Abstract
KD-247, a humanized monoclonal antibody to an epitope of gp120-V3 tip, has potent cross-neutralizing activity against subtype B primary human immunodeficiency virus type 1 (HIV-1) isolates. To assess how KD-247 escape mutants can be generated, we induced escape variants by exposing bulked primary R5 virus, MOKW, to increasing concentrations of KD-247 in vitro. In the presence of relatively low concentrations of KD-247, viruses with two amino acid mutations (R166K/D167N) in V2 expanded, and under high KD-247 pressure, a V3 tip substitution (P313L) emerged in addition to the V2 mutations. However, a virus with a V2 175P mutation dominated during passaging in the absence of KD-247. Using domain swapping analysis, we demonstrated that the V2 mutations and the P313L mutation in V3 contribute to partial and complete resistance phenotypes against KD-247, respectively. To identify the V2 mutation responsible for the resistance to KD-247, we constructed pseudoviruses with single or double amino acid mutations in V2 and measured their sensitivity to neutralization. Interestingly, the neutralization phenotypes were switched, so that amino acid residue 175 (Pro or Leu) located in the center of V2 was exchanged, indicating that the amino acid at position 175 has a crucial role, dramatically changing the Env oligomeric state on the membrane surface and affecting the neutralization phenotype against not only anti-V3 antibody but also recombinant soluble CD4. These data suggested that HIV-1 can escape from anti-V3 antibody attack by changing the conformation of the functional envelope oligomer by acquiring mutations in the V2 region in environments with relatively low antibody concentrations.
The envelope protein (Env) of human immunodeficiency virus type 1 (HIV-1) presents on the virus surface as “spikes” composed of trimers comprising three gp120-gp41 complexes (6, 32, 33). Among the regions that induce the neutralization antibody (NAb) response, the third variable domain (V3 loop) of gp120 is considered one of the major targets of the host immune response (23, 69). It has been estimated that as much as half of the antibody response against HIV-1 Env in patient serum is directed against the V3 region (43). A recent crystallographic study revealed that the V3 loop contains features that are essential for coreceptor binding and that the extended nature and antibody accessibility of V3 are associated with its immunodominance (20).
HIV-1 primary isolates are relatively resistant to neutralization by NAbs and recombinant soluble CD4 (rsCD4) compared with variants selected for growth in permanent cell lines (42, 52, 55). Studies addressing differences between neutralization-sensitive and -resistant variants have revealed the involvement of several mechanisms that underlie the neutralization resistance of primary isolates, including the occlusion of epitopes within the oligomer, extensive glycosylation, and extension of variable loops from the surface of the complex, as well as steric and conformational blocking of receptor binding sites (7, 12, 32, 38, 49, 50, 54, 62). The structural features of gp120 tolerate a vast array of mutations that permit the selection of neutralization escape variants, as has been previously demonstrated in culture assays, animal models, and infected individuals (24).
Although there are ample data showing that NAbs can protect against HIV-1 infection in vitro and in animal models in vivo, activity in infected humans remains controversial (3, 4, 9, 14, 22, 40, 48, 58). Studies addressing NAbs in primary infections have suggested that most recently infected individuals mount a vigorous antibody response against autologous viruses. However, the rapid evolution of HIV in the presence of NAbs results in the emergence of escape mutants. As a consequence, at any time during an early stage of the HIV disease, NAbs are more likely to recognize earlier autologous viruses than contemporaneous ones. Despite evidence of phenotypic resistance, the genetic basis of the mechanism allowing primary viruses to escape from NAbs is poorly understood. Wei et al. found that glycosylation in the envelope plays an important role in allowing escape from neutralization (62). In contrast, in a recent study Frost et al. found that viral escape from NAbs is correlated with the rate of amino acid substitution rather than changes in glycosylation or insertions or deletions in the envelope (14). Because of the polyclonal nature of NAbs in patient sera, it is difficult to clarify the genetic mechanism responsible for neutralization escape.
Neutralization escape from anti-V3 monoclonal antibodies (MAbs) has been induced in T-cell-line-adapted viruses in several experiments and associated with amino acid substitution within the epitope in the V3 loop (8, 37, 65). However, Park et al. showed that human sera with neutralizing antibodies that contained polyclonal antibodies directed at the V3 region induced neutralization-resistant variants without V3 amino acid substitution (46). Neutralization studies using anti-V3 antibodies against primary isolates suggest that the neutralization resistance phenotype is associated with changes in the sequences outside V3, rather than variation within the V3 epitope (29, 62). However, the contribution of each change in the envelope to the emergence of escape mutants remains unclear because they are not selected under neutralizing MAb pressure.
Recently, we described a humanized MAb, KD-247, that displayed cross-neutralizing activity against HIV-1 clade B isolates (11). The epitope of KD-247 was mapped to six amino acids around the PGR core sequence at the tip of the V3 loop. The shortest reactive peptide recognized by KD-247 was determined to be IGPGR, which is shared by 49% of HIV-1 isolates in clade B (35). In addition, complete protection from challenge infection by a pathogenic strain of simian-human immunodeficiency virus 89.6 was observed when a high concentration of the antibody was used in an animal model (10). A molecularly cloned CCR5-tropic HIV-1 strain, JR-FL, which is relatively resistant to neutralization (15, 50), was exposed to KD-247 to obtain a neutralization escape mutant (67). Induction of the neutralization-resistant virus with a mutation in the V3 tip was observed in the presence of a high concentration of KD-247, and the escape variant was found to be more sensitive to CCR5 inhibitors and rsCD4 than the original strain (67).
The present study sought to understand how virus mutation impacts the activity of an anti-V3 MAb, KD-247. For this we subjected a primary R5 virus, MOKW, to selection pressure by KD-247. The present data suggested that it is necessary to pass a phased step so that the escape mutant against the anti-V3 antibody can emerge. Neutralization escape variants with V2 mutations in gp120 could be selected at relatively low KD-247 pressures, but high concentrations of KD-247 were required for induction of a completely resistant variant with amino acid substitution in the epitope. Moreover, we present evidence suggesting that some V2 mutations change the tertiary or quaternary structure of the envelope trimers on the viral surface that are involved in the neutralization resistance of the primary isolate. Clarification of the mechanisms responsible for this neutralization resistance may provide important insight into possible methods for the induction of potent and cross-neutralizing antibody responses capable of neutralizing various primary isolates.
(This work was presented in part at the 13th Conference on Retroviruses and Opportunistic Infection, Denver, CO, 5 to 8 February 2006 [55a].)
MATERIALS AND METHODS
Cells, culture conditions, reagents, and viruses.
PM1/CCR5 cells (68) were maintained in RPMI 1640 medium (Sigma) supplemented with 10% heat-inactivated fetal calf serum (FCS; HyClone Laboratories, Logan, UT), 50 U/ml of penicillin, 50 mg/ml of streptomycin, and 100 μg/ml of G418 (Sigma). 293T cells were maintained in Dulbecco's modified Eagle medium (Sigma) supplemented with 10% heat-inactivated FCS. The CD4- and CCR5-expressing human osteogenic sarcoma cell line GHOST-hi5 was maintained in Dulbecco's modified Eagle medium supplemented with 10% FCS, G418 (200 μg/ml), hygromycin B (100 μg/ml; Sigma), and puromycin (1 μg/ml; Sigma).
KD-247, an anti-gp120-V3 antibody, was produced as previously described (11). 17b, a monoclonal antibody against the CD4-induced epitope, and immunoglobulin Gb12 (IgGb12), a monoclonal antibody against the CD4-binding epitope, were provided by the National Institutes of Health AIDS Research and Reference Reagent Program. 447-52D, an anti-gp120 V3 MAb, was a gift from Suzan Zolla-Pazner (New York University School of Medicine). 2D7, an anti-CCR5 MAb, and RPA-T4, an anti-CD4 MAb, were purchased from BD Biosciences Pharmingen (San Jose, CA). Human rsCD4 was purchased from R&D Systems, Inc. (Minneapolis, MN). TAK-779, a CCR5 inhibitor, was kindly provided by Takeda Chemical Industries, Ltd. (Osaka, Japan). AK-602, a CCR5 inhibitor, was gifted by Ono Pharmaceutical Co., Ltd. (Osaka, Japan).
The R5 primary HIV-1, MOKW virus, was isolated from a drug-naïve Japanese patient (36). This virus was passaged in phytohemagglutinin-activated peripheral blood mononuclear cells (PBMCs), and the culture supernatant was stored at −80°C until use.
Isolation of a KD-247-resistant mutant from MOKW virus in vitro.
The selection of KD-247 escape variants from MOKW virus was performed as previously described (67). Briefly, MOKW virus was preincubated in the presence of KD-247 for 30 min at 37°C, and then PM1/CCR5 cells (4 × 104) were exposed to 500 times the 50% tissue culture infective dose (TCID50) of the preincubated MOKW. After incubation for 5 h at 37°C, cells were pelleted down and resuspended in RPMI 1640 medium supplemented with 10% FCS without KD-247. Viral replication was monitored by observation of the cytopathic effects in PM1/CCR5 cells. The culture supernatant was harvested on day 7 and used to infect fresh PM1/CCR5 cells for the next round of culture in the presence of increasing concentrations of KD-247. When the virus began to propagate rapidly in the presence of KD-247, the MAb concentration was further increased. After the virus was passaged in the presence of up to 2,000 μg/ml KD-247 in PM1/CCR5 cells, a KD-247-resistant virus, MOKW9p(2000), was recovered from the cell culture supernatant. MOKW virus was also passaged for the same time period in PM1/CCR5 cells in the absence of KD-247, and the resulting virus was designated MOKW9p(−).
