Abstract
Herpesvirus saimiri (HVS) is the prototype gamma-2 herpesvirus, which naturally infects the squirrel monkey Saimiri sciureus, causing an asymptomatic but persistent infection. The latent phase of gamma-2 herpesviruses is characterized by their ability to persist in a dividing cell population while expressing a limited subset of latency-associated genes. In HVS only three genes, open reading frame 71 (ORF71), ORF72, and ORF73, are expressed from a polycistronic mRNA. ORF73 has been shown to be the only gene essential for HVS episomal maintenance and can therefore be functionally compared to the human gammaherpesvirus latency-associated proteins, EBNA-1 and Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen (LANA). HVS ORF73 is the positional homologue of KSHV LANA and, although it shares limited sequence homology, has significant structural and functional similarities. Investigation of KSHV LANA has demonstrated that it is able to mediate KSHV episomal persistence by tethering the KSHV episome to host mitotic chromosomes via interactions with cellular chromosome-associated proteins. These include associations with core and linker histones, several bromodomain proteins, and the chromosome-associated proteins methyl CpG binding protein 2 (MeCP2) and DEK. Here we show that HVS ORF73 associates with MeCP2 via a 72-amino-acid domain within the ORF73 C terminus. Furthermore, we have assessed the functional significance of this interaction, using a variety of techniques including small hairpin RNA knockdown, and show that association between ORF73 and MeCP2 is essential for HVS chromosomal attachment and episomal persistence.
Herpesvirus saimiri (HVS) is the prototype gamma-2 herpesvirus in the Rhadinovirus genus and as such shares significant homology with the human gammaherpesviruses Kaposi's sarcoma-associated herpesvirus (KSHV) and Epstein-Barr virus (EBV) (2, 4, 34). All gamma-2 herpesviruses share a colinear genomic organization, where homologous genes are found in similar locations and orientations. However, genes specific to each virus can be found interspersed throughout their respective genomes. The genomes of HVS and KSHV share particular similarity in that of the 81 open reading frames (ORFs) encoded by KSHV, at least 66 share significant homology with ORFs found within HVS (34).
A characteristic possessed by all rhadinoviruses is their ability to cause a latent, persistent infection in lymphoid cell populations. In particular, HVS establishes an asymptomatic, lifelong infection in T cells of the squirrel monkey Saimiri sciureus. Throughout latent infection the virus is maintained in a dividing cell population as a high-copy-number circularized episome, without chromosome integration. To sustain a latent episomal infection in dividing cells, each gammaherpesvirus must retain the ability to replicate and faithfully segregate viral episomes to each progeny cell. For the gamma-2 herpesviruses, this is achieved through the expression of a limited subset of latency-associated genes. Analysis of HVS latent gene expression has shown that only three genes, ORF71, ORF72, and ORF73, are expressed during latency (19). Each of these genes is also expressed during KSHV latency (16, 39), and in both viruses, ORF71 and ORF72 encode cellular homologues of an antiapoptotic FLICE inhibitory protein (40) and a cyclin D homologue (11), respectively. However, the ORF73 proteins in each virus have no known cellular homologues.
KSHV ORF73, which is often referred to as the latency-associated nuclear antigen (LANA), shares limited sequence homology with HVS ORF73. However, the proteins encoded by the two genes possess significant structural similarity. Each protein contains a small N-terminal domain and a larger C terminus separated by a central, repetitive acidic region (20). Most significantly, functional characterization of each protein has shown that they share several common features. First, in vitro immunofluorescent analysis of HVS ORF73 and KSHV LANA has demonstrated that each associates with host mitotic chromosomes during cell division (5, 9, 14, 41). Furthermore, each protein has the ability to bind its respective viral episome through association with the viral terminal repeat (TR) domains (5, 12-14, 21, 41). Specific interaction between KSHV LANA and the KSHV TR DNA has been shown to be mediated by a 13- to 20-bp imperfect palindrome located within nucleotides 603 to 622 of the TR sequence (6, 15). Similarly, it has been suggested that HVS TR DNA contains two potential ORF73 binding sites, which possess 64% and 53% identity with the well-characterized KSHV TR binding sequences (41).
Functional analysis of HVS episomal persistence using mutants lacking all latency-associated genes (i.e., ORF71, -72, and -73) demonstrated that these genes are required for the ability of HVS to sustain episomal maintenance (8). Moreover, replacement of ORF73 alone rescued HVS episomal persistence to wild-type levels (8). This clearly indicates that ORF73 is the only protein essential for the maintenance of HVS episomal persistence (8). Hence, like EBV nuclear antigen-1 (EBNA-1) and KSHV LANA, HVS ORF73 is the key latency-associated protein, required for efficient long-term latent persistence.
Analysis of both KSHV LANA and EBNA-1 has shown that each protein is able to direct chromosomal colocalization through interactions with cellular chromosome-associated proteins (7, 14, 22, 26, 37, 38). The amino terminus of the EBNA-1 protein, particularly two domains located between amino acids 40 and 89 and amino acids 328 and 377, has been shown to be responsible for its chromosome association (28). Substantial evidence suggests that the EBNA-1 protein mediates chromosomal association via an interaction with the cellular nuclear protein EBP-2 (EBNA-1 binding protein 2) (23-25, 38), although it has also been shown that the cellular DNA binding proteins HMGA1a and histone H1 can functionally replace the amino terminus of EBNA-1 (22, 35).
Multiple interactions between KSHV LANA and chromosome-associated proteins have also been reported. Initially, Cotter and Robertson reported that LANA interacts with the linker histone H1 (14). Shinohara et al. also demonstrated that an N-terminal deletion of LANA caused a loss of chromosomal association; however, replacement of this domain with histone H1 restored its ability to support KSHV episomal persistence (37). In addition, recent analysis has shown that the N terminus of LANA is sufficient for docking onto chromosomes via an interaction with the folded region of the core histones H2A-H2B (7). However, evidence also suggests that interactions between the C terminus of LANA and cellular bromodomain-containing proteins also play a role in KSHV chromosomal association (32, 42, 45). Furthermore, Krithivas et al. have demonstrated that LANA interacts with the cellular proteins methyl CpG binding protein 2 (MeCP2) and DEK (26). DEK is a ubiquitous nuclear protein of approximately 43 kDa and has been found to be predominately associated with chromatin (43). Likewise, MeCP2 is an abundant nuclear protein of approximately 75 to 80 kDa that preferentially binds to methylated CpG dinucleotides, although it has also been shown to posses AT hook binding domains (3, 27, 31). Extensive research suggests that it plays a key role in the regulation of gene transcription, possibly through its ability to facilitate heterochromatin compaction (17, 30).
