (H-series) and defined six major regions, themselves subdivided into many subgroups. Region D1 (residues 31 to 220, domain VI), D2 (residues 303 to 487, domains I and II), D3a (residues 488 to 505, hinge between domains II and III), and Dd5a (residues 630 to 720, domains IV and V) are the target of neutralizing MAbs, but it is unclear whether they require complement (33, 37). mar mutations for MAbs H126, H233, H1375, and H1435 (D2a) map to domain I (19). Together, these previous studies, as well as those reported here, highlight the importance of domain I and confirm that multiple domains contribute to gB function.
In addition, several MAbs of the H series that bind to amino acids 31 to 50 show neutralizing activity, suggesting that the putative domain VI is also functionally important. We narrowed down the epitope for H1817 to residues 31 to 43 that constitute FR4. We are currently testing the function of the N terminus by mutagenesis.
Recent work on HSV-2 gB showed that insertion of two amino acids after residues Pro354 and Ser399 (number starting at the first Met) had little effect on gB2 overall conformation and cell surface expression, although it inhibited gB2 fusion activity (25). The corresponding conserved residues in HSV-1 gB, Pro358 and Ser405, are exposed and lie at the end of domain I (FR1) and in domain II (FR2), respectively. Given the high degree of homology between gB1 and gB2, we hypothesize that these gB2 mutants define the equivalent of our gB1 FR1 and FR2 in gB2.
Final observations.
The crystal structure established with the soluble truncated form of gB, gB730t, revealed structural similarities with the proposed postfusion of VSV G protein (15, 39, 43). This information suggests but does not prove that the crystal structure of gB730t represents a postfusion form of gB. Most of the MAbs of the present study that bind the truncated form of gB also bind gB expressed on the surface of infected cells. Future studies are planned to examine the binding of the MAbs to the virion surface since the virion contains the prefusion form of gB. Thus far, our results indicate that the MAbs studied here recognized both pre- and postfusion gB. Whether some MAbs bind better with a form or the other is a possibility we are investigating.
Domain I and domain II of gB bear a remarkable structural similarity to plekstrin homology (PH) domains (15). Interestingly, PH domains are found in cytosolic proteins involved in intracellular signaling, where they mediate both protein-protein and protein-lipid interactions (24). Recently, we found that gB binds to lipid rafts during virus entry (4). One hypothesis is that the PH domains of gB interact with host cell membranes during virus entry. The fact that the epitopes of several neutralizing MAbs to gB map to domains I and II support the notion that they play an important role in HSV entry.
Our earlier structural studies and the functional map presented here suggest that gB functions as a fusion protein in HSV entry. However, HSV fusion is a complex event that also requires gD, a gD receptor, and the heterodimer gH/gL (36, 44). It may also require a gB receptor (3). Conformational changes occurring in gD as a consequence of receptor binding are likely to induce downstream events affecting in turn the conformation of gB and gH/gL (22). Therefore, changes in gB and gH/gL, as well as the contributions of these glycoproteins to fusion, are the subjects of intensive ongoing studies. In addition, attempts to identify a gB receptor are under way.
Acknowledgments
This study was supported by Public Health Service grant NS-36731 from the National Institute of Neurological Disease and Stroke to R.J.E. and grants AI-056045, AI-18289, and AI-065886 from the National Institute of Allergy and Infectious Disease to R.J.E., G.H.C., and E.E.H., respectively.