Amplification of viral cDNA and nucleotide sequencing.
Viral RNA was extracted from cell culture supernatants with several concentrations of KD-247 using a QIAamp viral RNA kit (QIAGEN). Viral RNAs were reverse transcribed using a High Capacity cDNA Archive Kit (Applied Biosystems) with specific antisense primer ENVN (5′-CTGCCAATCAGGGAAGTAGCCTTGTGT-3′). Nested PCR was performed to amplify the gp120 C1 to C4 coding region as described previously (60). The primers used were as follows: for the first-step PCR, 1B (5′-AGAAAGAGCAGAAGACAGTGGCAATGA-3′) and H (5′-TAGTGCTTCCTGCTGCTCCCAAGAACCC-3′); for the second-step PCR, 2B (5′-AGCAGAAGACAGTGGCAATGAGAGTGA-3′) and F (5′-ATATAATTCACTTCTCCAATTGTCCCTCAT-3′). The products of the nested PCR were inserted in the TA vector (Invitrogen) and sequenced using a Big Dye Terminator, version 1.1 (Applied Biosystems), in accordance with the manufacturer's instructions.
MTT assay.
The neutralization-sensitivities of each passaged MOKW virus to KD-247 were determined as previously described (67). Briefly, PM1/CCR5 cells (2 × 103 cells/well) were exposed to 100 TCID50 of each passaged virus in the presence of various concentrations of KD-247 in 96-well round-bottom microculture plates and incubated at 37°C for 7 days. After removal of 100 μl of medium from each well, 10 μl of MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide) solution (7.5 mg/ml) in phosphate-buffered saline (PBS) was added to each well, and the plate was incubated at 37°C for 3 h. After incubation, 100 μl of acidified isopropanol containing 4% (vol/vol) Triton X-100 was added to each well to dissolve the formazan crystals. The optical density (wavelength, 570 nm) was measured using a microplate reader. Assays were performed in duplicate or triplicate.
Construction of mutant envelope expression vectors.
Proviral DNA was extracted from each batch of passaged MOKW virus-infected PM1/CCR5 cells using a QIAamp DNA blood mini kit (QIAGEN). For the construction of each passaged envelope expression vector, we used pCXN2, which has a chicken actin promoter. Briefly, we amplified MOKW gp160 regions using LA Taq (Takara) with primers ENVA (5′-GGCTTAGGCATCTCCTATGGCAGGAAGAA-3′) and ENVN (see above). The products of the PCR were inserted into pCR-XL-TOPO (Invitrogen). The sequences of the amplified env region of MOKW virus were confirmed using an ABI377 automated DNA sequencer. The EcoRI fragment of pCR-XL-MOKW containing the entire env region was ligated into pCXN2 to give pCXN-MOKW-RDP (the last three letters of the MOKW virus constructs represent the amino acids at positions 166, 167, and 175 in the V2 region), pCXN-MOKW-KNL/C3m, and pCXN-MOKW-KNL/V3m. The pCXN-MOKW-KNL vector was constructed by replacing the StuI-Bsu36I fragment of pCXN-MOKW-KNL/C3m with a corresponding MOKW-RDP fragment. The pCXN-MOKW-RDP/V3m and pCXN-MOKW-RDP/C3m vectors were constructed by replacing the StuI-Bsu36I fragment of pCXN-MOKW-RDP with the corresponding pCXN-MOKW-KNL/V3m or pCXN-MOKW-KNL/C3m fragments, respectively. pCXN-MOKW-KNP, pCXN-MOKW-RDL, pCXN-MOKW-KDL, and pCXN-MOKW-RNL were generated by site-directed mutagenesis using a QuickChange Site-Directed Mutagenesis Kit (Stratagene) in accordance with the manufacturer's instructions.
Pseudovirus preparation.
Five micrograms of pNL4-3.luc.R−E− and 0.5 μg of pRSV-Rev (18), supplied by the NIH AIDS Research and Reference Reagent Program, and 4.5 μg of the MOKW Env-expressing pCXN2 were cotransfected into 293T cells using Effectene transfection reagent (QIAGEN). At 24 h after the transfection, the pseudovirus-containing supernatants were harvested, filtered through a 0.2-μm-pore-size filter, and stored at −80°C. To measure the pseudovirus activity, a luminescence assay using GHOST-hi5 cells was used as previously described (60).
Neutralization assays.
A single-cycle infectivity assay was used to measure the neutralization of MOKW pseudoviruses as described previously (60). Briefly, MAbs at various concentrations and a pseudovirus suspension corresponding to 100 TCID50 were preincubated on ice for 15 min. The virus-antibody mixtures were added to GHOST-hi5 cells, which on the preceding day had been seeded in a 96-well plate (1.5 × 104 cells/well). Cultures were incubated for 2 days at 37°C, washed with PBS, and lysed with lysis buffer (Luc PGC-50; PicaGene). Following transfer of the cell lysates to luminometer plates (Coastar 3912), the luciferase activity (in relative light units) in each well was measured using luciferase substrate (100 μl/well; PicaGene) in a TR717 microplate luminometer (Applied Biosystems). The reduction in infectivity was determined by comparing the relative light units in the presence and absence of MAbs and was expressed as the percentage of neutralization. The same assay was repeated two to three times.
In vitro binding assay to the MOKW envelope-expressing cell surfaces.
In vitro binding assays were performed as previously described (53, 67). EcoRI fragments of MOKW env genes from pXL-MOKWs were subcloned into the corresponding sites in pDNR-1r (Clontech). The vectors were sequenced to confirm the presence of the desired env gene and the absence of other changes. The env gene fragments were then subcloned into pLP-IRES2-EGFP (Clontech) using Cre-recombinase (Clontech) in accordance with the manufacturer's instructions. 293T cells were cotransfected with pRSV-Rev (0.5 μg) and pLP-IRES2-EGFP-MOKW (9.5 μg) using the Effectene transfection reagent. After 36 h, the cells were harvested, incubated with each anti-HIV-1 MAb in combination with biotin-conjugated anti-human IgG and peridinin chlorophyll protein-conjugated streptavidin (BD Pharmingen), gated for the green fluorescent protein (GFP)-positive area, and analyzed using a FACSCalibur flow cytometry system.
MAb-gp120 binding assay.
Culture supernatants containing the pseudotyped viruses were treated with 1% nonionic Nonidet P-40 to provide a source of gp120. Binding assays for MAbs to gp120 were then performed essentially as described elsewhere (41, 59). Briefly, gp120 proteins from transfected culture supernatants, diluted in Tris-buffered saline containing 10% FCS and 1% Nonidet P-40, were captured onto solid phase via their carboxyl termini using sheep polyclonal antibody D7324 (Aalto Bioreagents, Dublin, Ireland). MAb was added in PBS containing 10% FCS and 0.1% nonionic detergent Tween 20, and bound MAb was detected with alkaline phosphatase-conjugated goat anti-human IgG (Sigma) followed by the addition of phosphatase substrate (Sigma). A405 measurements were taken using a microplate reader.
Construction of chimeric NL4-3/MOKW env proviruses.
Chimeric proviruses were constructed from the pNL4-3 proviral plasmid (AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases) by overlapping PCR as previously described, with minor modifications (31). Briefly, the gp160 coding sequences were amplified from the cloning vectors using the primers EnvFv (5′-AGCAGAAGACAGTGGCAATGAGAGCGAAG-3′) and EnvR (5′-TTTTGACCACTTGCCACCCATCTTATAGC-3′). A portion of the NL4-3 provirus from nucleotides 5284 to 6232 was amplified with primers NL(5284)F (5′-GGTCAGGGAGTCTCCATAGAATGGAGG-3′) and NL(6232)Rv (5′-CTTCGCTCTCATTGCCACTGTCTTCTGCT-3′). This fragment encompasses the unique EcoRI restriction site in pNL4-3. Another fragment from the NL4-3 provirus spanning nucleotides 8779 to 9045 was amplified using the primers NL(8779)F (5′-GCTATAAGATGGGTGGCAAGTGGTCAAAA-3′) and NL(9045)R (5′-GATCTACAGCTGCCTTGTAAGTCATTGGTC-3′). This fragment includes the unique XhoI restriction site in pNL4-3. Overlapping PCR was used to join the gp160 coding sequence from the desired clone to the fragment encompassing bases 8779 to 9045 that had been amplified from pNL4-3. The resulting fragment was then similarly joined to the amplified fragment encompassing bases 5284 to 6232 from pNL4-3. The product was digested with EcoRI and XhoI and subcloned into the corresponding site in pBluescript SK(+) (Stratagene) for sequencing and subsequent manipulation. The EcoRI-XhoI fragment for each env gene was then subcloned back into pNL4-3. The results were proviral plasmids that differed from each other only in the env gene. The resulting plasmids were designated pNL-MOKW-RDL and pNL-MOKW-KNL.