Our studies suggest that HVS ORF73, like KSHV LANA, also binds to several chromosome-associated proteins, including histones and several members of the bromodomain protein family (unpublished data). However, in this report we demonstrate a specific association between HVS ORF73 and MeCP2, mediated through ORF73 C-terminal amino acids 324 to 395. Moreover, we have demonstrated, using a variety of functional assays, including small hairpin RNA (shRNA) knockdown, that this interaction is critical for the maintenance of latent HVS infection in a dividing cell population.
MATERIALS AND METHODS
Plasmid constructs.
C-terminal ORF73 (ORF73C) was PCR amplified with primers pET1 and pET2 (see Fig. 3a and b). These oligonucleotides incorporated BamHI and XhoI restriction sites, respectively, to assist cloning of the PCR product. PCR (35 cycles, where 1 cycle consists of 30 s at 94°C, 40 s at 54°C, and 40 s at 68°C) was performed using 2.5 U of Platinum Pfx DNA polymerase (Invitrogen). The product was inserted upstream and in frame of the histidine-tagged ORF in the prokaryotic expression vector pET21(b) (Novagen) to produce pET21(b)-73C. The ORF73C deletion series was constructed using the primers detailed in Fig. 3b. Oligonucleotides labeled F1-F4 and R1-R3 incorporated EcoRI and XhoI restriction sites, respectively. PCR amplification and incorporation into pET21b were performed as described above, producing vectors pET21b-73CΔ1 through pET21b-73CΔ10.
FIG. 3.
(a) Schematic representation of the ORF73 C-terminal deletion series. CAS 1 and CAS 2, described previously, are indicated (9). (b) Primers used during cloning of ORF73C fragments into the bacterial expression construct pET21b are listed and the sequences described.
The vector encoding an ORF73 C-terminal green fluorescent protein (GFP)-tagged fusion protein was constructed by PCR amplification (as described above) of ORF73C with primers incorporating SacI and KpnI restriction enzyme sites (5′-CGCGAGCTCGGACCAAGTACTCCACGTTTACCA-3′ and 5′-CGGGGTACCTTCTATAGGCAGGCTTTTGCTAAA-3′). The PCR fragment was then cloned into the pEGFP-C3 vector (Clontech) in frame and downstream of the GFP gene.
The plasmid encoding FLAG-tagged MeCP2 (pFLAG-MeCP2) was kindly provided by D. Hayward (Johns Hopkins School of Medicine, Baltimore, MD). pFLAG-DEK was supplied by Gerard Grosveld (St. Jude Children's Research Hospital, Memphis, TN). pcDNA3.1-FLAG (Invitrogen) was used as an empty-vector negative control.
In order to select cells expressing MeCP2, the human MeCP2 sequence was PCR amplified from the pFLAG-MeCP2 vector using primers incorporating EcoRI and XbaI restriction enzyme sites (5′-CCGGAATTCAGTAGCTGGGATGTTAGGGCT-3′ and 5′-CTAGTCTAGATCAGCTAACTCTCTCGGTCA-3′). The MeCP2 sequence was then cloned into the neomycin-resistant mammalian expression vector pFLAG-CMV (Sigma) downstream and in frame of the FLAG tag sequence to produce pFLAG-neo-MeCP2.
Cell culture, transfection, and viruses.
HeLa, 293T, NIH 3T3, SW480, and A549 cells were maintained in Dulbecco modified Eagle medium (GIBCO) supplemented with 10% fetal calf serum (FCS) and 5 U/ml of penicillin and streptomycin (Invitrogen). Cells were seeded at approximately 5 × 105 per 35-mm-diameter petri dish 24 h prior to transfection. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol, with 4 μg of the appropriate DNAs.
HVS-bacterial artificial chromosome (BAC)-GFP (strain A11) (titer, 1 × 106 PFU/ml) was used throughout; its construction has been described previously (44). Routinely, approximately 1 × 106 cells of each type per 35-mm-diameter petri dish were infected with 1 × 105 PFU/ml of HVS-BAC-GFP.
The 293T persistently infected stable cell line (293T-HVS-GFP) was constructed by infection of 293T cells and selection of cells harboring HVS-BAC-GFP by using 50 μg/ml hygromycin. Latently infected cells were maintained by regular passage for more than 8 weeks. Low-molecular-weight episomal DNA was extracted from the cells as previously described (44). The DNA was subsequently transformed into electrocompetent Escherichia coli DH10B and plated onto LB agar plates containing 12.5 μg/ml chloramphenicol.
The NIH-MeCP2 stable cell line was created by transfection of NIH 3T3 cells with the pFLAG-neo-MeCP2 vector. Cells expressing the neomycin resistance gene encoded by pFLAG-neo-MeCP2 were selected with 800 μg/ml of Geneticin (Invitrogen). Selected cells were maintained for several weeks by routine passage. Expression of MeCP2 in the selected cells was confirmed by immunofluorescence microscopy using a monoclonal anti-FLAG primary antibody (Sigma), diluted 1:500.
Cell culture episomal maintenance assays.
Infected SW480, NIH 3T3, and NIH-MeCP2 cells were maintained in 200 μg/ml hygromycin for 24 h, followed by trypsinization and dilution to a cell density of approximately 1 × 102/ml (with hygromycin selection maintained as appropriate). 293T-HVS-GFP latently infected cells were maintained in 50 μg/ml hygromycin and diluted to a cell density of approximately 5 × 102/ml. The cells were then seeded at a density of approximately 10 (SW480 and NIH 3T3) or 50 (293T-HVS-GFP and NIH-MeCP2) cells per single 6.4-mm-diameter well in a 96-well plate and incubated at 37°C under 5% CO2 for 1 week. The plates were then analyzed for colony formation and scored as the percentage of wells positive for colony outgrowth. All assays were performed at least in duplicate.
In vitro binding analysis.
pET21b-73C expression vectors were transformed into competent E. coli BL21 and grown in culture. Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) before pelleting and solubilizing in solubilization buffer (50 mM Tris-HCl [pH 8], 1.5 mM NaCl, 1% Triton X-100, Complete protease inhibitors [Roche]). Cells were sonicated prior to retention of the soluble fraction. Each soluble fraction was incubated with nickel-nitrilotriacetic acid agarose beads (QIAGEN), immobilizing the histidine-tagged 73C proteins. Unbound proteins were removed by washing the beads with wash buffer (50 mM NaH2PO4, 150 mM NaCl, 10 mM imidazole, 0.05% Triton X-100 [pH 8]). Binding and purification of ORF73 proteins were confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis. HeLa cells were transfected with 4 μg of the appropriate DNA. At 24 h posttransfection, the cells were harvested and lysed with lysis buffer (300 mM NaCl, 50 mM Tris-HCl [pH 8], 1% Tween 20, 1 mM EDTA, Complete protease inhibitors [Roche]). Where appropriate, lysed cell extracts were treated with 4 U of DNase I (Sigma) per cell extract. To ensure efficient digestion of the cellular DNA, EDTA was omitted from the lysis buffer (see above) during DNase analysis. Each cell extract was subsequently incubated with bead-bound, purified ORF73C protein and washed three times with lysis buffer (including EDTA in all cases). The immobilized proteins were resolved on 9% SDS-polyacrylamide gels before being transferred to nitrocellulose membranes. Following transfer, the membranes were blocked in 2% (wt/vol) nonfat milk powder-0.1% Tween 20-phosphate-buffered saline (PBS) solution prior to incubation with a 1:1,000 dilution of the anti-FLAG antibody M2 (Sigma) and a 1:1,000 dilution of a horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (DAKO). The nitrocellulose membranes were developed using electrochemiluminescent (ECL) detection reagents (Amersham). Complete DNA digestion with DNase I was confirmed by PCR analysis. Cell extracts that were either left untreated or treated with DNase I were used as templates in a PCR using primers directed against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (primer 1, 5′-CCACCCATGGCAAATTCCATGGCA; primer 2, 5′-TCTAGACGGCAGGTCAGGTCCACC).