We thank Becton, Dickinson and Company for their cooperation in supplying hybridomas, which were critical in this work. We are grateful to L. Pereira, J. Glorioso, P. Spear, and F. Tufaro for reagents. We thank M. Berne of the Tufts Protein Chemistry Facility for N-terminal sequencing. We also thank all of the members of the Cohen and Eisenberg laboratories for critical advice and helpful discussions.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Aldaz-Carroll, L., J. C. Whitbeck, M. Ponce de Leon, H. Lou, L. Hirao, S. N. Isaacs, B. Moss, R. J. Eisenberg, and G. H. Cohen. 2005. Epitope-mapping studies define two major neutralization sites on the vaccinia virus extracellular enveloped virus glycoprotein B5R. J. Virol. 79:6260-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banfield, B. W., Y. Leduc, L. Esford, K. Schubert, and F. Tufaro. 1995. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J. Virol. 69:3290-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, F. C., J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2005. Herpes simplex virus glycoprotein B binds to cell surfaces independently of heparan sulfate and blocks virus entry. J. Virol. 79:11588-11597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, F. C., J. C. Whitbeck, M. Ponce de Leon, H. Lou, R. J. Eisenberg, and G. H. Cohen. 2003. Specific association of glycoprotein B with lipid rafts during herpes simplex virus entry. J. Virol. 77:9542-9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cairns, T. M., D. J. Landsburg, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2005. Contribution of cysteine residues to the structure and function of herpes simplex virus gH/gL. Virology 332:550-562. [DOI] [PubMed] [Google Scholar]
- 6.Cairns, T. M., M. S. Shaner, Y. Zuo, M. Ponce-de-Leon, I. Baribaud, R. J. Eisenberg, G. H. Cohen, and J. C. Whitbeck. 2006. Epitope mapping of herpes simplex virus type 2 gH/gL defines distinct antigenic sites, including some associated with biological function. J. Virol. 80:2596-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 8.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 76:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durrer, P., Y. Gaudin, R. W. Ruigrok, R. Graf, and J. Brunner. 1995. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J. Biol. Chem. 270:17575-17581. [DOI] [PubMed] [Google Scholar]
- 10.Fujinaga, S., T. Sugano, Y. Matsumoto, Y. Masuho, and R. Mori. 1987. Antiviral activities of human monoclonal antibodies to herpes simplex virus. J. Infect. Dis. 155:45-53. [DOI] [PubMed] [Google Scholar]
- 11.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 12.Gianni, T., C. Forghieri, and G. Campadelli-Fiume. 2006. The herpesvirus glycoproteins B and H*L are sequentially recruited to the receptor-bound gD to effect membrane fusion at virus entry. Proc. Natl. Acad. Sci. USA. [DOI] [PMC free article] [PubMed] [Retracted]
- 13.Handler, C. G., G. H. Cohen, and R. J. Eisenberg. 1996. Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J. Virol. 70:6076-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Handler, C. G., R. J. Eisenberg, and G. H. Cohen. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J. Virol. 70:6067-6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 16.Highlander, S. L., W. H. Cai, S. Person, M. Levine, and J. C. Glorioso. 1988. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J. Virol. 62:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Highlander, S. L., D. J. Dorney, P. J. Gage, T. C. Holland, W. Cai, S. Person, M. Levine, and J. C. Glorioso. 1989. Identification of mar mutations in herpes simplex virus type 1 glycoprotein B which alter antigenic structure and function in virus penetration. J. Virol. 63:730-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isola, V. J., R. J. Eisenberg, G. R. Siebert, C. J. Heilman, W. C. Wilcox, and G. H. Cohen. 1989. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J. Virol. 63:2325-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kousoulas, K. G., B. Huo, and L. Pereira. 1988. Antibody-resistant mutations in cross-reactive and type-specific epitopes of herpes simplex virus 1 glycoprotein B map in separate domains. Virology 166:423-431. [DOI] [PubMed] [Google Scholar]
- 20.Krummenacher, C., I. Baribaud, M. Ponce de Leon, J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2000. Localization of a binding site for herpes simplex virus glycoprotein D on herpesvirus entry mediator C by using antireceptor monoclonal antibodies. J. Virol. 74:10863-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krummenacher, C., A. Carfi, R. J. Eisenberg, and G. H. Cohen. 2007. Entry of herpesviruses into cells: the enigma variations. In G. Simmons (ed.), Viral entry into host cells, in press. Landes Bioscience, Austin, TX. [Online.] http://www.eurekah.com/chapter/3035. [DOI] [PubMed]
- 22.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laquerre, S., R. Argnani, D. B. Anderson, S. Zucchini, R. Manservigi, and J. C. Glorioso. 1998. Heparan sulfate proteoglycan binding by herpes simplex virus type 1 glycoproteins B and C, which differ in their contributions to virus attachment, penetration, and cell-to-cell spread. J. Virol. 72:6119-6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemmon, M. A. 2003. Phosphoinositide recognition domains. Traffic 4:201-213. [DOI] [PubMed] [Google Scholar]
- 25.Li, W., T. J. Minova-Foster, D. D. Norton, and M. I. Muggeridge. 2006. Identification of functional domains in herpes simplex virus 2 glycoprotein B. J. Virol. 80:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marlin, S. D., S. L. Highlander, T. C. Holland, M. Levine, and J. C. Glorioso. 1986. Antigenic variation (mar mutations) in herpes simplex virus glycoprotein B can induce temperature-dependent alterations in gB processing and virus production. J. Virol. 59:142-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, C. G., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and N. W. Fraser. 2001. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol. Ther. 3:160-168. [DOI] [PubMed] [Google Scholar]
- 28.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 29.Muggeridge, M. I., S. R. Roberts, V. J. Isola, G. H. Cohen, and R. J. Eisenberg. 1990. Herpes simplex virus, p. 459-481. In A. R. Neurath (ed.), Immunochemistry of viruses, vol. II. The basis for serodiagnosis and vaccines. Elsevier Biochemical Press, Amsterdam, The Netherlands.