Virus preparation and viral replication assay in PM1/CCR5 cells.
Three micrograms of pNL-MOKW-RDL or pNL-MOKW-KNL was transfected into 293T cells using the Effectene transfection reagent. At 24 h after transfection, the virus-containing supernatants were harvested, filtered through a 0.2-μm-pore-size filter, and frozen in aliquots at −150°C. Viral yields were quantified by a RETROtek HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) kit (ZeptoMetrix). PM1/CCR5 cells (1 × 104) were exposed to NL4-3/MOKW env chimeric viruses corresponding to 10 ng of p24 and then preincubated for 4 h at 37°C. After incubation, cells were pelleted down and resuspended in RPMI 1640 medium supplemented with 10% FCS. Viral replication was monitored by measuring p24 kinetics in duplicate.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the DNA Data Bank of Japan under accession numbers AB262847 to AB262951 (passaged samples) and AB262952 to AB262961 (env expression vectors).
RESULTS
Anti-HIV-1 activity of KD-247 for the primary R5 isolate, MOKW virus.
KD-247 recognized an epitope that contains the IGPGR sequence located at the tip of V3 and neutralized primary HIV-1 in clade B with matching sequence motifs (11). To study how bulked primary R5 virus can escape from anti-V3 antibody, we selected a genetically heterogenous HIV-1 primary isolate, MOKW, rather than a molecular clone to allow escape mutants to be selected from a quasi-species pool as well as to be generated de novo. MOKW virus was isolated by standard PBMC culture from a drug-naïve Japanese patient infected with HIV-1 by heterosexual contact (36). The isolate was sensitive to neutralization by KD-247 with a 50% inhibitory concentration (IC50) of 3.4 μg/ml, which is comparable to the IC50 values of the Ba-L, JR-FL, and 89.6 viruses (data not shown).
Selection of KD-247 escape mutants from MOKW virus.
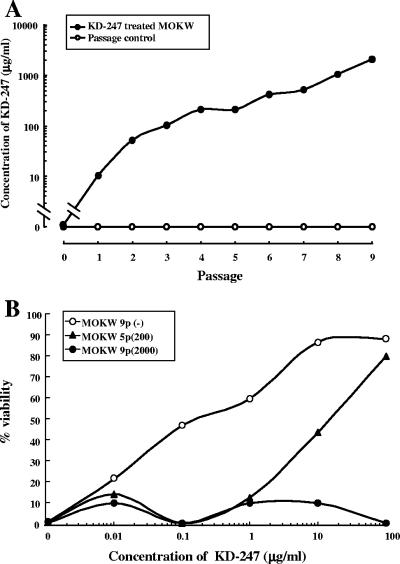
To select an HIV-1 variant that could escape neutralization by KD-247 in vitro, we exposed PM1/CCR5 cells to MOKW virus and serially passaged the virus in the presence of increasing concentrations of KD-247. PM1/CCR5 cells were highly sensitive to both X4 and R5 HIV infection and were accompanied by prominent syncytia (68). As a control, MOKW virus was passaged under the same conditions but without KD-247 to allow us to monitor spontaneous changes that occurred in the virus during prolonged PM1/CCR5 cell passaging. The selected virus was initially propagated in the presence of 10 μg/ml KD-247, and during the course of the selection procedure, the MAb concentration was increased to 2,000 μg/ml (Fig. 1A). After five rounds of passaging, a viral variant, designated MOKW5p(200), arose that replicated in the presence of 200 μg of KD-247 per ml. Moreover, after nine rounds of passaging, a viral variant, designated MOKW9p(2000), arose that infected PM1/CCR5 cells efficiently in the presence of 2,000 μg/ml KD-247 (Fig. 1A). We harvested each passaged virus and a passaged control virus, designated MOKW9p(−), and evaluated their sensitivity to KD-247 using the MTT assay (Fig. 1B). The IC50 values of KD-247 against the MOKW9p(−), MOKW5p(200), and MOKW9p(2000) viruses were 0.15 μg/ml, 16 μg/ml, and >100 μg/ml, respectively, indicating that MOKW virus had acquired a resistance phenotype against KD-247 during the in vitro selection.
FIG. 1.
Selection of neutralization-resistant virus variants against KD-247. (A) The selection was carried out in PM1/CCR5 cells, as described in Materials and Methods. (B) Sensitivity of MOKW5p(200) and MOKW9p(2000) virus to KD-247 as determined by MTT assay. PM1/CCR5 cells (2 × 103) were exposed to 100 TCID50 of MOKW9p(−), MOKW5p(200), or MOKW9p(2000) virus and were cultured in the presence of various concentrations of KD-247. The IC50 values were determined by MTT assay on day 7 of culture. The assay was conducted in duplicate. The values shown are representative of three separate experiments.
Sequences of the envelope region of the KD-247 escape mutants.
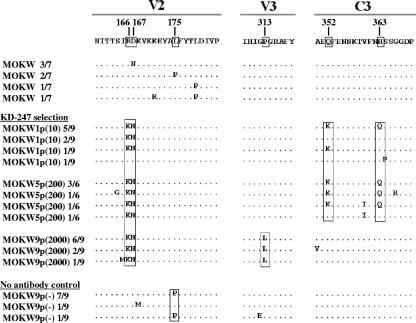
To determine the genetic basis underlying the resistance of the variant MOKW strains, the env gene was amplified and sequenced. The C1 to C4 regions of the envelope were sequenced after cloning the PCR product for each region using cDNAs synthesized from viral RNAs obtained from the supernatants of infected cells, as previously described (67). A total of six to nine clones for each sample from PCR products from the passaged viruses were isolated and sequenced.
Before selection by KD-247 was begun, the V2 regions of MOKW env had variable amino acid sequences (Fig. 2). In the first passage, two amino acid mutations in the V2 region and two amino acid mutations in the C3 region (five of nine clones) appeared, and after the fifth passage, the ratios of V2 and C3 mutated variants had further increased (seven of eight clones). However, after the ninth passage, the C3 mutations had completely disappeared, and a Pro-to-Leu substitution (P313L) in the V3 tip emerged in addition to the mutations in the V2 region (Fig. 2). The appearance of escape mutants with a V3 tip mutation was anticipated, because prior studies on the profile of KD-247 binding to peptides suggested that amino acid substitution at the V3 tip abrogates MAb binding (11). Some changes in the envelope sequence in other regions, including C1, V1, C2, V4, and C4, of the escape mutant were found even at early time points in the presence of the selective pressure. It is possible that these mutations also confer resistance to KD-247 but lead to viruses with decreased fitness, and thus they did not expand in the subsequent passage (Fig. 2 and data not shown). The virus passaged in PM1/CCR5 cells without KD-247 did not show the P313L substitution at passage 9 (zero of nine clones) (Fig. 2). However, accumulation of a mutation of leucine to proline at position 175 (L175P) in the V2 region was observed in the culture without KD-247. This mutation was not found in any passaged variants with KD-247.
FIG. 2.
Amino acid sequences of gp120 from the supernatants of MOKW-infected PM1/CCR5 cells passaged in the presence or absence of KD-247. Viral RNA from the cell culture supernatants at several concentrations of KD-247 was reverse transcribed. After the obtained cDNAs were subjected to PCR amplification and cloning, the env regions in the viruses passaged in the presence or absence of KD-247 were sequenced. The V2, V3, and C3 regions are indicated. The top amino acid sequence represents one of the major sequences from supernatants of MOKW-infected PBMCs. The locations and numbers of specific amino acids, based on the HXB2 sequence, are shown above the consensus line. The number of clones with the listed sequence among the total number of clones tested is given after each designation. For each set of clones, the deduced amino acid sequence of the gp120 was aligned by the single amino acid code. Dots denote sequence identity.
Neutralization sensitivities of mutated MOKW pseudoviruses.