Coimmunoprecipitation.
Approximately 1 × 107 293T cells were transfected with pEGFP-ORF73C and incubated for 36 h. The cells were then collected and lysed with 500 μl of radioimmunoprecipitation assay buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% sodium deoxycholate, Complete protease inhibitors [Roche]). The cells were then precleared with protein A-agarose (GE Healthcare) prior to incubation with a polyclonal anti-MeCP2 antibody (Abcam) for 2 h at 4°C. As a negative control, cell extracts were also incubated with a polyclonal anti-ORF57 antibody (as described by Goodwin et al. [18]). In each case, the immunocomplex was captured using protein A-agarose before being washed in PBS and resuspended in lysis buffer. Each sample was then loaded onto a 10% SDS-polyacrylamide gel before being transferred to a nitrocellulose membrane. Following blocking with a 5% (wt/vol) nonfat milk powder-0.1% Tween 20-PBS solution, ORF73C protein precipitated in association with MeCP2 was detected with a monoclonal anti-GFP antibody (BD Living Colors, diluted 1:1,000). A HRP-conjugated anti-mouse secondary antibody (diluted 1:1,000; DAKO) was then detected using ECL detection reagents (Amersham).
Confocal microscopy.
Approximately 5 × 105 HeLa cells were cotransfected with pEGFP-ORF73 and pFLAG-MeCP2 expression plasmids as described above. Transfected cells were serum starved in a medium containing 2% FCS prior to induction of cell division by incubation with a medium containing 10% FCS. Induced cells were fixed with 4% formaldehyde and permeabilized with 0.5% Triton X-100. Fixed cells were labeled with a 1:1,000 dilution of a monoclonal anti-FLAG antibody (Sigma), followed by staining with a 1:250 dilution of Texas red-conjugated anti-mouse immunoglobulin G, allowing visualization of the FLAG-MeCP2 fusion proteins. Cellular DNA was identified by staining fixed cells with 0.1 μM TO-PRO-3 iodide (Molecular Probes). Prepared slides were visualized on an upright Zeiss LSM 510 Meta confocal microscope system.
Reverse transcription-PCR (RT-PCR) analysis.
Total RNA was prepared from SW480, A549, and NIH 3T3 cells using TRIzol purification (Invitrogen) according to the manufacturer's instructions. The RNA was reverse transcribed by Superscript III reverse transcriptase (Invitrogen) for 1 h at 50°C using an oligo(dT) primer. cDNA was amplified by PCR primers directed against GAPDH (described above) and MeCP2 (primer 1, 5′-GGGATGTTAGGGCTCAGGGA; primer 2, 5′-AACCTTCAGGCAAGGTGGGG). MeCP2 primers were specifically designed to recognize sequences homologous to mice and humans.
shRNA knockdown of MeCP2 expression in 293T-HSV-GFP latently infected cells.
293T-HVS-GFP latently infected cells were transfected with shRNA vectors (Invivogen) directed against MeCP2 (psiRNA-MeCP2-custom.gcc) and luciferase protein (psiRNA-LucGL3-custom) as a negative control. Each vector also encoded a zeocin resistance gene. Following transfection, all cells were treated with 150 μg/ml of zeocin (Invitrogen) and incubated for 24, 48, and 72 h, at which time the cells were harvested and cell pellets frozen at −20°C. Cell pellets were resuspended in lysis buffer and loaded in duplicate onto 9% SDS-polyacrylamide gels. Following transfer to nitrocellulose membranes and blocking with a 5% (wt/vol) nonfat milk powder-0.1% Tween 20-PBS solution, alternative gels were incubated with either an anti-MeCP2 (Abcam) or an anti-GAPDH (Biodesign) antibody and then incubated with HRP-conjugated rabbit or mouse secondary antibodies (diluted 1:1,000; DAKO) before visualization with ECL detection reagents (Amersham). Cells treated with each shRNA vector were also plated onto 96-well microtiter plates at 0 and 72 h posttransfection for episomal maintenance analysis as described above.
RESULTS
ORF73C interacts with MeCP2 but not DEK.
We and others have previously demonstrated that ORF73 is essential and sufficient for HVS latent episomal persistence (8, 12, 13, 41). Furthermore, ORF73C has been shown to colocalize with host mitotic chromosomes during cell division (9, 41). The structural and functional similarities between HVS ORF73 and KSHV LANA led us to postulate that an analogous mechanism of chromosomal association may be used by these virus proteins. Data suggest that KSHV LANA interacts with multiple chromosome-associated proteins (7, 14, 26, 37). These include the cellular chromosome-associated proteins MeCP2 and DEK (26). Therefore, we set out to assess if HVS ORF73C also associated with these cellular proteins and, moreover; whether they are required for HVS episomal persistence.
Initially, HVS ORF73C (amino acids 241 to 407) was cloned into a bacterial expression vector in frame with a histidine epitope tag, producing pET21b-73C. The histidine-tagged ORF73C protein was subsequently expressed and bound to nickel-conjugated agarose beads (Fig. 1a). FLAG epitope-tagged DEK and MeCP2 proteins were expressed in HeLa cells and the cell extracts incubated with the ORF73C-conjugated beads. After washing, proteins bound to the ORF73C-conjugated beads were separated by SDS-PAGE and detected by Western blotting using an anti-FLAG antibody. The results indicate that DEK does not associate with ORF73C (Fig. 1bi). In contrast, repeated analysis indicates that ORF73C specifically bound the chromosomal protein MeCP2 (Fig. 1bii).
FIG. 1.
The HVS ORF73 C terminus associates with MeCP2 but not DEK during in vitro binding assays. (a) Bacterially expressed ORF73C was bound to nickel-agarose beads. (b) The ORF73C-conjugated beads were then incubated with HeLa cell extracts previously transfected with a plasmid expressing an empty FLAG vector (−ve), FLAG-tagged DEK, or FLAG-tagged MeCP2. Proteins attached to the ORF73C-conjugated beads were analyzed by Western blotting using a monoclonal anti-FLAG antibody as a primary antibody. Total-cell extracts representing 5% of total input were run as positive controls (+ve).