- 30.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 31.Ohlin, M., V. A. Sundqvist, M. Mach, B. Wahren, and C. A. Borrebaeck. 1993. Fine specificity of the human immune response to the major neutralization epitopes expressed on cytomegalovirus gp58/116 (gB), as determined with human monoclonal antibodies. J. Virol. 67:703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Para, M. F., M. L. Parish, A. G. Noble, and P. G. Spear. 1985. Potent neutralizing activity associated with anti-glycoprotein D specificity among monoclonal antibodies selected for binding to herpes simplex virions. J. Virol. 55:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pereira, L., M. Ali, K. Kousoulas, B. Huo, and T. Banks. 1989. Domain structure of herpes simplex virus 1 glycoprotein B: neutralizing epitopes map in regions of continuous and discontinuous residues. Virology 172:11-24. [DOI] [PubMed] [Google Scholar]
- 34.Pereira, L., D. Dondero, B. Norrild, and B. Roizman. 1981. Differential immunologic reactivity and processing of glycoproteins gA and gB of herpes simplex virus types 1 and 2 made in Vero and HEp-2 cells. Proc. Natl. Acad. Sci. USA 78:5202-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Romero, P., A. Perez, A. Capul, R. Montgomery, and A. O. Fuller. 2005. Herpes simplex virus entry mediator associates in infected cells in a complex with viral proteins gD and at least gH. J. Virol. 79:4540-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 37.Qadri, I., C. Gimeno, D. Navarro, and L. Pereira. 1991. Mutations in conformation-dependent domains of herpes simplex virus 1 glycoprotein B affect the antigenic properties, dimerization, and transport of the molecule. Virology 180:135-152. [DOI] [PubMed] [Google Scholar]
- 38.Rey, F. A. 2006. Molecular gymnastics at the herpesvirus surface. EMBO Rep. 7:1000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 40.Showalter, S. D., M. Zweig, and B. Hampar. 1981. Monoclonal antibodies to herpes simplex virus type 1 proteins, including the immediate-early protein ICP 4. Infect. Immun. 34:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 42.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Investig. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steven, A. C., and P. G. Spear. 2006. Biochemistry Viral glycoproteins and an evolutionary conundrum. Science 313:177-178. [DOI] [PubMed] [Google Scholar]
- 44.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Utz, U., W. Britt, L. Vugler, and M. Mach. 1989. Identification of a neutralizing epitope on glycoprotein gp58 of human cytomegalovirus. J. Virol. 63:1995-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 47.Watson, R. J., J. H. Weis, J. S. Salstrom, and L. W. Enquist. 1984. Bacterial synthesis of herpes simplex virus types 1 and 2 glycoprotein D antigens. J. Investig. Dermatol. 83:102s-111s. [DOI] [PubMed] [Google Scholar]
- 48.Whitley, R. J., and B. Roizman. 2001. Herpes simplex virus infections. Lancet 357:1513-1518. [DOI] [PubMed] [Google Scholar]