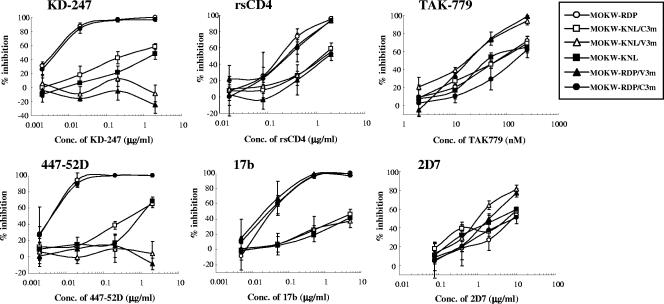
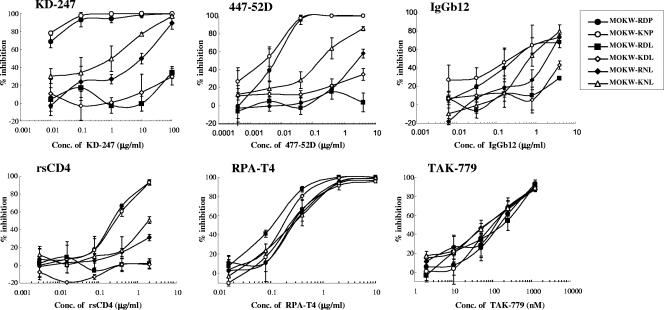
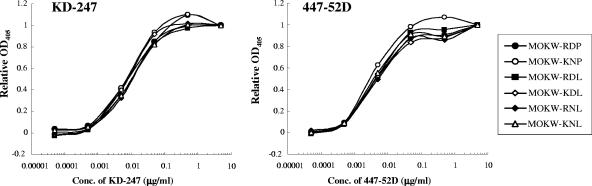
To determine which substitutions were responsible for KD-247 resistance, we constructed luciferase-reporter viruses which were pseudotyped with the representative envelopes of MOKW5p(200), MOKW9p(2000), and passaged viruses without KD-247 [MOKW9p(−)], and were designated MOKW-KNL/C3m, MOKW-KNL/V3m, and MOKW-RDP virus, respectively (Fig. 3). Chimeric envelopes were constructed by replacing the mutated-region (V2, V3, or C3) with a corresponding MOKW-RDP virus (designated MOKW-KNL, MOKW-RDP/V3m, and MOKW-RDP/C3m, respectively), and then sensitivity was compared with that of the passaged virus without KD-247. As shown in Fig. 4, the V3-tip-mutated pseudoviruses, MOKW-KNL/V3m and MOKW-RDP/V3m, were completely resistant to KD-247 (>25,000-fold), whereas V2-mutated viruses, MOKW-KNL/C3m and MOKW-KNL, were only partially resistant (125-fold and 500-fold, respectively) (Fig. 4 and Table 1). The involvement in neutralization resistance of mutation in the V1/V2 region has been reported by number of researchers (12, 49, 50, 54). Our results show that the MOKW variants that had V2 mutations and a resistance phenotype against KD-247 were selected under pressure from relatively low concentrations of KD-247 (10 to 200 μg/ml) and that evolution of fully resistant variants with a mutation in the V3 tip was observed under pressure from high concentrations of the antibody.
FIG. 3.
Schematic representation of recombinant MOKW env genes used for analysis of the genetic basis for resistance to KD-247. MOKW-RDP, MOKW-KNL/C3m, and MOKW-KNL/V3m env genes were amplified from passaged MOKW virus-infected PM1/CCR5 cells in the absence or presence of KD-247. MOKW-KNL, MOKW-RDP/V3m, and MOKW-RDP/C3m env genes were constructed by replacing each region of MOKW-RDP with the corresponding MOKW-KNL/C3m or MOKW-KNL/V3m sequence. MOKW-KNP, MOKW-RDL, MOKW-KDL, and MOKW-RNL viruses were constructed by site-directed mutagenesis. Construction of the clones and mutagenesis procedures are described in Materials and Methods. The locations and numbers of specific amino acids, based on the HXB2 sequence, are shown above the MOKW-RDP sequence.
FIG. 4.
Neutralization sensitivities of pseudoviruses with env genes from passaged MOKW viruses to MAbs, rsCD4, and CCR5 inhibitors. Pseudoviruses with the envelope sequences listed on the figure were prepared as described in Materials and Methods. KD-247, 447-52D, rsCD4, and 17b were preincubated with 100 TCID50 of each MOKW pseudotype virus for 15 min, followed by the addition of the mixtures to the target cells (GHOST-hi5). The target cells were treated with TAK-779 and 2D7 for 15 min, followed by inoculation of the pseudotype clones. The inhibitory effects were determined by measuring the luciferase activities on day 2 of culture. Conc, concentration.
TABLE 1.
Anti-HIV-1 activities of various MAbs and inhibitors toward MOKW pseudoviruses
| Class | Compound | IC50 (μg/ml) of the indicated virus (relative IC50)a
|
|||||
|---|---|---|---|---|---|---|---|
| MOKW-RDP | MOKW-KNL/C3m | MOKW-KNL/V3m | MOKW-RDP/C3m | MOKW-KNL | MOKW-RDP/V3m | ||
| V3 MAbs | KD-247 | 0.004 (1) | 0.5 (125) | >100 (>25,000) | 0.005 (1.3) | 2 (500) | >100 (>25,000) |
| 447-52D | 0.004 (1) | 0.5 (125) | >2 (>500) | 0.004 (1) | 0.8 (200) | >2 (>500) | |
| CD4-induced MAb | 17b | 0.035 (1) | >5 (>143) | >5 (>143) | 0.03 (0.86) | >5 (>143) | 0.02 (0.57) |
| CD4 | rsCD4 | 0.18 (1) | 1.3 (7.22) | 1.5 (8.33) | 0.24 (1.33) | 1.8 (10) | 0.24 (1.33) |
| CCR5 MAb | 2D7 | 8 (1) | 6.8 (0.85) | 1 (0.13) | 8 (1) | 3.2 (0.4) | 2 (0.25) |
| CCR5 inhibitor | TAK-779 | 63 (1) | 63 (1) | 18 (0.29) | 140 (2.22) | 65 (1) | 18 (0.29) |
| CD4 MAb | RPA-T4 | 0.4 (1) | 0.26 (0.65) | 0.22 (0.55) | 0.5 (1.25) | 0.22 (0.55) | 0.44 (1.1) |
GHOST-hi5 cells were exposed to 100 TCID50 of each MOKW pseudovirus and then cultured in the presence of various concentrations of MAb or inhibitors. The IC50 values were determined using the luciferase reporter assay on day 2 of culture. All assays were conducted in triplicate. The value in parentheses is the ratio of the IC50 of the compound to the IC50 of the MOKW-RDP virus. Values for the compound TAK-779 are nanomolar concentrations. Data shown are representative of two or three separate experiments.
We then determined whether the KD-247 escape variants remained sensitive to other neutralizing antibodies (447-52D and 17b), rsCD4, anti-CCR5 antibody (2D7), anti-CD4 antibody (RPA-T4), and the small-molecule CCR5 inhibitor (TAK-779) (Fig. 4 and Table 1). The KD-247 escape variants with the P313L mutation, MOKW-KNL/V3m and MOKW-RDP/V3m, were also resistant to another anti-V3 MAb, 447-52D, and V2-mutated viruses without V3 mutation, MOKW-KNL/C3m and MOKW-KNL, were partially resistant (the same as for KD-247). In contrast, the V2-mutated viruses (MOKW-KNL/C3m, MOKW-KNL/V3m, and MOKW-KNL) showed resistance to rsCD4 and 17b (a MAb to the CD4-induced epitope; CD4i) compared with the pseudoviruses without V2 mutations, i.e., MOKW-RDP, MOKW-RDP/C3m, and MOKW-RDP/V3m. Moreover, the pseudoviruses with V3 tip mutations, MOKW-KNL/V3m and MOKW-RDP/V3m, became significantly more sensitive to TAK-779 and 2D7 compared with pseudoviruses without the P313L mutation (Fig. 4 and Table 1). No significant differences with respect to sensitivity to RPA-T4 were observed between any pseudoviruses (Table 1). These data indicate that V3 tip and V2 mutations confer neutralization resistance against anti-V3 antibodies and that these mutations affect viral sensitivity to neutralizing antibodies recognizing different epitopes and anti-CCR5 antibody/agents.
Binding affinity of neutralizing antibodies to MOKW Env proteins on the cell surface.
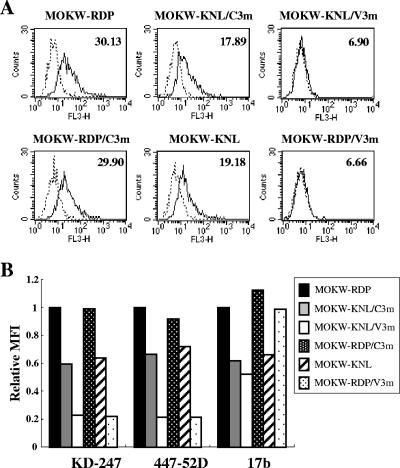
To elucidate the mechanism by which escape virus variants with V3 tip and V2 mutations become less sensitive to neutralizing antibodies, MOKW Env-expressing 293T cells were established by transfection with each Env expression plasmid and then stained with the MAbs. Binding of KD-247, 447-52D, and 17b to the surface-expressed Env proteins was assayed using fluorescence-activated cell sorter (FACS) analysis. As shown in Fig. 5A, the mean fluorescence intensities (MFIs) of KD-247 binding to the Env proteins without either V2 or V3 mutations (the MOKW-RDP and MOKW-RDP/C3m cells) were 30.13 and 29.20, respectively. However, the corresponding values for the V3-tip-mutated Env-expressing cells (MOKW-KNL/V3m and MOKW-RDP/V3m) were almost the same as negative controls (6.90 and 6.66, respectively). The MFI of the V2-mutated Env-expressing cells (MOKW-KNL/C3m and MOKW-KNL) indicated a lower binding affinity (17.89 and 19.18, respectively) than for Env proteins without V2 and V3 mutations. The binding pattern of 447-52D to these Env-expressing cells was similar to that of KD-247 (Fig. 5B). However, reduction in the binding of 17b was observed for strains with V2-mutated Env proteins (MOKW-KNL/C3m, MOKW-KNL/V3m, and MOKW-KNL), whereas no difference in 17b binding was noted for the V3-mutants without V2 mutations (Fig. 5B). These findings are consistent with the results of a single-round neutralization assay (Fig. 4). Taken together, these data suggest that the mutations in V2 have a significant influence on access by antibodies to V3 as well as to the CD4i epitope. This is because access by antibodies to the epitopes of the functional envelope is related to neutralization sensitivity or resistance.