Confirmation of ORF73C and MeCP2 interaction using in vivo coimmunoprecipitation.
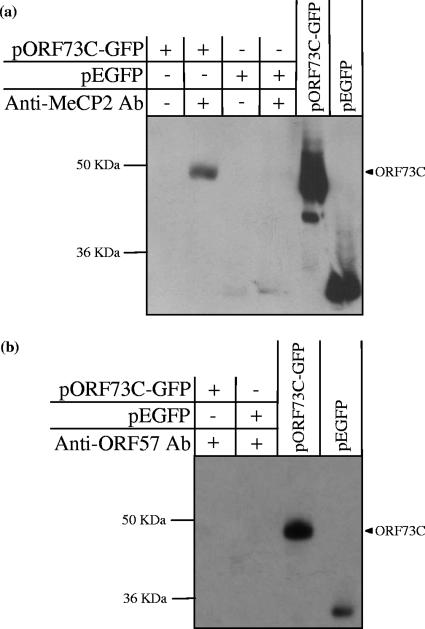
To confirm the interaction between ORF73C and MeCP2, in vivo coimmunoprecipitation was performed using an antibody directed against endogenous MeCP2. In order to allow detection of ORF73C, 293T cells were transfected with a vector expressing ORF73C as a GFP-tagged fusion protein (pEGFP-73C). An empty GFP vector (pEGFP-C3) was also transfected as a negative control. An anti-MeCP2 antibody was then used to immunoprecipitate MeCP2 protein, and an interaction with ORF73C was detected by Western blotting with an anti-GFP antibody (Fig. 2a). ORF73C was clearly coimmunoprecipitated in association with MeCP2; however, to ensure that this interaction was specific, the assay was repeated using an unrelated polyclonal antibody directed against HVS ORF57 (Fig. 2b). ORF73C-GFP was not immunoprecipitated by the anti-ORF57 antibody, confirming that the previously observed in vitro interaction between MeCP2 and ORF73C also occurs in vivo.
FIG. 2.
ORF73C is coimmunoprecipitated with MeCP2 in vivo. (a) 293T cells were transfected with a vector expressing the ORF73C-GFP fusion protein (pORF73C-GFP) and an empty EGFP vector (pEGFP). Cells were harvested, lysed, and incubated with or without a polyclonal anti-MeCP2 antibody (Ab). Immunocomplexes were captured using protein A agarose, washed, and separated on an SDS-PAGE gel. Analysis by Western blotting using an anti-GFP antibody indicates that ORF73C is immunoprecipitated in association with MeCP2. (b) To ensure that ORF73C is specifically immunoprecipitated by the anti-MeCP2 antibody, the immunoprecipitation was repeated using an alternative polyclonal antibody directed against HVS ORF57. Following Western blotting with an anti-GFP antibody, no interaction was observed, indicating that ORF73C is specifically immunoprecipitated in association with MeCP2. For both panels, 10% of the pORF73C-GFP and pEGFP total-cell extract was run as a positive control.
Mapping of the ORF73C domain responsible for association with MeCP2.
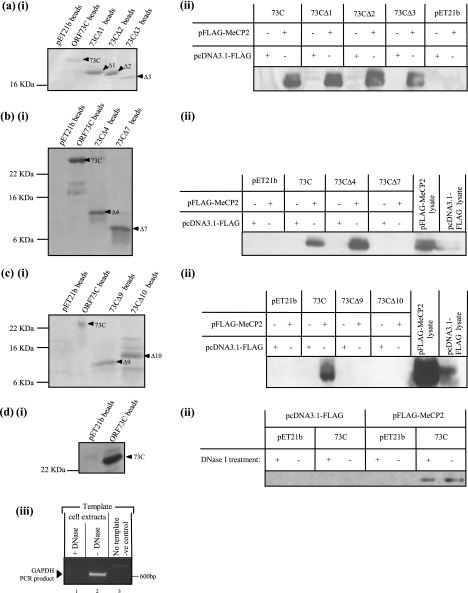
To map the domain within ORF73C required for association with MeCP2, a series of ORF73C deletions were cloned and expressed as recombinant histidine-tagged proteins (Fig. 3). Previous analysis using confocal microscopy suggested that removal of the first 62 amino acids of ORF73C (up to and including chromosomal association site 1 [CAS 1]) or 12 amino acids from the C terminus (CAS 2) (Fig. 3) results in loss of ORF73C chromosomal colocalization (9). Therefore, to assess if deletion of these ORF73C regions also results in loss of ORF73C-MeCP2 association, similar deletion constructs were expressed as histidine-tagged recombinant proteins (Fig. 4ai) and used in the in vitro binding analysis as described above. Analysis by Western blotting indicated that each of the truncated proteins retains the ability to interact with MeCP2 (Fig. 4aii).
FIG. 4.
In vitro binding assays using ORF73 C-terminal deletion proteins (a, b, and c) and DNase I-treated cellular extracts (d). pET21b-73C deletion proteins were expressed and bound to nickel-agarose beads (ai, bi, ci, and di) prior to incubation with HeLa cell extracts previously transfected with pFLAG-MeCP2 or an empty FLAG vector (pcDNA3.1-FLAG). Proteins associated with ORF73C-conjugated beads were analyzed by Western blotting using an anti-FLAG primary antibody (aii, bii, cii, and dii). Deletion analyses shown in panels aii, bii, and cii indicate that only full-length 73C and deletion proteins through 73CΔ4 are able to associate with MeCP2. Five to 10% of total pFLAG-MeCP2 and pcDNA3.1-FLAG cell lysates was run as a positive control where shown. Note that a small amount of MeCP2 lysate control has spilled over in the input lanes of cii; MeCP2 is not expressed in the empty FLAG control. PCR analysis using primers directed against GAPDH indicates that DNase I treatment has successfully digested cellular DNA in the treated cell extracts (diii). Panel dii demonstrates that DNase I treatment does not affect the ability of ORF73C and MeCP2 to associate, indicating that the interaction is not dependent on the presence of cellular DNA.
Subsequently, further deletion constructs were designed to identify the minimal ORF73C domain required for association with MeCP2 (Fig. 3). Each deletion construct was expressed as a recombinant histidine-tagged protein, although, as shown in Fig. 3, several of the proteins were insoluble and could not be used in the assay. However, in vitro binding analysis using the remaining soluble proteins demonstrated that ORF73C amino acids 324 to 395 (i.e., deletion protein 73CΔ4) are sufficient for MeCP2 binding (Fig. 4bii and cii).
ORF73 and MeCP2 interact in the absence of DNA.
To ensure that the interaction between ORF73 and MeCP2 was not due to a common affinity for DNA, in vitro binding assays as described above were repeated following treatment of the mammalian cell extracts with DNase I. To confirm that DNase I treatment had been successful, treated and untreated cell extracts were used as templates in a PCR utilizing primers directed against the housekeeping gene encoding GAPDH. This analysis clearly demonstrated that complete digestion of cellular DNA had occurred following addition of DNase I (Fig. 4diii, compare lanes 1 and 2). Subsequently, the treated cell extracts were incubated with ORF73C-conjugated agarose beads, washed, and analyzed by Western blotting using an anti-FLAG antibody. As Fig. 4dii shows, digestion of cellular DNA had no effect on ORF73C-MeCP2 binding, confirming that their association is mediated by a direct protein-protein interaction.
ORF73 colocalizes with MeCP2 on mitotic chromosomes.
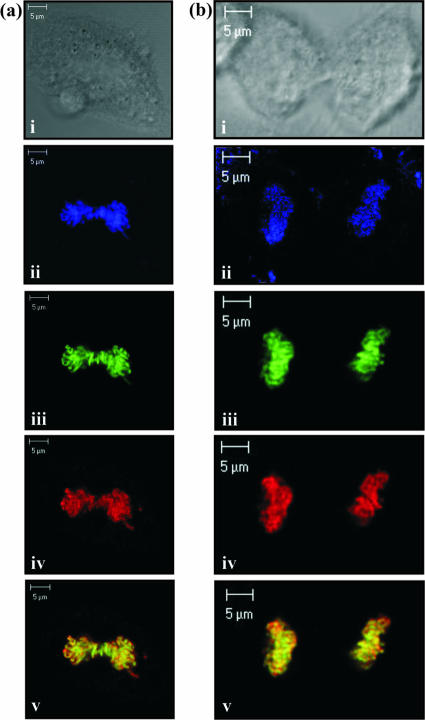
To analyze the cellular localization of ORF73 and MeCP2, indirect immunofluorescence was utilized. HeLa cells were transfected with ORF73-GFP and MeCP2-FLAG expression constructs and after 24 h were induced into mitosis by the addition of 10% serum following overnight serum starvation. The cells were then fixed and observed using laser scanning confocal microscopy. Examples of mitotic cells in the metaphase and telophase stages of division, stained with TO-PRO-3 iodide nucleic acid stain, are shown in Fig. 5a and b, respectively. HVS ORF73 was directly visualized using GFP fluorescence (Fig. 5aiii and biii), and MeCP2 was identified using anti-FLAG and Texas red antibodies (Fig. 5aiv and biv). Overlay of these images suggests that ORF73 and MeCP2 are bound to similar regions on the mitotic chromosomes, consistent with their association during cell division (Fig. 5av and bv).
FIG. 5.
ORF73 and MeCP2 colocalize on mitotic chromosomes during the metaphase (a) and telophase (b) stages of mitosis. HeLa cells were serum starved, induced to divide by the replenishment of serum, stained with anti-FLAG and Texas red-conjugated antibodies, and visualized by laser scanning confocal microscopy. Images show bright field (i), TO-PRO-3 iodide-stained nucleic acid (ii), ORF73C-GFP (iii), and FLAG-MeCP2 protein (iv) at a magnification of ×400 (a) or ×630 (b). (av and bv) merged GFP and Texas red fluorescent staining, highlighting ORF73 and MeCP2 colocalization on the cellular DNA.
MeCP2 expression is required for HVS episomal persistence.
During investigation of the association of KSHV with MeCP2, Krithivas et al. noted that GFP-LANA did not associate with NIH 3T3 mouse chromosomes; however, association was restored upon heterologous expression of human MeCP2 (26). They postulated that the lack of association could be due to poor expression of MeCP2 in NIH 3T3 cells, or that LANA attaches to MeCP2 through nonconserved regions in the MeCP2 murine and human homologues (sequence analysis indicates 71% homology). To assess the level of MeCP2 expression in NIH 3T3 cells, RT-PCR analysis was performed using primers directed against MeCP2. Analysis confirmed that MeCP2 is at best poorly expressed in NIH 3T3 cells compared with SW480 and A549 human carcinoma epithelial cells, which are known to support HVS episomal persistence (Fig. 6a, lanes 10 to 12).
FIG. 6.
HVS cannot persist in NIH 3T3 cells when virus persistence is selected. (a) Mouse NIH 3T3 cells do not express MeCP2 at detectable levels when analyzed by RT-PCR. (b) (i) NIH 3T3 and SW480 cells infected with HVS-BAC-GFP successfully form colonies following episomal persistence analysis without virus selection. (ii) NIH 3T3 cells are unable to form colonies when virus persistence is selected with hygromycin. (c) Expression of heterologous MeCP2 in the NIH-MeCP2 stable cell line rescues the ability of NIH 3T3 cells to maintain HVS episomal persistence. (i) Immunofluorescent microscopy indicates that approximately 50 to 60% of NIH-MeCP2 cells express heterologous MeCP2. (ii) Colony formation analysis indicates that heterologous expression of MeCP2 in NIH-MeCP2 cells is able to rescue hygromycin-resistant colony formation.
Therefore, the abilities of HVS to persist episomally in NIH 3T3 (low-MeCP2-expressing) and SW480 (high-MeCP2-expressing) cells were compared using cell culture maintenance assays. HVS-BAC-GFP, previously engineered to constitutively express the enhanced green fluorescent protein (EGFP) as well as a hygromycin resistance gene (44), was used to infect SW480 and NIH 3T3 cells. Following successful infection of both cell types (data not shown), the ability of each cell type to maintain the virus was analyzed by plating the infected cells on a 96-well microtiter plate. Following incubation under hygromycin selection, the numbers of wells positive for hygromycin-resistant colony outgrowth were quantified and compared (Fig. 6bii). To ensure that the results generated were due solely to the lack of HVS episomal persistence, cells were also incubated in the absence of virus selection (Fig. 6bi). Figure 6b demonstrates that although both cell types are able to form colonies when infected without hygromycin selection, only SW480 cells are able to maintain the HVS episome when viral persistence is selected for by using hygromycin.
To ensure that the inability of NIH 3T3 cells to maintain HVS episomal persistence was a consequence of low MeCP2 expression, NIH 3T3 cells were transfected with a plasmid expressing human MeCP2, and a stable cell line was produced. Analysis by immunofluorescent microscopy confirmed that approximately 50 to 60% of the NIH-MeCP2 cell line expressed high levels of heterologous MeCP2 (Fig. 6ci). Subsequently the ability of the NIH-MeCP2 cells to maintain HVS episomal persistence was compared with that of native NIH 3T3 cells by cell culture maintenance assays, as described above. Following successful infection of both the NIH 3T3 and NIH-MeCP2 cells with HVS-BAC-GFP, the cells were plated into 96-well plates in the presence of hygromycin selection. As shown in Fig. 6cii, stable expression of human MeCP2 in the NIH 3T3 cell line resulted in a statistically significant increase in hygromycin-resistant colony formation. These results confirm that low expression of MeCP2 significantly hinders HVS episomal persistence in dividing cells.
shRNA knockdown of MeCP2 expression inhibits HVS episomal persistence in a cell line latently infected with HVS.