FIG. 5.
Comparison of antibody binding to cell surface-expressed MOKW Env proteins. (A) 293T cells transfected with MOKW Env expression vectors were harvested at 24 h posttransfection and stained with KD-247. Flow cytometry data for binding of the KD-247 (black lines) to cell surface MOKW Env proteins are shown among GFP-gated 293T cells along with the control antibody (normal human IgG; dotted lines). The number at the top right of each graph is the MFI. (B) Each bar indicates the relative binding of KD-247, 447-52D, and 17b to MOKW Env-expressing cell surfaces. Data were normalized to each antibody's MFI for MOKW-RDP virus. FL3-H, relative fluorescence.
Identification of the V2 region site responsible for the neutralization resistance phenotype by site-directed mutagenesis of specific residues.
Because KD-247 recognizes an epitope containing the IGPGR amino acid sequence in the V3 tip, the MAb could not bind the V3 tip of mutated Env proteins. Consequently, KD-247 does not neutralize V3-tip-mutated virus strains (67). However, the mechanism of neutralization resistance associated with V2 mutations is not known. To clarify the responsible mutation in the V2 region that confers the escape phenotype with respect to KD-247, we introduced V2 amino acid changes individually and in combination into the MOKW-RDP Env expression vector (Fig. 3) and measured the sensitivities of pseudoviruses with these envelopes to KD-247. As shown in Fig. 6, the R166K/D167N double mutant, MOKW-KNP virus, showed almost the same neutralization sensitivity as MOKW-RDP virus against KD-247. Surprisingly, a single amino acid change (P175L in MOKW-RDL) was sufficient to confer >10,000-fold resistance upon MOKW-RDP virus, with an IC50 of >100 μg/ml. R166K/P175L (MOKW-KDL) mutations also conferred resistance. Both the MOKW-RDL and MOKW-KDL viruses were much more resistant than the fully V2-mutated virus, MOKW-KNL (>100-fold and >10-fold resistance, respectively) (Fig. 6). The D167N/P175L (MOKW-RNL) mutant was more resistant than MOKW-KNL virus (10-fold) but less resistant than the MOKW-RDL and MOKW-KDL viruses. We also constructed a V2 mutant of JR-FL and confirmed that JR-FL with an amino acid substitution of Leu to Pro at position 175 became highly sensitive to KD-247 compared with JR-FL with Leu at position 175 in Env (data not shown). These results suggest that residue 175 (Pro or Leu) is the crucial amino acid for determining neutralization sensitivity against KD-247 and that the phenotypic influence of the R166K and D167N changes is strictly context dependent, requiring the presence of Leu at residue 175.
FIG. 6.
Neutralization sensitivities of pseudoviruses with env genes from MOKW9C(−) virus with selected V2 mutations to MAbs, rsCD4, and CCR5 inhibitors. Pseudoviruses that have envelope sequences with the selected V2 mutations listed on Fig. 4 were prepared as described in Materials and Methods. KD-247, 447-52D, rsCD4, and IgGb12 were preincubated with 100 TCID50 of each MOKW pseudotype virus for 15 min, followed by addition of the mixtures to the target cells (GHOST-hi5). Target cells were treated with TAK-779 and RPA-T4 for 15 min, followed by inoculation of the pseudotype clones. Inhibitory effects were determined by measuring the luciferase activities on day 2 of culture. Conc, concentration.
We then determined whether these pseudoviruses with various V2 mutations remained sensitive to other neutralizing antibodies (447-52D and IgGb12), rsCD4, 2D7, RPA-T4, and TAK-779 (Fig. 6). MOKW-RDL and MOKW-KDL viruses were also resistant to another anti-V3 MAb, 447-52D, CD4 binding site MAb, IgGb12, and rsCD4. MOKW-RNL was partially resistant compared with MOKW-KNL but less resistant than MOKW-RDL and MOKW-KDL against 447-52D, IgGb12, and rsCD4. These results were similar to those for KD-247. All V2-mutated clones were sensitive to TAK-779 and 2D7, as was MOKW-RDP virus (Fig. 6 and data not shown). However, the anti-CD4 MAb RPA-T4 neutralized both the MOKW-RDL and MOKW-KDL viruses at an approximately threefold lower concentration than other viruses (Fig. 6). These results suggest that the amino acids at positions 166 and 167 (with RD and KN sequences) may help compensate for any reduced fitness of viruses with Leu at residue 175. On the other hand, Pro at position 175 in MOKW-RDP virus might be accumulated because it confers better fitness to replicate on PM1/CCR5 cells in the absence of KD-247 pressure.
Binding affinity of MAbs against monomeric or cell surface-expressed gp120 with mutations in V2.
To determine the difference in binding of MAbs to monomeric gp120 of MOKW Env with V2 mutations relative to that of MOKW-RDP virus, we performed MAb binding assays. Monomeric gp120 was prepared from pseudoviruses that had a series of V2 mutations and was captured on an ELISA plate, followed by detection by MAbs. No difference was noted in the binding activity of KD-247 or 447-52D to monomeric gp120 from V2-mutated and MOKW-RDP envelopes (Fig. 7). These results suggest that V2-mutated Env proteins retain the neutralizing epitope at least in monomeric gp120.
FIG. 7.
Binding affinity of anti-V3 MAbs to monomeric gp120. Viral lysates for each MOKW pseudovirus were used. gp120 was captured onto microtiter wells using a sheep polyclonal antibody specific for the C terminus of gp120. Serial dilutions of KD-247 or 447-52D were tested for binding by ELISA. Because of differences in the amount of bound gp120, optical density at 405 nm (OD405) values were normalized to saturating levels of antibody (5 μg/ml) for comparison. Conc, concentration.
In contrast to the monomeric form, gp120 expressed on the cell surface contained, to a certain degree, functional envelope oligomers that were directly related to the infectivity and neutralization sensitivity of the virus. To compare the binding activity of MAbs for the surface-expressed Env with the results obtained for monomeric gp120, 293T cells transfected with MOKW-RDP and MOKW-KNP viruses, a V2 mutant strain, were subjected to FACS analysis. As shown in Fig. 8A and B, the relative binding of KD-247, 447-52D, and IgGb12 to Env expressed on the cell surface was no different than for MOKW-RDP and MOKW-KNP.
FIG. 8.
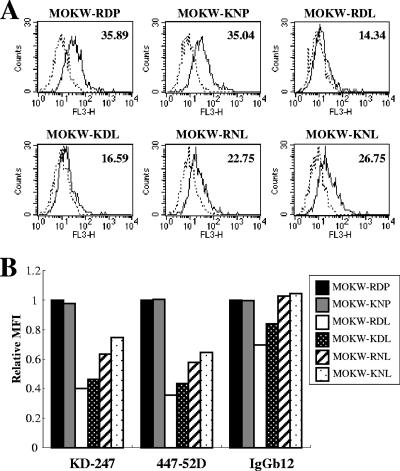
Comparison of antibody binding to cell surface-expressed MOKW Env proteins with V2 mutations. (A) 293T cells transfected with MOKW Env-expression vectors were harvested at 24 h posttransfection and stained with KD-247. Flow cytometry data for binding of the KD-247 (black lines) to cell surface MOKW Env proteins are shown for GFP-gated 293T cells along with data for the control antibody (normal human IgG; dotted lines). The number at the top right of each graph is the MFI. (B) Each bar indicates relative binding of KD-247, 447-52D, and IgGb12 to MOKW Env-expressing cell surfaces. Data were normalized to each antibody's MFI for MOKW-RDP virus. FL3-H, relative fluorescence.
Consistent with the results of the single-round neutralization assay shown in Fig. 6, MOKW-RDL virus had the lowest binding affinity for all tested MAbs. To determine which mutations (166K or 167N) further influence binding affinity, in addition to the MOKW-RDL background, we constructed MOKW-KDL and MOKW-RNL Env proteins and measured the binding affinity by FACS. The MOKW-KDL Env was found to have a slightly greater binding affinity for KD-247, 447-52D, and IgGb12 than MOKW-RDL Env. But cell surface binding of all tested MAbs to MOKW-RNL was better than for MOKW-KDL. The strain with a fully V2-mutated Env, MOKW-KNL, had a binding profile that was intermediate between single- or double-mutated Env proteins and nonmutated Env, but in the case of IgGb12, the binding affinity of MAbs for MOKW-KNL was comparable to that for MOKW-RDP. These data were consistent with the results obtained from the neutralizing assay using a high concentration of each MAb (Fig. 6).
Comparison of replication kinetics between the NL-MOKW-RDL and NL-MOKW-KNL viruses.