Although KSHV LANA and MeCP2 have been shown to interact and this interaction was suggested to be important for episomal persistence, no MeCP2 depletion assay was performed to confirm the requirement for MeCP2. Therefore, to truly assess the importance of MeCP2 to HVS episomal persistence, we used shRNA to knock down MeCP2 expression in a latently infected cell line. 293T cells were infected with HVS-BAC-GFP, which constitutively expresses the genes encoding GFP as well as chloramphenicol and hygromycin resistance (Fig. 7a). Infected cells were selected using 50 μg/ml of hygromycin and maintained for more than 8 weeks in cell culture. As shown with other latently HVS infected cell lines, to ensure that the HVS genome was maintained in the 293T cells as a nonintegrated episome; episome recovery assays were performed as previously described (44). Low-molecular-weight DNA was extracted from the cells and transformed into electrocompetent E. coli DH10B. Following transformation, colonies were found on LB plates containing chloramphenicol, indicating that recovery of circularized HVS episomal DNA had been successful. This confirms that HVS is maintained as a nonintegrated latent episome in the 293T-HVS-GFP cells.
FIG. 7.
Analysis of HVS episomal persistence following shRNA knockdown of MeCP2 expression in a latently infected cell line. (a) 293T-HVS-GFP latently infected cells constitutively express GFP as well as hygromycin and chloramphenicol resistance. The HVS genome can be maintained indefinitely as a nonintegrated, circularized episome in these cells. (b) Vectors expressing shRNA directed against MeCP2 or luciferase protein (control) were transfected into 293T-HVS-GFP cells. The cells were incubated for 0, 24, 48, and 72 h before harvest and analysis by Western blotting. Analysis of GAPDH expression indicates that all samples were loaded equally. Detection of MeCP2 with an anti-MeCP2 antibody indicates that the MeCP2 shRNA specifically inhibits MeCP2 expression after 48 and 72 h posttransfection. (c) (i) Analysis of colony formation in the absence of virus selection using hygromycin indicates that the shRNA treatment alone does not inhibit episomal colony formation. (ii) In the presence of virus selection, inhibition of MeCP2 expression in the 293T-HVS-GFP latently infected cells correlates with a substantial decrease in episomal colony formation in a 96-well episomal persistence assay.
The latently infected 293T-HVS-GFP cells were subsequently transfected with a vector encoding shRNA targeted against MeCP2. To ensure that specific knockdown of MeCP2 had occurred, we also transfected the 293T-HVS-GFP cells with a vector containing shRNA directed against luciferase. Cells successfully transfected with each shRNA vector were selected for using zeocin and subsequently incubated for 0, 24, 48, and 72 h. Following incubation, the cells were harvested and MeCP2 protein expression analyzed by Western blotting using antibodies directed against MeCP2, as well as GAPDH as a loading control. As shown in Fig. 7b, specific knockdown of MeCP2 was achieved at 48 and 72 h posttransfection, in contrast to the control vector. Once MeCP2 knockdown had been confirmed, cells transfected with both control luciferase shRNA and MeCP2 shRNA were analyzed by a colony formation assay at 0 and 72 h after transfection of the shRNA vectors. To ensure that any reduction in colony formation was not due to inhibition of cell growth following transfection with the shRNA vectors, the 96-well colony-forming assay was performed without virus selection using hygromycin. As shown in Fig. 7ci, shRNA treatment did not inhibit 293T-HVS-GFP colony formation. However, in the presence of virus selection, loss of MeCP2 expression in the 293T-HVS-GFP latently infected cells significantly inhibited HVS episomal persistence. This is illustrated in Fig. 7cii, where a dramatic decrease in the number of wells containing hygromycin-resistant colonies was seen 72 h after transfection of the MeCP2 shRNA. These data confirm the importance of MeCP2 to HVS latent episomal persistence.
DISCUSSION
Maintenance of the HVS episome during the latent phase of the virus life cycle is fundamental to its ability to persist in a dividing cell population. We have previously shown that ORF73 is the sole viral gene required for HVS episomal persistence (8) and as such is functionally analogous to the related gammaherpesvirus genes encoding EBNA-1 and KSHV LANA. Furthermore, similarities between the genomic locations and protein structures of KSHV LANA and HVS ORF73, as well as their significant functional similarities, led us to postulate that these proteins may mediate episomal persistence by similar mechanisms. Both KSHV LANA and HVS ORF73 have been shown to colocalize with host mitotic chromosomes (5, 9, 14, 41), as well as possessing an ability to bind their respective episomal genomes via the TR DNA (5, 12-14, 21, 41). Hence, both proteins are thought to mediate latent persistence in a dividing cell population by tethering the virus episome to host mitotic chromosomes; ensuring that the virus genome is carried to each daughter cell. Following these observations, we set out to determine the mechanism by which HVS is able to attach itself to the host chromosome during cell division.
To date, KSHV LANA has been shown to interact with multiple cellular proteins, including several host chromosome-associated proteins, proposed to mediate its chromosomal localization. These include components of the nucleosome complex (i.e., histones H2A-H2B) as well as the linker histone H1 and, most recently, the bromodomain protein Brd4 (7, 14, 32, 37, 45). Additionally, Krithivas et al. demonstrated that the chromosome-associated proteins DEK and MeCP2 were able to target KSHV LANA to cellular chromosomes (26). Due to the distinct similarities between KSHV LANA and HVS ORF73, we are currently investigating whether HVS ORF73 uses similar protein interactions to mediate chromosomal association. Preliminary data suggest that HVS ORF73 also interacts with multiple host chromosome-associated proteins, and our investigations of these interactions are currently ongoing. Here, however, we have presented data demonstrating an interaction between ORF73C and the cellular protein MeCP2 as well as the functional significance of this interaction to the HVS latent state. This is the first time that depletion of MeCP2 by shRNA knockdown has shown that MeCP2 is essential for episome persistence of a gamma-2 herpesvirus.
Previous immunofluorescent analysis of ORF73 chromosomal colocalization demonstrated that the C terminus is sufficient for its chromosome association (9). Therefore, we used ORF73C to identify possible interactions with chromosome-associated proteins. We were able to demonstrate by a variety of assays that HVS ORF73C associates with MeCP2 via a direct protein-protein interaction, which occurs both in vitro and in vivo. Furthermore, deletion analysis of ORF73C demonstrated that a 72-amino-acid domain toward the C terminus of the protein (amino acids 324 to 395) is sufficient for MeCP2 association. A significant finding during this deletion analysis was that removal of the previously defined CASs (CAS 1 and CAS 2) had no effect on ORF73C-MeCP2 association. Previous analysis of the CAS 1 domain suggested that it contains a chromatin binding motif made up of a proline followed by two lysines (PKK) (9). A similar motif is also found within the DNA binding domains of the histone H1 and H2 proteins, as well as within a number of herpesvirus ORF73 homologues and EBP-2 (9). Mutation of the CAS1 PKK domain resulted in loss of ORF73 chromosome association, suggesting that this motif assists in ORF73 DNA binding. Furthermore, it was shown that loss of CAS 2 (but not CAS 1) correlated with loss of ORF73 self-association as well as chromosome localization, suggesting that ORF73 multimerization is required for chromosomal tethering (9). Therefore, although these domains were not required for the interaction of ORF73 with MeCP2, it is possible that the CAS 1 and CAS 2 domains may have a role in assisting ORF73 chromosomal association throughout its latent phase.
MeCP2 is a ubiquitously expressed nuclear protein whose abundant chromosomal localization is mediated through its high affinity for methylated CpG dinucleotides (31). A typical diploid nucleus contains approximately 107 methyl-CpGs, while quantitative Western blotting suggests that there are approximately 106 molecules of MeCP2 per nucleus (30), theoretically allowing every MeCP2 molecule to bind to the chromosome. Although the structure and composition of eukaryotic heterochromatin have yet to be fully understood, it is clear that chromosome-associated proteins play a vital role in the continual and essential modification of the chromosomal environment. Due to the requirement for constant modification, it is not surprising that the majority of chromosome-associated proteins are in a dynamic state of chromosome association and dissociation (1, 33). For example, MeCP2 can displace histone H1 in a Xenopus oocyte extract (30), and it has been shown that MeCP2 and histone H1 bind to chromatin via similar mechanisms (10). However, it was also noted that the ability of MeCP2 to compete with histone H1 chromosome localization ceased after approximately 40% of resident histone H1 had been replaced (30). This suggests that although these proteins bind to similar locations within chromatin, each has sufficient binding opportunities to maintain abundant and constant association with heterochromatin. Therefore, an ability to bind to an abundant and tightly associated chromosomal protein, such as MeCP2, would assist HVS chromosome association in the dynamic chromosomal environment throughout cell division.
Investigation of the related gammaherpesvirus latency-associated protein EBNA-1 primarily implicates the cellular protein EBP-2 in EBNA-1 chromosome association (38). The amino terminus of EBNA-1 is sufficient for its chromosomal localization, and two domains have been identified as responsible for this interaction (amino acids 33 to 89 and 328 to 378) (29). Although interaction with EBP-2 is clearly important to EBNA-1 chromosome association, it has also been shown that the amino-terminal domain of EBNA-1 can be functionally replaced by the cellular DNA binding proteins HMGA1a and histone H1 (22, 35). Interestingly, Sears et al. suggest that in addition to EBP-2 binding, EBNA-1 may also possess intrinsic DNA binding activity mediated by AT hook binding domains akin to those found in cellular DNA binding proteins (36). This is particularly noteworthy in the context of our study, since MeCP2 also contains an AT hook DNA binding domain (3).
Although the association of HVS ORF73 with MeCP2 has been clearly identified, it is evident that gammaherpesvirus chromosome attachment is likely to be facilitated by several alternative interactions. This leads to the question, what is the functional significance of ORF73's association with MeCP2? Here we demonstrate that although MeCP2 may not be the sole target to which ORF73 binds, the interaction between ORF73 and MeCP2 is essential for HVS episomal persistence. Initially we investigated the ability of HVS to persist in the low-MeCP2-expressing cell line NIH 3T3. Previous investigation of KSHV LANA chromosome association suggested that NIH 3T3 cells do not express a sufficient concentration of MeCP2 to enable LANA chromosomal association (26). We were able to confirm this observation following RT-PCR analysis of MeCP2 expression in NIH 3T3 cells compared with that in two cell lines known to support HVS episomal persistence. The data clearly indicated that NIH 3T3 cells expressed undetectable levels of MeCP2 compared with both SW480 and A549 cells, confirming that NIH 3T3 cells provide a good in vitro model for study of the role of HVS ORF73C-MeCP2 association. Previous analysis of HVS episomal persistence by a colony maintenance assay with the human carcinoma epithelial cell line SW480 has shown that this protocol provides a consistent and reliable method, which can be used to assess the ability of HVS to persist episomally (8). Therefore, the abilities of wild-type HVS to persist in NIH 3T3 (low-MeCP2-expressing) and SW480 (high-MeCP2-expressing) cells were compared using this assay. Our results indicate that the low-MeCP2-expressing NIH 3T3 cells were unable to maintain viral persistence when virus-infected cells were selected with hygromycin. Additionally, overexpression of MeCP2 in NIH 3T3 cells rescued episomal persistence and resulted in a significant increase in hygromycin-resistant colony formation. Since immunofluorescent microscopy indicated that only 50 to 60% of the stable NIH-MeCP2 cells successfully expressed heterologous MeCP2, these results strongly suggest that expression of MeCP2 is critical to HVS episomal persistence.
Although studies with NIH 3T3 cells indicate that low expression of MeCP2 hinders HVS episomal persistence, it is also possible that further differences in these cells may contribute to the inability of HVS to persist. Therefore, to conclusively prove that the interaction of ORF73 with MeCP2 was vital to HVS episomal persistence, we used shRNA to specifically reduce MeCP2 expression in a latently infected cell line. Upon successful knockdown of MeCP2 expression in the 293T-HVS-GFP latently infected cells, episomal persistence was drastically reduced under selective conditions. This indicates that in the absence of MeCP2 expression, the latently infected cells were unable to retain the virus episome and hence lost hygromycin resistance as the cells divided. Therefore, we believe that the association of ORF73 with MeCP2 is essential to the maintenance of HVS episomes in a dividing cell population. Although the specific role of MeCP2 has not been addressed in this article, the similar localizations of MeCP2 and ORF73 on mitotic chromosomes suggested that MeCP2 may contribute to the chromosomal attachment of ORF73. We have attempted to investigate this hypothesis further by assessing the localization of ORF73 in NIH 3T3 versus NIH-MeCP2 cells; however, no differences were observed. This leads to the possibility that other interactions are involved in chromosomal attachment and that the MeCP2 interaction may have an enhancing or stabilizing effect which is essential for the long-term persistence of the episome.
Therefore, in summary, while we do not exclude the possibility that HVS uses multiple interactions to secure lifelong latent infection, the data we present here have clearly demonstrated that HVS episomal persistence is functionally dependent on the interaction between ORF73 C-terminal amino acids 324 to 395 and the cellular chromosome-associated protein MeCP2.
Acknowledgments
We thank D. Hayward (Johns Hopkins School of Medicine, Baltimore, MD) and Gerard Grosveld (St. Jude Children's Research Hospital, Memphis, TN) for provision of the MeCP2 and DEK expression constructs and D. A. Mann (Newcastle University, Newcastle, United Kingdom) for the shRNA vectors.