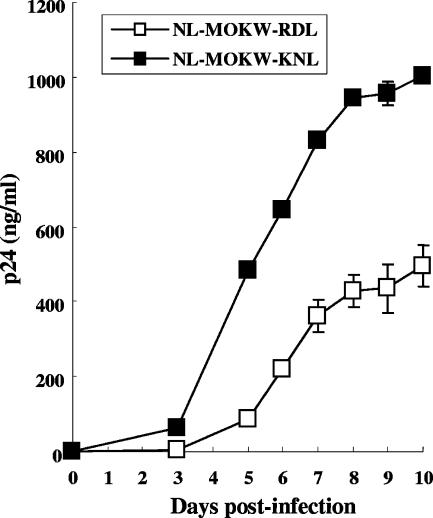
Although the MOKW-RDL variant was much more resistant against KD-247 than the MOKW-KNL variant (Fig. 6) and the RD sequence was more prevalent than KN at positions 166 and 167 in the V2 region before selection (Fig. 2), the MOKW variants with 166K/167N/175L were selected and outgrown under KD-247 pressure (Fig. 2). It was possible that the KN sequences at positions 166 and 167 are necessary to compensate for the fitness of the variants with 175L in PM1/CCR5 cells, as shown in Fig. 6. To clarify the role of KN at positions 166 and 167 in replication, we constructed replication-competent viruses with a MOKW Env with RD or KN in addition to 175L (NL-MOKW-RDL and NL-MOKW-KNL) and compared their replication kinetics. As shown in Fig. 9, NL-MOKW-KNL virus replicated faster than NL-MOKW-RDL virus in PM1/CCR5 cells. These data suggested that KN sequences at positions 166 and 167 with the 175L variant confer a replication advantage in PM1/CCR5 cells. Therefore, the intermediate-resistant variant MOKW virus with the KNL sequence in the V2 region might replicate more rapidly than the highly resistant variant MOKW virus with RDL against KD-247 in the course of selection.
FIG. 9.
Replication kinetics of infectious molecular clones NL-MOKW-RDL and NL-MOKW-KNL. PM1/CCR5 cells were exposed to NL-MOKW-RDL (open square) or NL-MOKW-KNL (filled square) and cultured for 10 days. Virus replication was monitored by measuring the amounts of p24 Gag protein produced in the culture supernatants. The data are representative of the results from two independent experiments.
DISCUSSION
Although an attack from the humoral immune response, especially anti-V3 NAb, is lasting against HIV-1 in vivo, it is not clear why the V3 tip sequence is conserved in the course of the infection. In the present study, by using a genetically heterogeneous HIV-1 primary R5 isolate, MOKW virus, we found that V2- and C3-mutated viruses expanded under conditions with a relatively low concentration of KD-247. Further, we found that the V3-tip-mutated virus was induced only under conditions with a high concentration of MAb (more than 500 μg/ml). Using region-swapping analysis, it was found that both V2 and V3 tip mutations can cause an escape phenotype against anti-V3 antibody. Neutralization escape variants with V2 mutations could be selected from quasi-species existing in the primary isolate at relatively low antibody pressures. On the other hand, highly resistant variants with amino acid substitutions in the V3 epitope emerged via evolution of the virus in the presence of a high concentration of the MAb.
The V1/V2 region of gp120 is highly diverse, not only in respect to virus subtypes but also in respect to intraspecies diversity in the same patient (16, 61, 66). The primary isolate, MOKW, also displayed diversity in the V2 region (Fig. 2), and the first-passaged virus already harbored mutations in the V2 region. Many researchers have reported that the V1/V2 domain strongly influences neutralization of the anti-V3 MAbs, MAbs to the other epitopes, and rsCD4 (12, 13, 25, 27, 30, 44, 49, 50). Moreover, structural models of the Env trimer have been proposed that place the base of the V1/V2 loop of one subunit in proximity to the V3 loop of a neighboring subunit (32, 34). In the present study we observed a reduction in the binding of anti-V3 MAbs to V2-mutated Env expressed on the cell surface, whereas mutations in V2 did not have an effect on the binding of the MAbs to monomeric gp120. These results suggest the association of V2 mutations with anti-V3 antibody accessibility in the context of the oligomeric conformation of the functional envelope. It has been proposed that the gp120 of T-cell-line-adapted strains forms a relatively open conformation and that the primary isolate trimeric complex has a more closed conformation (2, 51). These findings suggest that antibody-induced V2 mutations may affect envelope oligomers on the viral surface so that they form a more closed conformation; thus, neutralization epitopes become less accessible to antibodies. The essential amino acid residues responsible for neutralization resistance located at the center of the V2 region may have a role in the interaction with the V3 loop of neighboring gp120 molecules.
We previously described the in vitro selection and characterization of a KD-247-escape mutant of JR-FL (67). The amino acid substitution that was critical for the resistance phenotype was Gly to Glu at residue 314 (G314E) in the V3 tip region. The genetically engineered mutant was completely resistant to neutralization by KD-247. Other researchers have also reported the induction of V3-mutated viruses by strain-specific anti-V3 MAb in in vitro culture systems (8, 37, 65). In earlier studies, combinations of genetically cloned viruses and highly potent NAbs were used for in vitro selection (8, 37, 65). The escape mutants were induced in the presence of high concentrations of MAbs to acquire V3 tip mutation(s). In contrast to these observations in vitro, the Gly-Pro-Gly amino acid sequence in the V3 tip varies to a negligible extent in clinical isolates from HIV-1-infected patients (35). The important role of the V3 tip in forming the β-turn of the V3 loop and in interacting with chemokine receptors may partly explain the discrepancy between in vivo and in vitro studies (19). In addition to the V3 loop, variation in or near the V1/V2 region is known to contribute to coreceptor usage of HIV-1 (17, 21, 27, 28, 47, 56, 64, 66). However, it is possible that HIV-1 suffers critical damage with respect to replication and infectivity through mutation in the V3 region, especially in the tip region, because the V3 loop plays a major role in the interaction of gp120 with coreceptors. Thus, mutations in the V2 region may be important not only to avoid anti-V3 pressure but also to maintain replication efficiency at a suitable level.
In the present study, which used a MOKW primary virus for selection, the virus underwent acquisition of resistance via V2 mutations and then V2 plus V3 mutations in response to increases in the concentration of MAb. By contrast, no V2 mutations were selected in JR-FL by KD-247 pressure in our previous study (67). Because MOKW was a primary isolate, the viruses contained quasi-species of related but distinct viruses, and relatively resistant variants with mutations in V2 were easily selected for replication. Pinter et al. found that inherent neutralization resistance in JR-FL is mediated by the V1/V2 domain (50). It is therefore possible that the V1/V2 sequence (or the conformation of this sequence) in JR-FL already had a resistance phenotype against anti-V3 antibodies because the escape variant underwent mutation directly in the V3 tip of the KD-247-reacting epitope (67).
In the in vitro selection process, amino acid residue 175 (Pro or Leu) in the V2 region of MOKW virus played a crucial role in dramatically changing the oligomeric state of the envelopes. However, MOKW-RDP obtained by prolonged culture in vitro without KD-247 became neutralization sensitive compared with MOKW virus. Residue 175P was the amino acid responsible for the change to the neutralization-sensitive phenotype, whereas viruses with 175L became highly resistant to the MAbs and rsCD4. The same phenomenon was observed in the relatively resistant strain JR-FL. Residue 175L is highly conserved among HIV-1 strains and is located at the center of the V2 loop (35), and the V2 region also mediates gp41-independent intersubunit contact (5). It is therefore possible that the V2 region, including residue 175, by mediating changes in the conformation of the gp120 oligomer, contributes to resistance to neutralization by limiting the exposure of epitopes.
Although the MOKW-RDL virus had a highly resistant phenotype against KD-247, MOKW virus with R166K/D167N and P175L in the V2 region and with the C3 mutations, which were less neutralization resistant than MOKW-RDL virus, were expanded in in vitro selection. Substitutions at residues 165 to 167 during the adaptation of various HIV-1 strains to replication in vitro have been reported; the adaptation is associated with an increase of the positive charge of this amino acid motif (1, 13, 39, 52, 57, 63). In our present study, the amino acid change at 166/167 in the V2 region in passaged MOKW viruses with KD-247 was RD to KN, again increasing the positive charge. MOKW-RDL virus was partially sensitive to anti-CD4 MAb (RPA-T4) compared with MOKW viruses with 175P, MOKW-RDP, and other 166/167-mutated MOKW viruses. Pugach et al. also showed that charged amino acids at residues 165 to 167 with 175L in the V2 region emerged during in vitro replication and that these viruses also had their sensitivity to rsCD4 and resistance to the anti-CD4 antibody slightly changed by the charged amino acids at positions 165 to 167 (52). It is therefore possible that amino acid mutations at positions 166 and 167 are necessary to compensate variants with 175L for interactions with CD4 molecules on the target cell membrane. As shown in Fig. 9, KN sequences at positions 166 and 167 with the 175L variant confer replication advantage in PM1/CCR5 cells. Therefore, residues 166 and 167 may help compensate for the reduced fitness of the viruses with Leu at position 175 in PM1/CCR5 cells. C3 mutations may also be involved in a minor compensation effect in replication cycles under moderate selective pressure from KD-247 (45, 60).