This work was funded by the BBSRC.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Adkins, N. L., M. Watts, and P. T. Georgel. 2004. To the 30-nm chromatin fiber and beyond. Biochim. Biophys. Acta 1677:12-23. [DOI] [PubMed] [Google Scholar]
- 2.Albrecht, J., J. Nicholas, D. Biller, K. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. Craxton, H. Coleman, and B. Fleckenstein. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind, L., and D. Landsman. 1998. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 26:4413-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 6.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbera, A. J., J. V. Chodaparambil, B. Kelley-Clarke, V. Joukov, J. C. Walter, K. Luger, and K. M. Kaye. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856-861. [DOI] [PubMed] [Google Scholar]
- 8.Calderwood, M., R. E. White, R. A. Griffiths, and A. Whitehouse. 2005. Open reading frame 73 is required for herpesvirus saimiri A11-S4 episomal persistence. J. Gen. Virol. 86:2703-2708. [DOI] [PubMed] [Google Scholar]
- 9.Calderwood, M. A., K. T. Hall, D. A. Matthews, and A. Whitehouse. 2004. The herpesvirus saimiri ORF73 gene product interacts with host-cell mitotic chromosomes and self-associates via its C terminus. J. Gen. Virol. 85:147-153. [DOI] [PubMed] [Google Scholar]
- 10.Chandler, S. P., D. Guschin, N. Landsberger, and A. P. Wolffe. 1999. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry 38:7008-7018. [DOI] [PubMed] [Google Scholar]
- 11.Chang, Y., P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, K. Godden, H. Paterson, R. A. Weiss, and S. Mittnacht. 1996. Cyclin encoded by KS herpesvirus. Nature 382:410. [DOI] [PubMed] [Google Scholar]
- 12.Collins, C. M., M. M. Medveczky, T. Lund, and P. G. Medveczky. 2002. The terminal repeats and latency-associated nuclear antigen of herpesvirus saimiri are essential for episomal persistence of the viral genome. J. Gen. Virol. 83:2269-2278. [DOI] [PubMed] [Google Scholar]
- 13.Collins, C. M., and P. G. Medveczky. 2002. Genetic requirements for the episomal maintenance of oncogenic herpesvirus genomes. Adv. Cancer Res. 84:155-174. [DOI] [PubMed] [Google Scholar]
- 14.Cotter, M. A., and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 15.Cotter, M. A., C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 16.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgel, P. T., R. A. Horowitz-Scherer, N. Adkins, C. L. Woodcock, P. A. Wade, and J. C. Hansen. 2003. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 278:32181-32188. [DOI] [PubMed] [Google Scholar]
- 18.Goodwin, D. J., K. T. Hall, M. S. Giles, M. A. Calderwood, A. F. Markham, and A. Whitehouse. 2000. The carboxy terminus of the herpesvirus saimiri ORF 57 gene contains domains that are required for transactivation and transrepression. J. Gen. Virol. 81:2253-2265. [DOI] [PubMed] [Google Scholar]
- 19.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, I. M. Carr, A. J. Stevenson, A. F. Markham, and A. Whitehouse. 2000. Analysis of gene expression in a human cell line stably transduced with herpesvirus saimiri. J. Virol. 74:7331-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, A. F. Markham, and A. Whitehouse. 2000. Characterization of the herpesvirus saimiri ORF73 gene product. J. Gen. Virol. 81:2653-2658. [DOI] [PubMed] [Google Scholar]
- 21.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapoor, P., and L. Frappier. 2003. EBNA1 partitions Epstein-Barr virus plasmids in yeast cells by attaching to human EBNA1-binding protein 2 on mitotic chromosomes. J. Virol. 77:6946-6956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kapoor, P., B. D. Lavoie, and L. Frappier. 2005. EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases. Mol. Cell. Biol. 25:4934-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapoor, P., K. Shire, and L. Frappier. 2001. Reconstitution of Epstein-Barr virus-based plasmid partitioning in budding yeast. EMBO J. 20:222-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 28.Mackey, D., and B. Sugden. 1999. The linking regions of EBNA1 are essential for its support of replication and transcription. Mol. Cell. Biol. 19:3349-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marechal, V., A. Dehee, R. Chikhi-Brachet, T. Piolot, M. Coppey-Moisan, and J. C. Nicolas. 1999. Mapping EBNA-1 domains involved in binding to metaphase chromosomes. J. Virol. 73:4385-4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nan, X., F. J. Campoy, and A. Bird. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 31.Nan, X., P. Tate, E. Li, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ottinger, M., T. Christalla, K. Nathan, M. M. Brinkmann, A. Viejo-Borbolla, and T. F. Schulz. 2006. Kaposi's sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. J. Virol. 80:10772-10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phair, R. D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D. T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo, J. J., R. A. Bohenzky, M.-C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sears, J., J. Kolman, G. M. Wahl, and A. Aiyar. 2003. Metaphase chromosome tethering is necessary for the DNA synthesis and maintenance of oriP plasmids but is insufficient for transcription activation by Epstein-Barr nuclear antigen 1. J. Virol. 77:11767-11780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 78:11487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinohara, H., M. Fukushi, M. Higuchi, M. Oie, O. Hoshi, T. Ushiki, J. Hayashi, and M. Fujii. 2002. Chromosome binding site of latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus is essential for persistent episome maintenance and is functionally replaced by histone H1. J. Virol. 76:12917-12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 40.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517-521. [DOI] [PubMed] [Google Scholar]
- 41.Verma, S. C., and E. S. Robertson. 2003. ORF73 of herpesvirus saimiri strain C488 tethers the viral genome to metaphase chromosomes and binds to cis-acting DNA sequences in the terminal repeats. J. Virol. 77:12494-12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viejo-Borbolla, A., M. Ottinger, E. Bruning, A. Burger, R. Konig, E. Kati, J. A. Sheldon, and T. F. Schulz. 2005. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. J. Virol. 79:13618-13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waldmann, T., I. Scholten, F. Kappes, H. G. Hu, and R. Knippers. 2004. The DEK protein—an abundant and ubiquitous constituent of mammalian chromatin. Gene 343:1-9. [DOI] [PubMed] [Google Scholar]
- 44.White, R. E., M. A. Calderwood, and A. Whitehouse. 2003. Generation and precise modification of a herpesvirus saimiri bacterial artificial chromosome demonstrates that the terminal repeats are required for both virus production and episomal persistence. J. Gen. Virol. 84:3393-3403. [DOI] [PubMed] [Google Scholar]
- 45.You, J., V. Srinivasan, G. V. Denis, W. J. Harrington, Jr., M. E. Ballestas, K. M. Kaye, and P. M. Howley. 2006. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen interacts with bromodomain protein Brd4 on host mitotic chromosomes. J. Virol. 80:8909-8919. [DOI] [PMC free article] [PubMed] [Google Scholar]