The neutralization resistance of primary HIV-1 variants is considered instrumental for HIV-1 persistence in the presence of NAbs in vivo. Various immunological pressures always induce escape variants by eliciting appropriate mutation(s) (14, 60, 62). In the present study, we found that HIV-1 could escape from the broadly reactive anti-V3 MAb, KD-247, by stepwise mutation in the V2 and V3 regions. These observations strongly support the idea that the major problem facing the development of V3-based immunogens is not sequence variation within V3 but, rather, that access of most V3-directed antibodies to their epitopes in functional Env complexes is blocked, often by the V1/V2 domain (29, 50).
Our observations support the hypothesis that neutralization escape in a primary isolate is mainly mediated by amino acid substitutions in the V2 region in vivo, because only a moderate selective pressure by neutralizing antibodies against autologous viruses has been reported for infected individuals (9, 14, 26, 50, 62). The large sequence diversity observed for V2 in quasi-species existing in patients may represent the accumulation of escape mutants early in HIV-1 infection in response to NAb pressure. Our observations may also explain why the V3 sequence in quasi-species existing in patients is relatively conserved in the face of a vigorous antibody response, especially in early HIV-1 infection. A recent study by Deeks et al. has important implications for understanding the NAb response against autologous virus (9): although NAb responses against contemporaneous autologous viruses are absent in early HIV infection, they can be detected at low levels in some patients with chronic infection. These data suggest the existence of an NAb response that overcomes the emergence of escape mutants. Further characterization of the response in humans who have potent and broadly neutralizing activities not affected by V1/V2 blocking effects may allow the identification of additional neutralization sites in HIV-1 Env, which might allow new targets to be identified for vaccine development.
Acknowledgments
We are grateful to S. Zolla-Pazner of the New York University School of Medicine for providing MAb 447-52D and to Yasuhiro Kou for technical support. We also thank Yoko Kawanami for excellent technical assistance.
This work was supported in part by the Ministry of Health, Labor and Welfare of Japan (H-16-AIDS-001 and -012); a Grant-in aid for Scientific Research (C-18591119) from the Ministry of Education, Science and Culture of Japan; and the Cooperative Research Project on Clinical and Epidemiological Studies of Emerging and Re-emerging Infectious Diseases.
Footnotes
Published ahead of print on 24 January 2007.
REFERENCES
- 1.Bouma, P., M. Leavitt, P. F. Zhang, I. A. Sidorov, D. S. Dimitrov, and G. V. Quinnan, Jr. 2003. Multiple interactions across the surface of the gp120 core structure determine the global neutralization resistance phenotype of human immunodeficiency virus type 1. J. Virol. 77:8061-8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burton, D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, Y., L. Qin, L. Zhang, J. Safrit, and D. D. Ho. 1995. Virologic and immunologic characterization of long-term survivors of human immunodeficiency virus type 1 infection. N. Engl. J. Med. 332:201-208. [DOI] [PubMed] [Google Scholar]
- 4.Carotenuto, P., D. Looij, L. Keldermans, F. de Wolf, and J. Goudsmit. 1998. Neutralizing antibodies are positively associated with CD4+ T-cell counts and T-cell function in long-term AIDS-free infection. AIDS 12:1591-1600. [DOI] [PubMed] [Google Scholar]
- 5.Center, R. J., P. L. Earl, J. Lebowitz, P. Schuck, and B. Moss. 2000. The human immunodeficiency virus type 1 gp120 V2 domain mediates gp41-independent intersubunit contacts. J. Virol. 74:4448-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Center, R. J., R. D. Leapman, J. Lebowitz, L. O. Arthur, P. L. Earl, and B. Moss. 2002. Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J. Virol. 76:7863-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, K. S., J. D. Steckbeck, J. L. Rowles, R. C. Desrosiers, and R. C. Montelaro. 2004. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J. Virol. 78:1525-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Costa, S., K. S. Slobod, R. G. Webster, S. W. White, and J. L. Hurwitz. 2001. Structural features of HIV envelope defined by antibody escape mutant analysis. AIDS Res. Hum. Retrovir. 17:1205-1209. [DOI] [PubMed] [Google Scholar]
- 9.Deeks, S. G., B. Schweighardt, T. Wrin, J. Galovich, R. Hoh, E. Sinclair, P. Hunt, J. M. McCune, J. N. Martin, C. J. Petropoulos, and F. M. Hecht. 2006. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J. Virol. 80:6155-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eda, Y., T. Murakami, Y. Ami, T. Nakasone, M. Takizawa, K. Someya, M. Kaizu, Y. Izumi, N. Yoshino, S. Matsushita, H. Higuchi, H. Matsui, K. Shinohara, H. Takeuchi, Y. Koyanagi, N. Yamamoto, and M. Honda. 2006. Anti-V3 humanized antibody KD-247 effectively suppresses ex vivo generation of human immunodeficiency virus type 1 and affords sterile protection of monkeys against a heterologous simian/human immunodeficiency virus infection. J. Virol. 80:5563-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eda, Y., M. Takizawa, T. Murakami, H. Maeda, K. Kimachi, H. Yonemura, S. Koyanagi, K. Shiosaki, H. Higuchi, K. Makizumi, T. Nakashima, K. Osatomi, S. Tokiyoshi, S. Matsushita, N. Yamamoto, and M. Honda. 2006. Sequential immunization with V3 peptides from primary human immunodeficiency virus type 1 produces cross-neutralizing antibodies against primary isolates with a matching narrow-neutralization sequence motif. J. Virol. 80:5552-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etemad-Moghadam, B., Y. Sun, E. K. Nicholson, G. B. Karlsson, D. Schenten, and J. Sodroski. 1999. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J. Virol. 73:8873-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Follis, K. E., M. Trahey, R. A. LaCasse, and J. H. Nunberg. 1998. Continued utilization of CCR5 coreceptor by a newly derived T-cell line-adapted isolate of human immunodeficiency virus type 1. J. Virol. 72:7603-7608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost, S. D., T. Wrin, D. M. Smith, S. L. Kosakovsky Pond, Y. Liu, E. Paxinos, C. Chappey, J. Galovich, J. Beauchaine, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Neutralizing antibody responses drive the evolution of human immunodeficiency virus type 1 envelope during recent HIV infection. Proc. Natl. Acad. Sci. USA 102:18514-18519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorny, M. K., K. Revesz, C. Williams, B. Volsky, M. K. Louder, C. A. Anyangwe, C. Krachmarov, S. C. Kayman, A. Pinter, A. Nadas, P. N. Nyambi, J. R. Mascola, and S. Zolla-Pazner. 2004. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J. Virol. 78:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groenink, M., R. A. Fouchier, S. Broersen, C. H. Baker, M. Koot, A. B. van't Wout, H. G. Huisman, F. Miedema, M. Tersmette, and H. Schuitemaker. 1993. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science 260:1513-1516. [DOI] [PubMed] [Google Scholar]
- 17.Harrowe, G., and C. Cheng-Mayer. 1995. Amino acid substitutions in the V3 loop are responsible for adaptation to growth in transformed T-cell lines of a primary human immunodeficiency virus type 1. Virology 210:490-494. [DOI] [PubMed] [Google Scholar]
- 18.Hope, T. J., X. J. Huang, D. McDonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, Q., J. O. Trent, G. D. Tomaras, Z. Wang, J. L. Murray, S. M. Conolly, J. M. Navenot, A. P. Barry, M. L. Greenberg, and S. C. Peiper. 2000. Identification of ENV determinants in V3 that influence the molecular anatomy of CCR5 utilization. J. Mol. Biol. 302:359-375. [DOI] [PubMed] [Google Scholar]
- 20.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253:71-74. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson, J. M., N. Colman, N. A. Ostrow, R. W. Simson, D. Tomesch, L. Marlin, M. Rao, J. L. Mills, J. Clemens, and A. M. Prince. 1993. Passive immunotherapy in the treatment of advanced human immunodeficiency virus infection. J. Infect. Dis. 168:298-305. [DOI] [PubMed] [Google Scholar]
- 23.Javaherian, K., A. J. Langlois, C. McDanal, K. L. Ross, L. I. Eckler, C. L. Jellis, A. T. Profy, J. R. Rusche, D. P. Bolognesi, S. D. Putney, et al. 1989. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc. Natl. Acad. Sci. USA 86:6768-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, W. E., and R. C. Desrosiers. 2002. Viral persistance: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499-518. [DOI] [PubMed] [Google Scholar]
- 25.Kang, S. M., F. S. Quan, C. Huang, L. Guo, L. Ye, C. Yang, and R. W. Compans. 2005. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology 331:20-32. [DOI] [PubMed] [Google Scholar]
- 26.Kimura, T., K. Yoshimura, K. Nishihara, Y. Maeda, S. Matsumi, A. Koito, and S. Matsushita. 2002. Reconstitution of spontaneous neutralizing antibody response against autologous human immunodeficiency virus during highly active antiretroviral therapy. J. Infect. Dis. 185:53-60. [DOI] [PubMed] [Google Scholar]
- 27.Koito, A., G. Harrowe, J. A. Levy, and C. Cheng-Mayer. 1994. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J. Virol. 68:2253-2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koito, A., L. Stamatatos, and C. Cheng-Mayer. 1995. Small amino acid sequence changes within the V2 domain can affect the function of a T-cell line-tropic human immunodeficiency virus type 1 envelope gp120. Virology 206:878-884. [DOI] [PubMed] [Google Scholar]
- 29.Krachmarov, C., A. Pinter, W. J. Honnen, M. K. Gorny, P. N. Nyambi, S. Zolla-Pazner, and S. C. Kayman. 2005. Antibodies that are cross-reactive for human immunodeficiency virus type 1 clade A and clade B V3 domains are common in patient sera from Cameroon, but their neutralization activity is usually restricted by epitope masking. J. Virol. 79:780-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krachmarov, C. P., W. J. Honnen, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. 2006. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J. Virol. 80:7127-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhmann, S. E., P. Pugach, K. J. Kunstman, J. Taylor, R. L. Stanfield, A. Snyder, J. M. Strizki, J. Riley, B. M. Baroudy, I. A. Wilson, B. T. Korber, S. M. Wolinsky, and J. P. Moore. 2004. Genetic and phenotypic analyses of human immunodeficiency virus type 1 escape from a small-molecule CCR5 inhibitor. J. Virol. 78:2790-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 33.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwong, P. D., R. Wyatt, Q. J. Sattentau, J. Sodroski, and W. A. Hendrickson. 2000. Oligomeric modeling and electrostatic analysis of the gp120 envelope glycoprotein of human immunodeficiency virus. J. Virol. 74:1961-1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber. 2005. HIV sequence compendium 2005, LA-UR 06-0680. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 36.Maeda, K., K. Yoshimura, S. Shibayama, H. Habashita, H. Tada, K. Sagawa, T. Miyakawa, M. Aoki, D. Fukushima, and H. Mitsuya. 2001. Novel low molecular weight spirodiketopiperazine derivatives potently inhibit R5 HIV-1 infection through their antagonistic effects on CCR5. J. Biol. Chem. 276:35194-35200. [DOI] [PubMed] [Google Scholar]
- 37.Masuda, T., S. Matsushita, M. J. Kuroda, M. Kannagi, K. Takatsuki, and S. Harada. 1990. Generation of neutralization-resistant HIV-1 in vitro due to amino acid interchanges of third hypervariable env region. J. Immunol. 145:3240-3246. [PubMed] [Google Scholar]
- 38.McCaffrey, R. A., C. Saunders, M. Hensel, and L. Stamatatos. 2004. N-Linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mo, H., L. Stamatatos, J. E. Ip, C. F. Barbas, P. W. Parren, D. R. Burton, J. P. Moore, and D. D. Ho. 1997. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. off. J. Virol. 71:6869-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montefiori, D. C., T. S. Hill, H. T. Vo, B. D. Walker, and E. S. Rosenberg. 2001. Neutralizing antibodies associated with viremia control in a subset of individuals after treatment of acute human immunodeficiency virus type 1 infection. J. Virol. 75:10200-10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore, J. P., Y. Cao, D. D. Ho, and R. A. Koup. 1994. Development of the anti-gp120 antibody response during seroconversion to human immunodeficiency virus type 1. J. Virol. 68:5142-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, J. P., and D. D. Ho. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67:863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morikita, T., Y. Maeda, S. Fujii, S. Matsushita, K. Obaru, and K. Takatsuki. 1997. The V1/V2 region of human immunodeficiency virus type 1 modulates the sensitivity to neutralization by soluble CD4 and cellular tropism. AIDS Res. Hum. Retrovir. 13:1291-1299. [DOI] [PubMed] [Google Scholar]
- 45.Otto, C., B. A. Puffer, S. Pohlmann, R. W. Doms, and F. Kirchhoff. 2003. Mutations in the C3 region of human and simian immunodeficiency virus envelope have differential effects on viral infectivity, replication, and CD4-dependency. Virology 315:292-302. [DOI] [PubMed] [Google Scholar]
- 46.Park, E. J., L. K. Vujcic, R. Anand, T. S. Theodore, and G. V. Quinnan, Jr. 1998. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J. Virol. 72:7099-7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pastore, C., R. Nedellec, A. Ramos, S. Pontow, L. Ratner, and D. E. Mosier. 2006. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J. Virol. 80:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pilgrim, A. K., G. Pantaleo, O. J. Cohen, L. M. Fink, J. Y. Zhou, J. T. Zhou, D. P. Bolognesi, A. S. Fauci, and D. C. Montefiori. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176:924-932. [DOI] [PubMed] [Google Scholar]
- 49.Pinter, A., W. J. Honnen, P. D'Agostino, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2005. The C108g epitope in the V2 domain of gp120 functions as a potent neutralization target when introduced into envelope proteins derived from human immunodeficiency virus type 1 primary isolates. J. Virol. 79:6909-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poignard, P., E. O. Saphire, P. W. Parren, and D. R. Burton. 2001. gp120: biologic aspects of structural features. Annu. Rev. Immunol. 19:253-274. [DOI] [PubMed] [Google Scholar]
- 52.Pugach, P., S. E. Kuhmann, J. Taylor, A. J. Marozsan, A. Snyder, T. Ketas, S. M. Wolinsky, B. T. Korber, and J. P. Moore. 2004. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology 321:8-22. [DOI] [PubMed] [Google Scholar]
- 53.Sakaguchi, N., T. Kimura, S. Matsushita, S. Fujimura, J. Shibata, M. Araki, T. Sakamoto, C. Minoda, and K. Kuwahara. 2005. Generation of high-affinity antibody against T cell-dependent antigen in the Ganp gene-transgenic mouse. J. Immunol. 174:4485-4494. [DOI] [PubMed] [Google Scholar]
- 54.Saunders, C. J., R. A. McCaffrey, I. Zharkikh, Z. Kraft, S. E. Malenbaum, B. Burke, C. Cheng-Mayer, and L. Stamatatos. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79:9069-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawyer, L. S., M. T. Wrin, L. Crawford-Miksza, B. Potts, Y. Wu, P. A. Weber, R. D. Alfonso, and C. V. Hanson. 1994. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J. Virol. 68:1342-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Shibata, J., K. Yoshimura, A. Honda, A. Koito, T. Murakami, and S. Matusushita. 2006. A role of mutations in non-V3 envelope regions for escape from a broad neutralizing anti-V3 monoclonal antibody, KD-247, during in vitro selection, abstr. 415. 13th Conf. Retrovir. Opportunistic Infect., Denver, CO, 5 to 8 February 2006.
- 56.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1992. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:9434-9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sullivan, N., M. Thali, C. Furman, D. D. Ho, and J. Sodroski. 1993. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trkola, A., H. Kuster, P. Rusert, B. Joos, M. Fischer, C. Leemann, A. Manrique, M. Huber, M. Rehr, A. Oxenius, R. Weber, G. Stiegler, B. Vcelar, H. Katinger, L. Aceto, and H. F. Gunthard. 2005. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nat. Med. 11:615-622. [DOI] [PubMed] [Google Scholar]
- 59.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, F. X., T. Kimura, K. Nishihara, K. Yoshimura, A. Koito, and S. Matsushita. 2002. Emergence of autologous neutralization-resistant variants from preexisting human immunodeficiency virus (HIV) quasi species during virus rebound in HIV type 1-infected patients undergoing highly active antiretroviral therapy. J. Infect. Dis. 185:608-617. [DOI] [PubMed] [Google Scholar]
- 61.Wang, N., T. Zhu, and D. D. Ho. 1995. Sequence diversity of V1 and V2 domains of gp120 from human immunodeficiency virus type 1: lack of correlation with viral phenotype. J. Virol. 69:2708-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 63.Wrin, T., T. P. Loh, J. C. Vennari, H. Schuitemaker, and J. H. Nunberg. 1995. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J. Virol. 69:39-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida, K., M. Nakamura, and T. Ohno. 1997. Mutations of the HIV type 1 V3 loop under selection pressure with neutralizing monoclonal antibody NM-01. AIDS Res. Hum. Retrovir. 13:1283-1290. [DOI] [PubMed] [Google Scholar]
- 66.Yoshimura, K., S. Matsushita, A. Hayashi, and K. Takatsuki. 1996. Relationship of HIV-1 envelope V2 and V3 sequences of the primary isolates to the viral phenotype. Microbiol. Immunol. 40:277-287. [DOI] [PubMed] [Google Scholar]
- 67.Yoshimura, K., J. Shibata, T. Kimura, A. Honda, Y. Maeda, A. Koito, T. Murakami, H. Mitsuya, and S. Matsushita. 2006. Resistance profile of a novel broadly neutralizing anti-HIV monoclonal antibody, KD-247, that shows favorable synergism with anti-CCR5 inhibitors in vitro. AIDS 20:2065-2073. [DOI] [PubMed] [Google Scholar]
- 68.Yusa, K., Y. Maeda, A. Fujioka, K. Monde, and S. Harada. 2005. Isolation of TAK-779-resistant HIV-1 from an R5 HIV-1 gp120 V3 loop library. J. Biol. Chem. 280:30083-30090. [DOI] [PubMed] [Google Scholar]
- 69.Zolla-Pazner, S. 2004. Identifying epitopes of HIV-1 that induce protective antibodies. Nat. Rev. Immunol. 4:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]